BRCA1 expression can be lost through a germline or somatic mutation or through promotor hypermethylation. Detection of BRCA1 loss in ovarian cancer has potential prognostic and therapeutic significance. Our data indicate that BRCA1 immunohistochemistry alone is not sufficient to screen for BRCA1 germline mutations but is a reproducible and accurate means of identifying BRCA1 loss by any mechanism.
Keywords: BRCA1, immunohistochemistry, genetic testing, ovarian cancer, biomarkers, hypermethylation
Abstract
Background
BRCA1 expression can be lost by a variety of mechanisms including germline or somatic mutation and promotor hypermethylation. Given the potential importance of BRCA1 loss as a predictive and prognostic biomarker in high-grade serous ovarian cancer, we sought to evaluate the utility of BRCA1 immunohistochemistry (IHC) in screening for BRCA1 loss by germline, somatic, and epigenetic mechanisms.
Patients and methods
Patients with advanced high-grade serous ovarian cancer who had previously undergone germline BRCA1 testing were identified. Samples from each tumor were stained for BRCA1 and reviewed independently by two pathologists blinded to BRCA status. Tumors with abnormal BRCA1 IHC and wild-type germline testing underwent further evaluation for somatic BRCA1 mutations and promoter hypermethylation. McNemar's test was used to determine the association of BRCA1 IHC with germline BRCA1 mutations and BRCA1 loss through any mechanism. Kaplan–Meier methods were used to estimate overall survival (OS), and the log-rank test was used to assess differences between groups.
Results
Inter-rater reliability between the two pathologists on BRCA IHC interpretation was very good (kappa coefficient 0.865, P = 0.16; McNemar's test). BRCA1 IHC was abnormal in 36% (48/135) of cases. When compared with germline BRCA1 status, BRCA1 IHC had a high negative predictive value (95.4%) but a low positive predictive value (PPV, 52.1%). When accounting for promoter hypermethylation and somatic mutations as alternative methods of BRCA1 loss, the PPV rose to 87.5%. Five-year OS rate was 49.6% [95% confidence interval (CI) 26.3% to 69.3%] for patients with germline BRCA1 mutations, 50.4% (95% CI 27.5% to 69.5%) for germline wild-type BRCA1 and abnormal IHC, and 52.1% (95% CI 38.4% to 64.2%) for germline wild-type BRCA1 and normal IHC (P = 0.92).
Conclusions
BRCA1 IHC interpretation was a highly reproducible and accurate modality for detecting germline, somatic, or epigenetic mechanisms of BRCA1 loss. These results support further development of BRCA1 IHC as a potential biomarker for BRCA1 loss in high-grade serous ovarian cancer.
introduction
BRCA1 germline mutations confer an improved prognosis in high-grade serous ovarian cancer through a variety of mechanisms [1, 2]. The absence of intact homologous recombination DNA repair due to loss of BRCA1 function renders these cancers sensitive to agents that cause double-stranded DNA breaks, such as platinum compounds [3–5]. More recently, poly(ADP) ribose polymerase (PARP) inhibitors have also shown promising activity in BRCA1 germline mutant tumors [6, 7]. It is not known, however, whether other common mechanisms of BRCA1 loss, including somatic mutation or promoter hypermethylation [8], confer a similarly favorable prognosis or improved sensitivity to DNA damaging agents or PARP inhibitors.
