Background: Treg cell stability is ensured by a CNS2-Cbfβ-Runx1-Foxp3 feedback loop.
Results: Activation of the COT/Tpl2-MEK-ERK pathway decreases the DNA binding activity of Foxp3 and down-regulates Treg cell function and Foxp3 expression in vivo.
Conclusion: The COT/Tpl2-MEK-ERK pathway modulates Treg cell identity.
Significance: Therapeutic interventions targeting the COT/Tpl2-MEK-ERK pathway might contribute to treating autoimmune diseases and optimize human Treg cell therapy.
Keywords: Autoimmunity; Cell Signaling; Immunology; Inflammation; T Cell; Treg Cell, Instability, Foxp3, COT, MEK, ERK
Abstract
The instability of regulatory T (Treg) cells is involved in the pathogenesis of autoimmune diseases and also highlights safety concerns with regard to clinical Treg cell therapy. Cell-intrinsic molecular events linked to this Treg cell instability in vivo cells, which leads to safety concerns regardingare still obscure. Here we developed a novel luciferase-based reporter system and performed an unbiased screening for kinases that potentially modulate Foxp3 function. We found that the active form of COT/Tpl2 specifically inhibits the DNA binding activity of Foxp3 through a MEK-ERK-dependent pathway. Moreover, Treg cell-specific expression of activated MEK1 led to dysregulation of Treg function and instability of Foxp3 expression in vivo. Our results support the hypothesis that outside inflammatory signals act through the COT/Tpl2-MEK-ERK signaling pathway to destabilize the Treg lineage.
Introduction
T cells confer antigen-specific immune responses and immunological memory to protect the body from pathogens and cancer cells. However, misguided or excessive T cell responses targeting self-antigens can cause autoimmune diseases, such as type I diabetes and multiple sclerosis (1). Foxp3+ regulatory T (Treg)2 cells play central roles in maintaining immune homeostasis and self-tolerance, which is essential for the control of pathogenic T cell responses (2, 3). Recently, Treg cells have demonstrated promising potential in clinical therapy for autoimmune diseases (4, 5).
Foxp3, a member of the forkhead transcription factor family, is associated with CD4+CD25+ T cells, acting as the most reliable marker for the Treg cell lineage (6). Moreover, Foxp3 is indispensable for the development and function of Treg cells, as exemplified by mice harboring the scurfy mutation, germ line deletion, or conditional deletion of the Foxp3 gene. They develop severe autoimmune diseases because of defects in the development and/or functions of Treg cells (7–10). Notably, scurfy mice can be rescued by Foxp3 transgene expression (6), and ectopic expression of Foxp3 confers partial surface phenotypes and suppressive function of Treg cells to non-Treg cells in vitro (11), indicating that Foxp3 directly programs the development and function of Treg cells.
It has been demonstrated that a fraction of Foxp3+ Treg cells can lose Foxp3 expression in vivo, especially in lymphopenic and inflammatory settings (12–16). These unstable Treg cells usually acquire effector Th cell-like phenotypes and turn into pathogenic cells, which leads to safety concerns regarding human Treg cell therapy (13, 15, 16). Considerable evidence supports the hypothesis that Treg cell instability is associated with both external signals and cell-intrinsic events, including proinflammatory cytokines (17), strong engagement with autoantigens (13, 15), IL-2 depletion (15), epigenetic modification of the Foxp3 locus (18), Foxp3 protein stability (19), and modulation of microRNAs (20), but it is still not clear how cell-intrinsic signaling pathways are linked to Treg cell instability.
Stable Foxp3 expression in the progeny of Treg cells is ensured by a positive feedback loop comprising the CNS2 (also known as TSDR) region in the Foxp3 gene locus, the Cbfβ-Runx1 transcription factor, and Foxp3 itself, in which CNS2, Cbfβ-Runx1, and Foxp3 bind to each other to form a transcription complex (7, 21–24). Treg cells lacking CNS2, Cbfβ, or Runx1 gradually lose or down-regulate Foxp3 expression, indicating that defects in this positive feedback loop promote Treg cell instability (21, 22). The formation of this feedback loop is largely dependent on the methylation status of the CNS2 region and the DNA binding activity of the Cbfβ-Runx1-Foxp3 complex. Demethylated CNS2 in Treg cells favors the recruitment of the Cbfβ-Runx1-Foxp3 complex to CNS2, whereas methylated CNS2 in conventional T cells and TGF-β-induced Treg cells does not (22). Consistent with this, the DNA methyltransferase family promotes Treg cell instability by increasing the level of CpG methylation in the CNS2 region (18).
Attenuating the DNA binding activity of Foxp3 potentially breaks the CNS2-Cbfβ-Runx1-Foxp3 feedback loop, resulting in Treg cell instability. As a transcription factor, Foxp3 binds target gene loci through its forkhead/winged helix (FKH) domain, which is critical to Foxp3 function. Of great significance, most IPEX patients carry genetic mutations in the FKH domain (25).
