Abstract
Bile acids (BAs) are known to regulate their own homeostasis, but the potency of individual bile acids is not known. This study examined the effects of cholic acid (CA), chenodeoxycholic acid (CDCA), deoxycholic acid (DCA), lithocholic acid (LCA) and ursodeoxycholic acid (UDCA) on expression of BA synthesis and transport genes in human primary hepatocyte cultures. Hepatocytes were treated with the individual BAs at 10, 30, and 100μM for 48 h, and RNA was extracted for real-time PCR analysis. For the classic pathway of BA synthesis, BAs except for UDCA markedly suppressed CYP7A1 (70–95%), the rate-limiting enzyme of bile acid synthesis, but only moderately (35%) down-regulated CYP8B1 at a high concentration of 100μM. BAs had minimal effects on mRNA of two enzymes of the alternative pathway of BA synthesis, namely CYP27A1 and CYP7B1. BAs increased the two major target genes of the farnesoid X receptor (FXR), namely the small heterodimer partner (SHP) by fourfold, and markedly induced fibroblast growth factor 19 (FGF19) over 100-fold. The BA uptake transporter Na+-taurocholate co-transporting polypeptide was unaffected, whereas the efflux transporter bile salt export pump was increased 15-fold and OSTα/β were increased 10–100-fold by BAs. The expression of the organic anion transporting polypeptide 1B3 (OATP1B3; sixfold), ATP-binding cassette (ABC) transporter G5 (ABCG5; sixfold), multidrug associated protein-2 (MRP2; twofold), and MRP3 (threefold) were also increased, albeit to lesser degrees. In general, CDCA was the most potent and effective BA in regulating these genes important for BA homeostasis, whereas DCA and CA were intermediate, LCA the least, and UDCA ineffective.
Keywords: Human primary hepatocyte cultures, bile acids, CYP7A1, FGF19, BSEP, OSTα/β
Abbreviations
- CYP
Cytochrome P450
- FXR
farnesoid X receptor
- SHP
small heterodimer partner
- CA
cholic acid
- CDCA
chenodeoxycholic acid
- DCA
deoxycholic acid
- LCA
lithocholic acid
- UDCA
ursodeoxycholic acid
- FGF19
fibrosis growth factor 19
- NTCP
Na+-taurocholate co-transporting ploypeptide
- OATP1B3
organic anion-transporting polypeptide 1B3
- BSEP
bile salt export pump
- Ostα/β
organic solute transporter α/β
- MRP
multidrug-resistant protein
Bile acids (BAs) are physiological detergents that regulate bile flow and facilitate the intestinal absorption of lipids, nutrients, and lipid-soluble vitamins (Chiang, 2009). For the BA-biosynthetic enzymes in liver, cholesterol 7α-hydroxylase (CYP7A1) is the rate-limiting enzyme in the classic pathway of BA synthesis. In humans, further hydroxylation at the 12 position of the steroid nucleus by CYP8B1 leads to one of the primary BAs, cholic acid (CA); chenodeoxycholic acid (CDCA) is another primary BA (Vlahcevic et al., 1999). In addition, derivatives of cholesterol, namely the oxysterols, can also be catabolized to BAs through another BA synthetic pathway, often referred to as the alternative pathway. Two critical enzymes responsible for the synthesis of BAs in the alternative pathway are CYP27A1 and CYP7B1 (Chiang, 2009; Russell, 2009; Vlahcevic et al., 1999).
In humans, the primary BAs are CA and CDCA. The two major secondary BAs, namely deoxycholic acid (DCA) and lithocholic acid (LCA), are formed by 7-dehydroxylation of CA and CDCA, respectively, by intestinal bacteria (Zhang and Klaassen, 2010). Ursodeoxycholic acid (UDCA) is a primary BA in some mammals (e.g., bear) and has been used to treat cholesterol gallstones, primary biliary cirrhosis (PBC), and cholestasis (Hofmann and Hagey, 2008). Individual BAs differ markedly in their potency to produce biological effects (Parks et al., 1999). For example, CDCA is more effective than UDCA in feedback regulation of BA synthesis (Ellis et al., 2003), and is more toxic than UDCA (Hillaire et al., 1995). Our laboratory has recently shown that the hepatotoxicity of feeding BAs in mice varies among different BAs, LCA is the most toxic whereas UDCA is the least toxic BA (Song et al., 2011).
Various transporters are important in mediating the disposition of BAs in the liver and intestine (Klaassen and Aleksunes, 2010). In liver, BAs are actively taken up into hepatocytes by the sodium-dependent taurocholate co-transporting polypeptide (NTCP/SLC10A1), which is located on the sinusoidal membrane of hepatocytes. The organic anion transporting peptides (OATP/SLCO transporters) are involved in sodium-independent uptake of unconjugated BAs into liver (Csanaky et al., 2011). BAs in hepatocytes are excreted into bile at the canalicular membrane of hepatocytes by an ATP-dependent transporter, the bile salt export pump (BSEP/ABCB11) (Plass et al., 2002). The heterodimer ABCG5/ABCG8, which is located on the canalicular membrane of hepatocytes, mediates biliary excretion of cholesterol (Yu et al., 2002). BA-efflux transporters located at the basolateral membrane of hepatocytes include the multidrug resistance-associated proteins (MRP3/ABCC3) and the heterodimer organic solute transporter (OSTα/OSTβ), transport BAs, and other solutes from hepatocytes back into the systemic circulation (Ballatori et al., 2005).
BAs are thought to regulate their own homeostasis via a BA sensor, the farnesoid X receptor (FXR/NR1H4) (Makishima et al., 1999; Parks et al., 1999). Activation of FXR in the liver increases the expression of its target gene SHP/NROB2, which down-regulates the key BA-biosynthetic enzyme CYP7A1 (Goodwin et al., 2000). This is often referred to as the hepatic FXR-SHP signaling pathway. A second mechanism is fibroblast growth factor 19 (FGF19), a hormone that is released into blood and decreases the synthesis of BAs in the liver. Both the FXR-SHP pathway and FXR-FGF19 pathway are important in regulating BA biosynthesis (Chiang, 2009; Kong et al., 2012).
There are species differences in BA biosynthesis, BA signaling, BA transport, and BA toxicity (Handschin et al., 2005; Li et al., 2011; Marion et al., 2012; Song et al., 2007, 2009, 2011). This highlights the need for a good in vitro human model. Isolated primary human hepatocytes have the capacity to synthesize normal conjugated BAs at a rate similar to that in vivo (Ellis and Nilsson, 2010). Human primary hepatocyte cultures have been shown to be a good model to study BA homeostasis and regulation, including the FXR-SHP signaling pathway (Li and Chiang, 2005; Jahan and Chiang, 2005), and the FXR-FGF19 signaling pathway (Song et al., 2009). BA homeostasis is also regulated by hepatocyte growth factor (Song et al., 2007), glucagon, cAMP, and forkhead box transcription factor O1 (Li et al., 2006; Song and Chiang, 2006), as well as by vitamin D receptor signaling (Han and Chiang, 2009). Thus, human hepatocyte primary culture appears to be a good model to study BA homeostasis and signaling pathways.
