Dear Editor,
Plants grown in close proximity experience a change in light quality, and respond by reallocating energy resources from storage organs to stem-like organs. This adaptive response, called the shade-avoidance syndrome (SAS), allows the shaded plant to grow and compete effectively against its neighbors. SAS is initiated upon detection by the phytochrome photoreceptors of a lowering of the ratio of red to far-red light (R/FR), leading to the synthesis of plant hormones and a transcriptional cascade that targets genes involved in growth. Among these genes is PIL1 (PHYTOCHROME INTERACTING FACTOR 3-LIKE 1), which encodes a bHLH transcription factor, whose expression is induced by up to 100-fold within 30min of exposure to shade (Salter et al., 2003); yet, PIL1’s precise role in shade avoidance is unknown. The Salter paper concluded that PIL1 worked with TOC1 to restrict growth to a particular time of day, and that PIL1 is necessary for the normal display of the rapid elongation response to shade (Salter et al., 2003). Later, Roig-Villanova and colleagues (2006) showed that PIL1 is a negative regulator of the SAS with only phenotype of pil1-4 and pil1-4phyB without any mechanism. To further understand the function of PIL1 in transducing phytochrome signals during the shade-avoidance response, we examined phenotypes of PIL1 loss- and gain-of-function mutants in simulated shade and proposed three possible modes of PIL1 action based on its protein stability and interaction with DNA and PIFs to regulate gene expression.
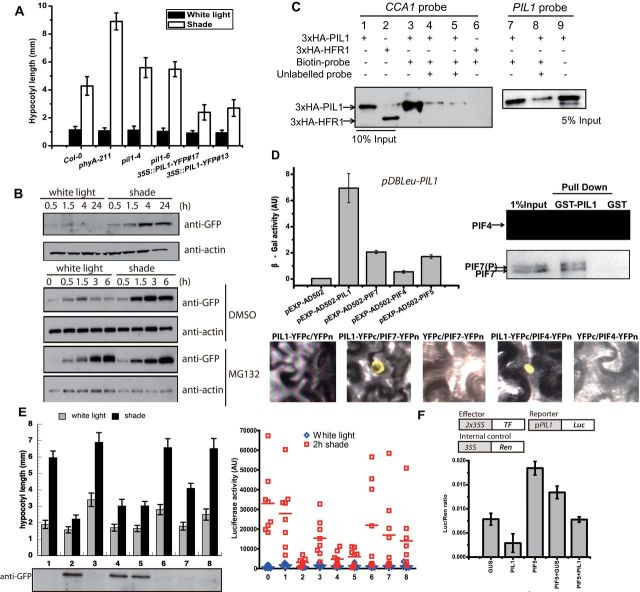
We first obtained two Arabidopsis mutants with T-DNA insertions (Salk_043937C termed pil1-4 and Salk_025598C termed pil1-6) in the PIL1 coding region and also generated plants that stably overexpressed a PIL1–YFP fusion protein under the CaMV 35S promoter (35S::PIL1–YFP #17 and #13). Consistently with a previous report (Roig-Villanova et al., 2006), pil1-4 and pil1-6 mutant seedlings had slightly longer hypocotyls under shade conditions (Figure 1A). Furthermore, hypocotyls of PIL1-overexpressing lines were ~50% shorter than wild-type under shade (Figure 1A). This observation suggests that PIL1 plays a role as a decelerator of growth during early shade avoidance.
Figure 1.
PIL1 Participates in a Negative Feedback Loop that Regulates Its Own Gene Expression in Response to Shade.
(A) Quantification of hypocotyl lengths of pil1 mutants and PIL1 overexpression line.
(B) PIL1 is degraded by the 26S proteasome in white light and accumulated under shade. DMSO or 50 μM MG132 was added 1 h before exposure to the different light conditions. Anti-GFP antibody was used to detect PIL1–YFP.
(C) DNA-binding activity of PIL1 by in vitro DNA-binding assay. Lanes 4, 5 (the same as 4), and 8 were used unlabeled probes to compete with a biotin-labeled probe. Anti-HA antibody was used to detect protein.
(D) PIL1 can interact with PIFs. In the left panel, pDBleu–PIL1 as bait was transformed with pEXP–AD502, pEXP–AD502–PIL1, pEXP–AD502–PIF7, pEXP–AD502–PIF4, and pEXP–AD502–PIF5 in yeast. In the right panel, proteins bound to GST–PIL1 were detected by immunoblotting using anti-myc antibody. In the bottom panel, BiFC assays show interaction between PIL1 and PIF7/PIF4.
(E) Negative regulation of shade-induced PIL1 transcriptional activation by PIL1 overexpression. Top panel shows hypocotyl length and luciferase activity in white light or shade from eight independent lines harboring a 35S::PIL1–CFP transgene in the pPIL1::LUC background. Average luciferase activities after shade treatment are shown as bars. ‘0’ presents LUC acitivity of pPIL1::LUC without transgene. Various PIL1–CFP protein levels are shown at the bottom.
