Abstract
Ubiquitination of the αN-terminus of protein substrates has been reported sporadically over the past twenty years. However the identity of an enzyme responsible for this unique ubiquitin (Ub) modification has only recently been elucidated. We show the ubiquitin-conjugating enzyme (E2) Ube2w employs a novel mechanism to facilitate the specific ubiquitination of the α-amino group of its substrates that involves recognition of backbone atoms of intrinsically disordered N-termini. We present the NMR-based solution ensemble of full-length Ube2w that reveals a structural architecture unlike any other E2, in which its C-terminus is partly disordered and flexible to accommodate variable substrate N-termini. Flexibility of the substrate is critical for recognition by Ube2w and point mutations in, or removal of, the flexible C-terminus of Ube2w inhibits substrate binding and modification. Mechanistic insights reported here provide guiding principles for future efforts to define the N-terminal-Ubiquitome in cells.
Introduction
The attachment of ubiquitin (Ub) to cellular proteins is a highly regulated process that requires three enzyme activities. First, an E1 ubiquitin-activating enzyme forms a thioester bond between its active site cysteine and the Ub C-terminus in an ATP-dependent reaction. Second, Ub undergoes a transthiolation reaction with the active site cysteine of an E2 ubiquitin-conjugating enzyme forming an E2~Ub conjugate. Third, E2~Ub interacts with an E3 ubiquitin ligase to modify protein targets via a RING-type, HECT-type, or RING-between-RING-type (RBR-type) mechanism. A distinguishing feature of RING-type mechanisms is that the E3 activates the E2~Ub conjugate to transfer Ub directly from the E2 active site to the substrate1. Thus, in RING-type mechanisms, the E2 plays a direct role in interacting with substrate and dictating the final ubiquitinated product. The diversity of products depends on the enzymes involved and the biological context and may include the addition of a single Ub onto a substrate lysine or the synthesis of poly-ubiquitin chains built from any of Ub’s seven lysine residues. To generate such diversity, there are ~40 human E2s that have presumably evolved disparate functions. Some E2s are specific for a single chain type, such as the Ubc13/Mms2 complex (K63-linked chains)2 or Ube2k (K48-linked chains)3, while others such as UbcH5c are promiscuous and can build Ub chains of multiple linkages 4. Some E2s such as Ube2e1 and Ube2t add only a single Ub to their target substrate5,6. There is some evidence that certain E2s may transfer Ub to non-canonical amino acids such as serine, threonine, and cysteine7,8. The E2 Ube2w was recently reported to attach mono-Ub to the αN-terminus of substrates rather than to the εNH2 side chain group of lysine residues9,10.
Here we show that Ube2w specifically mono-ubiquitinates the αN-terminus of diverse substrates by recognizing backbone atoms of disordered N-termini. The solution ensemble of Ube2w reveals a novel UBC (“Ubiquitin Conjugating”) domain architecture (Supplementary Results, Supplementary Fig. 1). Though the first 118 residues adopt a canonical E2 fold, the Ube2w C-terminal region is partially unstructured and can occupy multiple positions near the active site. Removal of the final twenty C-terminal residues or a single point mutation within this region abrogates Ube2w ubiquitin transfer activity and impacts recognition and binding of multiple substrates. Furthermore, N-terminal substrate recognition and subsequent Ub transfer catalyzed by Ube2w are intimately dependent on the non-canonical arrangement of Ube2w C-terminal residues relative to its active site.
Results
Ube2w adds mono-Ub to intrinsically disordered N-termini
RNA Polymerase Subunit 8 (RPB8) is a highly conserved subunit of RNA polymerases I, II, and, III that is ubiquitinated in cells by the RING E3 ligase BRCA1/BARD1 following UV-induced DNA damage11. To identify the E2 – BRCA1/BARD1 pair(s) that can ubiquitinate RPB8, in vitro ubiquitination assays were performed using the minimal RING heterodimer of BRCA1/BARD1 (BC112/BD115), and E2s that had previously been shown to interact with BRCA1/BARD1: Ube2w, UbcH5c, UbcH7, and Ube2e112. Although RPB8 contains eight lysines, only Ube2w modifies RPB8 with Ub in the presence of BC112/BD115 (Fig. 1a, Supplementary Fig. 2a). Ube2w also exhibits E3-independent modification, though with substantially lower activity (Supplementary Fig. 2a, Supplementary Figure 3). Mass spectrometry analysis of the mono-ubiquitinated RPB8 product confirmed that the Ub is attached to RPB8’s αN-terminus (Supplementary Fig. 4). It should be noted that an initial mono-Ub attached by Ube2w can serve as a primer for poly-Ub chain synthesis by another E2 such as Ubc13/MMS2 or Ube2k10,12,13.
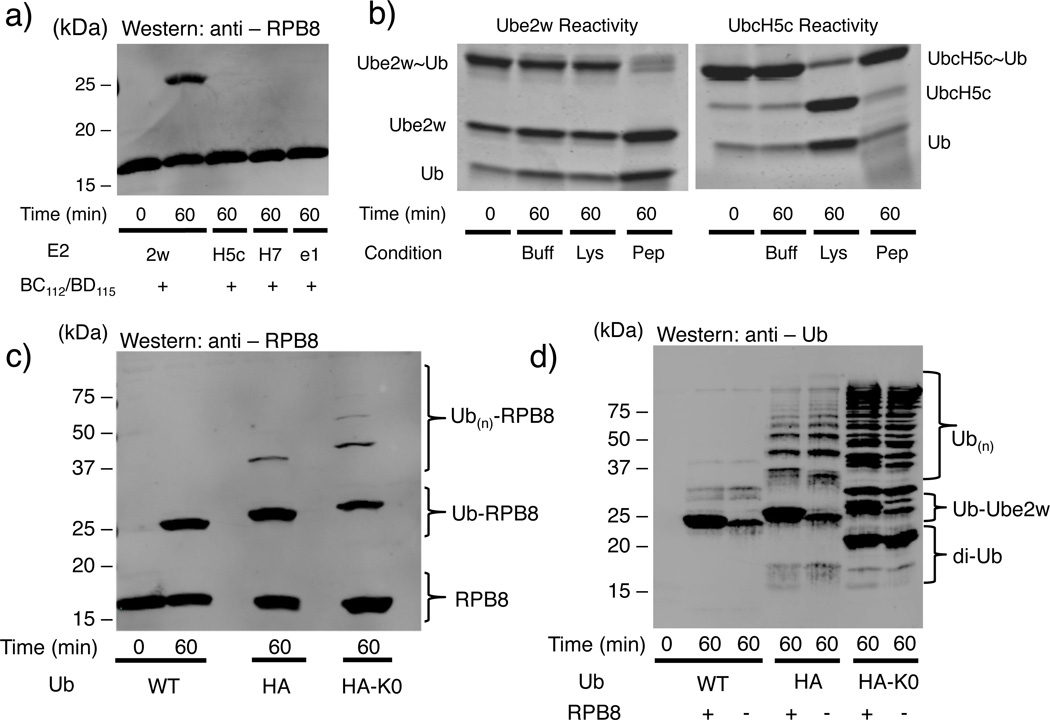
Figure 1.
Ube2w has distinct E2 activity. (a) Ube2w transfers a single Ub to RPB8 in vitro while other BRCA1-interacting ubiquitin conjugating enzymes UbcH5c (“H5c”), UbcH7 (“H7”), and Ube2e1 (“e1”) do not (Supplementary Fig. 2a). (b) Left A nucleophile reactivity assay reveals Ube2w has intrinsic activity with αNH2 groups of a peptide with a free NH2 group at its N-terminus (NH2-A-G-G-S-Y-COO-; 50 mM) but not the εNH2 groups of lysine. Right, Identical reactions with UbcH5c~Ub conjugates confirm the previously reported lysine reactivity of UbcH5c and reveal it to be unreactive towards the peptide (Supplementary Fig. 2b). (c) Products generated on RPB8 depend on the Ub species in the reaction. Lanes 1 and 2: a single Ub is attached to RPB8 in a reaction with WT-Ub. Lane 3: Attachment of an additional Ub is detected in a reaction HA-Ub, which contains a 13-residue tag at the N-terminal end of Ub; Lane 4: Reaction carried out with lysine-less HA-Ub (HA-Ub(K0)) confirms that Ube2w builds linear Ub chains (i.e., attaches the C-terminus of one Ub to the N-terminus of another) on RPB8 with HA-Ub (Supplementary Fig. 2d). (d) Reactions shown in Panel (c) were blotted for Ub, revealing that Ube2w builds linear poly-Ub chains only when Ub harbors an N-terminal HA-tag (Supplementary Fig. 2e).
