Abstract
Genome-wide association studies (GWAS) have identified hundreds of genetic susceptibility loci for cancers and other complex diseases. However, the public health and clinical relevance of these discoveries is unclear. Evaluating the combined associations of genetic and environmental risk factors, particularly those that can be modified, will be critical in assessing the utility of genetic information for risk stratified prevention. In this commentary, using breast cancer as a model, we show that genetic information in combination with other risk factors can provide levels of risk stratification that could be useful for individual decision-making or population-based prevention programs. Our projections are theoretical and rely on a number of assumptions, including multiplicative models for the combined associations of the different risk factors, which need confirmation. Thus, analyses of epidemiological studies with high-quality risk factor information, as well as prevention trials, are needed to empirically assess the impact of genetics in risk stratified prevention.
With an increasing number of genetic susceptibility loci being identified for complex diseases, it is now important to examine whether genetic information could have utility in public health applications. Possible applications include risk stratified prevention and screening strategies targeted to susceptible subgroups of the population at elevated risk, or conversely defining subgroups at low risk that would benefit least from interventions. The value of genetic information for population-based screening programs has been recently discussed (1,2). The purpose of this commentary is to assess the potential utility of genetic information in primary prevention, either at the individual or the population level (1–3), for instance, by providing more accurate measures of individual risk to help make informed decisions on life-style changes or taking medications; or by targeting individuals at elevated risk for risk factor modification and chemoprevention programs. We are using breast cancer as an example, because this is a malignancy with a large number of known susceptibility loci, established modifiable risk factors such as menopausal hormone therapy (MHT), options for chemoprevention (ie, endocrine therapies to prevent estrogen receptor positive [ER+] breast cancer [4]), and screening strategies for early detection. Our model-based analyses illustrate the potential impact of genetic information for breast cancer prevention, and highlight the need for more empirical assessment of cancer risks for the combined associations of genetic and environmental risk factors (broadly defined as life-style, endogenous factors, and external environment).
Utility of Polygenic Risk Information to Improve Breast Cancer Risk Stratification
From a broad perspective, the utility of risk factor information for disease prevention critically depends on whether this information can provide sufficient “risk stratification,” i.e., its ability to define several population strata with sufficient differences in absolute risks to alter the risk-benefit tradeoff for an intervention. In the past, the impact of adding new risk factors into existing risk prediction models has been often assessed by changes in the area under the receiver operating characteristic curve (AUC) of the model. The AUC, which is a measure of discriminatory accuracy of a model, is defined as the probability that the predicted risk is higher for a case than a noncase (range of 0.5 for no discrimination to 1.0 for perfect discrimination). This measure, which depends only on relative risk (RR) parameters, cannot capture the degree of stratification of absolute risk that a new risk factor can add to a model. In particular, when the baseline risk of a disease is high, overall or in certain subgroups of the population defined by existing risk factors, a polygenic risk score even with modest ability to increase AUC (5–7) could substantially add to risk stratification. We recently demonstrated an example of such risk-stratification by a polygenic risk score (PRS) for smoking-related risk of bladder cancer (8). Calibration, ie, how well the predicted risks of developing disease agree with the actual observed risk, is another important measure of risk model performance that requires data from prospective cohort studies.
GWAS coupled with very large scale replication studies have identified 76 single nucleotide polymorphisms (SNP) for breast cancer at genome-wide significance levels (9–13). These SNPs explain about 15% of the familial risk (assuming a sibling relative RR of 2.0), with a further approximately 15% attributed to additional low and intermediate risk variants that did not reach genome-wide significance levels, according to the recent Collaborative Oncological Gene-Environment Study (COGS) Project (9,14). Although each of these loci is responsible for a small increase in risk, when combined, they could provide levels of risk stratification useful for the prevention of breast cancer, particularly in conjunction with information on family history and other risk factors.
To assess the extent that polygenic risk information can add to risk stratification by itself or in conjunction with other risk factors, we estimated the expected RR distribution and corresponding absolute risks of invasive breast cancer according to different sets of risk factors in the United Kingdom (UK) population. Simulation-based models were used to calculate the expected RR distribution that was calibrated to UK cancer rates to obtain estimates of absolute risk for women aged 40 and 50 years (see Supplementary Methods, available online). Models were based on published allele frequencies of SNPs, joint distribution of environmental risk factors from UK population–based surveys or published reports, and RR estimates from previous publications, adjusted by confounders when available (Supplementary Table 1, available online) (15–27).