The lack of widely available means of identifying BRCA1 somatic mutations and promoter hypermethylation in the clinic has hampered the ability to understand the clinical significance of these alterations. Even as somatic tumor sequencing becomes more commonplace, predicting the functional impact of low-frequency mutations on BRCA1 protein expression can be challenging. A straightforward method to directly identify BRCA1 protein loss would be potentially useful in prognostication, stratification for clinical trials, and selection among standard and investigational therapies for recurrent ovarian cancer. To address this unmet need, we evaluated the utility of BRCA1 immunohistochemistry (IHC), an inexpensive and widely available technique, in screening for BRCA1 loss by germline, somatic, and epigenetic mechanisms.
methods
patient selection
Institutional Review Board approval was obtained for this analysis. Eligible patients were seen at Memorial Sloan Kettering Cancer Center (MSK) between 1 August 1996 and 1 August 2010 for newly diagnosed stage III or IV high-grade serous ovarian, fallopian tube, or primary peritoneal cancer. All diagnoses were confirmed by two expert gynecologic pathologists. All patients had consented to BRCA1 mutation testing on one of two IRB-approved studies being conducted by the Clinical Genetics Service [9]. Patients who did not have tumor specimens available at MSK for analysis were excluded. To minimize ascertainment bias, patients whose first visit to MSK occurred >6 months after their date of diagnosis were also excluded from the survival analysis. All BRCA1 mutations were predicted to be deleterious. Patients with variants of unknown significance were considered to be BRCA1 wild-type.
BRCA immunohistochemistry
A triplicate tissue microarray containing samples from each tumor was prepared and stained for BRCA1 using previously published methods [10]. MS110 from Calbiochem (EMD Millipore, Billerica, MA), Catalog number OP92, was used. The epitope for this antibody is 304 amino acids from the N-terminus of BRCA1. Each IHC stain was reviewed independently by two pathologists blinded to BRCA status of each tumor, and scored as absent, equivocal, or retained using the following criteria:
Loss: <5% of tumor nuclei staining, positive internal control (Figure 1A).
Equivocal: 5%–10% of tumor cell nuclei staining, less intense compared with the positive internal control (Figure 1B).
Retained: >10% of tumor cell nuclei staining, positive internal control or >5% when staining intensity of tumor cell nuclei is similar to the internal control (Figure 1C).
Figure 1.

Representative BRCA1 IHC stains and Interpretation. Low and high power images of loss (A), equivocal (B), and retained (C) BRCA1 IHC stains based on percent of tumor nuclei staining (green arrow). Loss and equivocal samples were considered abnormal and retained samples normal. Stroma (red arrow) and tumor infiltrating lymphocytes (black arrow) were used as positive controls.
The stromal cells serve as an internal positive control of the validity of the stain, since they retain a normal copy of BRCA1 even if tumor cells do not. If BRCA1 was retained, no further sections were obtained. If BRCA1 was scored absent or equivocal, a whole section was prepared and a final score obtained using the same procedure. This was done to avoid false negatives associated with patchy BRCA1 IHC staining. Of note, even tumors with retained staining do not typically show diffuse staining of strong intensity; rather, staining is often of moderate to strong intensity and limited in distribution to 30%–60% of tumor cells, usually in a patchy rather than a geographic pattern.
Tumors with retained staining were considered ‘normal’ and those with absent or equivocal staining were considered ‘abnormal’. To evaluate whether BRCA1 IHC may have utility as a dynamic biomarker, BRCA1 IHC stains were repeated in recurrent tissue (when available) from patients with abnormal BRCA1 IHC in pretreatment tissue and correlated to platinum sensitivity status.
BRCA1 somatic sequencing and promoter hypermethylation testing
Tumors with abnormal BRCA1 IHC and wild-type germline testing underwent further testing to assess for somatic BRCA1 mutations and BRCA1 DNA promoter hypermethylation. DNA was extracted from microdissected formalin-fixed, paraffin-embedded tumor and normal tissues according to standard laboratory protocols. All tumors contained a minimum of 50% tumor cell nuclei. We used a custom target capture deep sequencing assay to perform massively parallel sequencing across the entire coding region of BRCA1. Paired normal and tumor samples were sequenced to a median depth of 272× with 96.5% of the targeted sequence in BRCA1 covered at 100× or greater. Custom oligonucleotide probes were designed to capture all protein-coding exons and splice regions. Captured regions were sequenced on an Illumina HiSeq 2000 (Illumina, San Diego, CA) and reads were aligned to the reference human genome (hg19). All candidate mutations were manually reviewed using the Integrative Genomics Viewer [11].
