Abstract
Motivation: cddApp is a Cytoscape extension that supports the annotation of protein networks with information about domains and specific functional sites from the National Center for Biotechnology Information’s conserved domain database (CDD). CDD information is loaded for nodes annotated with NCBI numbers or UniProt identifiers and (optionally) Protein Data Bank structures. cddApp integrates with the Cytoscape apps structureViz2 and enhancedGraphics. Together, these three apps provide powerful tools to annotate nodes with CDD domain and site information and visualize that information in both network and structural contexts.
Availability and implementation: cddApp is written in Java and freely available for download from the Cytoscape app store (http://apps.cytoscape.org). Documentation is provided at http://www.rbvi.ucsf.edu/cytoscape, and the source is publically available from GitHub http://github.com/RBVI/cddApp.
Contact: scooter@cgl.ucsf.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The functional annotation of proteins is critical to understanding biology. In most proteins, units of biological function are domains that contain key active sites, critical structural components or interaction sites. A single protein may contain one or more domains, and the acquisition of domains can provide new interaction partners or new functions. One approach to inferring basic protein function is to characterize the functional domains that the protein contains. The National Center for Bioinformatics Information’s (NCBI) conserved domain database (CDD) (Marchler-Bauer et al., 2011) is a repository of manually curated and computationally derived protein domain family models that are searchable through a Web interface or Web services. The conserved domains (CDs) in the CDD database are organized into families of CDs related by common evolutionary ancestry. Structurally and functionally important regions of the domain are annotated based on literature and structural evidence wherever possible in each of the CD. This resource is a valuable tool for researchers looking to identify or assign functions of individual proteins.
Another commonly used tool for inference and analysis of biological function is Cytoscape (Cline et al., 2007). Cytoscape is an open-source desktop application for the visualization and analysis of biological networks, including pathways, protein–protein interaction networks and protein–protein similarity networks. Cytoscape includes an extension mechanism that allows developers to add ‘apps’ to Cytoscape that enhance its analytical or visualization capabilities (Lotia et al., 2013).
Cytoscape and the CDD are complementary, but no mechanism exists to allow using these tools together. We embarked on a collaborative effort to link Cytoscape’s network visualization and analysis features with the CDD domain annotations to support researchers interested in exploring functional annotation of proteins in a network context.
2 DESCRIPTION
The cddApp provides three main functions: using the CDD Web services to search for domains in proteins in the current network, visualizing the domain annotations and linking the domain annotations to the structure through a companion app.
cddApp adds a new cddApp menu in Cytoscape’s top-level Apps menu that has four submenus: Load CDD Domains for Network, Load CDD Domains for selected Node(s), Show CDD Domain Panel and Hide CDD Domain Panel. It also adds a submenu in the Apps node context menu to allow loading annotations for a single node and showing the domain architecture diagram for that node. When users select one of these menu items, they are asked for the column containing the node identifier (CDD accepts GI numbers, UniProt identifiers and protein sequences) and, optionally, a column containing the identifiers of any Protein Data Bank (PDB) (Berman et al., 2007) structures.
Once the results are available from the CDD, cddApp adds a new Results Panel tab, CDD Domains, with a table of information about the annotated domains and sites in a table (right side of Fig. 1). For each domain, there is a link to open up its detailed information at the CDD Web site. If a PDB column was selected as part of the load and a PDB structure was annotated for a node, the Results Panel will also contain a checkbox to open that structure in UCSF Chimera (Pettersen et al., 2004). This option will only appear if the structureViz2 (Morris et al., 2007) app is also loaded. The CDD Domains panel also has checkboxes to use the enhancedGraphics app, if loaded, to paint pie charts for domains and features onto nodes.