Eight different models were developed for sets of risk factors that require different sources of information: Model 1 is based only on questionnaire-based risk factors (age, age at menarche, number of births, age at first live birth, oral contraceptive use [for women age 40 years] and combined MHT use [for women age 50 years], body mass index [BMI], alcohol, smoking, personal history of benign breast disease [BBD], and family history of breast cancer in first-degree relatives); Model 2 is based on questionnaire-based risk factors plus measurements of mammographic density (i.e., risk assessment requires a previous mammogram); Model 3 is based on a PRS (i.e., risk assessment requires a genetic test) as a measure of the combined associations of 76 currently known SNPs explaining approximately15% of the familial risk (referred to as “76-SNP PRS”) (see Supplementary Methods, available online); Model 4 is based on questionnaire-based risk factors plus 76-SNP PRS; Model 5 is based on all factors combined; and Models 6–8 are the same as Models 3–5 but with a different PRS explaining approximately 30% of the familial risk, referred to as “improved PRS” (see Supplementary Methods, available online).
Table 1 shows the percentage of the population and the percentage of cancers in subgroups of the population at moderate risk (RR > 2.0–3.0) and high risk (RR > 3.0) of invasive breast cancer, as defined by the above eight sets of risk factors. RRs are reported with respect to the average risk in the population (ie, lifetime risk from age 20 to 79 for a woman in the UK of 10.2%, accounting for competing mortality). The risk categories are similar to those used by the UK’s National Institute for Health and Clinical Excellence (NICE) guidelines on familial breast cancer for recommendations on the use of enhanced surveillance and chemoprevention (ie, lifetime risk of 17- <30% and ≥30% for moderate and high risk categories, respectively) (28).
Table 1.
Identification of women age 50 years in a UK population at moderate and high-risk of invasive breast cancer*
| Parameter | Model 1: Qx risk factors† | Model 2: Qx risk factors + density | Model 3: 76-SNP PRS |
Model 4: Qx risk factors
+ 76-SNP PRS |
Model 5: Qx risk factors
+ density + 76-SNP PRS |
Model 6: Improved PRS |
Model 7: Qx risk factors
+ improved PRS |
Model 8: Qx risk factors
+ density + improved PRS |
|---|---|---|---|---|---|---|---|---|
| AUC | 0.618 | 0.635 | 0.624 | 0.670 | 0.680 | 0.672 | 0.703 | 0.708 |
| % Population (% cases) at different risk thresholds‡ | ||||||||
| Moderate risk (RR > 2.0 - 3.0) | 3.8 (8.8) | 4.3 (10.0) | 3.6 (8.2) | 5.5 (13.0) | 5.8 (13.7) | 6.0 (14.1) | 6.0 (14.4) | 6.1 (14.7) |
| High risk (RR > 3.0) | 1.0 (3.7) | 1.3 (4.8) | 0.4 (1.4) | 2.4 (9.3) | 2.7 (10.8) | 2.1 (8.1) | 3.8 (16.0) | 4.1 (17.5) |
* Defined as relative risk (RR) > 2.0–3.0 and RR > 3.0, respectively, compared with the population average and for different combinations of risk factors and two polygenic risk scores (PRS). The following parameters are shown for eight risk prediction models: AUC, the % of the population found at moderate and high levels of risk according to the different models, and the % of cases in the population expected to occur among women at these levels of risk. AUC = area under the receiver operating characteristic curve; PRS = polygenic risk score; Qx = questionnaire; RR = relative risk; SNP = single nucleotide polymorphism.
† Questionnaire-based risk factors include age at menarche, parity, age at first birth, combined MHT, body mass index, benign breast disease, alcohol intake, smoking, and family history of breast cancer in first-degree relatives.
‡ Lifetime risk (from age 20 to 80 years), 10-year and five-year risk thresholds corresponding to RRs of 2.0 and 3.0 for a woman age 50 years: RR = 2.0: 19.4% lifetime risk, 5.2% 10-year risk and 2.6% five-year risk; RR = 3.0: 27.5% lifetime risk, 7.8% 10-year risk and 3.9% five-year risk.