The present study has utilized human primary hepatocyte cultures, obtained from the University of Pittsburgh through the Liver Tissue Cell Distribution System, to simultaneously study regulatory effects of five BAs, i.e., the primary BAs CA and CDCA, the secondary BAs DCA and LCA, as well as UDCA. The effects of BAs on expression of genes important in BA biosynthesis, BA signaling pathway activation, and BA transporters were investigated.
MATERIALS AND METHODS
Human primary hepatocyte cultures
Human primary hepatocyte cultures were obtained from the Liver Tissue and Cell Distribution System of the National Institute of Diabetes and Digestive and Kidney Diseases (LTCDS; N01-DK-7-0004/HHSN267200700004C, S. Strom, University of Pittsburgh, PA). The hepatocytes were from six donors: HH1850, HH1849, HH1649, HH1479, HH1426, and HH11425. Hepatocytes were maintained in William E medium supplemented with 10% fetal bovine serum, antibiotics, and hepatocyte growth supplements, and cultured in 5% CO2 and 50% humidity for 48 h before addition of BAs.
BA treatments
CA, CDCA, DCA, LCA, and UDCA were purchased from Sigma-Aldrich Chemical Co. (St Louis, MO), and the 10, 30, and 100mM stock solutions were prepared in dimethyl sulfoxide (DMSO), and added to cultures to the final concentrations of 10, 30, and 100μM. The BAs’ concentration selection was based on CDCA (1–100μM) used in human hepatocytes (Song et al., 2009). The final DMSO concentration in the BA-treated groups and DMSO control group was 0.1%. Forty-eight hours after cell treatment, cells were harvested in RNA-Bee (Tel-Test Inc., Friendswood, TX) and frozen at −80°C prior to experiments.
RNA isolation
Total RNA was isolated using RNA-Bee reagent according to the manufacturer's instructions. RNA quantity was determined by the 260/280 ratio (>1.8) and quality by formaldehyde-agarose gel electrophoresis for visualization of 18S and 28S rRNA bands after ethidium bromide staining. Total RNA was diluted with diethyl pyrocarbonate-treated deionized water to a concentration of 100 ng/μl.
Real-time RT-PCR analysis
Total RNA was purified with RNeasy columns (Qiagen, Valencia, CA) and reverse transcribed with Multiscript reverse transcriptase using a High Capacity RT kit from Applied Biosystems (Foster City, CA). Primers were designed with Primer3 software (version 4), and are listed in Supplementary table 1. The Power SYBR Green Master Mix (Applied Biosystems) was used for real-time reverse-transcription polymerase chain reaction (RT-PCR) analysis. Differences in gene expression between groups were calculated using cycle threshold (Ct) values, which were normalized with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) of the same sample, and relative transcript levels were calculated with vehicle controls of each donor set as 100%.
Heatmap
A one-way hierarchical clustering dendrogram was generated by standarding the data by the formula of Xi-standardized (Z score) = (Xi − Xmean)/SD, followed by importing the data into JMP v. 11.0 (SAS, Cary, NC) to determine the expression patterns of the genes important for BA biosynthesis, BA signaling, and BA transport following culture with BAs at various concentrations.
Statistical analysis
The data were calculated as mean ± SEM. For comparisons among three or more groups, data were analyzed using a one-way analysis of variance (ANOVA), followed by Duncan's multiple range test. The significant level was set at p < 0.05 in all cases.
RESULTS
Human hepatocyte cultures were obtained from six donors over a three-year period. Although there were significant individual variations in response to BA treatments, the trend of alterations was similar. The data obtained from each donor was considered as n = 1. For the expression of key genes (CYP7A1, FXR, SHP, FGF19, BSEP, OSTα, and OSTβ), all six donors were examined; for other genes, 3–5 donors were used.
Effect of Various BAs on BA Synthesis Genes
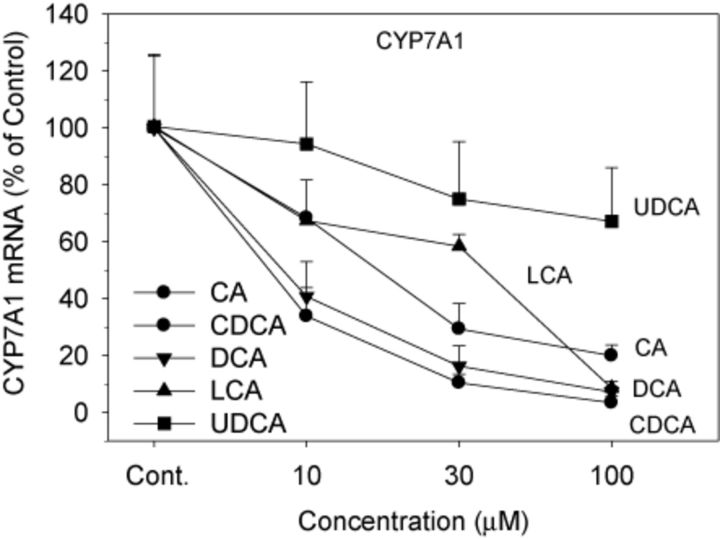
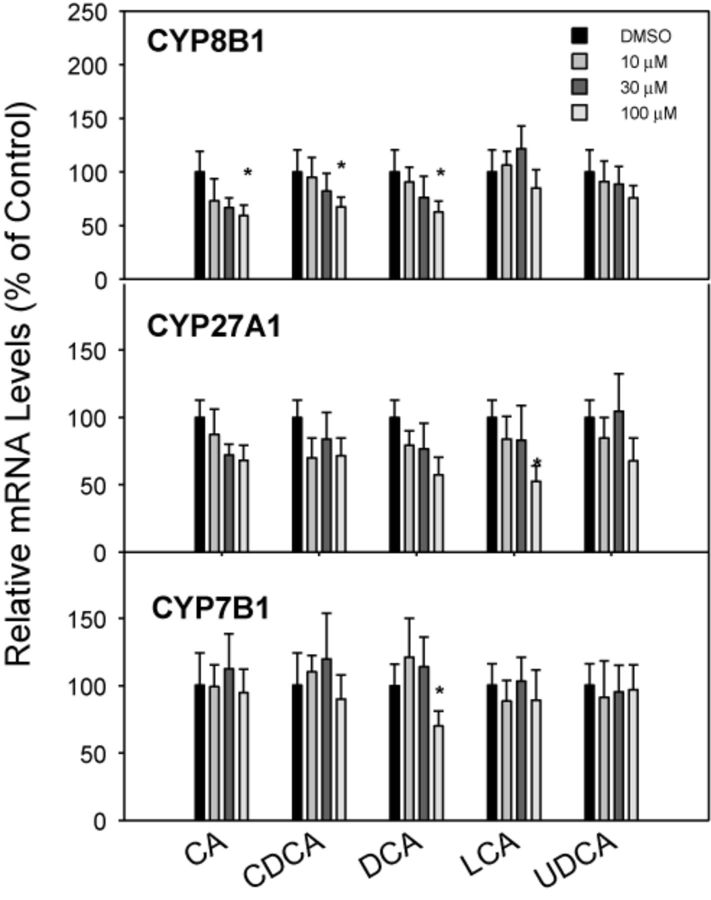
The mRNA expression of genes responsible for BA synthesis by the classic (CYP7A1 and CYP8B1) and alternative (CYP27A1 and CYP7B1) pathways is shown in Figures 1 and 2. CA, DCA, CDCA, and LCA dramatically and dose-dependently decreased the expression of CYP7A1, the rate-limiting enzyme for BA biosynthesis (Chiang, 2009). The four BAs dose-dependently decreased CYP7A1 mRNA 75–90%, but UDCA had no effect on CYP7A1 mRNA levels (Fig. 1). CDCA and DCA were more effective than CA and LCA in decreasing CYP7A1 at 10μM. In comparison, CYP8B1 mRNA was decreased only at the high concentration of 100μM of CA, CDCA, and DCA (Fig. 2). Regarding genes encoding the alternative pathway of BA biosynthesis (CYP27A1 and CYP7B1), the BAs had little or no effect; only the highest concentration of LCA decreased the mRNA of CYP27A1, and DCA at the highest concentration decreased CYP7B1 (Fig. 2).
