Abstract
Background. Differentiation between gametocyte-producing Plasmodium falciparum clones depends on both high levels of stage-specific transcripts and high genetic diversity of the selected genotyping marker obtained by a high-resolution typing method. By analyzing consecutive samples of one host, the contribution of each infecting clone to transmission and the dynamics of gametocyte production in multiclone infections can be studied.
Methods. We have evaluated capillary electrophoresis based differentiation of 6 length-polymorphic gametocyte genes. RNA and DNA of 25 µL whole blood from 46 individuals from Burkina Faso were simultaneously genotyped.
Results. Highest discrimination power was achieved by pfs230 with 18 alleles, followed by pfg377 with 15 alleles. When assays were performed in parallel on RNA and DNA, 85.7% of all pfs230 samples and 59.5% of all pfg377 samples contained at least one matching genotype in DNA and RNA.
Conclusions. The imperfect detection in both, DNA and RNA, was identified as major limitation for investigating transmission dynamics, owing primarily to the volume of blood processed and the incomplete representation of all clones in the sample tested. Abundant low-density gametocyte carriers impede clone detectability, which may be improved by analyzing larger volumes and detecting initially sequestered gametocyte clones in follow-up samples.
Keywords: Plasmodium falciparum, transmission dynamics, gametocytes, genotyping, capillary electrophoresis, pfg377, pfs230
Malaria infection and transmission dynamics both describe the appearance, loss or persistence of genotypes of Plasmodium parasites in a given host. Although infection dynamics describe longitudinal changes among asexual parasite clones, the focus of transmission dynamics lies on the sexual stages, gametocytes. To answer gaps in our knowledge on parasite reproduction and transmission, both the sexual and asexual stages, concurrently present in a host, need to be analyzed by genotyping. Examples of specific research questions are: Do all concurrent P. falciparum clones contribute to gametocyte production? Do drug-resistant clones contribute more to transmission? Does within-host competition between clones or other environmental factors affect the start and duration of gametocyte production?
Superinfections of already infected hosts and a high number of concurrent infections are common in areas of high malaria transmission. Polymorphic molecular markers are amplified to differentiate concurrent clones. The number of clones per blood sample (multiplicity) varies according to the transmission intensity; mean multiplicity of infection (MOI) was 2 in Papua New Guinea (PNG) and almost twice as much in Tanzania [1]. MOI is age-dependent and peaks in highly endemic settings in the age range of 5–9 year-olds [2]. Human and rodent models suggested that clone multiplicity affects transmission stages [3–5]. Antimalarials also were found to affect transmission. Residual submicroscopic parasitemia after ACT treatment was associated with a higher transmission in Kenyan children [4], but the individual clones within the infection were not differentiated in this study. A further determinant of transmission is the quantity and duration of gametocyte production. Asexual P. falciparum clones can persist in a host for many months as asymptomatic infections [6]. From this observation the question arises whether gametocytes are produced continuously by each clone, and whether gametocyte production is up-regulated or suppressed by concurrent clones of the same or other Plasmodium species.
Genotyping of gametocytes depends on high stage-specific expression and high genetic diversity of the chosen genotyping marker in the study area. Classical length-polymorphic markers for differentiation of gametocytes are pfs230 and pfg377. Pfs230 was first observed as a potential transmission-blocking antigen in 1988 and thereafter characterized by several immunological studies [7–10]. Williamson and co-workers first described 2 polymorphic repeat regions in pfs230 by comparing 5 cultured parasite lines [11]. A separate polymorphic, glutamate-rich region within pfs230 was described, but diversity was limited [12].
Another frequently used genotyping marker, pfg377, is specifically expressed in female gametocytes. Transcripts are detectable from gametocyte stage III onward [13]. Menegon and co-workers developed 4 pfg377 gametocyte genotyping assays [14]. The first longitudinal monitoring of gametocyte-producing clones was conducted in samples from Sudan. Results indicated that gametocytes were present for up to 8 months of dry season and thus were considered the most probable source of malaria outbreaks in the following rainy season [5, 15]. Gametocytes from multiclone P. falciparum infections persisted 3 times longer than those from single-clone infections; thus multiplicity of infection may promote either longer persistence or continuous production of gametocytes [5]. Feeding experiments in the Gambia confirmed that gametocytes from coinfecting clones were simultaneously transmitted to mosquitoes [16]. Despite a lower multiplicity of gametocyte clones compared to asexual MOI, it was found that clones not detected on RNA level still produced gametocytes and nevertheless contributed to transmission [16]. Of all asexual clones detected in Thai patients, 25% had no corresponding pfg377 transcript and thus no molecularly detectable level of gametocytes [17].
These previous studies have provided relevant information on malaria epidemiology and transmission dynamics but were hampered by the limited resolution of the available gametocyte-genotyping methods. Size-polymorphic diversity of molecular markers used in these earlier studies was maximal 7 for pfg377 and 4 for pfs230 [12, 16]. To improve the discriminatory power of markers pfg377 and pfs230, we created amplicons spanning several polymorphic domains of these genes and increased accuracy of fragment sizing by replacing gel-based sizing by capillary electrophoresis (CE). In addition, we screened the gametocyte transcriptome [18] for tandem repeat-containing genes expressed only in gametocytes and evaluated these in search for novel high-resolution markers. Our assays were applied to asexual parasites by targeting genomic DNA (gDNA) from field samples and in parallel to gametocytes from the same blood samples by targeting RNA. Our aim was to employ high-resolution typing to gain a clearer picture on how many coinfecting asexual clones simultaneously produce gametocytes.