Results in Table 1 show substantial improvements in risk stratification when adding information on polygenic risk to other known risk factors, as determined by the eight risk prediction models. For 50-year old women, questionnaire-based risk factors and mammographic breast density could identify 5.6% of the population at moderate-to-high risk that would account for 14.9% of the cases in the population. The 76-SNP PRS by itself could identify 4.0% of the population at moderate-to-high risk, capturing 9.6% of the cancers. When combining the 76-SNP PRS with questionnaire-based risk factors and density, one could identify 8.5% of the population capturing 24.5% of the cancers. This could increase to 10.2% of the population capturing 32.2% of cases for the improved PRS (Table 1). A similar level of risk stratification can be obtained for women at age 40 years, with 34.4% of the cases occurring in 10.2% of the population at moderate-to-high risk (Supplementary Table 2, available online). It should be noted that substantial improvements in risk stratification can be obtained even when the AUC improvements are relatively small (Table 1).
The percentage of the population crossing a threshold for moderate-to-high levels of risk only increases moderately when including more risk factors in the models (eg, an additional 5.4% of women are found to be at moderate-to-high risk when comparing Model 1 [4.8%] with Model 8 [10.2%]). However, the percentage of cases captured in this risk group increases substantially (eg, an additional 19.7% of cases are captured when comparing Model 1 [12.5%] with Model 8 [32.2%]), which is the desirable impact as risk models improve. Finally, contrary to previous suggestions based on information on only seven known SNPs at the time (6), the addition of mammographic breast density results in a smaller improvement in risk prediction than adding information on polygenic risk (now based on 76-SNPs). For instance, adding density to a model with questionnaire-based risk factors results in an additional 2.4% of cases being captured by the moderate-to-high risk groups; whereas addition of the 76-SNP PRS results in an additional 9.8% of cases captured and the improved PRS in an additional 17.9% of cases. This is consistent with estimates from a Swedish study that observed a larger improvement in the discriminatory accuracy of the Gail model when adding 18 SNPs than when adding breast density and BMI (29).
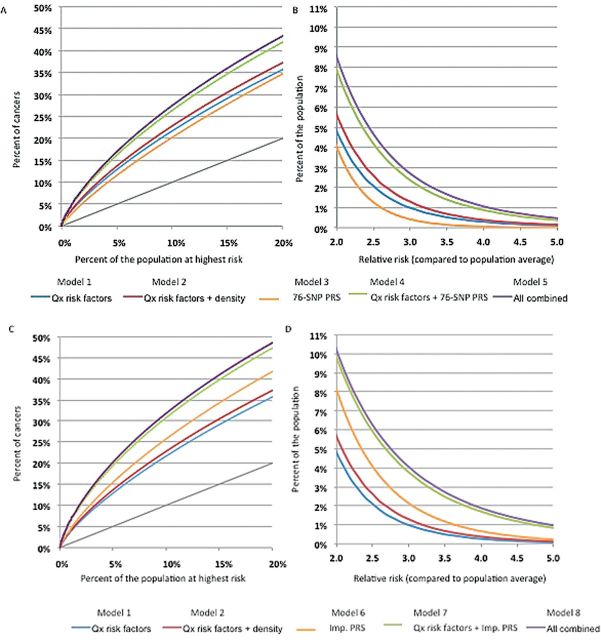
Figure 1 illustrates graphically the ability to discriminate risk in a population of women aged 50 years in the UK, expressed as the percentage of cancers expected to occur in different groups of the population identified at highest risk according to different sets of risk factors (panels A and C) and the percent of the population crossing different RR thresholds compared with the population average risk (panels B and D). This figure indicates that addition of polygenic information has a substantial impact in identifying subsets of the population more likely to develop breast cancer. For instance, if one were to stratify the population according to a risk score based on questionnaire-based risk factors alone, about 22% of breast cancers in the population would be expected to occur in the top 10% of the population identified as having the highest risk. The percentage of cancers would increase from 22% to 27% with the addition of information on the 76-SNP PRS and to 32% with the addition of the improved PRS. It should also be noted that the improved PRS could have stronger risk discrimination ability by itself than questionnaire-based risk factors.
Figure 1.
Partial Receiver Operating Curves (ROC) showing the percentage of cases of breast cancer expected to occur in groups of the population at highest predicted risk (A, C), and graphs for the percentage of the population crossing breast cancer relative risk (RR) thresholds (compared with the average risk in the population) (B, D). Estimates are for a UK population of women aged 50 years, for eight risk prediction models, including different sets of risk factors and two polygenic risk scores (PRSs): the 76–single nucleotide polymorphism (SNP) PRS based on currently known SNPs explaining 15% of the familial risk (A, B) and an improved PRS explaining 30% of the familial risk (C, D). PRS = polygenic risk score; Qx = questionnaire; SNP = single nucleotide polymorphism.