FIG. 1.
Effects of BAs on CYP7A1 mRNA levels. Human primary hepatocyte cultures were treated with individual BAs at concentrations of 10, 30, and 100μM for 48 h, and the expression of CYP7A1 was determined by real-time RT-PCR analysis. Data are mean + SEM (n = 6).
FIG. 2.
Effects of BAs on BA-metabolism enzyme mRNA levels. Human primary hepatocyte cultures were treated with individual BAs at concentrations of 10, 30, and 100μM for 48 h, and the expression of CYP8B1, CYP27A1, and CYP7B1 was determined by real-time RT-PCR analysis. Data are mean + SEM (n = 5–6). *Significantly different from DMSO-treated controls, p < 0.05.
Effect of Various BAs on FXR-SHP and FXR-FGF19 Pathways
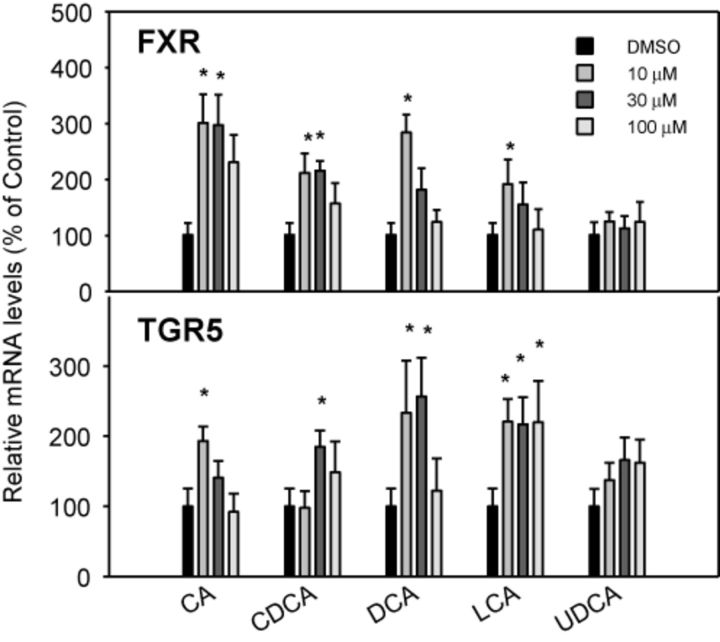
BAs are physiological ligands of FXR, a nuclear receptor (Parks et al., 1999), and TGR5, a membrane receptor (Pols et al., 2011; Sato et al., 2008). The effects of BAs on the mRNA levels of FXR and TGR5 are shown in Figure 3. CA, CDCA, DCA, and LCA increased the mRNA of FXR, but no dose-response was evident. At the highest dose (100μM), the expression of FXR was not induced by the BAs. UDCA was ineffective in increasing the mRNA of FXR. Similarly, BAs, except for UDCA, increased mRNA levels of the BA sensor TGR5 2–3-fold, without clear dose-responses (Fig. 3).
FIG. 3.
Effects of BAs on the mRNA levels of BA sensors FXR and TGR5. Human primary hepatocyte cultures were treated with individual BAs at concentrations of 10, 30, and 100μM for 48 h, and the expression of FXR and TGR5 was determined by real-time RT-PCR analysis. Data are mean + SEM (n = 6). *Significantly different from DMSO-treated controls, p < 0.05.
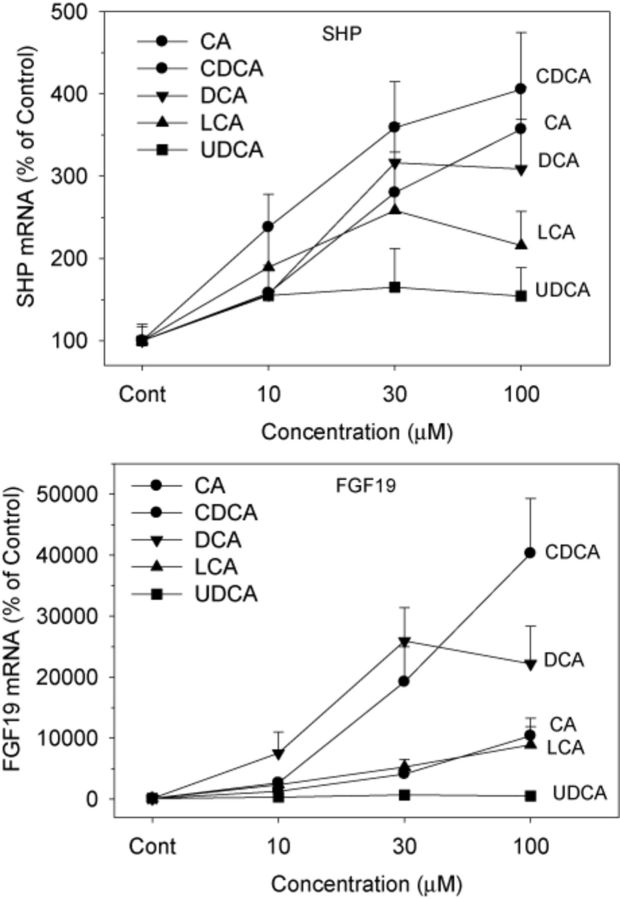
Both FXR-SHP and FXR-FGF19 pathways are major BA homeostasis regulators (Chiang, 2009). In contrast to modest induction of FXR and TGR5, BAs markedly induced small heterodimer partner (SHP) (up to fourfold) and FGF19 (up to 400-fold), with clear dose-responses (Fig. 4). All BAs except UDCA increased the mRNA expression of SHP, and CDCA was the most effective, followed closely by CA and DCA. In comparison, all BAs increased the mRNA of FGF19. For example, CDCA increased FGF19 17-fold at 10μM, and 400-fold at 100μM. Even UDCA increased FGF19 by sixfold at 100μM. The rank orders of FGF19 induction were CDCA ≈ CA > DCA ≈ LCA > UDCA.