To explore the links among cell-intrinsic signaling pathways, the DNA binding activity of Foxp3, and Treg cell instability, we performed an unbiased screen for kinases that modulate the DNA binding activity of Foxp3 using a novel luciferase-based reporter system. We found that activation of the COT/Tpl2-MEK-ERK signaling pathway inhibited the DNA binding activity of Foxp3 and promoted Treg cell instability in vivo. Our results support the hypothesis that external inflammatory signals modulate Treg cell stability through the COT/Tpl2-MEK-ERK signaling pathway, which provides a potential target for therapeutic intervention to treat autoimmune diseases and optimize human Treg cell therapy.
EXPERIMENTAL PROCEDURES
Plasmids
DNA elements with six tandem copies of an optimized FOXP3 binding motif (5′-gtaaacaagagtaaacaagtcacgatcgcaaggtaaacaagacaacacgattgtaaacaagtccgaattccaaggtaaacaagagtaaaca-3′) or five tandem copies of the Gal4DB binding motif (5′-cggagtactgtcctccgagcggagtactgtcctccgac tcgagcggagtactgtcctccgatcggagtactgtcctccgcgaattccggagtactgtcctccg-3′) were inserted into the NheI/BglII sites of the vector pGL3-promoter (Promega) to generate pFOXP3Luc or pGal4Luc, respectively. Plasmid pVP16-FL (full-length) was engineered by inserting a recombinant fragment, VP16-FOXP3, that contains the activation domain of herpes simplex virion protein 16 (VP16) and full-length human FOXP3 into the NotI/SalI sites of p3×FLAGcmv7.1 (Sigma). On the basis of pVP16-FL, other plasmids encoding truncated and/or mutated versions of VP16-FOXP3 or VP16-Gal4 were generated. Recombinant fragments HA-FOXP3DelN (amino acids 1–181 truncated), VP16-DelN, and VP16-Gal4 were also cloned into the NotI/SalI sites of pMSCV-IRES-Thy1.1. A library of 192 human kinases and kinase-related ORFs with a myristoylation sequence and FLAG epitope tag was obtained from Addgene (Addgene collection 1000000012) (26). pWZL-Neo-Myr-FLAG-EGFP was engineered by inserting the recombinant fragment myr-FLAG-EGFP into the EcoRI/SalI sites of pWZL-Neo-Myr-DEST. pEGFP-FOXP3DelN was engineered by inserting FOXP3DelN (amino acids 1–181 truncated) into the SalI/BamH I sites of pEGFP-C1. pLEX-DUSP6 was obtained from Addgene (Addgene plasmid 27975) (27). pcDNA3.1-MEK1DD and pcDNA3.1-MEK1mut (A-loop truncated) were provided by Prof. Y. H. Peng (Peking University, Beijing, China).
Luciferase Assay
To characterize the reporter system, 48-well tissue culture plates were seeded with 1 × 105 HEK293T cells/well 6 h before transfection using Lipofectamine 2000 (Invitrogen). Type A DNA mixtures (p3×FLAGcmv7.1-based construct:pFOXP3Luc:pRL-TK = 75:25:1) were introduced into HEK293T cells according to the specifications of the manufacturers. Similarly, type B DNA mixtures (kinase construct:pMSCV-VP16-DelN:pFOXP3Luc:pRL-TK = 45:45:10:1) were prepared for the screening of the kinase library and the following experiments. U0126 was added to the cell culture 6 h after transfection at final concentrations of 0, 20, and 40 μm. pLEX-DUSP6 was cotransfected with COT constructs at a ratio of 2. The dose effects of kinase were analyzed by serial dilution of the kinase construct in the type B DNA mixtures. For all of the experiments described above, empty vectors, GAL4-derived constructs, or EGFP constructs were used for control groups. Cells were washed with 1× PBS and lysed with 1× passive lysis buffer 24 h post-transfection (Promega, catalog no. E1941). Dual luciferase activity was measured using the Dual-Luciferase® reporter assay system (Promega, catalog no. E1960) and GloMax®20/20 luminometers according to the specifications of the manufacturers. Experiments were conducted in triplicate. Data were normalized to control groups. Differences in relative light units or fold activation were analyzed for statistical significance using unpaired Student's t test.
Nucleotide Pulldown and Western Blot Assays
To test the DNA binding activity of various versions of FOXP3, 6-well tissue culture plates were seeded with 4 × 105 HEK293T cells/well 6 h before transfection. The p3×FLAGcmv7.1-based constructs were introduced into HEK293T cells according to the specifications of the manufacturers. Similarly, DNA mixtures (kinase construct:pVP16-DelN = 2:1) were introduced into HEK293T cells. Twenty-four hours post-transfection, cells were washed with 1× PBS and lysed with Nonidet P-40 lysis buffer containing 150 mm NaCl, 50 mm Tris (pH 7.4), 1% Nonidet P-40, 1 mm PMSF, and protease inhibitors (Beyotime, China, catalog no. P0013F). The expression of versions of FOXP3 protein in cell lysates was confirmed by Western blotting using anti-FLAG antibodies. Properly diluted lysates were incubated with 10 μg of poly deoxyinosinic-deoxycytidylic acid (Sigma) and 40 μl of streptavidin-agarose beads (Sigma) coated with 5′-biotinylated FOXP3 binding oligonucleotide (5′-CAAGGTAAACAAGAGTAA ACAAAGTC-3′) overnight at 4 °C on a roller. The beads were washed three times with 500 μl of ice-cold wash buffer (1× PBS, 1 mm EDTA, 1 mm PMSF, and 0.1% Nonidet P-40), resuspended in 40 μl of SDS sample loading buffer, heated at 95 °C for 10 min, and analyzed by Western blotting using anti-FLAG antibody. The protein degradation assay was performed by introducing mixtures (kinase construct:pMSCV-HA-FOXP3DelN = 1:1) into HEK293T cells. Cycloheximide (200 μg/ml, Sigma) was added to the cell culture 24 h after transfection. Following incubation for 0, 0.5, 1, 2, and 4 h, cells were harvested and lysed for Western blotting assays using anti-HA and anti-β-actin antibodies.