A direct measure of the intrinsic aminolysis activity of an E2~Ub conjugate is its reactivity towards the εNH2 group of free lysine. Many E2s, such as UbcH5c, that transfer Ub to lysine side chains of protein substrates readily transfer Ub to free lysine14. As shown in Fig. 1b (left panel), the Ube2w~Ub conjugate remained intact in the presence of free lysine but reacted completely with a peptide containing a free N-terminal amino group and no lysine (NH2-Ala-Gly-Gly-Ser-Tyr-COO-) (Fig. 1b left, Supplementary Fig 2b,c,5). In contrast, UbcH5c~Ub reacted completely with free lysine but did not react with the peptide substrate (Fig. 1b right panel). Thus, Ube2w’s intrinsic aminolysis reactivity is limited to αNH2 groups in the context of a polypeptide, as it did not transfer Ub to the αNH2 group of a free amino acid, while UbcH5’s intrinsic aminolysis reactivity is limited to εNH2 groups of lysine residues.
In addition to RPB8, full-length CHIP (Carboxy terminus of HSP70-interacting protein), the minimal U-box domain of CHIP, small ubiquitin-like modifier 2 (SUMO-2), tau, ataxin-3, FANCL, FANCD2, and Ube2w have been reported to be ubiquitinated by Ube2w9,10,15,16. In all cases, a single mono-Ub is attached, implying the N-terminus of the attached Ub cannot also serve as a Ube2w substrate. Serendipitously, we discovered that Ub harboring a 13-residue N-terminal Human influenza hemagglutinin-tag (HA-tag) is a robust Ube2w substrate in vitro. Unlike with WT-Ub, Ube2w adds multiple HA-Ubs onto RPB8 and this activity is retained with a lysine-less version, HA-Ub(K0), consistent with each attachment occurring through the αNH2 group (Fig. 1c, Supplementary Fig. 2d). Notably, the major species formed under the reaction conditions used here (i.e., HA-Ub in excess over RPB8) are free poly-HA-Ub chains, as observed in an anti-Ub immunoblot of the same reaction (Fig. 1d, Supplementary Fig. 2e). {1H – 15N} NOE (hetNOE) values of HA-Ub, which are sensitive to high frequency motions on the pico to nano-second timescales, confirm that the additional residues in the HA tag are highly mobile as shown by their small and/or negative hetNOE values (Fig. 2a, Supplementary Fig. 6). In contrast, hetNOE values for Ub residues 1-72 show that the N-terminus of WT-Ub is well ordered17 as expected from Ub crystal structures (Fig. 2a,b). The ability of HA-Ub but not WT-Ub to serve as substrate illustrates that addition of a disordered segment to its natively structured N-terminus is sufficient to convert Ub into a Ube2w substrate. We note that all currently identified targets of Ube2w-dependent ubiquitination (see above) have or are predicted to have a disordered segment at their N-termini18–20 (Supplementary Fig. 7).
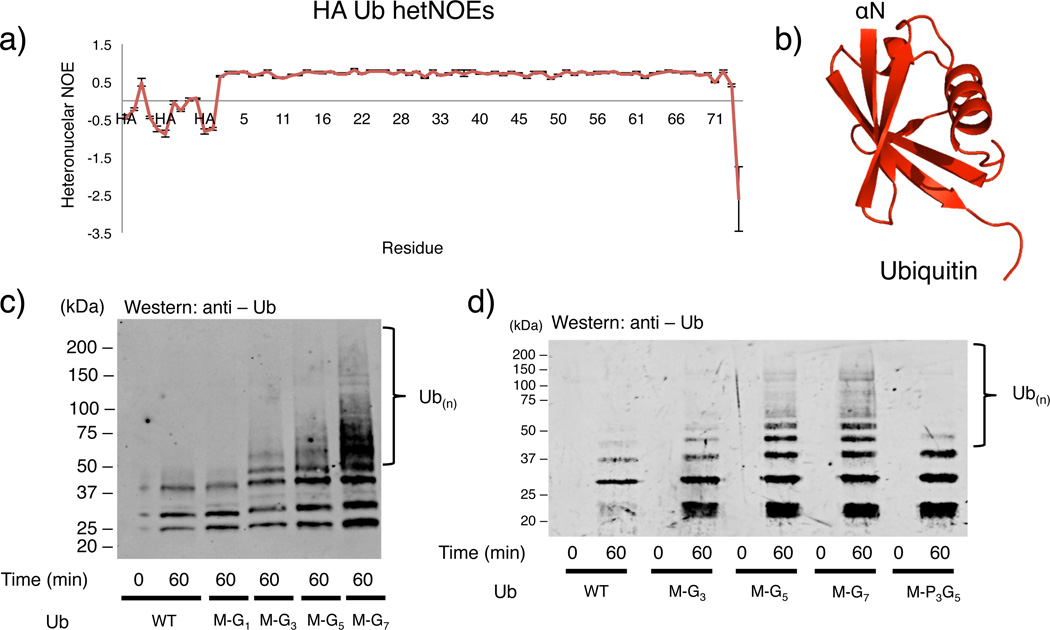
Figure 2.
Ube2w transfers Ub to flexible/disordered N-termini. (a) Negative {1H – 15N} hetNOE values for residues derived from the HA-tag are indicative of highly flexible amino acids. Errors bars represent the standard error from the mean. (Resonances from this tag are not assigned and are not plotted sequentially. They are labeled simply as “HA”). (b) Consistent with the {1H – 15N} hetNOE data, the crystal structure of Ub (PDB: 1UBQ) is ordered at its αN-terminus and immediately forms a β -strand with residue Met1. (c) Ub to which two N-terminal amino acids have been added at the N-terminus (Met-Gly-Ub) is not incorporated into chains by Ube2w and displays similar activity to WT Ub. Four N-terminal residues (Met-Gly3-Ub) are sufficient to induce Ube2w activity towards Ub. Addition of six (Met-Gly5-Ub) or eight (Met-Gly7-Ub) residues increases Ube2w N-terminal ubiquitination activity (Note: bands below 37kDa are consistent with auto-ubiquitinated E2 and E3) (Supplementary Fig. 2f). (d) N-terminal backbone amide groups are necessary for Ube2w-dependent ubiquitination. Ube2w shows increased activity with the addition of disordered N-terminal amino acids, (Lanes 1-8). Proline at positions 2-4 (Lanes 9,10) inhibits Ube2w chain-building activity to levels similar to WT-Ub (Supplementary Fig. 2g).
Ube2w recognizes the backbone atoms of its substrates
Diverse N-terminal sequences in protein substrates suggest two mechanistic possibilities: 1) Ube2w preferentially recognizes backbone atoms over amino acid side chains, and 2) regular N-terminal secondary structure elements (α -helices and β-sheets) would inhibit necessary contacts with Ube2w. To further test these hypotheses, we chose to add glycine residues (lacking side chains) to the N-terminus of Ub. Addition of two residues (Met-Gly-Ub) did not result in Ube2w-dependent modification, whereas addition of four N-terminal residues (Met-Gly3-Ub) results in modest activity, and additional glycine residues (Met-Gly5- or Met-Gly7-Ub) result in a further increase in Ube2w activity (Fig 2c, Supplementary Fig. 2f). The results show that in vitro, a disordered polypeptide chain composed of a methionine and three glycine residues is sufficient for Ube2w to recognize a protein as a potential ‘substrate’ and that Ube2w activity increases as additional disordered N-terminal glycines are present. Replacement of glycine residues with prolines disrupts the ability of Ube2w to use N-terminally tagged Ub as a substrate. Ub harboring three N-terminal prolines (Met-Pro3-Gly5-Ub) is incapable of forming large poly-Ub chains and forms similar products to WT-Ub indicating amide groups at positions two through four are necessary for Ube2w-dependent N-terminal ubiquitination (Fig. 2d, Supplementary Fig. 2g). Furthermore, a methionine at position 1 is not necessary for N-terminal ubiquitination by Ube2w (Supplementary Table 1). Altogether, the results are consistent with a model whereby Ube2w recognizes substrates through backbone carbonyl and amide groups rather than side chain atoms.