A CLIA-approved pyrosequencing assay was used for the simultaneous analysis and quantification of the degree of methylation at 11 CpG sites in the established promoter region of BRCA1 [12]. Bisulfite treatment of genomic DNA samples was used in the hydrolytic deamination of nonmethylated cytosines to uracils, whereas methylated cytosines are resistant to conversion. After a PCR, the methylation status at a given position is manifested in the ratio C (former methylated cytosine) to T (former nonmethylated cytosine) translating epigenetic information into sequence information and can be analyzed in the bisulfite-treated DNA. With each run of the BRCA1 methylation assay, positive, negative, and no template controls are included. The assay was performed once and not in replication. The degree of methylation is calculated as allele frequency using the following formula:
The average methylation fraction across 11 CpG sites is reported as positive if the mean methylation is between 10% and 99% and negative if it is <10%. The percent of methylation in normal and hypermethylated cases is provided in supplementary Figure S1, available at Annals of Oncology online.
statistical methods
McNemar's test was used to determine the association of BRCA1 IHC with BRCA1 germline test results as well as the association of BRCA1 IHC with BRCA1 loss through any mechanism. The agreement between pathologists was assessed via the kappa statistic. Overall survival (OS) was defined from the diagnosis date to the last follow-up date or the death date for all patients. Kaplan–Meier methods were used to estimate OS and the log-rank test was used to assess differences between patients (i) with germline BRCA1 mutations and (ii) without germline mutations who had (i) normal and (ii) abnormal IHC. Variables were regarded as significant at a level of 0.05.
results
Patient and disease characteristics describing the 135 patients analyzed are reported in Table 1. All patients had stage III or IV high-grade serous disease. Just over 20% harbored BRCA1 germline mutations. Approximately 90% of tumor specimens were from initial surgical resection. Of these cases, 14% had received neoadjuvant chemotherapy. The ovary or fallopian tube specimens were used in 84% of cases and a variety of metastatic sites for the remainder.
Table 1.
Patient and disease characteristics (N = 135)
| Characteristic | Number | Percent |
|---|---|---|
| Age at diagnosis | ||
| Median (mean) | 59 (57.69) | |
| Range | 32–82 | |
| Stage | ||
| IIIB | 5 | 3.7 |
| IIIC | 99 | 73.3 |
| IV | 31 | 23 |
| Optimally debulked | ||
| Yes | 102 | 77.9 |
| No | 29 | 22.1 |
| Intraperitoneal chemo | ||
| Yes | 60 | 47.2 |
| No | 67 | 52.8 |
| BRCA1 germline | ||
| Wild-type | 106 | 78.5 |
| Mutant | 29 | 21.5 |
| Tissue stained | ||
| Primary tumor, chemo-naïve | 103 | 76.3 |
| Primary tumor, chemo-treated | 17 | 12.6 |
| Recurrent tumor | 15 | 11.1 |
| Tumor site stained | ||
| Ovary/tube | 114 | 84.4 |
| Other | 21 | 15.6 |
| BRCA1 IHC result | ||
| Abnormal | 48 | 35.6 |
| Normal | 87 | 64.4 |
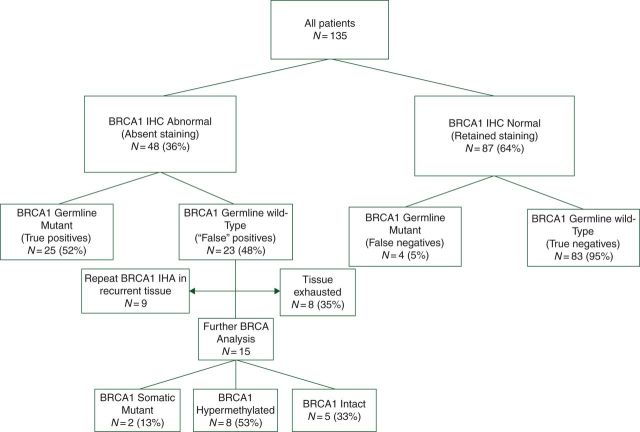
Inter-rater reliability between the two pathologists on final BRCA IHC interpretation was very good (kappa coefficient 0.865, P = 0.16; McNemar's test). The results of BRCA1 IHC testing are shown in Figure 2, and the performance of BRCA1 IHC testing with respect to BRCA1 germline status and BRCA1 loss by any mechanism is shown in Table 2. When compared with germline BRCA1 status, BRCA1 IHC had a high negative predictive value (95.4%) but a low positive predictive value (PPV, 52.1%). However, when accounting for promoter hypermethylation and somatic mutations as alternative mechanisms of BRCA1 loss, the PPV of BRCA1 IHC rose to 87.5% and the overall correct classification rate was 92.7%. BRCA1 staining characteristics were similar in germline and somatic mutant patients.