FIG. 4.
Effects of BAs on the mRNA levels of SHP and FGF19. Human primary hepatocyte cultures were treated with individual BAs at concentrations of 10, 30, and 100μM for 48 h, and the expression of SHP and FGF19 was determined by real-time RT-PCR analysis. Data are mean + SEM (n = 6).
Effects of BAs on BA Efflux Transporters
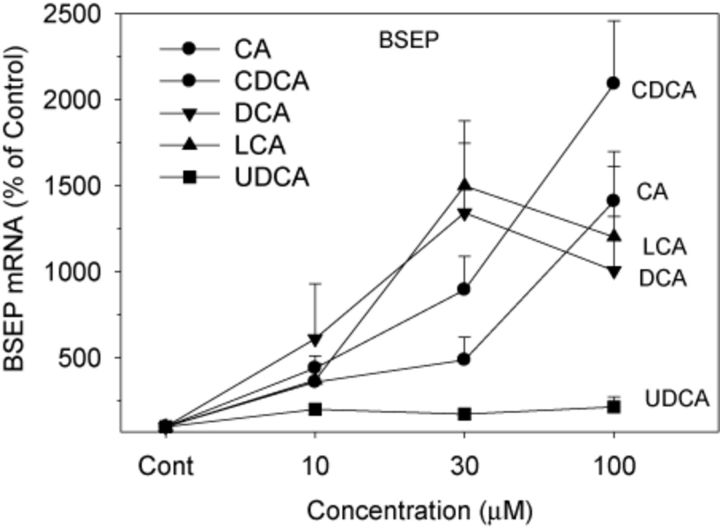
BSEP is a major BA efflux transporter responsible for the efflux of BAs into bile canaliculi, and thus for the prevention of cellular BA accumulation and toxicity (Marion et al., 2012; Ogimura et al., 2011). The expression of BSEP was increased 10–20-fold by all BAs except for UDCA (Fig. 5). CDCA was most effective in induction of BSEP at 100μM, followed by CA. Compared with CDCA and CA, both LCA and DCA were more effective at 30μM, but were less effective at 100μM. UDCA was ineffective in induction of BSEP at all concentrations.
FIG. 5.
Effects of BAs on mRNA levels of BSEP. Human primary hepatocyte cultures were treated with individual BAs at concentrations of 10, 30, and 100μM for 48 h, and the expression of BSEP was determined by real-time RT-PCR analysis. Data are mean + SEM (n = 5). *Significantly different from DMSO-treated controls, p < 0.05.
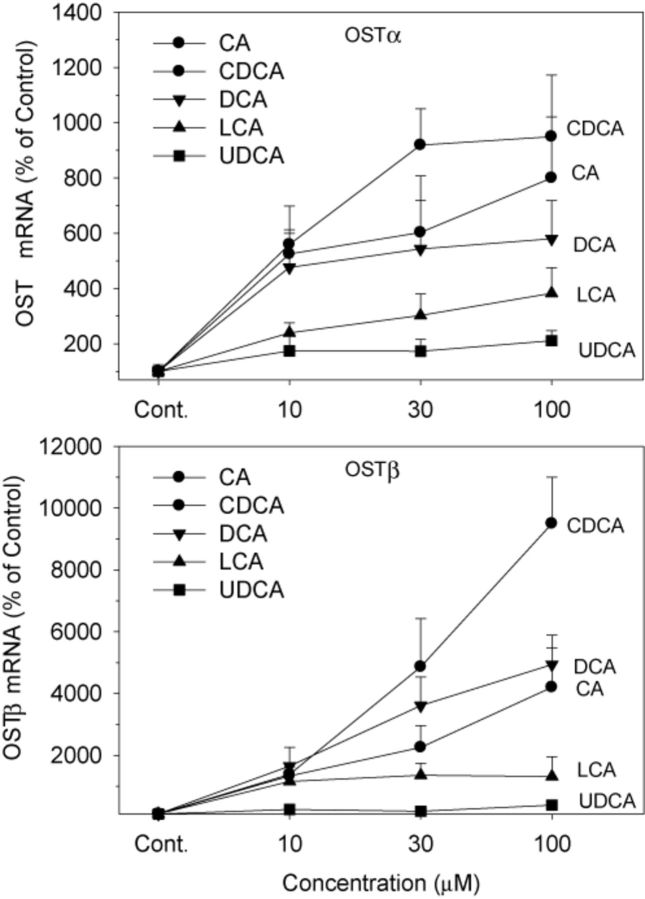
OSTα and OSTβ form heterodimers for the efflux of BAs from the liver back into the circulation (Ballatori et al., 2005). Treatment with CDCA, CA, LCA, and DCA increased the mRNA of OSTα 3–10-fold, and CDCA was the most effective (Fig. 6). UDCA was ineffective in induction of OSTα at any concentration. In comparison, CDCA at 100μM markedly increased OSTβ 95-fold, DCA and CA increased OSTβ 40–50-fold, and LCA 13-fold. UDCA at the highest concentration of 100μM increased OSTβ fourfold (Fig. 6).
FIG. 6.
Effects of BAs on mRNA levels of OSTα and OSTβ. Human primary hepatocyte cultures were treated with individual BAs at concentrations of 10, 30, and 100μM for 48 h, and the expression of OSTα and OSTβ was determined by real-time RT-PCR analysis. Data are mean + SEM (n = 5).