METHODS
Study Population and Ethics
The diversity of genotyping markers was determined in 111 archived anonymized DNA samples collected in Madang province, PNG, from April 2004 to February 2005 [19]. Scientific approval and ethical clearance was obtained from the Medical Research and Advisory Committee of the Ministry of Health in PNG (MRAC no. 09.24). Informed consent was obtained from parents or legal guardians prior to sampling. In addition, 46 archived anonymous RNA samples collected in the course of a cluster-randomized trial in Saponé, Burkina Faso [NCT01256658] [20] were used for evaluation of gametocyte detection assays. Ethical clearance was obtained from the National Ethical Committee for Health Research of Burkina Faso (no. 2013-3-019).
Nucleic Acid Extraction
DNA samples from PNG, stored at −20°C, had been extracted previously using QIAamp DNA Blood Kit (Qiagen) [19]. Total RNA of Burkina Faso samples was extracted from 25 µL whole blood stored with 125 µL RNAprotect Cell reagent (Qiagen). RNeasy Plus 96 kit (Qiagen) was used as previously described [21]. RNA was eluted in 50 µL water. The gDNA was eluted simultaneously from the gDNA elimination column (provided by the kit) using the QIAamp 96 Blood DNA Kit (Qiagen) protocol from the column washing step onwards. The gDNA was eluted twice in 50 µL of 40°C prewarmed AE elution buffer (Qiagen) following 30 minutes incubation. RNA and gDNA samples were stored at −20°C.
Validation of Genotyping Assays and Determination of Allelic Diversity of Markers
Diversity of 6 genotyping markers was determined in 111 gDNA samples from PNG. Primer sequences for pfg377 (PF3D7_1250100), pfs230 (PF3D7_0209000), pf11.1 (PF3D7_1038400), PF11_0214 (PF3D7_1120700), PFI1210w (PF3D7_0924600), and PFL0545w (PF3D7_1211000) are given in Table 1. Composition of reaction mixes and thermo profiles are shown in Supplementary Table 1. For CE sizing the products were diluted in water according to their agarose gel band intensity. Samples were analyzed by ABI3130xl using GS500LIZ as size standard. Electropherograms were analyzed using GeneMapper Software version 3.7. A cutoff set at 250 fluorescence units (FU) defined the minimal required peak height. In samples containing dominant peaks of >10 000 FU, the cutoff was increased to 500 FU. Stutter peaks (defined by accompanying peaks with a regular pattern of >6 bp and heights <20% of the main peak) were censored. A bin width of 3 bp was defined for each allele to accommodate small variations in fragment sizing. To test whether a size standard containing larger fragments would provide more accurate CE sizing, a subset of 13 pfg377 fragments were simultaneously sized by CE using GS1200LIZ (Applied Biosystems). The expected heterozygosity (HE) was calculated as published elsewhere [22].
Table 1.
Primary and Nested Primer Sequences for Gametocyte Genotyping Markers
| Marker | Primer | Sequences (5′->3′) |
|---|---|---|
| Pfs230 | ||
| Primary | Pfs230_PF | AAG ACA TGT CGC CCA GGG ATA |
| Pfs230_PR | TTC TTC TTC ATC ACC AAA TGG ATA T | |
| Nested | Pfs230_NF | VIC - CAG GGA TAA TTT TGT AAT RGA TGA TGa |
| Pfs230_NR | ACC TTG CCT TTC TTT TTC ATC TAC A - tail | |
| Pfg377 | ||
| Primary | Pfg377_PF | CAC AAC GAA GGT TAT ATA CCT CAT AC |
| Pfg377_PR | TCC ATT CTT CTT TAA GGT TCG CTT C | |
| Nested | Pfg377_NF | 6FAM - GAA GAT GAC GAA GGG GAT GAA G |
| Pfg377_NR | CTG TAA GAA TTG GTT ATT ACT TTT GTG G - tail | |
| PF11.1 | ||
| Primary | Pf11.1_PF1b | GAT ATA TTC TAA TAA T|TG TTC CAA TGG |
| Pf11.1_PF2 | AAG TGC AGG GGA TAG TGC AG | |
| Pf11.1_PR | CGG TAA TAC CAT AAG CTC CTC CT | |
| Nested | Pf11.1_NF | 6FAM - GGA ATA AGG ATG ATG ATG ACG AA |
| Pf11.1_NR | AAC CTT CAA ATT CTT TGT CTC TTT C - tail | |
| PF11_0214 | ||
| Primary | PF11_0214_PF | TCG AGA CAA ATT GAA AAG TTA TGG |
| PF11_0214_PR | TTA GTG GAT AAA TGA ATA TCT ACC G | |
| Nested | PF11_0214_NF | 6FAM - AAT GAT ACA GAT TGT GAA GAA TGG T |
| PF11_0214_NR | TGA GGA ATA TCG TTT TGT ATA AAT GTT - tail | |
| PFI1210w | ||
| Primary | PFI1210w_PF | TTG ATA AGG GAT ATA TAC ACA ACC ATA |
| PFI1210w_PR | TTC CCG TTG TGT ATT TAA GTA GAA T | |
| Nested | PFI1210w_NF | 6FAM - TGT TTC AAT TTA CCA TCT TTC TTT TC |
| PFI1210w_NR | GTT TTT CAA TTT TTA TGT TGT TCT CCA - tail | |
| PFL0545w | ||
| Primary | PFL0545w_PF | GGA AGG AAA CGA AGA AGA AAC A |
| PFL0545w_PR | AAA GAT TGA AAT GGA GAT TCA CCT | |
| Nested | PFL0545w_NF | VIC - TGA CAA AGG GCA CTT TAT TAT TT |
| PFL0545w_NR | TTT CTT CAA CAG CAT TTT GCA T – tail | |
a Primer sequence contains a wobble: R = A/G.
b Pf11.1_PF1 is spanning an intron boundary indicated by “|”.