Figure 2.
Consort diagram.
Table 2.
Performance of BRCA IHC by mechanism of loss
|
BRCA1 loss (germline only) |
BRCA1 loss (germline/somatic/methylation) |
|||
|---|---|---|---|---|
| No | Yes | No | Yes | |
| BRCA1 IHC results | ||||
| Normal | 83 | 4 | 83 | 4 |
| Abnormal | 23 | 25 | 5 | 35 |
| Rate | 95% CI | Rate | 95% CI | |
| BRCA1 IHC performance | ||||
| Sensitivity | 86.2% | 73.7% to 98.8% | 89.7% | 80.2% to 99.3% |
| Specificity | 78.3 | 70.5% to 86.2% | 94.3% | 89.5% to 99.2% |
| PPV | 52.1% | 38.0% to 66.2% | 87.5% | 77.3% to 97.8% |
| NPV | 95.4% | 91.0% to 99.8% | 95.4% | 91.0% to 99.8% |
| OCCR | 80.0% | 73.3% to 86.8% | 92.9% | 88.5% to 97.4% |
PPV, positive predictive value; NPV, negative predictive value; OCCR, overall correct classification rate.
Figure 3 depicts the result of BRCA1 IHC testing by location and mechanism of BRCA1 loss. There was no observable relationship between the location of BRCA1 mutation and the BRCA IHC result. Recurrent tissue was available for nine germline wild-type patients who had abnormal BRCA IHC staining in the primary tumor specimen. Seventy-eight percent (7/9) had reverted to normal BRCA1 IHC staining in the recurrent specimen and 86% (6/7) of these cases were resistant to platinum therapy (platinum-free interval ≤12 months).
Figure 3.
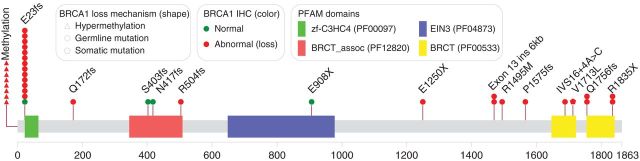
BRCA1 mutation and methylation map. E23fs is the 185delAG founder mutation. Q1756fs is the 5382/5385 insC founder mutation. One patient with multiple somatic BRCA1 mutations was not mapped.