Effects of BA Treatments on OATP1B3 and MRP3 mRNA Expression
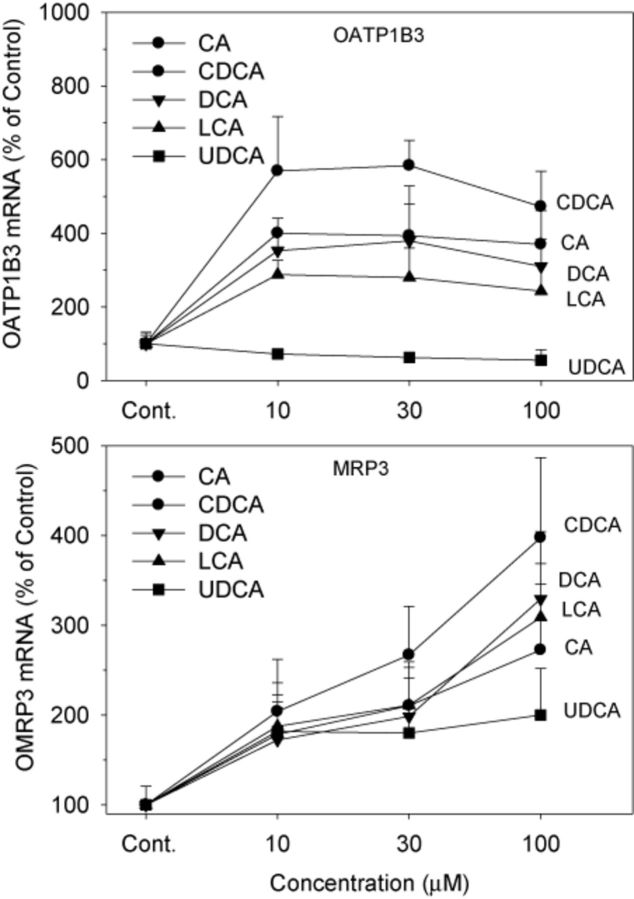
Both OATP1B1 and OATP1B3 are responsible for hepatic uptake of xenobiotics (Klaassen and Aleksunes, 2010), and Oatp1b2-null mice are deficient in hepatic uptake of unconjugated BAs (Csanaky et al., 2011). In human hepatocytes, OATP1B3 is a transporter of BAs (Briz et al., 2006; Mahagita et al., 2007). In the present study, BAs, except for UDCA, increased mRNA expression of OATP1B3 3–6-fold (Fig. 7). CDCA was the most effective, CA and DCA intermediate, followed by LCA. UDCA did not increase mRNA levels of OATP1B3; instead, it slightly decreased OATP1B3 expression by 25–40%.
FIG. 7.
Effects of BAs on the mRNA levels of OATP1B3 and MRP3. Human primary hepatocyte cultures were treated with individual BAs at concentrations of 10, 30, and 100μM for 48 h, and the expression of OATP1B3 and MRP3 was determined by real-time RT-PCR analysis. Data are mean + SEM (n = 4).
MRP3 is a transporter that effluxes BAs from liver into blood (Klaassen and Aleksunes, 2010). In the present study, all BAs increased mRNA expression of MRP3 in a concentration-dependent manner (Fig. 7). Again, CDCA was the most effective, and UDCA was the least effective.
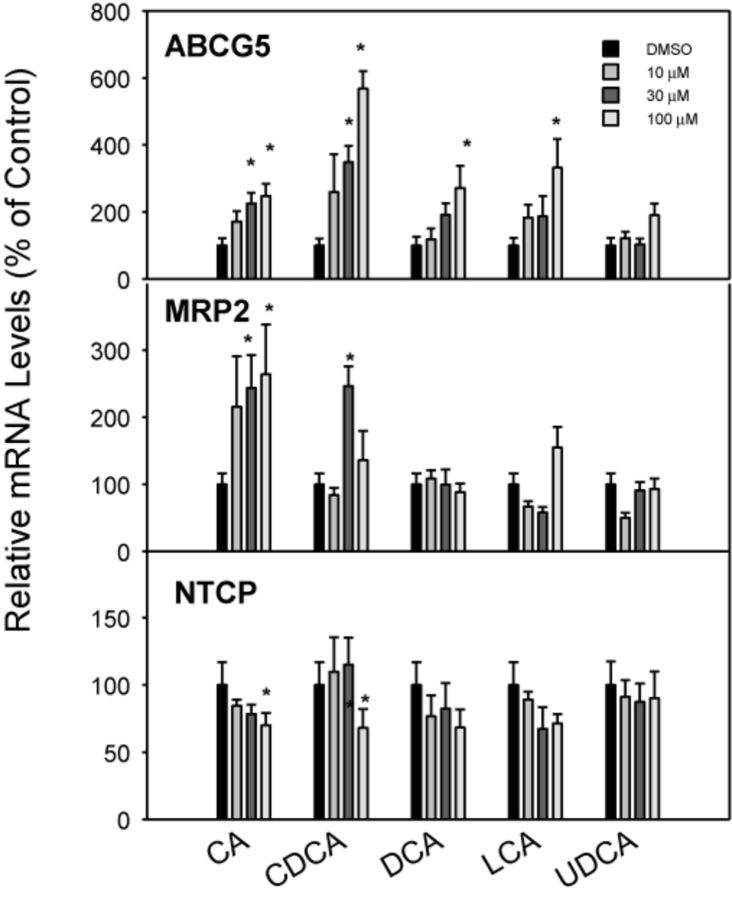
Effects of BA Treatments on ABCG5, MRP2, and NTCP mRNA Expression
The ABCG5 and ABCG8 genes encode half-transporter proteins that heterodimerize to form a transporter for efflux of plant sterols and BAs (Dieter et al., 2004; Klaassen and Aleksunes, 2010). All BAs except UDCA increased the expression of the efflux transporter ABCG5 by 2–5-fold, and CDCA was the most effective (Fig. 8). The effects of BA treatments on hepatic efflux transporter MRP2 are shown in the middle panel of Figure 8. Only the primary BAs (CA and CDCA) moderately increased mRNA expression of MRP2 at high concentrations (30–100μM) by about threefold, whereas DCA, LCA, and UDCA were ineffective. NTCP is thought to be the main uptake transporter in the liver for conjugated BAs (Csanaky et al., 2011). The mRNA expression of NTCP was slightly decreased at a high concentration (100μM) of CDCA and CA, and was not significantly altered by any concentration of DCA, LCA, or UDCA (Fig. 8).
FIG. 8.
Effects of BAs on mRNA levels of ABCG5, MRP2, and NTCP. Human primary hepatocyte cultures were treated with individual BAs at concentrations of 10, 30, and 100μM for 48 h, and the expression of ABCG5, MRP2, and NTCP was determined by real-time RT-PCR analysis. Data are mean + SEM (n = 3–4). *Significantly different from DMSO-treated controls, p < 0.05.