Ub transfer via aminolysis likely has differing requirements for an α-amino group as opposed to the ε-amino group of a lysine side chain21. The pKa of an N-terminal amino group is 7.7 ± 0.5 whereas lysine side chains have pKa values around 10.5 ± 1.122. Thus, at physiological pH, a larger proportion of α N-termini will be deprotonated, bypassing a need to deprotonate the incoming nucleophile. The pKa of an N-terminal amino group depends on the identity of the sidechain at position 1. However, our results indicate that Ube2w is capable of ubiquitinating N-termini over a wide pKa range (pKa = ~7.3 to ~9.1), indicating that the nucleophile’s pKa is not the determining factor for Ube2w’s special reactivity (Supplementary Fig. 2h,8)
Ube2w has a non-canonical UBC domain
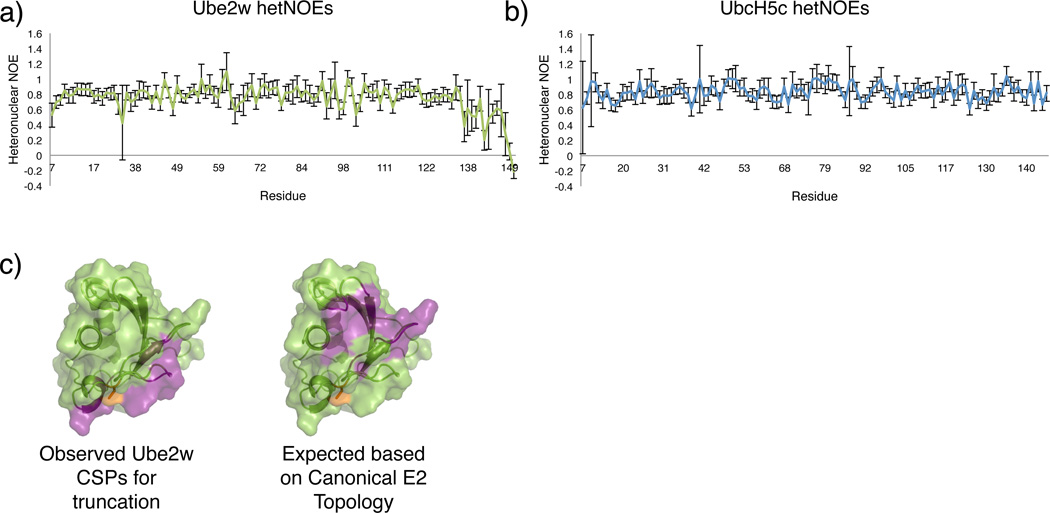
For insights into Ube2w’s unique Ub transfer specificity, we characterized the dynamic and structural properties of the E2 using NMR. Though a crystal structure exists (PDB: 2A7L), it is for a truncation of Ube2w that lacks residues Ser117- Cys15123. A similar truncated version of Ube2w can form an activated thioester with Ub (Supplementary Fig. 2i,9), but does not transfer Ub onto a substrate (data not shown) and thus lacks the structural features needed to understand Ube2w substrate selectivity. {1H – 15N} hetNOE values indicate that the final three residues of full-length Ube2w are highly flexible (negative hNOE values) and that residues 135-151 undergo higher frequency motions (small positive hetNOE values) than the rest of the protein (Fig. 3a). An identical experiment conducted on UbcH5c reveals its C-terminus undergoes motions consistent with the core of the protein (Fig. 3b).
Figure 3.
The Ube2w C-terminus is flexible and occupies a non-canonical position. (a) Residues 7-132 of Ube2w have generally uniform and positive {1H – 15N} hetNOE values. Beginning at residue 137, values decrease and ultimately become negative at the extreme C-terminus, consistent with a region that undergoes motions at higher frequencies than the core of the protein. Errors bars represent the standard error from the mean. (b) For comparison, UbcH5c has positive hetNOE values throughout its entire protein sequence, even at the far C-terminus. (c) Left, Experimental CSP data based on comparing the (1H – 15N) – HSQC-TROSY spectra of Ube2w-KK and Ube2w-131Δ-KK reveals that removal of the C-terminus perturbs residues near the active site, in the 310-helix, and on the ‘backside’ β -sheet (purple). Right, If C-terminal helices were to reside in their canonical positions in Ube2w a surface consisting of loops 3 and 5 would be perturbed by removal of residues 132-151. Residues depicted to be perturbed are colored in purple, demonstrating that the C-terminal region of Ube2w is different from other E2s.
To understand the unique structural properties of Ube2w, further NMR experiments were conducted. NMR data were collected on a monomeric Ube2w mutant (V30K/D67K-Ube2w termed “KK”) that reduces self-association at high concentrations, but retains Ube2w activity24. We compared the {1H, 15N} – HSQC-TROSY spectra of full-length Ube2w-KK with a fragment that is missing the C-terminal residues following Tyr131 (Ube2w-131-KK). Ube2w resonances for residues S86, N87, and T96 (near the active site), in the 310-helix, and on the ‘backside’ β-sheet experience large chemical shift perturbations (CSPs) as a consequence of removal of the C-terminus (Fig. 3c left, Supplementary Fig. 10a,b). These observations are surprising in that comparisons with canonical E2 structures, such as UbcH5c, predict a different set of CSPs would be observed, particularly with respect to the E2 active site and loops 3 and 5 (Fig. 3c right and Supplementary Fig. 1 for nomenclature)25.
Based on these results we sought to characterize the structure of full-length Ube2w. A conventional de novo NOE-based structure determination was precluded by a paucity of NOE crosspeaks (Supplementary Fig. 11). Therefore, we pursued an NMR-driven solution structure using alternative parameters. NMR chemical shifts (HN, N, C, Cα, Cβ) for 137 residues, 109 RDC values for NH pairs, and the CSPs between full-length and truncated Ube2w were initially input into the Rosetta-based algorithm, Chemical-Shift-ROSETTA (CS-ROSETTA) a program that utilizes chemical shift values and other experimentally-driven NMR restraints to generate solution ensembles26,27 (Supplementary Table 2,3, Supplementary Dataset 1, Supplementary Fig. 10c,12). This initial computational stage produced 16,000 structures that were further filtered using two spin label positions (C91 and C135) and SAXS to generate the final ensemble.
An ensemble of the twenty lowest energy structures is shown in Fig. 4a. Ube2w has a well-defined core that closely resembles canonical UBC domains, such as UbcH5c (Fig. 4b). The average pairwise RMSD for residues R7–S118 for the twenty members of the ensemble is 1.35 Å, indicating that the available experimental observations used are sufficient to define the structure. Furthermore, the twenty-member ensemble reveals favorable Ramachandran statistics (Supplementary Table 2). The average pairwise RMSD for all backbone atoms across the Ube2w ensemble is 4.1 Å, consistent with {1H – 15N} hetNOE values that reveal a highly flexible C-terminus. Features conserved amongst canonical E2s in the Ube2w solution structure include helix-1, the 4-stranded ‘backside’ β-sheet, the structural architecture of the active site, and helix-3 (“crossover” helix) (Fig 4a, b).
Figure 4.