Mice
Foxp3-GFP-Cre×R26-loxp-stop-loxp-YFP (termed TregYFP in this study) reporter mice were crossed with wild-type C57BL/6 mice to create a mixed NOD×B6 background (13). Rosa26-loxp-stop-loxp-MEK1DD-IRES-EGFP mice were obtained from The Jackson Laboratory (catalog no. 012352, C57BL/6-Gt (ROSA) 26Sortm8 (Map2k1*, EGFP) Rsky/J) (28) and bred to Foxp3-GFP-Cre×R26-loxp-stop-loxp-YFP mice to create progenies in the mixed NOD×B6 background. All mice were housed and bred under the same environmental conditions at the Beijing Laboratory Animal Research Center in accordance with the guidelines for the care and use of laboratory animals established by the Beijing Association for Laboratory Animal Science. All animal procedures were conducted according to the regulations of the Institute of Microbiology, Chinese Academy of Sciences Research Ethics Committee. The protocol was approved by the Research Ethics Committee of the Institute of Microbiology, Chinese Academy of Sciences.
Inhibitor Treatment and RNA Interference
Axillary and inguinal lymph node (pLN) cells from TregYFP mice were activated by plate-bound anti-CD3 (2 μg/ml/day) and anti-CD28 (1 μg/ml/day) for 5 days in the presence of 200 units/ml IL-2. COT inhibitor (CAS 871307-18-5, Santa Cruz Biotechnology) or U0126 (Sigma) was added to the cell culture medium at day 2 to a final concentration of 5 μm. Foxp3 protein expression was assessed at day 5 using a Foxp3 staining kit (eBioscience). The retrovirus vector LMP-Thy1.1 was engineered by inserting the Thy1.1 ORF into the NcoI/SalI sites of MSCV-LTRmiR30-PIG (Open Biosystems). A double-stranded DNA cassette that targets the coding region of mouse Cot was cloned into LMP-Thy1.1 according to the protocol of the manufacturer. Retrovirus production was performed as described previously (29). Pooled splenocytes and pLN cells from TregYFP mice were activated by plate-coated anti-CD3/CD28 for 2 days in the presence of 200 units/ml IL-2 before FACS for YFP+ cells. Sorted YFP+ cells were then infected with retrovirus and continued to be stimulated with plate-bound anti-CD3/CD28 for 3 days in the presence of 1000 units/ml IL-2 before intracellular Foxp3 staining.
Antibodies and Flow Cytometry
Labeled anti-CD4 (GK1.5), anti-CD8 (53–6.7), anti-CD25 (PC61), anti-CD45R (RA3–6B2), anti-CD44 (IM7), anti-CD62L (MEL14), anti-Foxp3 (FJK-16s), anti-Helios (22F6), anti-IL-2 (JES6–5H4), and anti-IFN-γ (XMG1.2) antibodies and specific isotype-matched control antibodies were from BD Biosciences or eBioscience. For cytokine analysis, cells were incubated for 3–4 h at 37 °C with 0.5 mm ionomycin, 10 ng/ml phorbol 12-myristate 13-acetate, and 3 mm monensin. Cells with YFP or GFP expression were prefixed in 1% (w/v) paraformaldehyde for 2 min, fixed, made permeable using a Foxp3 staining kit (eBioscience), and stained for intracellular proteins. The stained cells were analyzed on a FACSCalibur flow cytometer followed by data analysis using FlowJo software.
Induction of EAE
Eight- to twelve-week-old mice were immunized subcutaneously at four sites (50 μl/site) with 200 μl of emulsified complete Freund adjuvant (Sigma) supplemented with 4 mg/ml Mycobacterium tuberculosis H37Ra (Difco) and 300 μg of MOG35–55 peptide (MEVGWYRSPFSRVVHLYR-NK, ChinaPeptides, China) and received intraperitoneal injections of 2 × 109 heat-killed Bordetella pertussis organisms (Beijing Tiantan Biological products Co., Ltd.) at the time of immunization and 48 h later. Clinical disease was assessed by the scoring of hind limb paralysis as follows: no signs, 0; flaccid tail, 1; hind limb weakness, 2; partial hind limb paralysis, 3; complete hind limb paralysis, 4; and moribund mouse, 5.