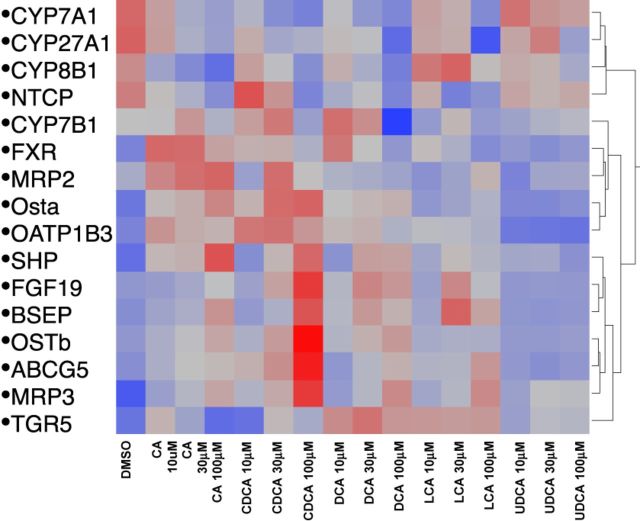
Hierarchical Clustering Dendrogram
Patterns of expression of mRNAs in response to BA treatment of human hepatocytes are shown in Figure 9. The 20 selected genes are important in BA biosynthesis (CYP7A1, CYP8B1, CYP27A1, and CYP7B1), BA homeostasis (FXR, SHP, FGF19, and TGR5), BA efflux (BSEP, OSTα, OSTβ, and MRP3), and other transporters (OATP1B3, ABCG5, MRP2, and NTCP). BAs are known to decrease BA synthesis and as noted in the top panel of Figure 9, DCA was the most potent and effective BA in decreasing the mRNA of CYP7A1, 27A1, and 8B1. Following DCA in potency and effectiveness was CDCA, followed by CA and LCA, and UDCA was relatively ineffective. The bottom portion of the figure indicates the mRNA of genes that were increased by BAs. It has been shown previously that BAs increase SHP and FGF19 (Inagaki et al., 2005), and CDCA was the most potent and effective in increasing these two genes, as well as BSEP, OSTβ, ABCG5, and MRP3. Whereas CDCA was the most potent and effective in increasing the mRNA of these genes, it was followed by DCA and CA, and UDCA was relatively ineffective in altering the expression of any of these genes.
FIG. 9.
The heatmap of BA treatment on BA homeostasis gene expression. Red indicates up-regulation and blue indicates down-regulation.
DISCUSSION
The present study demonstrates that treatment of primary human hepatocytes with individual BAs produces significant changes in mRNA expression of genes encoding BA synthesis, BA signaling, and BA transport. The primary purpose of this study is to simultaneously compare five different BAs, i.e., the primary BAs (CA and CDCA) and secondary BAs (DCA and LCA), and the therapeutic UDCA in altering the expression of these genes. Previous studies have shown that using cells derived from a money kidney (CV-1), FXR was activated the most by CDCA, and was followed by DCA > LCA > CA (Makishima et al., 1999; Parks et al., 1999).
CYP7A1 is the rate-limiting enzyme of BA synthesis. In the present study, CYP7A1 is markedly repressed by BA treatments of human hepatocytes. The present results (Fig. 1) are consistent with other reports that CDCA reduces CYP7A1 in human primary hepatocytes and in human cell lines (Han and Chiang, 2009, 2010; Li et al., 2006, 2011; Owsley and Chiang, 2003; Song et al., 2007, 2009). What is novel in the present study is that the CDCA reduction of CYP7A1 is simultaneously compared with the ability of CA, DCA, LCA, and UDCA at the same concentrations in six human donor livers to decrease CYP7A1. The results demonstrate that CDCA and DCA are very effective in suppression of CYP7A1, whereas CA and LCA have moderate effects, and UDCA is ineffective. Thus, individual BAs vary in their capability to decrease the mRNA of CYP7A1.
Other enzymes involved in BA synthesis, namely CYP27A1 and CYP8B1, were also categorized as mRNAs that decreased with increase in BA concentration (Fig. 9), however the decrease in expression of these two enzymes was much less than CYP7A1.
Whereas BAs can decrease the mRNA of their synthesis genes, they can also enhance the mRNA of some BA transporter genes. The primary transporter responsible for bile salt secretion into bile is the bile salt export pump (BSEP, ABCB11), a member of the ATP-binding cassette (ABC) superfamily, which is located at the bile canalicular domain of hepatocytes (Lam et al., 2010). Several in vitro studies have shown that CDCA up-regulates BSEP mRNA in HepG2 cells (Plass et al., 2002), and in human hepatocytes (Song et al., 2007). The present study extends these studies and demonstrates that BSEP is also induced by CA, DCA, CDCA, and LCA, but not UDCA. BSEP inhibitors increase cellular BA concentrations when exposed to BA mixtures in human and rat hepatocytes, and this inhibition is considered as a mechanism of drug-induced hepatotoxicity (Marion et al., 2012; Ogimura et al., 2011). Thus, increases in BSEP in human hepatocytes can be envisioned as an adaptive mechanism to reduce cellular BA burden to avoid BA toxicity.
Ostα/Ostβ heterodimers efflux BAs from the liver back into the blood (Ballatori et al., 2005; Boyer et al., 2006). The mRNA expression of the two genes is increased in patients with PBC, as well as in bile-duct ligated (BDL) rats (Boyer et al., 2006). In FXR-null mice, the mRNA expression of Ostα and Ostβ after BDL did not increase, suggesting that the induction of Ostα/Ostβ by BAs is FXR dependent (Boyer et al., 2006). The present data demonstrate that CDCA is the most effective inducer of Ostα (up to 10-fold) and Ostβ (up to 100-fold), followed by DCA and CA, and LCA being less effective, and UDCA the least effective. Drugs that inhibit BA transport in human hepatocytes also result in BA accumulation in rats, which corresponds to increased hepatotoxicity (Kostrubsky et al., 2003). A better understanding of BA regulation of transporters may aid the development of a therapeutic strategy for cholestasis.
MRP3/ABCC3 is also located on the sinusoidal membrane of hepatocytes, and it transports BAs out of hepatocytes back into the blood. Increased MRP3 is thought to be beneficial for elimination of increased BAs in hepatocytes during cholestasis (Klaassen and Slitt, 2005). Thus, the induction of the major BA efflux transporters (MRP2, MRP3, Ostα/Ostβ) by BAs may help to prevent BA-induced hepatotoxicity.
The major role of ABCG5 is to transport plant sterols out of cells (Dieter et al., 2004; Klaassen and Aleksunes, 2010). Feeding mice 1% CDCA increases hepatic expression of Abcg5/Abcg8 expression twofold (Dieter et al., 2004). Feeding mice BAs is likely to result in an accumulation of cholesterol and oxysterols, which are known activators of liver X receptor (LXR). The activation of LXR by oxysterols is known to up-regulate Abcg5/Abcg8 (Repa et al., 2002). In the present study with human hepatocytes, high concentrations (30–100μM) of CDCA, CA, DCA, and LCA increased ABCG5 3–5-fold. Therefore, BA-induced increase in ABCG5 mRNA expression may contribute to increased cholesterol efflux out of hepatocytes.
NTCP/SLC10A1 is considered the main transporter for uptake of conjugated BA into hepatocytes (Klaassen and Aleksunes, 2010). Down-regulation of Ntcp mRNA has been observed in several cholestatic animal models, including BDL rats (decreased 60%) (Slitt et al., 2007). However, whether this is due directly to BAs or hepatotoxicity is not known. In the present study, BAs have no appreciable effects on NTCP mRNA expression in cultured human hepatocytes. Only CDCA and CA at the highest concentration decreased NTCP mRNA levels.