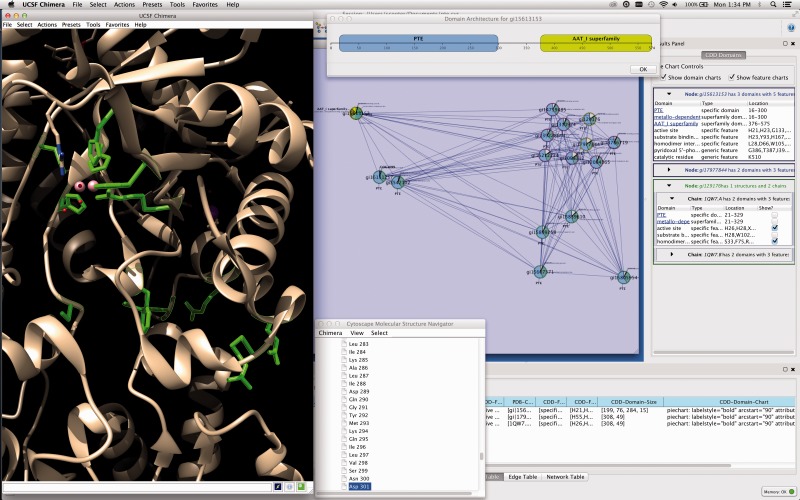
Fig. 1.
This screenshot of Cytoscape with cddApp installed shows the CDD domains panel in the Cytoscape Results Panel with three nodes selected. The bottom node is associated with a structure with two chains, and the top two nodes do not have PDB structures. The domain architecture diagram for the top node (gi 15613153) is shown. cddApp uses the Cytoscape apps enhancedGraphics and structureViz2 to paint pie charts on nodes and show the domain and site information on the structure, respectively. The active site and homodimer interface are shown in UCSF Chimera’s graphics window. A high-resolution export of the network image is provided as Supplementary File 1
All CDD annotations are stored in Cytoscape’s default node table and are saved as part of the session.
3 EXAMPLE USAGE
The structure–function linkage database (SFLD) (Akiva et al., 2014) is a data repository of highly curated functional annotations for a set of enzyme superfamilies. The SFLD supports the download of protein similarity networks (Atkinson et al., 2009) containing proteins annotated to specific functional families as well as those that have not yet been annotated.
The edges in these networks represent the similarity between a pair of proteins, generally as measured by the BLAST (Altschul et al., 1990) E-value. The small sample network in Figure 1 consists of a small number of proteins from the amidohydrolase superfamily that have been annotated by PFAM (Punta et al., 2012) as members of the phosphotriesterase (PTE) family. With cddApp, the network was annotated using name as the primary identifier column and pdbFilename as the column containing PDB IDs. Enabling the domain charts, one can quickly see that the protein (gi15613153) from Bacillus halodurans has an added domain (AAT_I superfamily) that is not present in the other PTE proteins. This may be further demonstrated by showing the domain architecture diagram (top of Fig. 1). Clicking on the link in the Results Panel to view the detailed information on the AAT_I superfamily fold, we learn that this domain is consistent with pyridoxal phosphate-dependent enzymes. It appears that this protein has gained a function not present in the other proteins in this network.
A step-by-step tutorial based on this example is available at http://www.rbvi.ucsf.edu/cytoscape/cddApp/tutorial.shtml.
4 SIGNIFICANCE AND CONCLUSION
CDD is a powerful tool in protein classification that allows users to analyze protein sequences in the context of domain families. The incorporation of CDD via cddApp into Cytoscape should likewise give the user additional insights into the domain compositions of proteins within his or her network. Additional decoration of each of the nodes via the enhancedGraphics app displays the protein’s domain architecture in a concise format and may uncover functional implications. The visualization of CDD annotations on the structures via the structureViz2 app will facilitate understanding the spatial relationships among key residues and lend insight into their functional roles. The links to the CDD Web site allow accessing the source data, including alignments, structure links, protein record links, literature citations and sequence trees. The cddApp connects Cytoscape and Chimera users to the wealth of resources available at CDD and NCBI, providing a new avenue to scientific discovery. By integrating with other apps, cddApp significantly expands functionality without duplication, leveraging existing knowledge. Together, these apps extend the utility of the Cytoscape environment for functional annotation.
Funding: This research was supported in part by the Intramural Research Program of the U.S. National Institutes of Health (NIH); National Library of Medicine; and by NIH National Institute of General Medical Science Grant [P41-GM103311].
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Akiva E, et al. The structure-function linkage database. Nucleic Acids Res. 2014;42:D521–D530. doi: 10.1093/nar/gkt1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Atkinson HJ, et al. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H, et al. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotia S, et al. Cytoscape app store. Bioinformatics. 2013;29:1350–1351. doi: 10.1093/bioinformatics/btt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JH, et al. structureViz: linking Cytoscape and UCSF Chimera. Bioinformatics. 2007;23:2345–2347. doi: 10.1093/bioinformatics/btm329. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.