Utility of Polygenic Risk Information for Targeted Breast Cancer Prevention
Although the above estimates show that we might be able to identify a relatively small percentage of the population at elevated risk that would capture a substantial proportion of cases occurring in the population, a majority of cases would still occur in women not identified as being at elevated risk. Therefore, although not a substitute for general prevention efforts applicable to the whole population (eg, maintaining a healthy weight, being physically active, drinking alcohol in moderation), targeting certain interventions to high-risk individuals could be more effective (30), for instance, when the intervention has associated harms (e.g., side effects of medication) and/or it is too expensive to be applied to the whole population.
Interventions targeted to susceptible individuals could have a stronger impact on disease absolute risk reduction when the combined associations of genetic (or other nonmodifiable risk factors) and modifiable risk factors are multiplicative. This is because under a multiplicative model for gene-environment joint associations, where RRs associated with a particular exposure remain the same for subjects in different polygenic risk categories, it is expected that risk differences (RDs) associated with the exposure will be larger for subjects with higher polygenic risk. Previous publications have shown that, with a few exceptions, most common susceptibility loci and environmental factors are likely to act multiplicatively on the risk of breast cancer (31–34). Below we illustrate how under the multiplicative model for gene-environment interaction, genetic risk-stratification can be useful for providing guidance to women about use of combined MHT and endocrine chemoprevention.
Combined Estrogen and Progesterone MHT Use
As the risk-benefit tradeoff for an intervention is critically related to the underlying risk differences, we assessed RD parameters associated with combined estrogen and progesterone MHT for menopausal symptoms for women in different categories of polygenic risk. Women with a family history of breast cancer are often advised to minimize the dose and duration of combined MHT use because of their underlying genetic risk (28). For women age 50 years under Model 8 in Table 1, we estimated that taking combined MHT increases the 10-year breast cancer risk by 2.5% (from 2.2% to 4.7%) for women without a family history, and by 4.4% (from 4.0% to 8.4%) for a women with a first-degree relative with breast cancer (Table 2). The stronger impact of MHT use on absolute risk of breast cancer for women with family history is the basis of the current recommendations.
Table 2.
Ten-year absolute risk of developing invasive breast cancer for a 50-year-old woman in the UK*
| Percentile | Women without a family history (~90% of the population) | Women with a family history (~10% of the population) | ||||||
|---|---|---|---|---|---|---|---|---|
| All | MHT never | MHT current | RD | All | MHT never | MHT current | RD | |
| All women | 2.5% | 2.2% | 4.7% | 2.5% | 4.4% | 4.0% | 8.4% | 4.4% |
| Stratified by percentiles of a 76-SNP PRS (15% of familial risk explained) | ||||||||
| <1% | 0.7% | 0.6% | 1.3% | 0.7% | 1.1% | 1.0% | 2.0% | 1.0% |
| 1–5% | 0.9% | 0.9% | 1.8% | 1.0% | 1.6% | 1.5% | 3.1% | 1.7% |
| >5–10% | 1.1% | 1.0% | 2.2% | 1.2% | 2.0% | 1.8% | 3.9% | 2.0% |
| >10–20% | 1.4% | 1.3% | 2.7% | 1.4% | 2.5% | 2.2% | 4.7% | 2.5% |
| >20–40% | 1.8% | 1.6% | 3.4% | 1.8% | 3.1% | 2.8% | 6.0% | 3.2% |
| >40–60% | 2.2% | 2.0% | 4.3% | 2.3% | 3.9% | 3.6% | 7.5% | 3.9% |
| >60–80% | 2.8% | 2.6% | 5.5% | 2.9% | 5.0% | 4.6% | 9.5% | 5.0% |
| >80–90% | 3.6% | 3.3% | 6.8% | 3.6% | 6.2% | 5.7% | 11.7% | 6.0% |
| >90–95% | 4.3% | 3.9% | 8.1% | 4.2% | 7.3% | 6.7% | 14.0% | 7.4% |
| >95–99% | 5.3% | 4.8% | 9.9% | 5.1% | 8.7% | 7.9% | 16.2% | 8.3% |
| >99% | 7.4% | 6.7% | 13.9% | 7.2% | 11.8% | 10.8% | 21.5% | 10.7% |
| Stratified by percentiles of an improved PRS (30% of familial risk explained) | ||||||||