The nuclear BA receptor FXR plays a central role in the feedback repression of CYP7A1 through the FXR-SHP (Chiang, 2009) and the FXR-FGF19 signaling pathways (Holt et al., 2003; Song et al., 2009), and is thought also to be important in the up-regulation of BA transporters. In the present study, the FXR target gene SHP was most effectively induced by CDCA, intermediate by CA and DCA, least by LCA, and not by UDCA. The ability of BAs to induce FGF19, the other major target gene of FXR, was led by CDCA and CA as the most effective, DCA and LCA intermediate, and DCA had no effect. Thus, the ability of individual BAs to activate FXR in cultured liver cells is quite different than reported previously using a kidney cell line where LCA was the second most effective BA to activate FXR, and CA was least effective (Makishima et al., 1999; Parks et al., 1999). This difference might be due to marked differences in the transport of BAs by the liver and kidney. It should be noted that the magnitude of FGF19 mRNA induction (up to 400-fold) is much higher than for SHP (fourfold). It was first shown that mouse Fgf15 (human homolog FGF19) was synthesized by the terminal small intestine and delivered to the liver to decrease BA synthesis (Inagaki et al., 2005). However, it was later shown (Song et al., 2009) that human hepatocytes synthesize FGF19, whereas it appears mouse liver does not synthesize Fgf15. In mice, it is thought that Fgf15 is more important than SHP down-regulation of BA synthesis (Kong et al., 2012; Song et al., 2009).
This study demonstrated that human primary hepatocyte cultures are a good model to study the potency of individual BA to regulate gene expression in human hepatocytes. The results obtained, together with our recent in vivo study in rats using these five individual BAs (Song et al., 2011), would greatly add to our understanding of physiological and toxicological effects of these five individual BAs, especially for CDCA, a primary BA with most potent effects on human hepatocytes (Song et al., 2009).
In summary, both the FXR-SHP and FXR-FGF19 signaling pathways are activated in cultured human hepatocytes by treatments with primary and secondary BAs. The classic pathway of (CYP7A1) BA synthesis is depressed more than the alternative pathway (CYP27A1). BA treatments dramatically activated the FXR-FGF19 pathway (100-fold), along with activation of the FXR-SHP pathway (fourfold). Notably, BA treatment of human hepatocytes also increased BA efflux transporters, namely BSEP, OSTα, and OSTβ. Among the BAs investigated, in general, CDCA is the most potent and effective, DCA and CA intermediate, LCA the least, whereas UDCA is generally ineffective in modulating these genes that regulate BA homeostasis.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Health (ES-013714, ES-09716, DK-081461, RR-021940).
Supplementary Material
Acknowledgments
The authors thank the students in Dr Klaassen's Lab for experimental help and critical review of this manuscript.
Footnotes
The authors certify that all research involving human subjects was done under full compliance with all government policies and the Helsinki Declaration.
REFERENCES
- Ballatori N., Christian W. V., Lee J. Y., Dawson P. A., Soroka C. J., Boyer J. L., Madejczyk M. S., Li N. OSTalpha-OSTbeta: A major basolateral bile acid and steroid transporter in human intestinal, renal, and biliary epithelia. Hepatology. 2005;42:1270–1279. doi: 10.1002/hep.20961. [DOI] [PubMed] [Google Scholar]
- Boyer J. L., Trauner M., Mennone A., Soroka C. J., Cai S. Y., Moustafa T., Zollner G., Lee J. Y., Ballatori N. Upregulation of a basolateral FXR-dependent bile acid efflux transporter OSTalpha-OSTbeta in cholestasis in humans and rodents. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:G1124–G1130. doi: 10.1152/ajpgi.00539.2005. [DOI] [PubMed] [Google Scholar]
- Briz O., Romero M. R., Martinez-Becerra P., Macias R. I., Perez M. J., Jimenez F., San Martin F. G., Marin J. J. OATP8/1B3-mediated cotransport of bile acids and glutathione: An export pathway for organic anions from hepatocytes. J. Biol. Chem. 2006;281:30326–30335. doi: 10.1074/jbc.M602048200. [DOI] [PubMed] [Google Scholar]
- Chiang J. Y. Bile acids: Regulation of synthesis. J. Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csanaky I. L., Lu H., Zhang Y., Ogura K., Choudhuri S., Klaassen C. D. Organic anion-transporting polypeptide 1b2 (Oatp1b2) is important for the hepatic uptake of unconjugated bile acids: Studies in Oatp1b2-null mice. Hepatology. 2011;53:272–281. doi: 10.1002/hep.23984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieter M. Z., Maher J. M., Cheng X., Klaassen C. D. Expression and regulation of the sterol half-transporter genes ABCG5 and ABCG8 in rats. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2004;139:209–218. doi: 10.1016/j.cca.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Ellis E., Axelson M., Abrahamsson A., Eggertsen G., Thörne A., Nowak G., Ericzon B. G., Björkhem I., Einarsson C. Feedback regulation of bile acid synthesis in primary human hepatocytes: Evidence that CDCA is the strongest inhibitor. Hepatology. 2003;38:930–938. doi: 10.1053/jhep.2003.50394. [DOI] [PubMed] [Google Scholar]
- Ellis E. C., Nilsson L. M. The use of human hepatocytes to investigate bile acid synthesis. Methods Mol. Biol. 2010;640:417–430. doi: 10.1007/978-1-60761-688-7_22. [DOI] [PubMed] [Google Scholar]
- Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- Han S., Chiang J. Y. Mechanism of vitamin D receptor inhibition of cholesterol 7alpha-hydroxylase gene transcription in human hepatocytes. Drug Metab. Dispos. 2009;37:469–478. doi: 10.1124/dmd.108.025155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C., Gnerre C., Fraser D. J., Martinez-Jimenez C., Jover R., Meyer U. A. Species-specific mechanisms for cholesterol 7alpha-hydroxylase (CYP7A1) regulation by drugs and bile acids. Arch. Biochem. Biophys. 2005;434:75–85. doi: 10.1016/j.abb.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Han S., Li T., Ellis E., Strom S., Chiang J. Y. A novel bile acid-activated vitamin D receptor signaling in human hepatocytes. Mol. Endocrinol. 2010;24:1151–1164. doi: 10.1210/me.2009-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillaire S., Ballet F., Franco D., Setchell K. D., Poupon R. Effects of ursodeoxycholic acid and chenodeoxycholic acid on human hepatocytes in primary culture. Hepatology. 1995;22:82–87. [PubMed] [Google Scholar]
- Hofmann A. F., Hagey L. R. Bile acids: Chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell. Mol. Life Sci. 2008;65:2461–2483. doi: 10.1007/s00018-008-7568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt J. A., Luo G., Billin A. N., Bisi J., McNeill Y. Y., Kozarsky K. F., Donahee M., Wang D. Y., Mansfield T. A., Kliewer S. A., et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17:1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Jahan A., Chiang J. Y. Cytokine regulation of human sterol 12alpha-hydroxylase (CYP8B1) gene. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;288:G685–G695. doi: 10.1152/ajpgi.00207.2004. [DOI] [PubMed] [Google Scholar]