NMR ensemble of Ube2w reveals a novel E2 architecture. (a) Solution ensemble of Ube2w derived from NMR restraints (backbone chemical shifts, CSPs, residual dipolar couplings (RDCs), paramagnetic spin-label data, and small-angle X-ray scattering (SAXS)) calculated with CS-Rosetta. The twenty lowest energy members of the ensemble are shown and reveal a well-defined core with high structural similarity to canonical E2s. The C-terminal region is partially disordered and occupies multiple positions near the Ube2w active site C91 (orange). (b) Similar views of a representative canonical UBC domain structure (UbcH5c; PDB 2FUH). (c) Helix-4 (penultimate helix) is in distinct positions in Ube2w (3 representatives of the 20-member ensemble are shown for clarity). A flexible loop emanating from helix-3 leads away from the protein core. Helix-4 is clustered in three distinct positions in the ensemble (Cluster 1, light gray; Cluster 2, gray; Cluster 3, dark gray). (d) Side-view of the full Ube2w ensemble looking down the helix-3 axis reveals the three clusters. (e) In all twenty members of the Ube2w ensemble residues N136-W145 occupy positions beneath the active site, C91 (orange). Residues 119-135 are not shown for clarity. No clustering is evident for this region and the Cβ atom of every residue is on average 14.5-17.5 Å away from the active site.
The first seven residues of Ube2w are not observed in our NMR spectra, indicating they undergo conformational exchange, most likely a helix-to-coil transition. However, due to the component of CS-Rosetta that utilizes SPARTA-based selection of protein fragments from the PDB, our ensemble contains an ordered N-terminus based on homology modeling. The most distinctive feature of the Ube2w ensemble is its C-terminal region (residues 127-151), which adopts multiple orientations near the active site. In the ensemble C-terminal positions are determined by the CSP restraints, the spin label effects at residues C91 and C135, and SAXS.
In Ube2w, a long disordered loop following the crossover helix leads away from the β -sheet and is followed by a single helix formed by residues 127-135. This helix does not appear to adopt a unique position but is located within 7-19 Å of the active site, C91, in all members of the ensemble (Fig. 4c). By contrast, in canonical UBC domains, a loop leads from the end of the crossover helix toward the protein core (near the ‘backside’ β -sheet) and is followed by two C-terminal helices, helix-4 (15.5 Å from active site) and helix-5 (Fig. 4b). Three distinct C-terminal clusters are evident in the ensemble, in which helix-4 occupies positions facing 1) closer to the 310-10 helix, 2) adjacent to the active site, and 3) closer to the ‘backside’ β -sheet (Fig 4c,d). In place of the final C-terminal helix present in canonical UBC folds, residues N136-W145 form a disordered region that occupies positions directly beneath the active site in all states of the ensemble, as revealed by the spin label attached to active-site C91 (Fig. 4e, Supplementary Fig. 13a). The final six amino acids are completely disordered and are not constrained to a particular region, consistent with {1H – 15N} hetNOE results.
The correlation between the experimental and back-calculated SAXS curves shows some disagreement (Supplementary Fig. 12f). Possible sources for the discrepancies are 1) presence of some aggregated material, 2) presence of low concentrations of dimeric Ube2w even with the dimer-disrupting mutation V30K/D67K, and 3) the aforementioned dynamics of the N-terminus. Nevertheless, the ensemble was generated from a combination of different experimental restraints and we believe it is an accurate representation of the predominant species in solution. In this respect, a recent crystal structure of an E2 from the fungi Agrocybe aegerita that shares 50% identity with Ube2w has both a non-canonical position for helix-4 that falls within our Ube2w ensemble and a disordered C-terminus (PDB: 3WE5)18. The average RMSD for all backbone atoms to the closest member of the Ube2w ensemble and this crystal structure is 2.55 Å over the entire protein sequence.
Ube2w C-terminus mediates substrate interactions
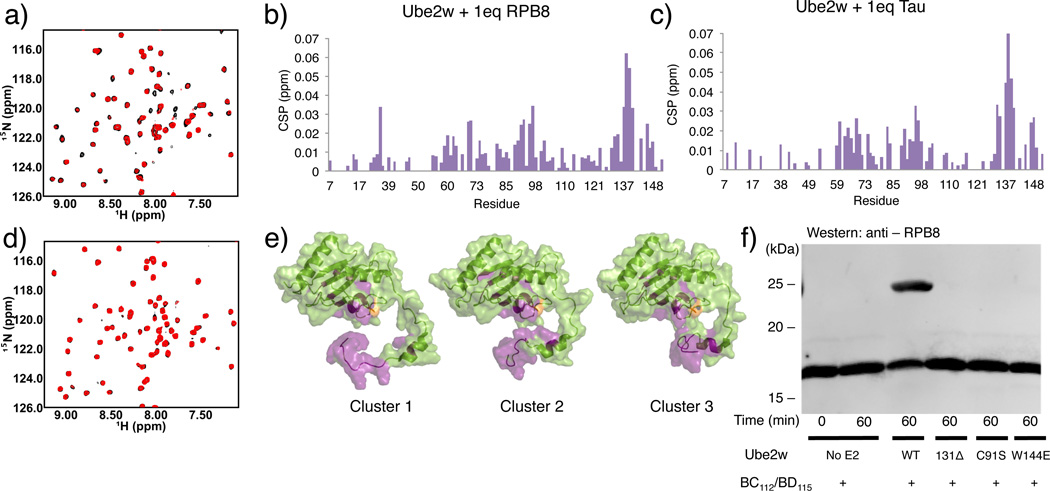
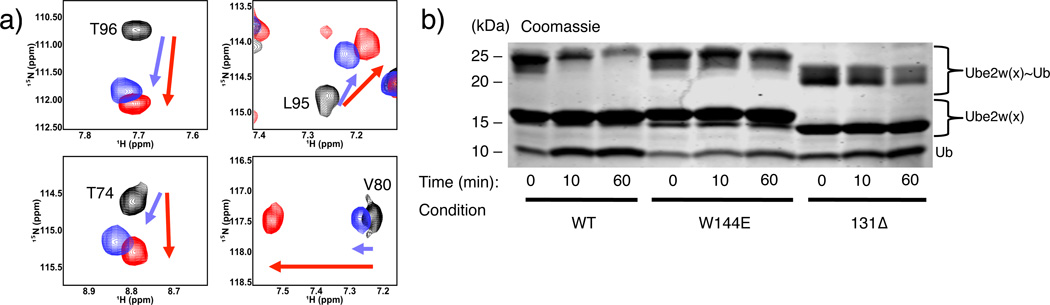
Multiple observations suggest that Ube2w plays a predominant role in mediating interactions with substrates. First, Ube2w shows robust Ub transfer activity (Fig. 1a left panel) in the presence of a minimal RING construct that offers no substrate binding functionality. Second, Ube2w adds Ub to the αN-terminus of substrates, in the absence of an E3, albeit at a slower rate (Supplementary Fig. 3) 9. RING-type E3s enhance the activity of other E2s by promoting closed E2~Ub conformations that promote Ub transfer21,28,29. Our results also demonstrate that Ube2w~Ub is allosterically activated by RING-type E3s to form closed conformations (Supplementary Fig. 2j,13). {1H – 15N} – HSQC-TROSY experiments capable of detecting low affinity binding interactions reveal peak broadening and chemical shift perturbations (CSPs) for a subset of 15N-Ube2w-KK resonances upon addition of the substrates RPB8 and tau (Fig. 5a,b,c Supplementary Fig. 14a). Based on the magnitude of the observed perturbations, these interactions are highly transient. Residues near the active site and within the C-terminal region are the most significantly perturbed upon addition of RPB8: Y131, K137, N138, K140, K141, K143, W144, and W145 in the C-terminus and S93, I94, L95, T96, and E97 in the 310-helix that immediately follows active site C91 (>1 standard deviation above/below mean for intensity loss/shifting, respectively). Intriguingly, all residues in the unstructured region positioned directly beneath the active site in the Ube2w ensemble (K137-W145) are significantly perturbed (Fig. 4e). C-terminally truncated Ube2w-131Δ-KK shows neither CSPs in NMR binding experiments with RPB8 nor Ub transfer activity, indicating that the C-terminus is essential for substrate recognition (Fig. 5d,e,f, Supplementary Fig. 2k). Finally, addition of tau, a Ube2w substrate with a different N-terminal sequence, perturbs a nearly identical set of residues in 15N-Ube2w-KK as does RPB8, suggesting side-chain identity plays, at most, a minimal role in Ube2w substrate recognition (Fig. 5c, Supplementary Fig. 14b,c).