| <1% | 0.4% | 0.3% | 0.7% | 0.4% | 0.6% | 0.6% | 1.3% | 0.7% |
| 1–5% | 0.6% | 0.5% | 1.2% | 0.6% | 1.0% | 1.0% | 2.1% | 1.1% |
| >5–10% | 0.8% | 0.7% | 1.5% | 0.8% | 1.4% | 1.3% | 2.7% | 1.4% |
| >10–20% | 1.0% | 0.9% | 2.0% | 1.1% | 1.8% | 1.7% | 3.5% | 1.8% |
| >20–40% | 1.5% | 1.3% | 2.8% | 1.5% | 2.6% | 2.3% | 5.0% | 2.6% |
| >40–60% | 2.0% | 1.9% | 3.9% | 2.1% | 3.6% | 3.2% | 6.9% | 3.6% |
| >60–80% | 2.9% | 2.6% | 5.5% | 2.9% | 4.9% | 4.5% | 9.4% | 4.9% |
| >80–90% | 3.9% | 3.6% | 7.5% | 4.0% | 6.8% | 6.2% | 12.8% | 6.6% |
| >90–95% | 5.1% | 4.6% | 9.7% | 5.1% | 8.5% | 7.8% | 15.7% | 7.9% |
| >95–99% | 6.8% | 6.2% | 12.7% | 6.5% | 10.8% | 9.9% | 19.7% | 9.8% |
| >99% | 10.9% | 10.0% | 20.2% | 10.2% | 15.9% | 14.7% | 29.2% | 14.5% |
* According to use of combined (estrogen and progesterone) menopausal hormone therapy (MHT) and familial risk, defined as family history in a first-degree relative and a polygenic risk score. PRS = polygenic risk score; RD = risk difference; SNP = single nucleotide polymorphism.
The above risk estimates are the average in the population for women with and without family history, and a PRS could allow finer risk stratification within these groups. For instance, the 10-year breast cancer risk for a woman without a family history taking MHT would range from 1.3% to 13.9% depending on whether she is in the lowest or highest one percentile of the 76-SNP PRS; and the corresponding range for a woman with a family history would be 2.0% to 21.5%. The RD estimates imply that 70 cases would be prevented by removing MHT use in 10000 women in the lowest one percentile of the 76-SNP PRS and negative family history, compared with 720 cases that would be prevented by removing MHT use in 10000 women in the highest one percentile of the PRS and positive family history. Of note, stratification with respect to the 76-SNP PRS indicates that women with positive family history in the lowest 20th percentile of polygenetic risk have similar or lower RDs associated with MHT than women without family history (Table 2). Similarly, women in the highest 10th percentile of polygenic risk but no family history have RDs similar or higher than women with a family history. Thus, polygenic risk could be useful for personalization of advice regarding use of MHT beyond family history.
While our analysis suggests that the combination of information on PRS and family history could provide substantial risk stratification in the population, the ultimate assessment of their utility for risk-benefit calculations needs to account for other risk factors for breast cancer, as well as those for other health effects from use of combined MHT, such as risk of endometrial cancer and osteoporosis.
Endocrine Chemoprevention
Selective estrogen receptor modulators (SERMs) are approved by the United States Food and Drug Administration for reducing breast cancer risk, and the recently published UK NICE guidelines for familial breast cancer recommend offering endocrine chemoprevention to women aged 40 to 50 years at high risk of breast cancer (ie, lifetime risk > 30%) without contraindications and considering this treatment for women at moderate risk (ie, lifetime risk of 17%-30%). However, the use of these drugs requires identification of women at those levels of risk. Our calculations based on all risk factors combined indicate that about 10% of women 50 years old found to be at moderate to high risk (capturing about 27% of cases in the population) could be identified as eligible for chemoprevention (Model 5 in Table 1), thus benefiting from a 38% reduction in risk (4). This could be improved to 11% of women capturing about 34% of cases when using the improved PRS (Model 8 in Table 1). However, preventive trials are needed to evaluate the joint associations of PRS and SERMs and assess the validity of the multiplicative joint effect assumption. In addition, introduction of such wide-scale preventive treatment would need to be closely monitored in the population, particularly in view of potentially severe side effects of treatment.