RESULTS
FOXP3Luc Indicates the DNA Binding Activity of FOXP3
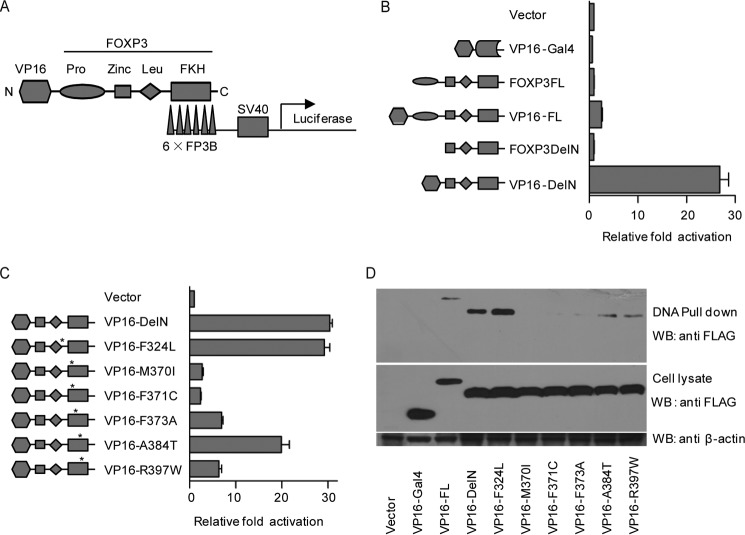
To study the DNA binding activity of FOXP3, we developed a novel luciferase-based reporter system (FOXP3Luc) in HEK293T cells. Briefly, FOXP3Luc consists of a reporter plasmid containing six tandem copies of an optimized FOXP3-binding DNA motif inserted upstream of the SV40-Luc cassette (pFOXP3Luc), a vector expressing full-length or truncated FOXP3 fused to the FLAG-tagged VP16 activation domain, and pRL-TK as an internal control (Fig. 1A). Theoretically, the VP16 domain can recruit additional transcriptional activators to enhance the transcription of the SV40-Luc gene upon FOXP3 binding to target sites through its FKH domain (30).
FIGURE 1.
FOXP3Luc reporter system for the DNA binding activity of FOXP3. A, schematic of the FOXP3Luc reporter system. Six tandem copies of an optimized FOXP3-binding motif (6×FP3B) were inserted upstream of the SV40-Luc cassette in the reporter plasmid (pFOXP3Luc), and a VP16 activation domain was fused to FOXP3 at its N terminus. The recruitment of VP16 domain to the 6×FP3B elements enhances the transcription of luciferase gene. B, readouts of FOXP3Luc composed of different versions of FOXP3. Vector, VP16-Gal4, FOXP3FL (full-length), VP16-FL, FOXP3DelN (amino acids 1–181 truncated), and VP16-DelN were coexpressed with pFOXP3Luc and pRL-TK in HEK293T cells, respectively. A Dual-Luciferase assay was performed 24 h after transfection. The ratio of firefly luciferase (Fluc)/Renilla luciferase (Rluc) was normalized to the vector group. C, readouts of FOXP3Luc composed of VP16-DelN harboring different IPEX mutations. VP16-DelN, VP16-F324L, VP16-M370I, VP16-F371C, VP16-F373A, VP16-A384T, and VP16-R397W were expressed and analyzed as in B. D, DNA pulldown assay for different versions of FOXP3. Streptavidin-agarose beads coated with 5′-biotinylated FOXP3 binding oligonucleotide were used to pull down different versions of FOXP3 from cell lysates. All experiments above were repeated independently at least three times. The error bars represent the mean ± S.D. WB, Western blot.
We first verified the function of the VP16 domain and the specificity of the FOXP3-binding elements by comparing the functions of different versions of FOXP3 in FOXP3Luc. Compared with the vector control, FOXP3FL (full-length) and FOXP3DelN (amino acids 1–181 truncated) had negligible effects on the readout of FOXP3Luc, whereas VP16-FL and VP16-DelN strikingly elevated the activity (∼3- and ∼30-fold, respectively) (Fig. 1B). Moreover, the optimized FOXP3-binding elements acted as specific targets of FOXP3 (but not the Gal4 DNA binding domain) in HEK293T cells, as shown by VP16-Gal4, which could not enhance the transcription of luciferase. It was striking that deletion of the N-terminal FOXP3 repressor domain (amino acids 1–181) increased the DNA binding activity of the FKH domain, which is consistent with gel shift findings reported previously (31).
Next we compared the DNA binding activities of VP16-DelN variants harboring different IPEX mutations. Compared with wild-type VP16-DelN, the F324L mutation had little effect on the function of VP16-DelN in FOXP3Luc. The A384T mutation caused a mild reduction in activity, and the M370I, F371C, F373A, and R397W mutations caused severe reductions, indicating that FOXP3Luc recapitulates the function of the FOXP3 FKH domain with high sensitivity and fidelity (Fig. 1C).