Figure 5.
The Ube2w C-terminus is required to interact with substrates. (a) {1H – 15N} – HSQC-TROSY spectrum of Ube2w-KK in the absence (black spectrum) and presence of 1 molar equivalent of RPB8 (red spectrum). Evidence for binding is seen as peak broadening (loss of intensity) and chemical shift perturbations of specific peaks in the Ube2w NMR spectrum. (b) A histogram showing chemical shift perturbations (CSPs) upon 1 molar equivalent of RP8 into Ube2w-KK. (c) Titration of 1 molar equivalent of tau into Ube2w-KK reveals very similar CSPs to addition of RPB8. (d) {1H – 15N} – HSQC-TROSY spectrum of Ube2w-131Δ-KK in the absence (black spectrum) and presence of 1 molar equivalent of RPB8 (red spectrum). Truncated Ube2w shows no interaction with RPB8 (e) Residues whose resonances have significant intensity losses and/or CSPs (> 1 standard deviation) are mapped in purple onto members of the Ube2w ensemble (one representative from each cluster). f) In an in vitro ubiquitination assay, Ube2w-131Δ does not transfer Ub to RPB8 after 1hr. Mutation of a single residue in the C-terminal region, W144E, also abrogates detectable activity. The loss of activity associated with the C-terminal region is equivalent to an active site-dead mutant, C91S (Supplementary Fig. 2k).
Although multiple sequence alignment of the Ube2w C-terminus with other E2s shows considerable divergence, the C-terminal region is strongly conserved among Ube2w orthologs. Even the slime mold Dictyostelium discoideum displays significant conservation of C-terminal amino acids (R133, N136, K137, P139, W144, H147, D148, D149) (Supplementary Fig. 15a). Our NMR experiments show residues N136 through W145 represent an important substrate-binding region. W144, which occupies positions as close as 7 Å (average distance is 16.7 Å) to the active site in the NMR ensemble of Ube2w, is one of the most highly conserved amino acids among Ube2w orthologs, is positioned in a highly disordered region of the Ube2w ensemble (Fig. 4e, Supplementary Fig. 15a, b), and shows significant perturbations upon substrate binding (Fig 5b,c,e). Substituting a glutamate (the corresponding residue in the UbcH5c sequence) for Ube2w Trp144 (W144E) completely abrogates Ube2w activity towards four different substrates: RBP8, ataxin-3, tau, and CHIP (Fig. 5f, Supplementary Fig. 16a-d). The mutation has a two-fold effect. First, substrate binding is inhibited, evidenced by decreased peak broadening upon addition of RPB8 to the mutant protein in an NMR binding experiment (Supplementary Fig. 16e). Second, the W144E mutation generates NMR CSPs in Ube2w active site resonances that resemble those seen in the truncated Ube2w-131 Δ mutant (Fig. 6a). To probe the role of the Ube2w C-terminus in N-terminal ubiquitination function, we employed our intrinsic reactivity assay. Ube2w~Ub readily transfers its Ub to the minimal peptide substrate in a 1hr reaction but Ube2w-131 Δ and Ube2w-W144E show almost no transfer to the peptide (Fig. 6b, Supplementary Fig. 2l). The slow loss of the Ube2w-131 Δ~Ub conjugate during the experiment is due to hydrolysis of Ub and not aminolysis with the substrate (Supplementary Fig. 2m, 15c). Thus, without an intact C-terminus, Ube2w does not transfer Ub to a free α-amino group.
Figure 6.
The Ube2w C-terminus facilitates α-amino group reactivity. (a) Selected {1H – 15N} resonances of residues near the Ube2w active site are compared in the spectra of full-length WT (black), W144E-Ube2w, (purple), and Ube2w-131Δ (red). Resonances move along similar trajectories as a result of the W144 mutation or C-terminal ablation, indicating similar chemical environments for the affected residues. (b) Mutation or ablation of the C-terminus affects the intrinsic aminolysis activity of Ube2w. In a 1hr reaction WT Ube2w~Ub shows robust transfer activity towards peptide (NH2-A-G-G-S-Y-COO-; 30 mM) as seen by increased amounts of free Ube2w and free Ub. Ube2w-W144E~Ub and the Ube2w-131Δ~Ub mutants show almost no Ub transfer activity to this minimal substrate over the same time period (Supplementary Fig. 2l).
Discussion
To date, Ube2w is the only E2 demonstrated to attach Ub directly and specifically to the N-terminus of proteins. While it shares certain mechanistic features with lysine-reactive E2s, our results indicate that Ube2w is uniquely adapted to facilitate selective α-amino ubiquitination. Similar to UbcH5c, binding to a RING E3 shifts the population of Ube2w~Ub toward a closed conformation that facilitates aminolysis of the E2~Ub thioester. However, a highly flexible C-terminal region allows Ube2w to bind and ubiquitinate a diverse set of disordered N-termini. The Ube2w C-terminal region adopts multiple orientations in proximity to the E2 active site cysteine. Most E2s, including those that transfer SUMO or Nedd8 to lysine sidechains have a conserved Asn residue (N77 in UbcH5) in the loop immediately preceding the active site that is proposed to play both catalytic and structural roles for transfer to substrate lysines. Asn77 is thought to stabilize the oxyanion intermediate formed following lysine attack of the E2~Ub thioester30 and likely performs a structural role in helping to stabilize the loop preceding the active site31. Notably, Ube2w contains a histidine at this position, H83. Mutation of H83 to Asn (H83N or H94N in human Ube2w isoform 2) results in a marked decrease in the E3-enhanced N-terminal ubiquitination activity of Ube2w9, highlighting structural adaptations for N-terminal ubiquitination. Notably, Ube2w is unique among the ~40 human E2s by containing a histidine at this key position, strongly implicating it as the sole E2 with N-terminal ubiquitination activity.
Our work shows that Ube2w’s non-canonical, flexible C-terminal structure provides a platform for recognition of diverse substrates. We propose a mechanism in which Ube2w recognizes and binds to backbone groups in substrates, explaining the observed requirement for N-terminal disorder in substrates. This mechanism is supported by observations that Ube2w can 1) transfer Ub to the α NH2 group of a 5 amino acid peptide but not to the αNH2 group of a free amino acid and 2) create linear Ub chains using Ub that has at least four unstructured residues (Met-Gly3-Ub) appended to its N-terminus but not if those residues lack amide groups (Met-Pro3-Gly5-Ub). (Fig. 1b left, Fig. 2e). Poly-glycine lacks a side chain that could serve as a basis for recognition, implicating interaction via backbone atoms. The N-terminal sequences of proteins targeted by Ube2w are diverse and we have not yet identified any Ube2w sequence preferences, but all the sequences are either known to be or predicted to be disordered18–20 (Supplementary Fig. 7). A mechanism that involves substrate backbone atoms would require those groups to be available; adoption of secondary structure such as α -helices and β -strands by N-terminal residues would place these groups in intramolecular hydrogen bonds, making them unavailable for formation of necessary contacts for Ube2w-dependent N-terminal ubiquitination.
Ube2w has evolved a unique C-terminus whose relative orientation to active site residues appears to be crucial for Ube2w α NH2 ubiquitination function, as its removal (Ube2w-131Δ) or alteration (Ube2w-W144E) reduces substrate binding towards disordered N-termini, affects the environment of active site residues, decreases intrinsic aminolysis activity, and consequently, inhibits ubiquitination of substrates (Fig. 5d, f, 6 and Supplementary Fig. 16). Based on our ensemble each individual member shows clear active site accessibility for an incoming substrate. Remarkably, C-terminal residues N136-W145, which can occupy multiple positions directly beneath the site of catalysis, represent the primary substrate recognition surface. Because Ube2w must ubiquitinate diverse substrate sequences, this region may have evolved to adopt multiple conformations to sterically accommodate unstructured N-termini that harbor unique side chain identities.