Concluding Remarks
Our estimates illustrate how information on polygenic risk, in addition to family history and other risk factors, could be used in primary prevention to identify subgroups of the population at different levels of risk, particularly those at elevated risk most likely to benefit from certain advice or an intervention. This is in spite of relatively small improvements in the AUC, indicating that using this as single measure of model performance can lead to erroneous conclusions about the clinical and public health utility of new risk predictors such as polygenic risk.
Our model-based estimates rely on RR estimates from large meta- or pooled analyses and population-based distributions. Although we used adjusted RR estimates and accounted for dependency of distribution of key risk factors when possible, because of the multiple sources of data used, we did not fully account for dependency and potential confounding effects of multiple risk factors. As for most existing risk prediction models, our model estimated the risk of developing invasive breast cancer; further work to develop models for both invasive and in situ breast cancer is needed. Our calculations also rely on the key assumption that genetic and environmental risks act multiplicatively. Although large studies are providing increasing support for multiplicative gene-environment associations (31-33,34), there is a need to continue to evaluate this assumption empirically and to obtain precise estimates of joint associations and absolute risk in subsets of population defined by the combination of environmental risk factors, chemo-preventive agents and polygenic risks. Such studies should include evaluation for additive interactions to directly evaluate if the expected risk reduction from removing an exposure differs by levels of genetic risk (8,35). This will require very large, studies with high quality information on risk factors. Calibration of risk models in prospective cohort studies to compare the predicted and observed risk will also be a critical part of the assessment of the performance of the models in providing accurate predictions of risk. Finally, easy-to-use risk prediction tools based on models validated in prospective cohorts will be needed for the adoption of risk-stratified strategies in the clinical practice.
Funding
MGC is funded by the Institute of Cancer Research and Breakthrough Breast Cancer, UK. NBG is funded by the Institute of Cancer Research, UK. NC is funded by Intramural Funds of the National Cancer Institute, National Institutes of Health.
Supplementary Material
The authors declare no conflict of interests.
References
- 1. Burton H, Chowdhury S, Dent T, Hall A, Pashayan N, Pharoah P. Public health implications from COGS and potential for risk stratification and screening. Nat Genet. 2013;45(4):349–351. [DOI] [PubMed] [Google Scholar]
- 2. Pashayan N, Pharoah P. Population-based screening in the era of genomics. Per Med. 2012;9(4):451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rose G. Sick Individuals and Sick Populations. Int J Epidemiol. 1985;14(1):32–38. [DOI] [PubMed] [Google Scholar]
- 4. Cuzick J, Sestak I, Bonanni B, et al. Selective oestrogen receptor modulators in prevention of breast cancer: an updated meta-analysis of individual participant data. Lancet. 2013;381(9880):1827–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wacholder S, Hartge P, Prentice R, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362(11):986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100(14):1037–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hüsing A, Canzian F, Beckmann L, et al. Prediction of breast cancer risk by genetic risk factors, overall and by hormone receptor status. J Med Genet. 2012;49(9):601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garcia-Closas M, Rothman N, Figueroa JD, et al. Common Genetic Polymorphisms Modify the Effect of Smoking on Absolute Risk of Bladder Cancer. Cancer Res. 2013;73(7):2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Michailidou K, Hall P, González-Neira A, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garcia-Closas M, Couch FJ, Lindstrom S, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45(4):392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bojesen SE, Pooley KA, Johnatty SE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45(4):371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. French JD, Ghoussaini M, Edwards SL, et al. Functional Variants at the 11q13 Risk Locus for Breast Cancer Regulate Cyclin D1 Expression through Long-Range Enhancers. Am J Hum Genet. 2013;92(4):489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mavaddat N, Antoniou AC, Easton DF, Garcia-Closas M. Genetic susceptibility to breast cancer. Mol Oncol. 2010;4(3):174–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. COGS Primer. http://www.nature.com/icogs/primer/common-variation-and-heritability-estimates-for-breast-ovarian-and-prostate-cancers/ Accessed May 30, 2013