Finally, to directly prove that FOXPLuc can indicate the DNA binding activity of FOXP3, we performed a DNA pulldown assay using streptavidin-agarose beads coated with 5′-biotinylated FOXP3-binding oligonucleotide. We found that VP16-Gal4 could not be detected in the pulldown products, whereas wild-type VP16-DelN yielded a strong signal in the same assay, indicating that VP16-DelN can specifically bind the 5′-biotinylated FOXP3-binding oligonucleotide. VP16-FL was poorly enriched by the beads, in line with its low activity in FOXP3Luc. Further, VP16-F324L showed comparable DNA binding activity to wild-type VP16-DelN, whereas the DNA binding activity of the VP16-M370I, VP16-F371C, VP16-F373A, and VP16-R397W mutants was severely attenuated. Moreover, VP16-A384T had moderately attenuated DNA binding activity (Fig. 1D). Taken together, the results of the DNA pulldown assays were consistent with the FOXP3Luc assay. Therefore, we suggest that FOXP3Luc can reliably represent the DNA binding ability of FOXP3. Similarly, we successfully constructed a Gal4Luc system, which had no mutual interference with FOXP3Luc (data not shown).
Screen for Kinases as Inhibitors of FOXP3Luc
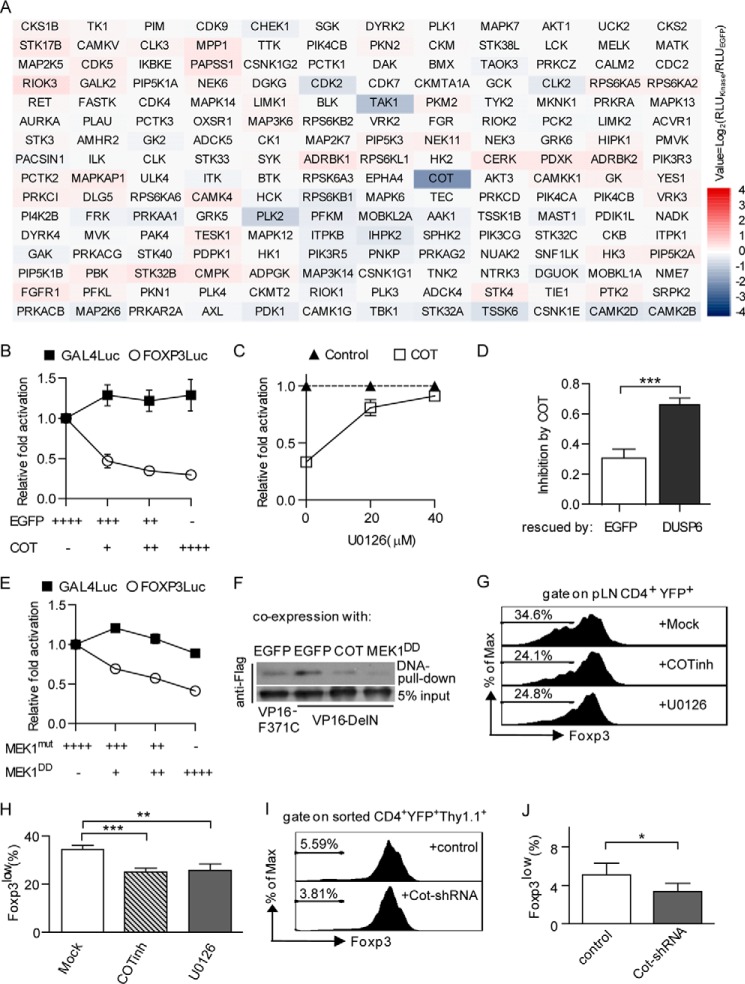
To investigate the cell-intrinsic signaling network linked to Treg cell instability, it is reasonable to study kinases that participate in diverse cellular signal transduction pathways. Therefore, we performed an unbiased screen for kinases that modulate the DNA binding activity of FOXP3 by utilizing a myr-kinase library in which the kinases are constitutively active in HEK293T cells because of membrane recruitment through their myristoylated N terminus. Moreover, the active forms of these kinases also represented their downstream signaling pathways.
For the unbiased screen, we used a defined FOXP3Luc system containing pMSCV-VP16-DelN, pFOXP3Luc, and pRL-TK. Myr-kinases were coexpressed with FOXP3Luc in HEK293T cells, and their effects on FOXP3Luc were normalized to the EGFP control. As shown in the heat map (Fig. 2A), COT (also called Tpl2 and MAP3K8) caused more than a 3-fold reduction in the readout of FOXP3Luc, whereas expression of the other kinases resulted in negligible effects on FOXP3Luc. Other members of the MAP kinase family in the library (e.g. MAP2K7-JNKs and MAP2K6-p38s) did not inhibit FOXP3Luc, with the exception of TAK1 (also called MAP3K7), which displayed a slight inhibitory effect. Next we found that COT inhibited FOXP3Luc in a dose-dependent manner. In contrast, COT had no inhibitory effect on Gal4Luc, indicating that COT specifically inhibited FOXP3Luc by affecting its DNA binding activity rather than the VP16 function (Fig. 2B).
FIGURE 2.