Sequencing of PCR Fragments of Single Clone Infections for Evaluating Fragment Sizing
Nucleotide sequences of 12 pfg377 and 10 pfs230 nested polymerase chain reaction (nPCR) fragments from single clone infections were determined in both directions for pfg377 and in one direction for pfs230 by direct Sanger sequencing. Sequences were analyzed with BioEdit version 7.3.2, and alignments were performed with T-Coffee multiple alignment server and BoxShade server version 3.21. Sequences were submitted to GenBank [KJ566743-KJ566764].
Evaluation of Sensitivity of Reverse Transcription (RT)-PCR
The detection limits of all nested RT-PCR assays were evaluated on a trendline of stage IV/V gametocyte in vitro culture of P. falciparum 3D7 as previously described [21]. RT of gene-specific complementary DNA (cDNA) was performed in a multiplex reaction using pfg377 and pfs230 primary reverse primers, 15 µL RNA and Superscript II (Invitrogen) according to the manufacturer's protocol. In a second multiplex RT reaction cDNA was reverse transcribed for pf11.1, PF10_0214, PFI1210w, and PFL0545w using the primary reverse primers (Table 1). In total, 4 µL of cDNA were added to the primary PCR (pPCR) mix and 2 µL of primary product to nPCR. The composition of reaction mixes and thermo profiles are shown in Supplementary Table 1. The nPCR products were run on a 2% agarose gel. The detection limit of each marker was compared to that of pfs25 qRT-PCR, which is highly sensitive and widely used [21].
Effects of DNase Treatment on RNA Quality
For marker pf11.1 an additional forward PCR primer was designed to span an exon-intron boundary (Table 1). Including this primer-binding site into an amplicon covering the polymorphic region of pf11.1 resulted in a 680 bp longer fragment. The sensitivity of both pf11.1 assays was assessed with 2 gametocyte trendlines that differed by omitting the DNase digest for the intron-spanning assay [21]. Assay conditions were identical except for a higher annealing temperature (58°C) for pPCR with the intron-spanning primer.
Evaluation of Gametocyte Genotyping Markers in Field Samples
The discrimination power for gametocyte clones in field samples was assessed for the 2 most diverse markers using 46 RNAs from Burkina Faso. Alleles detected on RNA level were compared to those found in gDNA of the same sample. In total, 5 µL RNA, equivalent to 2.5 µL whole blood, were reverse-transcribed and amplified for pfs230 and pfg377 by AffinityScript One-Step RT-PCR kit (Agilent Technologies) in simplex reactions. The nPCR was performed using 1 µL of primary product. Composition of reaction mixes and thermo profiles are shown in Supplementary Table 1. Reaction conditions were modified because the SuperScript II protocol (Invitrogen) used for work on parasite culture performed less well in field samples (data not shown).
The gDNA coextracted from the same blood samples was amplified for pfg377 and pfs230 as described above with the following modifications: an increased amount of 5 µL gDNA, equivalent to 1.25 µL whole blood, was added into a 30 µL reaction. Numbers of gametocytes originally present in whole blood samples were calculated by a conversion factor of 10−1.6225 × (copy number pfs25 transcripts/µL whole blood)0.8518 as described elsewhere [21]. Correlation between gametocyte density (pfs25 transcripts) and asexual density (S-type 18SrRNA copy numbers) with DNA or RNA-derived MOI was analyzed by Kendall rank correlation τ for nonparametric data.