- 15. Tice JA, Cummings SR, Smith-Bindman R, Ichikawa L, Barlow WE, Kerlikowske K. Using clinical factors and mammographic breast density to estimate breast cancer risk: development and validation of a new predictive model. Ann Intern Med. 2008;148(5):337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pharoah PD, Day NE, Duffy S, Easton DF, Ponder BA. Family history and the risk of breast cancer: a systematic review and meta-analysis. Int J Cancer. 1997;71(5):800–809. [DOI] [PubMed] [Google Scholar]
- 17. Nelson HD, MD MPH, et al. Risk Factors for Breast Cancer for Women Age 40 to 49: A Systematic Review and Meta-analysis. Ann Intern Med. 2012;9(156):635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McCormack VA, Santos Silva Dos I. Breast density and parenchymal patterns as markers of breast cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006;15(6):1159–1169. [DOI] [PubMed] [Google Scholar]
- 19. Hartmann LC, Sellers TA, Frost MH, et al. Benign breast disease and the risk of breast cancer. N Engl J Med. 2005;353(3):229–237. [DOI] [PubMed] [Google Scholar]
- 20. Byrne C, Schairer C, Brinton LA, et al. Effects of mammographic density and benign breast disease on breast cancer risk (United States). Cancer Causes Control. 2001;12(2):103–110. [DOI] [PubMed] [Google Scholar]
- 21. Reeves GK, Pirie K, Green J, Bull D, Beral V. Million Women Study Collaborators Comparison of the effects of genetic and environmental risk factors on in situ and invasive ductal breast cancer. Int J Cancer. 2012;131(4):930–937. [DOI] [PubMed] [Google Scholar]
- 22. Parkin DM. Is the recent fall in incidence of post-menopausal breast cancer in UK related to changes in use of hormone replacement therapy? Eur J Cancer. 2009;45(9):1649–1653. [DOI] [PubMed] [Google Scholar]
- 23. Gaudet MM, Gapstur SM, Sun J, Diver WR, Hannan LM, Thun MJ. Active smoking and breast cancer risk: original cohort data and meta-analysis. J Natl Cancer Inst. 2013;105(8):515–525. [DOI] [PubMed] [Google Scholar]
- 24. Cohort Fertility Tables in England and Wales. Office for National Statistics. http://data.gov.uk/dataset/cohort_fertility_england_and_wales Accessed June 19, 2013
- 25. Collaborative Group on Hormonal Factors in Breast Cancer Menarche, menopause, and breast cancer risk: individual participant meta-analysis, including 118 964 women with breast cancer from 117 epidemiological studies. Lancet Oncol. 2012;13(11):1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alcohol, tobacco and breast cancer – collaborative reanalysis of individual data from 53 epidemiological studies, including 58 515 women with breast cancer and 95 067 women without the disease. Br J Cancer. 2002;87(11):1234–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jacobi CE, Jonker MA, Nagelkerke NJ, van Houwelingen JC, de Bock GH.Prevalence of family histories of breast cancer in the general population and the incidence of related seeking of health care. J Med Genet. 2003;40(7):e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. NICE Familial Breast Cancer: Update of clinical guideline 14 and 41. http://www.nice.org.uk/nicemedia/live/13269/62338/62338.pdf. Accessed January 8, 2013.
- 29. Darabi H, Czene K, Zhao W, Liu J, Hall P, Humphreys K. Breast cancer risk prediction and individualised screening based on common genetic variation and breast density measurement. Breast Cancer Res. 2012;14(1):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rose G. The Strategy of Preventive Medicine. New York, NY: Oxford University Press; 1992. [Google Scholar]
- 31. Milne RL, Gaudet MM, Spurdle AB, et al. Assessing interactions between the associations of common genetic susceptibility variants, reproductive history and body mass index with breast cancer risk in the breast cancer association consortium: a combined case-control study. Breast Cancer Res. 2010;12(6):R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Campa D, Kaaks R, Le Marchand L, et al. Interactions Between Genetic Variants and Breast Cancer Risk Factors in the Breast and Prostate Cancer Cohort Consortium. J Natl Cancer Inst. 2011;103(16):1252–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Travis RC. Gene–environment interactions in 7610 women with breast cancer: prospective evidence from the Million Women Study. 2010. 1–13. [DOI] [PMC free article] [PubMed]
- 34. Nickels S, Truong T, Hein R, et al. Evidence of Gene-Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors. PLoS Genet. 2013;9(3):e1003284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Han SS, Rosenberg PS, Garcia-Closas M, et al. Likelihood Ratio Test for Detecting Gene (G)-Environment (E) Interactions Under an Additive Risk Model Exploiting G-E Independence for Case-Control Data. Am J Epidemiol. 2012;176(11):1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.