There are numerous reports implicating N-terminal ubiquitination of specific proteins in cells. Perhaps the best-studied example is myogenic transcriptional switch protein (MyoD). Mutation of all lysine residues to arginine did not abrogate the protein’s ubiquitination or its degradation in COS-7 cells, while specific chemical modification of the α -amino group through carbamylation32 inhibited ubiquitination of MyoD33. Other substrates identified by similar strategies are human papillomavirus 16 oncoprotein, E7 (E7-16), latent membrane protein 1, and inhibitor of differentiation 234. In each case, truncation of the N-terminal region inhibited ubiquitination and degradation implying a role for mobility/flexibility in the N-terminus34. The extracellular signal-regulated kinase 3 (ERK3) has been identified as a target for N-terminal ubiquitination in HEK-293 cells and an N-terminal Ub on ERK3 can be further modified to create Ub chains that signal for its degradation35. Bulky tags such as a Myc6 or EGFP on the N-terminus of WT-ERK3 (which contains 45 lysines) inhibited its degradation in a proteasome-dependent manner, whereas smaller tags, such as HA or His6-, had no such effect35. A crystal structure of ERK3 indicates that the first ~10 residues are disordered (PDB: 2I6L). The N-terminal regions of all the above proteins are either known to be or are predicted to be disordered, consistent with the notion that they could be cellular Ube2w substrates18–20.
Identification of cellular Ube2w substrates is a critical step towards understanding the function of N-terminal ubiquitination. In eukaryotic cells, protein N-termini appear to be an important site of regulation. N-terminal acetylation is a prevalent post-translational modification that likely plays a competing role with N-terminal ubiquitination, as acetylation is irreversible and would preclude further modification by Ub. Reports suggest that 60-90% of cytosolic proteins may harbor an acetyl moiety on their N-termini36. We note that roughly 25% of proteins in the human proteome are predicted to have at least ten N-terminal unstructured residues. Additionally, post-translational proteolysis may expose new disordered N-termini in protein substrates. An intriguing feature of the Ube2w C-terminus is that it harbors a nuclear localization signal suggesting that it could play a role in the regulation of nuclear proteins37. Indeed, a majority of N-terminally-ubiquitinated proteins identified thus far are nuclear proteins. Another intriguing possibility for Ube2w substrates include nascent polypeptide chains on stalled ribosomes that may not contain any lysine residues.
Identification of N-terminally ubiquitinated substrates using existing proteomics approaches poses challenges. Current Ub antibody enrichment methods that utilize the diGly-antibody do not recognize substrates ubiquitinated at their N-termini. N-terminal processing of proteins in cells means that an antibody derived towards a single Ub-amino acid linkage (for example, Gly-Gly-Met) will not be sufficient to recognize all potential substrates. Therefore, new methods to elucidate the N-terminal ubiquitome are required. As a caveat, future studies focused on Ube2w should avoid use of N-terminally tagged-Ub or substrates that introduce N-terminal disorder as these can induce non-native Ube2w activity. In sum, Ube2w is unique among members of the ubiquitin-conjugating enzyme family in both its structural and biochemical properties. Insights revealed here can guide future efforts to identify bona fide in vivo substrates for Ube2w and to further elucidate its distinct cellular function.
Methods
Plasmids, Protein Expression
Ube2w constructs (WT, V30K/D67K (KK), 131Δ, 131Δ-KK, W144E, W144E-KK L110Q, C91S, C91S-KK, C91S/C119S/C151S/KK) were expressed from the pET24 vector without affinity tags. Ube2w plasmids were transformed into Escherichia coli (BL21 DE3) cells and protein expression was induced with 0.5mM iso-propyl-β -D-thio-galactoside (IPTG) at OD600 of 0.6, followed by growth for 16hr at 16°C. Cells were lysed by French press in 25mM sodium phosphate (pH 7.0), 1mM EDTA for full-length constructs and 25mM 2-(N-morpholino) ethanesulfonic acid (pH 6.0), 1mM EDTA for 131 Δ constructs. Following centrifugal clarification, Ube2w was applied to a cation exchange column with an elution gradient of 0-0.5M NaCl. E2-rich fractions were identified by UV absorbance, concentrated, pooled, and further purified by size exclusion chromatography on a Superdex 75 (GE Healthcare) column equilibrated in 25mM sodium phosphate (pH 7.0), 150mM NaCl. This buffer was used for all NMR experiements.
RPB8 (1-150) was expressed from the pET28a vector with a thrombin cleavable HIS6-tag. RPB8 plasmids were transformed into Escherichia coli (BL21 DE3) cells and protein expression was induced with 0.5mM iso-propyl-β -D-thio-galactoside (IPTG) at OD600 of 0.6, followed by growth for 16hr at 16°C. Cells were lysed by French press in 25mM Tris-HCl (pH 7.6), 200mM NaCl, 10mM imidazole and applied to a Ni2+-NTA gravity flow column (Invitrogen) equilibrated in the same buffer. The column was washed with 5 column volumes of 25mM Tris-HCl (pH 7.6), 200mM NaCl, 50mM imidazole and eluted with 25mM Tris-HCl (pH 7.6), 200mM NaCl, 500mM imidazole. The elution was subject to cleavage at 4°C overnight in 25mM Tris-HCl (pH 7.6), 200mM NaCl with 1mg thrombin from bovine serum (Sigma-Aldrich). Following cleavage the dialyzed sample was re-applied to a Ni2+-NTA gravity flow column. The flow-through was collected, concentrated, and further purified by size exclusion chromatography on a Superdex 75 (GE Healthcare) column equilibrated in 25mM sodium phosphate (pH 7.0), 150mM NaCl.
Wheat Uba1 and Ub were cloned, expressed, and purified as previously described38. Identical purification protocols for WT-Ub were followed for, Met-Gly-Ub, Met-Gly3-Ub, Met-Gly5-Ub, Met-Gly7-Ub, Ub-I44A, HA-Ub and HA-Ub(K0).
BC112/BD115 was expressed from the pET28N and pCOT7N expression systems (generous gift of Dr. M. Wittekind, Bristol-Myers Squibb). Proteins were purified as previously described39. E2’s UbcH5c, Ube2e1, and UbcH7 were cloned, expressed, and purified as previously described12.
Ataxin-3 was cloned, expressed and purified as previously described40. Tau protein was cloned, expressed, and purified as previously described41.
Full length CHIP and CHIP U-box were cloned, expressed, and purified as previously described42,43.
In vitro ubiquitination assays
Ubiquitination assays involving RPB8 or HA-Ub were conducted at 37°C for 1hr in 25mM sodium phosphate (pH 7.0), 150mM NaCl. Protein concentrations were as follows: 1uM Uba1, 4uM E2 enzyme, 4uM RPB8, 2uM BC112BD115, 30uM Ub or HA-Ub, and 10mM MgCl2. Reactions were initiated by addition of 5mM ATP. Samples were boiled in SDS buffer, loaded onto an SDS-PAGE, and visualized by western blotting with appropriate antibodies; anti-RPB8 (Abnova), anti-Ub (Santa Cruz).
Ubiquitination assays for tau, ataxin-3, and CHIP were typically performed for 1-5 min at 37°C in 10 µl mixtures containing buffer A (50mM Tris [pH 7.5], 50mM KCl, 0.2mM DTT), Ubmix (2.5mM ATP, 5mM MgCl2, 50nM Ube1, and 250µM ubiquitin), 1µM indicated E2, 1µM CHIP, and 1µM of indicated substrate. For CHIP independent ubiquitination reactions CHIP was omitted from the reaction, substrate concentration was increased to 20uM, and E2 concentration was 9uM unless otherwise indicated. Reactions were stopped by addition of SDS-Laemmli buffer and boiling, followed by separation of proteins by SDS-PAGE and visualization by western blotting with appropriate antibodies.