Screening for FOXP3Luc inhibitors. A, effects of myr-kinases on FOXP3Luc. Myr-kinases and FOXP3Luc were coexpressed in HEK293T cells. A Dual-Luciferase assay was performed 24 h after transfection. The ratio of Fluc/Rluc was normalized to the EGFP control group. The binary logarithms of means are shown in a heat map. RLU, relative light units. B, effects of COT on FOXP3Luc and Gal4Luc. Serially diluted COT constructs were coexpressed with FOXP3Luc or Gal4Luc in HEK293T cells. Cells were analyzed as in A. C, U0126 antagonizes the effects of COT on FOXP3Luc. COT and FOXP3Luc were expressed as in A, U0126 was supplemented 6 h after transfection at final concentrations of 0, 20, or 40 μm. Cells were analyzed as in A. D, DUSP6 antagonizes the effects of COT on FOXP3Luc. DUSP6 and COT were coexpressed with FOXP3Luc in HEK293T cell. Cells were analyzed as in A. E, effects of MEK1DD on FOXP3Luc and Gal4Luc. Assays were performed as in B. F, DNA pulldown assays for VP16-DelN. EGFP, COT, and MEK1DD were coexpressed with VP16-DelN in HEK293T cells. Streptavidin-agarose beads coated with 5′-biotinylated FOXP3 binding oligonucleotide were used to pull down VP16-DelN from cell lysates. G, effects of COT and MEK1 inhibition on Foxp3 expression in Treg cells. pLN cells from TregYFP mice were activated by repetitive plate-bound anti-CD3/CD28 in the presence of 200 units/ml IL-2 for 5 days. COT inhibitor (5 μm) or U0126 (5 μm) were added at day2. H, quantification of frequency of Foxp3low cells among CD4+YFP+ cells described in G. I, effects of COT knockdown on Foxp3 expression in Treg cells. Pooled splenocytes and pLN cells from TregYFP mice were cultured as in F before FACS for YFP+ cells on day 2. Sorted YFP+ cells were infected with retrovirus packaging the LMP-Thy1.1-based vectors and activated by repetitive plate-bound anti-CD3/CD28 in the presence of 1000 units/ml IL-2 for 3 days. J, quantification of frequency of Foxp3low cells among CD4+YFP+Thy1.1 cells described in I. All experiments were repeated independently at least three times. The error bars represent the mean ± S.D. *, p < 0.05; **, p < 0.005; ***, p < 0.0005; two-tailed, paired Student's t test.
COT is an activator of ERK, which primarily depends on MEK, but not RAF (32), so we tested whether the inhibitory effect of COT was dependent on MEK-ERK signaling. We found that the inhibitory effect of COT on FOXP3Luc could be antagonized by either the MEK-specific inhibitor U0126 or the ERK-specific phosphatase DUSP6 (Fig. 2C, D). Moreover, overexpression of MEK1DD (constitutively active, rat S218D/S222D mutant) inhibited FOXP3Luc without affecting Gal4Luc (Fig. 2E). Further studies revealed that COT altered neither the protein degradation of HA-FOXP3DelN nor the nuclear transport of EGFP-FOXP3DelN in HEK293T cells (data not shown). Finally, DNA pull-down assays demonstrated that COT and MEK1DD decreased the DNA binding activity of Flag-VP16-DelN (Fig. 2F).
Attenuating the DNA binding activity of Foxp3 potentially breaks the CNS2-Cbfβ-Runx1-Foxp3 feedback loop for Foxp3 expression, resulting in Treg cell instability. To further determine the roles of COT/MEK activity in differentiated Treg cells, we cultured peripheral lymph node cells from “Treg fate mapping” mice (Foxp3-GFPCre×R26-loxp-stop-loxp-YFP, termed TregYFP mice in this study; they have Treg cells permanently labeled by YFP) in the presence of repetitive stimulation by CD3/CD28 antibodies. We found that the inhibition of either COT and MEK1/2 activity via a COT inhibitor (CAS 871307-18-5) or U0126 helped the YFP+ Treg cells to maintain stable Foxp3 expression (Fig. 2, G and H). Similarly, we observed that FACS-sorted Treg cells expressing Cot-specific shRNA displayed more stable Foxp3 expression than those expressing a scrambled shRNA under strong activation conditions (Fig. 2, I and J). Together, our results suggest that COT/Tpl2 specifically inhibits the DNA binding activity of FOXP3 through a MEK-ERK-dependent pathway, which may involve in the regulation of Treg cell lineage stability in vivo.