- Klaassen C. D., Aleksunes L. M. Xenobiotic, bile acid, and cholesterol transporters: Function and regulation. Pharmacol. Rev. 2010;62:1–96. doi: 10.1124/pr.109.002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen C. D., Slitt A. L. Regulation of hepatic transporters by xenobiotic receptors. Curr. Drug Metab. 2005;6:309–328. doi: 10.2174/1389200054633826. [DOI] [PubMed] [Google Scholar]
- Kong B., Wang L., Chiang J. Y., Zhang Y., Klaassen C. D., Guo G. L. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology. 2012;56:1034–1043. doi: 10.1002/hep.25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrubsky V. E., Strom S. C., Hanson J., Urda E., Rose K., Burliegh J., Zocharski P., Cai H., Sinclair J. F., Sahi J. Evaluation of hepatotoxic potential of drugs by inhibition of bile-acid transport in cultured primary human hepatocytes and intact rats. Toxicol. Sci. 2003;76:220–228. doi: 10.1093/toxsci/kfg217. [DOI] [PubMed] [Google Scholar]
- Lam P., Soroka C. J., Boyer J. L. The bile salt export pump: Clinical and experimental aspects of genetic and acquired cholestatic liver disease. Semin. Liver Dis. 2010;30:125–133. doi: 10.1055/s-0030-1253222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Chiang J. Y. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;288:G74–G84. doi: 10.1152/ajpgi.00258.2004. [DOI] [PubMed] [Google Scholar]
- Li T., Jahan A., Chiang J. Y. Bile acids and cytokines inhibit the human cholesterol 7 alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 2006;43:1202–1210. doi: 10.1002/hep.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Matozel M., Boehme S., Kong B., Nilsson L. M., Guo G., Ellis E., Chiang J. Y. Overexpression of cholesterol 7α-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology. 2011;53:996–1006. doi: 10.1002/hep.24107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahagita C., Grassl S. M., Piyachaturawat P., Ballatori N. Human organic anion transporter 1B1 and 1B3 function as bidirectional carriers and do not mediate GSH-bile acid cotransport. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293:G271–G278. doi: 10.1152/ajpgi.00075.2007. [DOI] [PubMed] [Google Scholar]
- Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- Marion T. L., Perry C. H., St Claire R. L., 3rd, Brouwer K. L. Endogenous bile acid disposition in rat and human sandwich-cultured hepatocytes. Toxicol. Appl. Pharmacol. 2012;261:1–9. doi: 10.1016/j.taap.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogimura E., Sekine S., Horie T. Bile salt export pump inhibitors are associated with bile acid-dependent drug-induced toxicity in sandwich-cultured hepatocytes. Biochem. Biophys. Res. Commun. 2011;416:313–317. doi: 10.1016/j.bbrc.2011.11.032. [DOI] [PubMed] [Google Scholar]
- Owsley E., Chiang J. Y. Guggulsterone antagonizes farnesoid X receptor induction of bile salt export pump but activates pregnane X receptor to inhibit cholesterol 7alpha-hydroxylase gene. Biochem. Biophys. Res. Commun. 2003;304:191–195. doi: 10.1016/s0006-291x(03)00551-5. [DOI] [PubMed] [Google Scholar]
- Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., et al. Bile acids: Natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Plass J. R., Mol O., Heegsma J., Geuken M., Faber K. N., Jansen P. L., Muller M. Farnesoid X receptor and bile salts are involved in transcriptional regulation of the gene encoding the human bile salt export pump. Hepatology. 2002;35:589–596. doi: 10.1053/jhep.2002.31724. [DOI] [PubMed] [Google Scholar]
- Pols T. W., Noriega L. G., Nomura M., Auwerx J., Schoonjans K. The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J. Hepatol. 2011;54:1263–1272. doi: 10.1016/j.jhep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa J. J., Berge K. E., Pomajzl C., Richardson J. A., Hobbs H., Mangelsdorf D. J. Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J. Biol. Chem. 2002;277:18793–18800. doi: 10.1074/jbc.M109927200. [DOI] [PubMed] [Google Scholar]
- Russell D. W. Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 2009;50:S120–S125. doi: 10.1194/jlr.R800026-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H., Macchiarulo A., Thomas C., Gioiello A., Une M., Hofmann A. F., Saladin R., Schoonjans K., Pellicciari R., Auwerx J. Novel potent and selective bile acid derivatives as TGR5 agonists: Biological screening, structure-activity relationships, and molecular modeling studies. J. Med. Chem. 2008;51:1831–1841. doi: 10.1021/jm7015864. [DOI] [PubMed] [Google Scholar]
- Slitt A. L., Allen K., Morrone J., Aleksunes L. M., Chen C., Maher J. M., Manautou J. E., Cherrington N. J., Klaassen C. D. Regulation of transporter expression in mouse liver, kidney, and intestine during extrahepatic cholestasis. Biochim. Biophys. Acta. 2007;1768:637–647. doi: 10.1016/j.bbamem.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Song K. H., Chiang J. Y. Glucagon and cAMP inhibit cholesterol 7alpha-hydroxylase (CYP7A1) gene expression in human hepatocytes: Discordant regulation of bile acid synthesis and gluconeogenesis. Hepatology. 2006;43:117–125. doi: 10.1002/hep.20919. [DOI] [PubMed] [Google Scholar]
- Song K. H., Ellis E., Strom S., Chiang J. Y. Hepatocyte growth factor signaling pathway inhibits cholesterol 7alpha-hydroxylase and bile acid synthesis in human hepatocytes. Hepatology. 2007;46:1993–2002. doi: 10.1002/hep.21878. [DOI] [PubMed] [Google Scholar]
- Song K. H., Li T., Owsley E., Strom S, Chiang J. Y. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009;49:297–305. doi: 10.1002/hep.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P., Zhang Y., Klaassen C. Dose-response of five bile acids on liver and serum bile acid concentrations and hepatoxicity in mice. Toxicol. Sci. 2011;123:359–367. doi: 10.1093/toxsci/kfr177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahcevic Z. R., Pandak W. M., Stravitz R. T. Regulation of bile acid biosynthesis. Gastroenterol. Clin. North Am. 1999;28:1–25. doi: 10.1016/s0889-8553(05)70041-8. [DOI] [PubMed] [Google Scholar]
- Yu L., Li-Hawkins J., Hammer R. E., Berge K. E., Horton J. D., Cohen J. C., Hobbs H. H. Overexpression of ABCG5 and ABCG8 promotes biliary cholesterol secretion and reduces fractional absorption of dietary cholesterol. J. Clin. Invest. 2002;110:671–680. doi: 10.1172/JCI16001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Klaassen C. D. Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J. Lipid Res. 2010;51:3230–3342. doi: 10.1194/jlr.M007641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.