RESULTS
High Allelic Diversity of Gametocyte Genotyping Markers in PNG
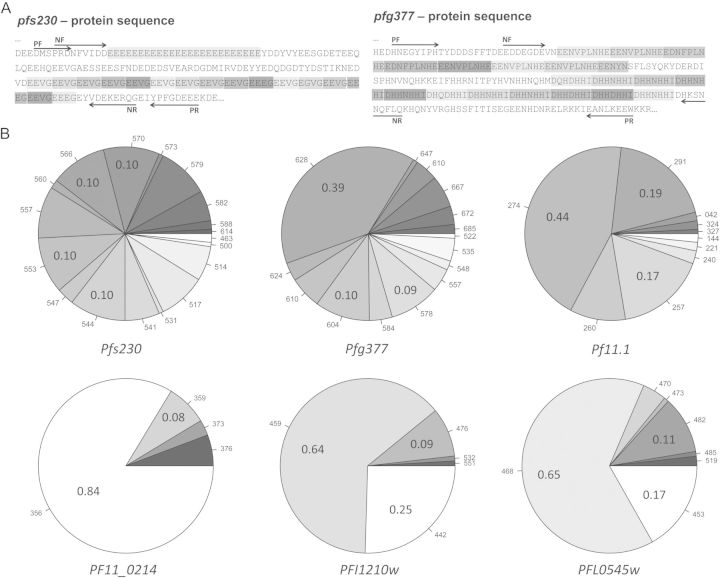
New length polymorphic and gametocyte specifically expressed genes were selected by screening publically available gametocyte transcriptome data [18] followed by tandem repeat detection using Tandem Repeats Finder [23]. Primers were designed to maximize size variation in amplified fragments. For pfg377 we combined polymorphic regions 2 and 3 described by Menegon into one larger amplicon [14]. Similarly, also our pfs230 amplicon spans 2 polymorphic regions (Figure 1A). Diversity of both markers in 111 gDNAs from PNG was highest with 18 pfs230 alleles (HE = 0.92) and 15 pfg377 alleles (HE = 0.81). The detection limit of each assay and parameters describing the genetic diversity and resolution of each marker are listed in Table 2. Allelic frequencies of the 6 gametocyte markers showed equal distribution for most of the pfs230 alleles, but for pfg377 a predominant allele (39%; Figure 1B). Sequence alignments of 12 pfg377 and 10 pfs230 nPCR products (Supplementary Figure 1) served for validating CE fragment sizing. Sizing of pfs230 fragments was more accurate than that of the larger pfg377 fragments. Comparison of 38 amplicons sized in parallel with GS500LIZ and GS1200LIZ size standards indicated that GS1200LIZ yielded better resolution for pfg377 with amplicon sizes >700 bp (Supplementary Figure 2). GS500LIZ worked well for pfs230 with amplicons <600 bp.
Figure 1.
Location of repeat regions within pfs230 and pfg377 amplicons and allelic diversity of 6 gametocyte markers. A, Markers pfs230 and pfg377 both span 2 distinct repeat regions. Individual repeat units are in shades of grey. Protein sequences of Plasmodium falciparum strain 3D7 were derived from PlasmoDB: PF3D7_0209000 (pfs230) and PF3D7_1250100 (pfg377). B, Allelic frequencies of 6 molecular markers for genotyping gametocytes determined in 111 cross-sectional samples from Papua New Guinea. Highest diversity was found for pfs230 (18 alleles) and pfg377 (15 alleles). The rounded average allele size is indicated for each allele in addition to the frequencies of the most frequent alleles of each marker in the study population. Abbreviations: NF, nested PCR forward primer; NR, nested PCR reverse primer; PCR, polymerase chain reaction; PF, primary PCR forward primer; PR, primary PCR reverse primer.
Table 2.
Resolution of 6 Polymorphic Gametocyte Markers in Comparison to Asexual Marker Msp2 in 111 P. falciparum Positive Cross-Sectional Samples From PNG
| Marker | Positive Samples | No. of Clones | No. of Alleles | CE-Product Size Range | Mean MOI | HE | In Vitro Detection Limit (Gametocyte/µL 3D7 Culture) |
|---|---|---|---|---|---|---|---|
| Msp2a | 111/111 | … | … | … | 1.56 | … | … |
| Pfs230 | 95/111 | 124 | 18 | 463–614 | 1.31 | 0.923 | 10 |
| Pfg377 | 97/111 | 117 | 15 | 521–695 | 1.21 | 0.816 | 1 |
| Pf11.1 | 100/111 | 125 | 10 | 143–327 | 1.25 | 0.734 | 1 if intron boundary 5 if no intron boundary |
| PF11_0214 | 100/111 | 104 | 4 | 355–376 | 1.04 | 0.293 | 10 |
| PFI1210w | 90/111 | 110 | 5 | 442–551 | 1.22 | 0.527 | 10 |
| PFL0545w | 95/111 | 113 | 7 | 453–519 | 1.19 | 0.546 | 10 |
Abbreviations: HE, heterozygosity; MOI, multiplicity of infection; Msp2, merozoite surface protein 2.
a Results from [19].
Detection Limits of Gametocyte Assays Assessed With a Trendline of Cultured Gametocytes
The limit of detection (LOD) of our gametocyte typing assays ranged from 1 (pfg377 and pf11.1 without DNase digestion) to 5 (pf11.1 with DNase digestion) and 10 gametocytes/µL culture (pfs230, PF11_0214, PFI1201w and PFL0545w; Table 2). These LODs were 50–500 times less sensitive than that of pfs25, a qRT-PCR assay able to detect 0.02 gametocytes/µL blood [21, 24]. Our pfg377 LOD was in line with earlier reports [14].
Quantifying the Gain in Sensitivity After Bypassing DNase Digestion
Using an intron-spanning marker permits omitting DNase digestion prior to reverse transcription but also increases amplicon length. By using alternative primers for marker pf11.1 we analyzed whether bypassing digestion would exceed the benefit of a smaller amplicon. Sensitivity was 5-fold higher for the 680 bp longer fragment not requiring DNase digestion (Table 2).