Nucleophile reactivity assays
Nucleophile reactivity assays were performed at 37°C in buffer containing 25mM sodium phosphate (pH 7.0), 150mM NaCl. Reactions contained 1uM Uba1, 20uM Ub, 20uM E2, 10mM MgCl2. Reactions were initiated with 5mM ATP and allowed for form E2~Ub conjugates for 30min. Nucleophile reactivity was induced by addition of 50mM free lysine or 50mM 5aa peptide (NH2-A-A-G-S-Y-COO-) and allowed to react for 1hr. Samples were collected in non-reducing SDS buffer and loaded onto a SDS-PAGE gel. Results were visualized by coommasie staining. Peptides were purchased (United Biosystems Inc).
Protein Identification by LC-MS/MS
Protein identification and ubiquitination sites on RPB8 were conducted based on previously described protocols9. Briefly, protein bands corresponding ubiquitinated substrate (as indicated) were excised and destained with 30% methanol for 4 h. Upon reduction (10 mm DTT) and alkylation (65 mm 2-chloroacetamide or iodoacetamide, with similar results) of the cysteines, proteins were digested overnight with sequencing grade, modified trypsin (Promega). Resulting peptides were resolved on a nano-capillary reverse phase column (Picofrit column, New Objective) using a 1% acetic acid/acetonitrile gradient at 300 nl/min and directly introduced into a linear ion-trap mass spectrometer (LTQ Orbitrap XL, Thermo Fisher). Data-dependent MS/MS spectra on the five most intense ions from each full MS scan were collected (relative Collision Energy 35%). Proteins were identified by searching the data against Human International Protein Index database (version 3.5) appended with decoy (reverse) sequences using the X!Tandem/Trans-Proteomic Pipeline (TPP) software suite. All peptides and proteins with a PeptideProphet and ProteinProphet probability score of >0.9 (false discovery rate <2%) were considered positive identifications and manually verified.
NMR Spectroscopy
All NMR samples were prepared in 25mM sodium phosphate (pH 7.0), 150mM NaCl using either 90% H2O/D2O or 100% D2O. Samples for Ube2w-KK utilized either uniformly 15N or 15N, 13C-labeled protein at concentrations from 400uM to 200uM. Titration experiments involving 15N – Ube2w-KK were performed by equimolar addition of unlabeled RPB8 or tau. The magnitude of chemical shift perturbations for each resonance was quantified in Hz according to the equation Δδj=((Δδj15N)2 + (Δδj 1H)2)1/2. Data collection for resonance assignments utilized standard three-dimensional NMR techniques44 collected on INOVA 600 and 800 MHz spectrometers (Varian) at Pacific Northwest National Labs (PNNL). All other NMR-based experiments (substrate titrations, {1H – 15N} hetNOEs, T1,T2 relaxation experiments, {1H – 15N} RDC measurements, paramagnetic spin labeling experiments) were collected on a 500 MHz Bruker Avance II (University of Washington). All spectra were collected at 25°C. Data were processed using NMR-Pipe/NMRDRaw45 and visualized with NMRView46.
Spin-Label Modification
A mutant form of Ube2w (C91S, C119S, C151S, V30K, D67K) was generated by site-directed mutagenesis to incorporate a single cysteine chemical modification at position C135. A similar mutant (C119S, C135S, C151S, V30K, D67K) was created for the C91 modification. The thiol-reactive relaxation probe 4-(2-iodoacetamido)-TEMPO (Sigma-Aldrich) was mixed at a 1:5, Ube2w:TEMPO molar ratio for 2hrs at 30°C. Reaction yields were quantified by MALDI-MS. Only those that reacted to completion (>95%) were utilized for spin-label experiments. Unreacted probe was cleared by dialysis overnight at 4°C. Identical {1H –15N} – HSQC-TROSY experiments were conducted in the presence and absence of the probe reducing agent, ascorbate.
Residual Dipolar Couplings (RDCs)
Pf1-phage (ALSA Biotech) was added to 20% in a solution containing 250uM 15N-Ube2w-KK in 25mM sodium phosphate (pH 7.0), 150mM NaCl. This resulted in precipitation of the protein around the phage. An additional 200mM NaCl was added in order dissolve the precipitant. The sample was centrifuged to remove bubbles. The final solution contained 10mg/ml phage and 350uM NaCl.
Small-Angle X ray Scattering (SAXS)
SAXS data were collected at Stanford Synchrotron Radiation Lightsource beamline 4-2. Data were collected for Ube2w at concentrations of 10, 5, and 0.5 mg/mL in 25mM sodium phosphate (pH 7.0), 150mM NaCl, and 2mM DTT at 25°C. SAXS statistics were calculated using the EMLB CRYSOL47 server.
CS-Rosetta structure calculation for Ube2w
Monomeric structures of Ube2w were generated from NMR data in combination with homologous structural information. The standard CS-Rosetta method is used to derive fragments from the sequence profile, predicted secondary structure, and backbone and Cβ chemical shift data24,25,48. Interatomic restraints are derived from alignments of Ube2w sequence to templates49. The computational protocol includes a low-resolution stage and a high-resolution stage. In the low-resolution step, side-chains are represented using a single, residue-specific pseudo-atom, positioned at the Cβ carbon. From an extended chain, structures are assembled by fragment insertion under a force field that favors compactness and formation of secondary structures. In the high-resolution stage, side-chains and hydrogen atoms are explicitly represented and the structures are optimized using the Rosetta full-atom energy function. In both stages of conformational sampling, Rosetta energy function is augmented with penalty term related to the homologous structural and experimental N-H RDC restraints. Backbone chemical shift perturbation data (residues 72, 78, 85, 86, 87, 95, 96, 97, 124 and 131) derived upon deletion of residues 132-151 was also included as ambiguous distance constraints during sampling and refinement. A total of 16,000 models are generated. Models with top 10% lowest Rosetta energy, and 25% lowest homologous constraint energy and 25% lowest RDC energy were selected for further analysis. Paramagnetic broadening data from spin labels at residues C91 and C135 and SAXS are used to rank the top models. Pearson correlation is calculated between experimental data and distance from paramagnetic center to HN atom. Distances were calculated by explicitly adding spin label to each model at residue 135 or 91. The top 20 models with best correlation with paramagnetic quenching and SAXS data were chosen as the final ensemble. Structural statistics were calculated using the Protein Structure Validation Server suite (PSVS)50. Favorable Ramachandran statistics were observed, with 88.9% of residues in most favored regions, 10.6% in additionally allowed regions, 0.5% in generously allowed regions, and 0% in disallowed regions.
Statistical Analysis
Pearson correlations were calculated by standard methods. The Pearson correlation is the standard product-moment correlation coefficient that describes the linear correlation between two variables.
Q factor for RDCs reflects the agreement between calculated Qcalc and experimental Qobs dipolar couplings as: Q = rms(Dcalc − Dobs)/{Da2[4 + 3R2]/5}1/2, where Da and R refer to the magnitude and rhombicity of the alignment tensor, respectively51.
SAXS agreement is calculated using the EMBL Crysol server, which evaluates X-ray solution scattering curves from atomic models. Chi describes the discrepancy between theoretical and experimental curves47.
Supplementary Material
Acknowledgements
We acknowledge D. Christensen and C. Eakin for their initial observations on Ube2w and J.N. Pruneda for collecting SAXS data on Ube2w. This work was supported by National Institute of General Medical Sciences grants R01 GM088055 (REK), R01 GM098503 (PSB), K99 NS073936 (KMS), R01 AG034228 (HLP), UW Hurd Fellowship Fund, and PHS NRSA 2T32 GM007270 (VV).
Footnotes
Author contributions
V.V., P.S.B. and R.E.K. conceived the experiments and wrote the manuscript. V.V performed the biochemical and structural experiments with help from K.M.S and E.D.D. D.M.W performed the initial characterization of Ube2w. V.B. and K.S.J.E.J. performed the mass spectrometry. L.S. and D.B. performed the structure calculations. H.J.P. provided guidance. R.E.K supervised the project.
Competing financial interests
The authors declare no competing financial interests.