(F) Transactivation activity of PIL1 promoter in tobacco using dual luciferase assay. PIL1 promoter activity expressed as a ratio of luciferase (Luc) to Renilla (Ren).
Although shade-induced accumulation of PIL1 transcripts is well documented, the regulation of PIL1 protein levels or activity has not been reported. Seedlings of line 35S::PIL1–YFP #17 were grown under continuous white light for 3 d, and then PIL1 protein levels were monitored over time from 0 to 24 h following transfer of seedlings from white light to shade. PIL1 protein gradually accumulated after seedlings were transferred to shade when compared with white light (Figure 1B). We then pretreated PIL1ox seedlings with 26S proteasome inhibitor MG132 or mock-treated with solvent control DMSO and then subjected the seedlings to shade or white-light conditions. MG132 treatment led to accumulation of PIL1 in white light. This indicates a white-light-dependent proteasome degradation of PIL1 protein (Figure 1B) which may explain why pil1 mutant and overexpression have no obvious phenotypes under white-light (high R/FR) conditions. To further examine light-mediated control of PIL1 stability, we measured protein accumulation in etiolated seedlings upon transfer to R or FR light. As shown in the Supplementary Data, long-term FR light treatment slightly stabilized PIL1 whereas R light had the opposite effect.
Previous studies have shown that the atypical HLH transcription factors HFR1, PAR1, and PAR2 are negative regulators of the shade-avoidance response (Hornitschek et al., 2009; Galstyan et al., 2011). These proteins do not directly bind DNA. Instead, they function through modulating the activity of other DNA-binding bHLHs, such as PIF4 and PIF5, by interactions through the HLH domain (Hornitschek et al., 2009; Galstyan et al., 2011; Hao et al., 2012). PIL1 is predicted to be a typical bHLH protein with H/K9–E13–R17 DNA-binding domain. We tested whether PIL1 can bind DNA by employing a previously described in vitro DNA-binding assay (Vert and Chory, 2006). 3xHA–PIL1 and 3xHA–HFR1 were synthesized in cell-free extracts to test binding to biotin-labeled dsDNA probes. We chose the G-box-containing region from CCA1 promoter (–301/–266) and PIL1 promoter (–1412/–1375) as probe. PIL1 pelleted readily with the G-box containing dsDNA probes (Figure 1C, lanes 3 and 7). This binding was effectively competed by an unlabeled DNA probe (Figure 1C, lanes 4, 5, and 8), indicating that PIL1 can directly bind to DNA similarly to PIFs whereas HFR1 (Figure 1C, lane 6) cannot.
In addition, we tested whether PIL1 is able to bind with PIFs. In yeast two-hybrid assays, the β-Gal reporter was activated when PIL1 was co-expressed with PIL1, PIF7, PIF4, and PIF5 (Figure 1D and Supplementary Data). A glutathione S-transferase (GST) pull-down assay was performed using GST–PIL1 purified from Escherichia coli which could pellet PIF4 and PIF7 from the extraction of seedlings overexpressing Flash-tagged (9Myc–6His–3Flag) PIF4 and PIF7 (Figure 1D and Supplementary Data). BiFC (Bimolecular Fluorescence Complementation) also confirmed the interaction between PIL1 and PIF7/PIF4, indicating that PIL1 is able to form a homodimer or heterodimers with PIFs in vivo.
To better understand how PIL1 gene expression is controlled and how it affects the shade-regulated transcription network, we constructed a transgenic line that fuses the PIL1 promoter to the firefly luciferase (pPIL1::LUC) reporter. The PIL1 from this transgenic line reported similar expression levels and responses to shade as the endogenous PIL1 locus. And the shade induction of LUC gene expression and activity in pil1-4 background was similar to that in wide-type (Supplementary Data). In contrast, PIL1–CFP overexpression in the pPIL1::LUC line reduced LUC activity and suppressed hypocotyl elongation. These phenotypes were associated with PIL1 overexpression, as they were only manifested in three independent lines (lines 2, 4, and 5) which succeeded in overexpression but not in the other five lines which failed to express the transgene (Figure 1E). To confirm the self-regulation directly, we conducted a transactivation assay in tobacco. We used the LUC reporter gene under the control of the 1.5-Kb region upstream from the translation initiation site of PIL1 as a reporter, and PIL1, PIF5, and control (GUS) were used as effectors under the control of a 2xCaMV 35S promoter. Lastly, a Renilla LUC gene was driven by the CaMV 35S promoter and used as a control for transformation efficiency (Figure 1F). Agrobacterium tumefaciens harboring this construct were infiltrated into tobacco and the ratio of Luc/Ren activity was measured in leaf punches for determining any effects on the PIL1 promoter. PIF5 has been shown as an activator of PIL1 transcription (Hornitschek et al., 2009) and, consistently, expression of PIF5 strongly increased the ratio of Luc/Ren, whereas expression of PIL1 reduced the ratio of Luc/Ren compared to the control samples (Figure 1F). When PIL1 and PIF5 were co-expressed at the same time, the ratio of Luc/Ren is between that from only PIL1 and only PIF5, and lower than co-expression of PIF5 and GUS. Besides PIL1 itself, we examined the expression level of YUCCA8, IAA5, and IAA29 by qRT–PCR in Col-0 and PIL1 overexpression lines treated by 1h of shade (Supplementary Data). Except a higher level of PIL1 in the PIL1 overexpression line, the expression of YUCCA8, IAA5, and IAA29 were lower than that in Col-0, which may explain the short hypocotyl length of PIL1 overexpression under shade.