Evaluation of Gametocyte Typing Markers in Field Samples
Markers pfs230 and pfg377 were genotyped in parallel in paired DNA/RNA samples coextracted from 46 blood samples from Burkina Faso, all of which had been gametocyte-positive by pfs25 qRT-PCR. RT-PCR was successful in 42/46 field samples for pfs230 and in 37/46 samples for pfg377. For pfs230 18 alleles were detected and 19 for pfg377. For each blood sample, genotypes detected in gDNA were compared with genotypes amplified from gametocyte transcripts. A higher concordance between DNA- and RNA-derived genotypes was observed for pfs230 (Figure 2). From the total of 93 pfs230 PCR fragments amplified from all DNA samples, 41 (44.1%) were not observed in the corresponding RNA fraction. These clones either did not produce gametocytes or were below the detection limit of RT-PCR. For pfg377 61.5% (48/78) of fragments were missed on RNA level (Table 3). Similarly, MOI on DNA level (MOIDNA) was higher for pfs230 with a mean of 2.21 [range 1–5] infections per carrier in contrast to 2.11 [range 1–4] for pfg377, also arguing for pfs230 as the more sensitive marker. When combining all DNA- and RNA-derived genotypes per sample, mean MOI of pfs230 and pfg377 increased to 3.14 [range 1–6] and 3.08 [range 1–5], respectively (Table 3). This combined MOI (MOIcombined) represents a more realistic, though still underestimated number of any stage of all coinfecting clones per sample. No correlation between MOIRNA, MOIDNA, MOIcombined, and gametocyte or asexual densities was found (Kendall rank correlation, all τ's > 0, all P-values > .07).
Figure 2.
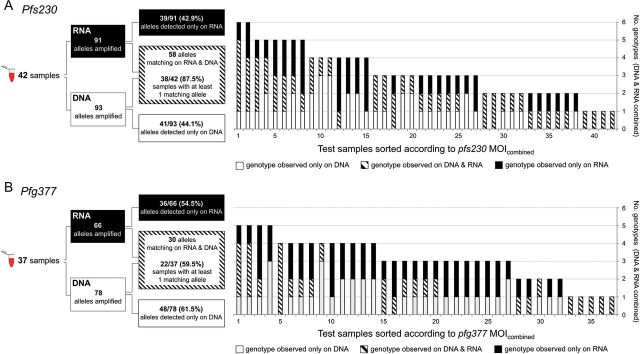
Schematic of analytical procedures (right panel) and overlap of genotypes detected simultaneously in RNA and DNA by blood sample (left panel). A, Pfs230, 42 paired RNA/DNA samples. B, Pfg377, 37 paired RNA/DNA samples. Abbreviation: MOI, multiplicity of infection.
Table 3.
Discrimination Power and Test Sensitivity of Gametocyte Typing Markers pfs230 and pfg377 in 46 Blood Samples From Burkina Faso
| Marker | pfs230 | pfg377 |
|---|---|---|
| No. of successful amplified samples | 42/46 | 37/46 |
| Detection limit in field samplesa | 2 gametocyte/µL WB | 3.5 gametocyte/µL WB |
| Median gametocyte counta | 17.0 gametocyte/µL WB | 17.9 gametocyte/µL WB |
| No. of different alleles | 18 | 19 |
| DNA/RNA sample pairs with at least 1 matching PCR fragment | 38/42 (85.7%) | 22/37 (59.5%) |
| Total no. of PCR fragments detected (DNA and RNA combined) | 132 | 114 |
| Proportion of DNA fragments not found on RNA level | 41/93 (44.1%) | 48/78 (61.5%) |
| Proportion of RNA fragments not found on DNA level | 39/91 (42.9%) | 36/66 (54.5%) |
| Combined mean MOI (DNA and RNA) | 3.14 [range 1–6] | 3.08 [range 1–5] |
| Mean MOI (DNA) | 2.21 [range 1–5] | 2.11 [range 1–4] |
| Mean MOI (RNA) | 2.17 [range 1–5] | 1.78 [range 1–4] |
Abbreviations: MOI, Multiplicity of infection; PCR, polymerase chain reaction; WB, whole blood.
a Determined by a conversion factor based on pfs25 transcripts copies/µL RNA [21].
DISCUSSION
Gametocyte typing depends on detection of transcripts from genes exclusively transcribed in gametocytes. In addition, extensive length polymorphism is required to permit tracking of gametocytes from multiclone infections. Multiple P. falciparum infections can coexist over weeks or months, but the variation in their relative densities and contribution to transmission over time has not yet been adequately quantified. The available data on gametocytes production of individual co-infecting clones were compromised by limited size-polymorphism in marker pfg377-R3 [5, 15–17, 25]. High-endemic settings are characterized by high MOI, where a limited marker resolution of ≥7 alleles will not adequately discriminate gametocytes of all clones present in a sample. By combining 2 repeat regions into 1 amplicon, we substantially improved the discriminatory power of both major markers. In 46 samples from Burkina Faso we detected 19 pfg377 and 18 pfs230 alleles by CE. A comparable diversity was observed in samples from PNG indicating that these markers may have sufficiently high diversity for genotyping in both African and non-African populations with different transmission intensity.