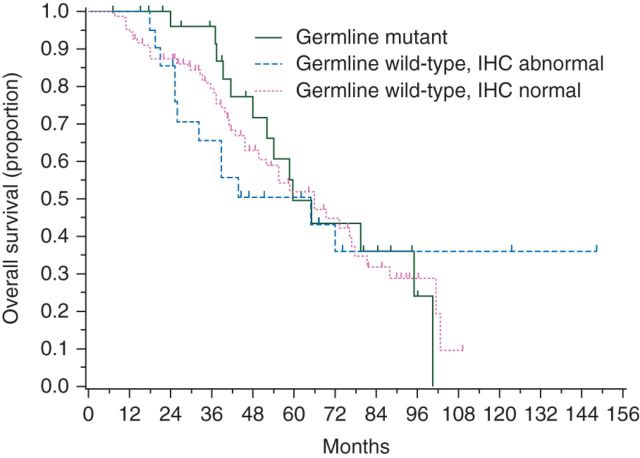
For the OS analysis, five patients were excluded because the interval from diagnosis to first evaluation at MSK was >6 months. The results for the OS analysis are shown in Table 3. Of the remaining 130 patients, 67 (51.5%) died of disease. The median duration of follow-up was 44.6 months (range: 7.2–148.3 months) for the 63 survivors. The 5-year OS rate was 49.6% [95% confidence interval (CI) 26.3% to 69.3%] for patients with germline BRCA1 mutations, 50.4% (95% CI 27.5% to 69.5%) for germline wild-type BRCA1 and abnormal IHC, and 52.1% (95% CI 38.4% to 64.2%) for germline wild-type BRCA1 and normal IHC. Differences between the three groups did not reach statistical significance (P = 0.92). The Kaplan–Meier curves for OS, stratified by BRCA1 germline and IHC status, are shown in Figure 4.
Table 3.
Overall survival (OS) by BRCA1 status
| BRCA1 status | N | Deaths | 5-year OS rate (95% CI) | Median OS months (95% CI) | HR (95% CI)* |
|---|---|---|---|---|---|
| Germline mutant | 28 | 14 | 49.6% (26.3% to 69.3%) | 59.5 (48.2–95.0) | Ref. level |
| Germline WT, IHC abnormal | 21 | 12 | 50.4% (27.5% to 69.5%) | 64.8 (25.9–NE) | 1.13 (0.52–2.46) |
| Germline WT, IHC normal | 81 | 41 | 52.1% (38.4% to 64.2%) | 65.9 (45.8–78.0) | 1.13 (0.62–2.08) |
*P = 0.918, obtained by using log-rank test.
Figure 4.
Overall survival by BRCA1 status.
discussion
In this comprehensive study of BRCA1 immunohistochemical testing in high-grade, advanced-stage, ovarian serous carcinomas, we found that this is an effective method to identify BRCA1 loss through both genetic and epigenetic mechanisms. When considering both mechanisms of BRCA1 loss, BRCA1 IHC correlation was excellent, with an overall correct classification rate of 93%. Our findings are consistent with, and expand upon, previous smaller case series of BRCA1 IHC testing performed to date [10, 13–15].
At present, BRCA1 germline testing is the only form of BRCA1 assessment routinely offered to patients with ovarian cancer. BRCA1 IHC identified 86% of patients with a BRCA1 germline mutation and therefore is not accurate enough to be used as prescreening before germline testing; however, our data indicate that in conjunction with routine BRCA1 germline testing, BRCA IHC may provide a reliable means of identifying patients with nongermline mechanisms of BRCA1 loss. As the clinical significance of somatic mutations and promoter hypermethylation is further ascertained, IHC could become useful as a companion to germline testing.
The results of BRCA1 staining were highly reproducible among pathologists blinded to the underlying BRCA1 status of each patient. Our two pathologists reached agreement in 126 of 134 (94%) cases. Moreover, in the eight cases where agreement was not reached, the disagreement was between absent versus equivocal staining. Therefore, none of the discrepancies involved scores that would have altered the final interpretation of the result (i.e. normal versus abnormal). Further studies will be necessary to determine whether similar inter-rater reliability would exist across institutions or among pathologists with less experience with BRCA1 IHC.