Accession Codes
PDB. The solution ensemble of full-length Ube2w was deposited under the code 2MT6. The NMR assignments were deposited to the BMRB under the accession code 25150.
References
- 1.Pickart CM. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:195–201. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 2.Deng L, et al. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z, Pickart CM. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. J. Biol. Chem. 1990;265:21835–21842. [PubMed] [Google Scholar]
- 4.Brzovic PS, Klevit RE. Ubiquitin Transfer from the E2 Perspective: why is UbcH5c so promiscuous? Cell Cycle. 2006;24:2867–2873. doi: 10.4161/cc.5.24.3592. [DOI] [PubMed] [Google Scholar]
- 5.Nuber U, Schwarz S, Kaiser P, Schneider R, Scheffner M. Cloning of human ubiquitin-conjugating enzymes UbcH6 and UbcH7 (E2-F1) and characterization of their interaction E6-AP and RSP5. J. Biol. Chem. 1996;271:2795–2800. doi: 10.1074/jbc.271.5.2795. [DOI] [PubMed] [Google Scholar]
- 6.Machida YJ, et al. UBE2T Is the E2 in the Fanconi Anemia Pathway and Undergoes Negative Autoregulation. Mol. Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 7.McDowell GS, Kucerova R, Philpott A. Non-canonical ubiquitylation of the proneural protein Ngn2 occurs in both Xenopus embryos and mammalian cells. Biochem. Biophys. Res. Commun. 2010;400:655–660. doi: 10.1016/j.bbrc.2010.08.122. [DOI] [PubMed] [Google Scholar]
- 8.Vosper JM, et al. Ubiquitylation on canonical and non-canonical sties targets the transcription factor neurogenin for ubiquitin-mediate proteolysis. J. Biol. Chem. 2009;284:15458–15468. doi: 10.1074/jbc.M809366200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scaglione KM, et al. The ubiquitin-conjugating enzyme (E2) Ube2w ubiquitinates the N terminus of substrates. J. Biol. Chem. 2013;288:18784–18788. doi: 10.1074/jbc.C113.477596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tatham MH, Plechanovová A, Jaffray EG, Salmen H, Hay RT. Ube2W conjugates ubiquitin to α-amino groups of protein N-termini. Biochem. J. 2013;453:137–145. doi: 10.1042/BJ20130244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu W, et al. BRCA1 ubiquitinates RPB8 in response to DNA damage. Cancer Res. 2007;67:951–958. doi: 10.1158/0008-5472.CAN-06-3187. [DOI] [PubMed] [Google Scholar]
- 12.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 13.Guzzo CM, et al. RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to site of DNA damage. Sci. Signal. 2012;5:1–7. doi: 10.1126/scisignal.2003485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. UBE2W interacts with FANCL and regulates the monoubiquitination of Fanconi anemia protein FANCD2. Mol. Cells. 2011;31:113–122. doi: 10.1007/s10059-011-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic Insight into Site-Resictricted Monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol. Cell. 2008;32:767–777. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Wand AJ, Urbauer JL, McEvoy RP, Beiber RJ. Internal dynamics of human ubiquitin revealed by 13C-relaxation studies of randomly fractionally labeled protein. Biochemistry. 1996;35:6116–6125. doi: 10.1021/bi9530144. [DOI] [PubMed] [Google Scholar]
- 18.Berman HM, et al. The Protein Data Bank. Nuc. Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelley LA, Sternberg MJ. E. Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 20.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 21.Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimsley GR, Scholtz JM, Pace CN. A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci. 2009;18:247–251. doi: 10.1002/pro.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheng Y, et al. A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Mol Cell Proteomics. 2012;11:329–341. doi: 10.1074/mcp.O111.013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vittal V, Wenzel DM, Brzovic PS, Klevit RE. Biochemical and structural characterization of the ubiquitin-conjugating enzyme UBE2W reveals the formation of a noncovalent homodimer. Cell Biochem. Biophys. 2013;67:103–110. doi: 10.1007/s12013-013-9633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laskowski RA. PDBsum new things. Nuc. Acids. Res. 2009;37:D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Y, et al. Consistent blind protein structure generation from NMR chemical shit data. Proc. Natl. Acad. Sci USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen Y, Vernon R, Baker D, Bax A. De novo protein structure generation from incomplete chemical shift assignments. J. Biol. NMR. 2009;43:63–78. doi: 10.1007/s10858-008-9288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruneda JN, et al. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu PY, et al. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berndsen CE, Wiener R, Yu IW, Ringel AE, Wolberger C. A conserved asparagine has a structural role in ubiquitin-conjugating enzyme. Nat. Chem. Biol. 2013;9 doi: 10.1038/nchembio.1159. 154-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hershko A, Heller H, Eytan E, Kaklij G, Rose IA. Role of the α-amino group of protein in ubiquitin-mediated protein breakdown. Proc. Natl. Acad Sci. USA. 1984;81:7021–7025. doi: 10.1073/pnas.81.22.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breitschopf K, Bengal E, Ziz T, Admon A, Ciechanover A. A novel site of ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 1998;17:5964–5973. doi: 10.1093/emboj/17.20.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. Review. [DOI] [PubMed] [Google Scholar]
- 35.Coulombe P, Rodier G, Bonneil E, Thibault P, Meloche S. N-Terminal ubiquitination of extracellular signal-regulated kinase 3 and p21 directs their degradation by the proteasome. Mol. Cell. Biol. 2004;24:6140–6150. doi: 10.1128/MCB.24.14.6140-6150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dormeyer W, Mohammed S, Breukelen B, Krijgsveld J, Heck AJ. Targeted analysis of protein termini. J. Proteome Res. 2007;6:4634–4645. doi: 10.1021/pr070375k. [DOI] [PubMed] [Google Scholar]
- 37.Yin G, et al. Cloning, characterization and subcellular localization of a gene encoding a human Ubiquitin-conjugating enzyme (E2) homologous to the Arabidopsis thaliana UBC-16 gene product. Front. Biosci. 2006;11:1500–1507. doi: 10.2741/1899. [DOI] [PubMed] [Google Scholar]
- 38.Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enyzmol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 39.Brzovic PS, et al. Binding and recognition oin the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. USA. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todi SV, et al. Cellular turnover of the polyglutamine disease protein ataxin-3 is regulated by its catalytic activity. J. Biol. Chem. 2007;282:29348–29358. doi: 10.1074/jbc.M704126200. [DOI] [PubMed] [Google Scholar]
- 41.Barghorn S, Biernat J, Mandelkow E. Amyloid Proteins: Methods and Protocols. Methods Mol. Biol. 2004;299:35–51. doi: 10.1385/1-59259-874-9:035. [DOI] [PubMed] [Google Scholar]
- 42.Winborn BJ, et al. The deubiquitinating denzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 2008;283:26436–26443. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Z, et al. Structure and interactions of the helical and U-box domains of CHIP, the C terminus of HSP70 interacting protein. Biochemistry. 2006;45:4749–4759. doi: 10.1021/bi0601508. [DOI] [PubMed] [Google Scholar]
- 44.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiements for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nuc. Mag. Res. Spec. 1999;34:93–158. [Google Scholar]
- 45.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 46.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 47.Svergun D, Barberato C, Koch MH. J. CRYSOL: A Program to Evaluate X-ray Solution Scatter of Biological Macromolecules from Atomic Coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 48.Raman S, et al. NMR structure determination for larger proteins using backbone-only data. Science. 2010;327:1014–1018. doi: 10.1126/science.1183649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson JM, et al. Accurate protein structure modeling using sparse NMR data and homologous structure information. Proc. Natl. Acad. Sci. USA. 2012;109:9875–9880. doi: 10.1073/pnas.1202485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhattacharya A, Tejero R, Gaetano MT. Evaluating protein structures determined by structural genomics consortia. Proteins. 66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 51.Ulmer TS, Ramirez BE, Delaglio F, Bax A. Evaluation of Backbone Proton Positions and Dynamics in a Small Protein by Liquid Crystal NMR Spectroscopy. J. Am. Chem. Soc. 125:9179–9191. doi: 10.1021/ja0350684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.