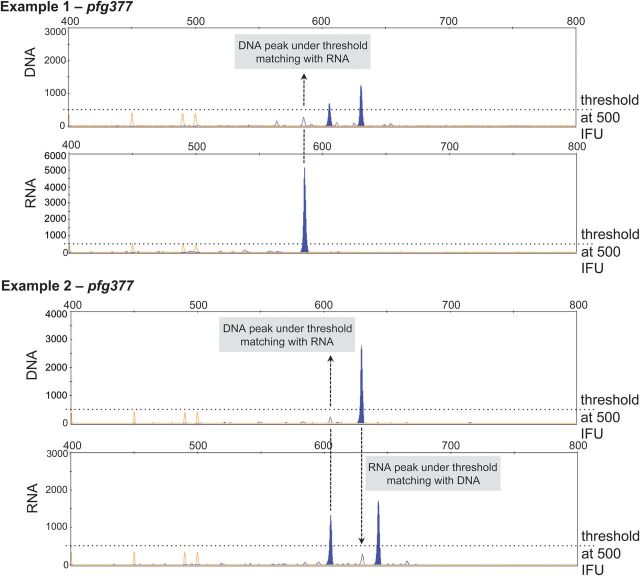
High MOI in the Burkina Faso study area [26] can contribute to discrepant results between RNA- and DNA-derived MOI. High MOI implies high clone competition in the host, resulting in turn in fluctuations in clone densities [27]. During PCR the presence of several templates of various concentrations may lead to template competition. Both effects of high MOI result in imperfect detectability [28]. This effect of competing templates, leading to lack of detection of genotypes either in DNA- or RNA-based detection, is enhanced by applying a cutoff for peak height in CE to separate background noise from real signals (Figure 3). In view of these inherent shortfalls, it seems essential to optimize sampling and preservation of both DNA and RNA to maximize the volume of template in PCR and RT-PCR in order to minimize the failures to detect all alleles present.
Figure 3.
Electropherograms of pfg377 fragments amplified by nested PCR from gDNA and nested RT-PCR from RNA coextracted from the same sample. Arrows indicate minority peaks, which had fallen below the cutoff, whereas matching fragments of significant peak height were present in the corresponding sample. Abbreviation: RT-PCR, reverse transcription polymerase chain reaction.
When comparing paired RNA- and DNA-derived fragments, 3 scenarios are expected: (I) RNA- and DNA-derived alleles match; here asexual stages and gametocytes of a clone are concurrently present in the blood sample, or DNA- and RNA- alleles both derive from gametocytes only. Yet, in multiclone infections a perfect match might be rare because the ratio of asexual vs sexual stages of each clone could differ considerably. (II) DNA-derived alleles exceed those obtained from RNA of the same sample. This is intuitively expected, because only some of the concurrent clones might produce gametocytes, as suggested by the frequent absence of gametocytes in some of the P. falciparum-positive blood samples despite molecular detection. (III) RNA-derived alleles are detected despite their absence on DNA level. This could occur when a gametocyte clone is still circulating while its asexual stages are already cleared by the immune system or below the detection limit. The gametocytes' nuclear DNA in this scenario remains below the detection limit of PCR or suffers from competition in multiclone infections.
In our study all 3 scenarios were seen, with scenario I predominant for pfs230 and scenario II for pfg377. An explanation for this discrepancy is offered by the differential performance of our 2 markers, which differed in their ability to detect a clone on both DNA- plus RNA level: pfs230 detected at least one matching genotype in >80% of samples, in contrast to only 60% for pfg377. Similarly, more RNA clones were missed by pfg377 (60%) than by pfs230 (45%). This argues for a higher sensitivity of pfs230 compared to pfg377 RT-PCR.
The imperfect detectability observed in asexual clones [6, 28] is aggravated in gametocyte detection, because gametocytes occur in densities about 100-fold lower than asexual stages [29]. Detection of gametocytes depends greatly on the blood volume processed, whereby a rare gametocyte clone might be present or absent by chance in the limited volume of blood processed. An additional limitation specific for Pfg377 consists in its expression restricted to female gametocytes. Our RT-PCR assays amplified gametocyte-specific transcripts in field samples that contained as little as 2 gametocytes/µL whole blood, as indicated by the LOD for pfs230 and is thus in the range of previously published assays [14, 30]. Even though this LOD permits detection of submicroscopic gametocytes, it does not reach the up to 100-fold higher sensitivity of pfs25 qRT-PCR [21]. This difference is mainly due to a lower expression rate of pfs230 compared to pfs25. Amplicon size and differential stability of the RNA may play an additional role as previously suggested [21].
We propose another strategy to address the problem of imperfect detectability of gametocyte clones: a longitudinal study design would permit to detect a particular genotype on RNA-level in a subsequent blood samples harboring higher gametocyte density. It is possible that sexual and asexual densities do not peak at the same time due to a 10 days maturation period of gametocytes. Therefore, a better match may be achieved by comparing results from consecutive bleeds. A gametocyte clone missed at an earlier sampling date might appear in the following sample. This approach parallels our strategy adapted to track asexual clones also fluctuating in their densities over time [28, 31, 32]. Nevertheless, even a longitudinal approach to gametocyte tracking will not overcome the imperfect detection of a gametocytemia that is persistently very low.
No other candidate of higher diversity and sensitivity than our CE-based pfg377 and pfs230 assays was found. Thus, length polymorphism of intragenic repeat regions in gametocyte-expressed genes seems to be less extensive than in genes expressed in asexual stages. For improving discrimination power beyond 18 alleles by pfs23-CE, alternative approaches could be investigated in, for example, future detection of single nucleotide polymorphisms by targeted next generation sequencing. However, the major challenges will likely persist, e.g. imperfect clone detectability in a limited blood volume from the field, expression levels of polymorphic gametocyte-specific genes, and assay sensitivity impaired by long amplicons.