We did not find a difference in OS between BRCA1 mutant, BRCA1 wild-type/IHC normal, and BRCA1 wild-type/IHC abnormal patients. Due to the small numbers of patients in each cohort and the convenience sample utilized, this analysis must be considered exploratory. The relatively small survival advantage conferred by BRCA1 germline mutations necessitates a very large cohort study to detect a statistically significant difference compared with sporadic ovarian cancers [1]. This suggests that, if BRCA1 somatic mutation or promoter hypermethylation conferred a similar survival advantage, a much larger population would be needed to detect this difference. Still, our results are consistent with a related analysis carried out and reported by The Cancer Genome Atlas [8]. Taken together, these data suggest that different mechanisms of BRCA1 loss may be associated with unique disease phenotypes. Although other investigators have reported an association between BRCA1 protein expression and outcome in ovarian carcinoma, these reports have been somewhat limited by the absence of universal BRCA1 germline testing and other assessments of BRCA1 inactivation [16–18].
Unlike BRCA1 germline sequencing, BRCA1 IHC may have utility as a dynamic biomarker throughout the disease course as methylation status changes or secondary gain-of-function mutations accumulate. Although our analysis was limited by the small number of recurrent samples, we observed that seven of eight patients—six of whom had developed at least intermediate resistance to platinum therapy—regained BRCA1 function in recurrent specimens, consistent with preclinical observations of restoration of BRCA1 function upon development of platinum resistance [3, 4]. This preliminary analysis suggests that BRCA1 IHC testing may be a useful biomarker for clinical trial selection or stratification, particularly in the setting of recurrent disease.
The importance of developing a real-time clinical test for nongermline mechanisms of BRCA1 loss is heightened by the recent development of PARP inhibitors, which have been shown to produce objective responses [6, 7] and improved progression-free survival in BRCA1/2 germline mutant ovarian cancers. However, the clinical benefit from PARP inhibitors is not limited to BRCA1 or BRCA2 germline mutated cancers. Recent investigations have identified deficiency in other key mediators of genomic stability including ATM [19], RAD51C [20], and MRE11 [21] that may also potentiate PARP inhibitor sensitivity. Importantly, preclinical data also indicate that BRCA1 promoter hypermethylation [22] and somatic mutation [23] may also be synthetically lethal with PARP inhibitors. Taken together, these data suggest that BRCA1 IHC may be useful not only as a screen for BRCA1 germline mutations but also as a predictive biomarker for PARP inhibitors in BRCA1 germline wild-type patients.
conclusion
Detection of BRCA1 loss in ovarian carcinomas has potential prognostic and therapeutic significance. Our data indicate that BRCA1 IHC might be an inexpensive, easy to implement, and reproducible means of identifying these patients in the clinic.
funding
Funded in part by the cancer center's core grant P30 CA008748. The core grant provides funding to institutional cores, such as Biostatistics and Pathology, which were used in this study. Also supported in part by a Stand Up to Cancer Dream Team Translational Research Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-DT0209), The Honorable Tina Brozman Foundation, and Be The Difference Foundation.
disclosure
The authors have declared no conflicts of interest.
Supplementary Material
references
- 1.Bolton KL, Chenevix-Trench G, Goh C, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307(4):382–390. doi: 10.1001/jama.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hyman DM, Zhou Q, Iasonos A, et al. Improved survival for BRCA2-associated serous ovarian cancer compared with both BRCA-negative and BRCA1-associated serous ovarian cancer. Cancer. 2012;118(15):3703–3709. doi: 10.1002/cncr.26655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Husain A, He G, Venkatraman ES, Spriggs DR. BRCA1 up-regulation is associated with repair-mediated resistance to cis-diamminedichloroplatinum(II) Cancer Res. 1998;58(6):1120–1123. [PubMed] [Google Scholar]