Compared to other negative regulators of SAS, unlike HFR1 and PAR1/2, PIL1 can bind DNA and regulate gene expression (Figure 1E). On the other hand, similarly to HFR1, PIL1 could form heterodimers with PIFs. The attenuation of reporter by introducing PIL1 expression (Figure 1F) could be caused by either a homo-dimerization of PIL1 itself or hetero-dimerization of PIL1 and other PIF, namely PIF5. Thus, two modes of PIL1 action are possible: (1) PIL1 may outcompete PIFs for binding DNA or PIL1/PIF heterodimers may reduce the growth promoting function of PIFs. (2) PIL1 may directly regulate gene expression in a PIF-independent manner through binding to different sites in promoters of downstream genes.
Another difference with HFR1, PIL1 contains an Active Phytochrome B-binding (APB) domain which is required for phyB-specific binding (Khanna et al., 2004). Despite the lack of evidence for full-length PIL1 interacting with phyB in vitro, the PIL1 APB motif has been shown to bind phyB Pfr in a photo-reversible manner (Khanna et al., 2004). The APB domain is required for PIF turnover (Khanna et al., 2004; Al-Sady et al., 2006; Lorrain et al., 2008; Bu et al., 2011). It is not sure whether APB of PIL1 affects the protein stability, while PIL1 is degraded in the light (Figure 1B) with similar kinetics to PIFs, which raises the third possibility that PIL1 may outcompete PIFs for binding to phyB.
How do these negative regulators cooperate during shade? HFR1 is induced by up to 4 d of shade treatment, whereas PIL1 is rapidly induced by 1 h of shade treatment and is self-limited. PIL1 accumulates rapidly and transiently in response to shade, which might be an early signal for Arabidopsis to ‘pause growth’, thereby allowing the plant to determine whether prolonged shading is imminent. This would slow down a commitment to the shade-avoidance lifestyle if it were unnecessary. Finally, a more robust and long-lasting negative feedback loop involves other negative regulators: HFR1, PAR1, and PAR2 (Hornitschek et al., 2009; Galstyan et al., 2011), which ensures that plants sense sustained shade conditions and make a ‘self-confident’ decision.
FUNDING
This work was funded by the School of Life Sciences of Fudan University and the State Key Laboratory of Genetic Engineering and Institute of Plant Biology, People’s Republic of China to L.L. and a grant from the NIH (RO1GM52413 to J.C.), by funding to J.C. from the Howard Hughes Medical Institute and by the Office of Science (BER) Department of Energy Grant DE-FG02-08ER64700DE to S.P.H. No conflict of interest declared.
Supplementary Material
REFERENCES
- Al-Sady B., Ni W., Kircher S., Schafer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell. 23, 439–446. [DOI] [PubMed] [Google Scholar]
- Bu Q., Zhu L., Dennis M.D., Yu L., Lu S.X., Person M.D., Tobin E.M., Browning K.S., Huq E. (2011). Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis . J. Biol. Chem. 286, 12066–12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galstyan A., Cifuentes-Esquivel N., Bou-Torrent J., Martinez-Garcia J.F. (2011). The shade avoidance syndrome in Arabidopsis: a fundamental role for atypical basic helix–loop–helix proteins as transcriptional cofactors. Plant J. 66, 258–267. [DOI] [PubMed] [Google Scholar]
- Hao Y., Oh E., Choi G., Liang Z., Wang Z.Y. (2012). Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant. 5, 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28, 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix–loop–helix transcription factors. Plant Cell. 16, 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323. [DOI] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou J., Sorin C., Devlin P.F., Martinez-Garcia J.F. (2006). Identification of primary target genes of phytochrome signaling: early transcriptional control during shade avoidance responses in Arabidopsis . Plant Physiol. 141, 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter M.G., Franklin K.A., Whitelam G.C. (2003). Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature. 426, 680–683. [DOI] [PubMed] [Google Scholar]
- Vert G., Chory J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature. 441, 96–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.