A major gap in our knowledge of P. falciparum transmission dynamics is the onset and duration of gametocytogenesis of each asexual clone in relation to coinfecting clones and the contribution of resistant clones to transmission. We envisage that the molecular description of clone transmission dynamics may yield molecular gametocyte-specific parameters similar to those used in the description of infection dynamics and complementing these, for example, the duration of gametocyte production or multiplicity of gametocyte clones. This will open up new investigations of clone interaction, within-host competition, and clonal fitness. So far, very little is known on gametocyte dynamics in natural infections, where concurrent clonal infections might contribute to transmission equally or in competition with each other. This determines parasite recombination in mosquitoes, which in turn has major consequences for development of multilocus drug resistance phenotypes or antigenic diversity.
In summary, we improved the resolution of existing markers for discriminating gametocyte clones, but were unable to find alternative polymorphic markers of higher diversity. Pfs230 emerged as the most sensitive and diverse marker. Detectability of minority clones was identified as a major problem for matching asexual clones with their gametocytes. The loss of minority clones seemed strongest in the high transmission setting with high mean MOI where about half of all clones were missed in either of the paired samples. Longitudinal analyses are needed to permit temporally staggered alignment of fragments to compensate imperfect detectability. This calls for longitudinal studies with short-term sampling intervals specifically designed for genotyping DNA and RNA targets in parallel.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank the study participants and their parents or guardians, and the field teams in PNG and Burkina Faso.
Author contribution. R. W. and I. F. designed and performed research and wrote the article; I. S., A. B. T., and P. S. contributed field samples, L. T., H. P. B., and I. M. contributed new reagents or analytical tools.
Financial support. This work was supported by Swiss National Science Foundation [grant 310030_134889], International Centers of Excellence in Malaria Research [grant U19 AI089686), National Health and Medical Research Council [grants GNT1021544 and GNT1043345] and Bill and Melinda Gates Foundation [grant OPP1034577].
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Schoepflin S, Valsangiacomo F, Lin E, Kiniboro B, Mueller I, Felger I. Comparison of Plasmodium falciparum allelic frequency distribution in different endemic settings by high-resolution genotyping. Malar J. 2009;8:250. doi: 10.1186/1475-2875-8-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith T, Beck H-P, Kitua A, et al. Age dependence of the multiplicity of Plasmodium falciparum infections and of other malariological indices in an area of high endemicity. Trans R Soc Trop Med Hyg. 1999;93(suppl 1):15–20. doi: 10.1016/s0035-9203(99)90322-x. [DOI] [PubMed] [Google Scholar]
- 3.Wargo AR, Huijben S, de Roode JC, Shepherd J, Read AF. Competitive release and facilitation of drug-resistant parasites after therapeutic chemotherapy in a rodent malaria model. Proc Natl Acad Sci U S A. 2007;104:19914–9. doi: 10.1073/pnas.0707766104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beshir KB, Sutherland CJ, Sawa P, et al. Residual Plasmodium falciparum parasitemia in Kenyan children after artemisinin-combination therapy is associated with increased transmission to mosquitoes and parasite recurrence. J Infect Dis. 2013;208:2017–24. doi: 10.1093/infdis/jit431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nassir E, Abdel-Muhsin AM, Suliaman S, et al. Impact of genetic complexity on longevity and gametocytogenesis of Plasmodium falciparum during the dry and transmission-free season of eastern Sudan. Int J Parasitol. 2005;35:49–55. doi: 10.1016/j.ijpara.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 6.Felger I, Maire M, Bretscher MT, et al. The dynamics of natural Plasmodium falciparum infections. PLoS ONE. 2012;7:e45542. doi: 10.1371/journal.pone.0045542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaslow DC, Quakyi IA, Syin C, et al. A vaccine candidate from the sexual stage of human malaria that contains EGF-like domains. Nature. 1988;333:74–6. doi: 10.1038/333074a0. [DOI] [PubMed] [Google Scholar]
- 8.Eksi S, Czesny B, Van Gemert GJ, Sauerwein RW, Eling W, Williamson KC. Malaria transmission-blocking antigen, Pfs230, mediates human red blood cell binding to exflagellating male parasites and oocyst production. Mol Microbiol. 2006;61:991–8. doi: 10.1111/j.1365-2958.2006.05284.x. [DOI] [PubMed] [Google Scholar]
- 9.Williamson KC, Keister DB, Muratova O, Kaslow DC. Recombinant Pfs230, a Plasmodium falciparum gametocyte protein, induces antisera that reduce the infectivity of Plasmodium falciparum to mosquitoes. Mol Biochem Parasitol. 1995;75:33–42. doi: 10.1016/0166-6851(95)02507-3. [DOI] [PubMed] [Google Scholar]
- 10.Pradel G. Proteins of the malaria parasite sexual stages: expression, function and potential for transmission blocking strategies. Parasitology. 2007;134(Pt. 14):1911–29. doi: 10.1017/S0031182007003381. [DOI] [PubMed] [Google Scholar]