Foxp3-specific Activation of MEK1 Promotes Treg Cell Instability in Vivo
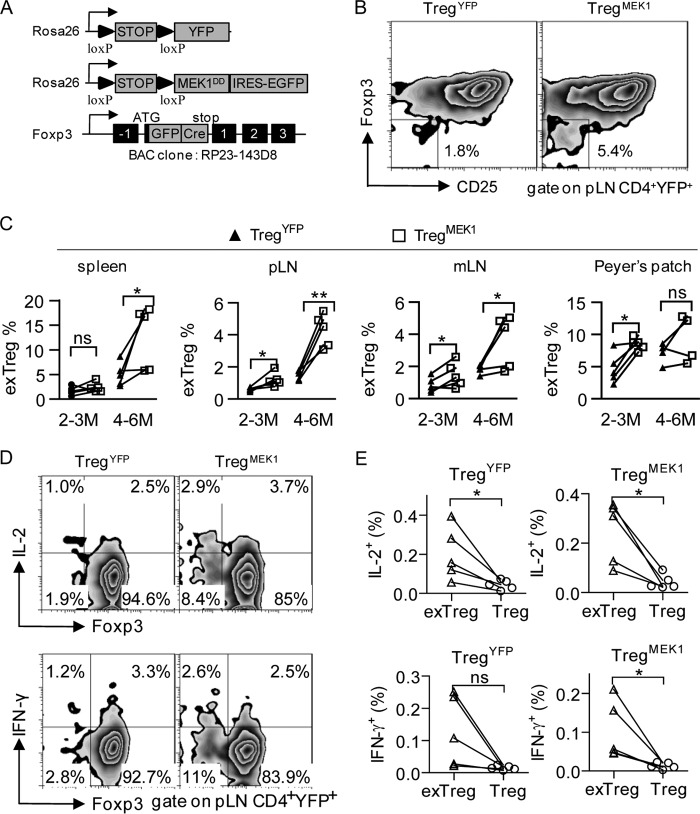
To investigate the roles of the MEK-ERK pathway in differentiated Treg cells in vivo, we crossed TregYFP mice with Rosa26-loxp-stop-loxp-MEK1DD-IRES-EGFP mice (The Jackson Laboratory, catalog no. 012352) to generate littermates containing Foxp3-specific expression of G/YFP.MEK1DD in a mixed NOD/B6 background, which were termed TregMEK1 mice in this study (Fig. 3A). The TregMEK1 mice specifically expressed the rat MEK1DD transgene in differentiated Treg cells after the “loxp-stop-loxp” cassette downstream of the Rosa26 promoter was excised by the Foxp3 BAC-driven GFP-Cre, which yielded a good model to study the MEK-ERK pathway in differentiated Treg cells in vivo. Most of the peripheral Foxp3-positive cells in TregYFP and TregMEK1 mice displayed Rosa26-YFP/GFP expression, indicating that the “Foxp3-GFP-Cre/loxp-stop-loxp-Rosa26” system worked well, as reported previously (20).
FIGURE 3.
Foxp3-specific activation of MEK1 promotes Treg cell instability in vivo. A, schematic of the transgene constructs used in this study. IRES, internal ribosome entry site. B, expression of surface CD25 and intracellular Foxp3 by gated CD4+YFP+ T cells from the pLN of 4- to 6-month-old TregYFP and TregMEK1 mice under homeostatic conditions. Numbers in and adjacent to the outlined areas indicate the percent of cells in each. C, frequency of CD25−Foxp3− exTreg cells among CD4+ YFP+ cells from the spleens, pLN, mLN, and Peyer's patches of 2- to 3-month-old and 4- to 6-month-old TregYFP and TregMEK1 mice under homeostatic conditions. D, expression of IL-2, IFN-γ, and intracellular Foxp3 by gated CD4+YFP+ T cells from pLN of TregYFP and TregMEK1 mice. Cells were incubated for 3–4 h at 37 °C with ionomycin, phorbol 12-myristate 13-acetate, and monensin for cytokine analysis. E, frequency of IL-2+ or IFN-γ+ cells among exTreg cells and Treg cells from pLN of TregYFP and TregMEK1 mice. Data are representative of at least five mice. *, p < 0.05; **, p < 0.005; ns, not significant; two-tailed, paired Student's t test.
To study the effects of MEK1 activation on the lineage stability of Treg cells, we analyzed the frequency of CD25−Foxp3−“exTreg” cells among CD4+YFP+ cells from TregYFP and TregMEK1 mice. Consistent with our previous reports, we found that a fraction of YFP+ Treg cells lost Foxp3 expression in both TregYFP and TregMEK1 mice under homeostatic conditions and that most such Foxp3− Treg cells were in the YFP+CD25− population (Fig. 3B). There was a correlation between the frequency of CD25−Foxp3− exTreg cells and the age of the mice, i.e. 4- to 6-month-old mice had a higher frequency of CD25−Foxp3− exTreg cells than 2- to 3-month-old mice (Fig. 3C). Importantly, compared with age-matched TregYFP mice, 2- to 3-month-old TregMEK1 mice had a significantly higher frequency of CD25−Foxp3− exTreg cells among CD4+YFP+ cells from the pLN, mLN, and Peyer's patch (pLN, p = 0.0247; mLN, p = 0.0391; Peyer's patch, p = 0.0157; Fig. 3C). Similarly, 4- to 6-month-old TregMEK1 mice had a significantly higher frequency of CD25−Foxp3− exTreg cells among CD4+YFP+ cells from the spleen, pLN, and mLN (spleen, p = 0.0483; pLN, p = 0.0031; mLN, p = 0.0487; Fig. 3C). These CD25−Foxp3− exTreg cells tended to produce IL-2 and IFN-γ upon phorbol 12-myristate 13-acetate/ionomycin stimulation (Fig. 3, D and E), which is consistent with the Th-like phenotypes of exTreg cells reported previously (13). Taken together, these data suggest that Foxp3-specific activation of MEK1 promotes Treg cell instability in vivo.