- 4.Sakai W, Swisher EM, Karlan BY, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451(7182):1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallagher DJ, Konner JA, Bell-McGuinn KM, et al. Survival in epithelial ovarian cancer: a multivariate analysis incorporating BRCA mutation status and platinum sensitivity. Ann Oncol. 2011;22(5):1127–1132. doi: 10.1093/annonc/mdq577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Audeh MW, Carmichael J, Penson RT, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376(9737):245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 7.Gelmon KA, Tischkowitz M, Mackay H, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12(9):852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheuer L, Kauff N, Robson M, et al. Outcome of preventive surgery and screening for breast and ovarian cancer in BRCA mutation carriers. J Clin Oncol. 2002;20(5):1260–1268. doi: 10.1200/JCO.2002.20.5.1260. [DOI] [PubMed] [Google Scholar]
- 10.Garg K, Levine DA, Olvera N, et al. BRCA1 immunohistochemistry in a molecularly characterized cohort of ovarian high-grade serous carcinomas. Am J Surg Pathol. 2013;37(1):138–146. doi: 10.1097/PAS.0b013e31826cabbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92(7):564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 13.Byrne TJ, Reece MT, Adams LA, et al. An antibody assay predictive of BRCA1 mutations in ovarian tumors and normal tissue. Oncol Rep. 2000;7(5):949–953. doi: 10.3892/or.7.5.949. [DOI] [PubMed] [Google Scholar]
- 14.Skytte AB, Waldstrom M, Rasmussen AA, et al. Identification of BRCA1-deficient ovarian cancers. Acta Obstet Gynecol Scand. 2011;90(6):593–599. doi: 10.1111/j.1600-0412.2011.01121.x. [DOI] [PubMed] [Google Scholar]
- 15.Vaz FH, Machado PM, Brandao RD, et al. Familial breast/ovarian cancer and BRCA1/2 genetic screening: the role of immunohistochemistry as an additional method in the selection of patients. J Histochem Cytochem. 2007;55(11):1105–1113. doi: 10.1369/jhc.7A7209.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carser JE, Quinn JE, Michie CO, et al. BRCA1 is both a prognostic and predictive biomarker of response to chemotherapy in sporadic epithelial ovarian cancer. Gynecol Oncol. 2011;123(3):492–498. doi: 10.1016/j.ygyno.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 17.Radosa MP, Hafner N, Camara O, et al. Loss of BRCA1 protein expression as indicator of the BRCAness phenotype is associated with favorable overall survival after complete resection of sporadic ovarian cancer. Int J Gynecol Cancer. 2011;21(8):1399–1406. doi: 10.1097/IGC.0b013e318227c990. [DOI] [PubMed] [Google Scholar]
- 18.Weberpals JI, Tu D, Squire JA, et al. Breast cancer 1 (BRCA1) protein expression as a prognostic marker in sporadic epithelial ovarian carcinoma: an NCIC CTG OV.16 correlative study. Ann Oncol. 2011;22(11):2403–2410. doi: 10.1093/annonc/mdq770. [DOI] [PubMed] [Google Scholar]
- 19.Gilardini Montani MS, Prodosmo A, Stagni V, et al. ATM-depletion in breast cancer cells confers sensitivity to PARP inhibition. J Exp Clin Cancer Res. 2013;32:95. doi: 10.1186/1756-9966-32-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min A, Im SA, Yoon YK, et al. RAD51C-deficient cancer cells are highly sensitive to the PARP inhibitor olaparib. Mol Cancer Ther. 2013;12:865–877. doi: 10.1158/1535-7163.MCT-12-0950. [DOI] [PubMed] [Google Scholar]
- 21.Koppensteiner R, Samartzis EP, Noske A, et al. Effect of MRE11 loss on PARP-inhibitor sensitivity in endometrial cancer in vitro. PLoS One. 2014;9:e100041. doi: 10.1371/journal.pone.0100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ibragimova I, Cairns P. Assays for hypermethylation of the BRCA1 gene promoter in tumor cells to predict sensitivity to PARP-inhibitor therapy. Methods Mol Biol. 2011;780:277–291. doi: 10.1007/978-1-61779-270-0_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drew Y, Mulligan EA, Vong WT, et al. Therapeutic potential of poly(ADP-ribose) polymerase inhibitor AG014699 in human cancers with mutated or methylated BRCA1 or BRCA2. J Natl Cancer Inst. 2011;103(4):334–346. doi: 10.1093/jnci/djq509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.