- 11.Williamson KC, Kaslow DC. Strain polymorphism of Plasmodium falciparum transmission-blocking target antigen Pfs230. Mol Biochem Parasitol. 1993;62:125–7. doi: 10.1016/0166-6851(93)90186-2. [DOI] [PubMed] [Google Scholar]
- 12.Niederwieser I, Felger I, Beck HP. Limited polymorphism in Plasmodium falciparum sexual-stage antigens. Am J Trop Med Hyg. 2001;64:9–11. doi: 10.4269/ajtmh.2001.64.9. [DOI] [PubMed] [Google Scholar]
- 13.Alano P, Read D, Bruce M, et al. COS cell expression cloning of Pfg377, a Plasmodium falciparum gametocyte antigen associated with osmiophilic bodies. Mol Biochem Parasitol. 1995;74:143–56. doi: 10.1016/0166-6851(95)02491-3. [DOI] [PubMed] [Google Scholar]
- 14.Menegon M, Severini C, Sannella A, et al. Genotyping of Plasmodium falciparum gametocytes by reverse transcriptase polymerase chain reaction. Mol Biochem Parasitol. 2000;111:153–61. doi: 10.1016/s0166-6851(00)00314-5. [DOI] [PubMed] [Google Scholar]
- 15.Abdel-Wahab A, Abdel-Muhsin AM, Ali E, et al. Dynamics of gametocytes among Plasmodium falciparum clones in natural infections in an area of highly seasonal transmission. J Infect Dis. 2002;185:1838–42. doi: 10.1086/340638. [DOI] [PubMed] [Google Scholar]
- 16.Nwakanma D, Kheir A, Sowa M, et al. High gametocyte complexity and mosquito infectivity of Plasmodium falciparum in the Gambia. Int J Parasitol. 2008;38:219–27. doi: 10.1016/j.ijpara.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Nakazawa S, Culleton R, Maeno Y. In vivo and in vitro gametocyte production of Plasmodium falciparum isolates from Northern Thailand. Int J Parasitol. 2011;41:317–23. doi: 10.1016/j.ijpara.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Young JA, Fivelman QL, Blair PL, et al. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol Biochem Parasitol. 2005;143:67–79. doi: 10.1016/j.molbiopara.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Marfurt J, Mueller I, Sie A, et al. Low efficacy of amodiaquine or chloroquine plus sulfadoxine-pyrimethamine against Plasmodium falciparum and P. vivax malaria in Papua New Guinea. Am J Trop Med Hyg. 2007;77:947–54. [PubMed] [Google Scholar]
- 20.Tiono AB, Ouedraogo A, Ogutu B, et al. A controlled, parallel, cluster-randomized trial of community-wide screening and treatment of asymptomatic carriers of Plasmodium falciparum in Burkina Faso. Malar J. 2013;12:79. doi: 10.1186/1475-2875-12-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wampfler R, Mwingira F, Javati S, et al. Strategies for detection of Plasmodium species gametocytes. PLoS ONE. 2013;8:e76316. doi: 10.1371/journal.pone.0076316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koepfli C, Ross A, Kiniboro B, et al. Multiplicity and diversity of Plasmodium vivax infections in a highly endemic region in Papua New Guinea. PLoS Negl Trop Dis. 2011;5:e1424. doi: 10.1371/journal.pntd.0001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–80. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider P, Schoone G, Schallig H, et al. Quantification of Plasmodium falciparum gametocytes in differential stages of development by quantitative nucleic acid sequence-based amplification. Mol Biochem Parasitol. 2004;137:35–41. doi: 10.1016/j.molbiopara.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 25.Gatei W, Kariuki S, Hawley W, et al. Effects of transmission reduction by insecticide-treated bed nets (ITNs) on parasite genetics population structure: I. The genetic diversity of Plasmodium falciparum parasites by microsatellite markers in western Kenya. Malar J. 2010;9:353. doi: 10.1186/1475-2875-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soulama I, Nebie I, Ouedraogo A, et al. Plasmodium falciparum genotypes diversity in symptomatic malaria of children living in an urban and a rural setting in Burkina Faso. Malar J. 2009;8:135. doi: 10.1186/1475-2875-8-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henning L, Schellenberg D, Smith T, et al. A prospective study of Plasmodium falciparum multiplicity of infection and morbidity in Tanzanian children. Trans R Soc Trop Med Hyg. 2004;98:687–94. doi: 10.1016/j.trstmh.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Koepfli C, Schoepflin S, Bretscher M, et al. How much remains undetected? Probability of molecular detection of human Plasmodia in the field. PLoS ONE. 2011;6:e19010. doi: 10.1371/journal.pone.0019010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouedraogo AL, Bousema T, de Vlas SJ, et al. The plasticity of Plasmodium falciparum gametocytaemia in relation to age in Burkina Faso. Malar J. 2010;9:281. doi: 10.1186/1475-2875-9-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeno Y, Nakazawa S, Dao LD, et al. A dried blood sample on filter paper is suitable for detecting Plasmodium falciparum gametocytes by reverse transcription polymerase chain reaction. Acta Trop. 2008;107:121–7. doi: 10.1016/j.actatropica.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Sama W, Owusu-Agyei S, Felger I, Vounatsou P, Smith T. An immigration-death model to estimate the duration of malaria infection when detectability of the parasite is imperfect. Stat Med. 2005;24:3269–88. doi: 10.1002/sim.2189. [DOI] [PubMed] [Google Scholar]
- 32.Bretscher MT, Valsangiacomo F, Owusu-Agyei S, Penny MA, Felger I, Smith T. Detectability of Plasmodium falciparum clones. Malar J. 2010;9:234. doi: 10.1186/1475-2875-9-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.