Foxp3-specific Activation of MEK1 Disrupts Treg Cell Function in EAE
Next, we tested whether the Treg cell instability induced by active MEK1 would accompany a functional defect in Treg cells. Superficially, we did not observe a significant difference between TregYFP and TregMEK1 mice in breeding and growth. Similarly, there were no statistically significant differences in overall immune homeostasis and Treg cell homeostasis in vivo under homeostatic conditions, except that the mean fluorescence intensity of Foxp3 staining in YFP+ Treg cells from the Peyer's patch of TregMEK1 mice was significant lower than that in TregYFP mice, which was probably linked to the enrichment of peripheral Treg cells and the immune activation environment in the intestine.
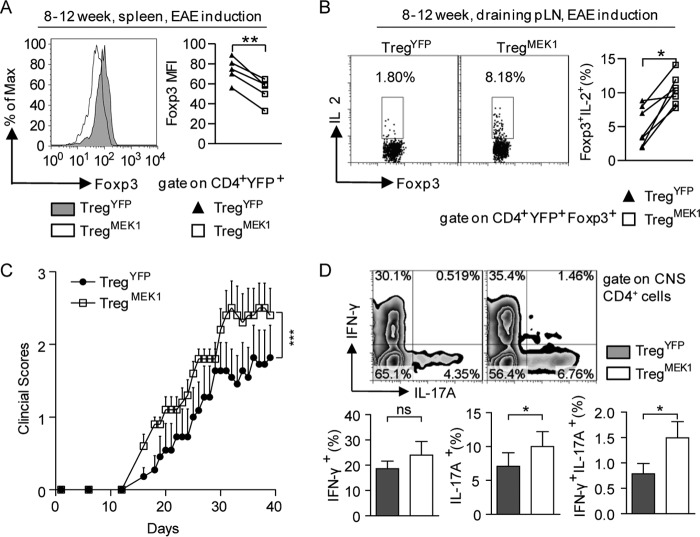
Recent studies have shown that CNS2 plays a critical role in maintaining FOXP3 expression in Treg cells and the knockout of CNS2 exacerbates experimental autoimmune encephalitis (EAE) development (33, 34). In addition, it has reported that MOG35–55 antigen-specific natural Treg cells are highly unstable in EAE (15). Therefore, we induced EAE in transgenic mice by using MOG35–55 peptide in complete Freund adjuvant and heat-killed B. pertussis. For strict comparison between individuals, mice were sacrificed and analyzed during the third week after immunization. Although most of the mice displayed no symptoms or only flaccid tails at the time of sacrifice, their immune systems were indeed activated by EAE induction, as shown by the enlarged spleens and higher frequencies of CD44hiCD62Llow T cells in EAE-induced mice compared with mice without EAE induction (data not shown). Under EAE-induced immune activation conditions, the mean fluorescence intensity of intracellular Foxp3 staining of splenic CD4+YFP+ cells from TregMEK1 mice was significantly lower than that from TregYFP mice (p = 0.0016, Fig. 4A). In addition, the frequency of IL-2+ cells among CD4+YFP+Foxp3+ cells from draining pLN of TregMEK1 mice was significantly higher than that in TregYFP mice (p = 0.0025, Fig. 4B). Importantly, TregMEK1 mice presented higher clinical scores and earlier onset of EAE disease than TregYFP mice, indicative of the functional defects of Treg cells in TregMEK1 mice (paired Student's t test, p < 0.0005, Fig. 4C). There were stronger Th17 responses in the CNS of TregMEK1 mice compared with those in TregYFP mice, as shown by the significantly higher frequency of IL-17A-producing and IFN-γ/IL-17A double-producing CD4+ T cells in CNS of TregMEK1 mice (Fig. 4D). Therefore, constitutive activation of MEK1 dysregulated the functions of differentiated Treg cells in EAE.
FIGURE 4.
Foxp3-specific activation of MEK1 dysregulates Treg cell function in EAE. A, MFI (mean fluorescence intensity) of intracellular Foxp3 of CD4+YFP+ cells from the spleens of TregYFP and TregMEK1 mice under EAE induction conditions. Mice were sacrificed and analyzed at the third week of EAE induction. B, frequency of IL-2+ cells among CD4+YFP+Foxp3+ cells from the draining pLN of TregYFP and TregMEK1 mice under EAE induction conditions. Cells were incubated for 3–4 h at 37 °C with ionomycin, phorbol 12-myristate 13-acetate, and monensin for cytokine analysis. C, clinical scores of EAE disease in TregYFP (n = 11) and TregMEK1 (n = 10) mice. D, frequency of IFN-γ+ or/and IL-17A+ cells among CD4+ cells from the CNS of TregYFP and TregMEK1 mice during the EAE peak phase. For all experiments, data are representative of and compiled from at least five mice. The numbers in the chart indicate the percentage of cells in each area. * p < 0.05; **, p < 0.005; ***, p < 0.0005; ns, not significant; two-tailed, paired Student's t test.
DISCUSSION
It is important to understand the cell-intrinsic signaling pathways linked to Treg cell instability in vivo, which provides targets for developing strategies to modulate Treg cell function and lineage stability in the clinic. In this study, we explored the roles of cell-intrinsic signaling pathways represented by ∼200 human kinases in the control of FOXP3 function. Our study provides a more comprehensive understanding of cell signal transduction in the generation of Treg cell instability and indicates that the COT/Tpl2-MEK-ERK signaling pathway promotes Treg cell instability in vivo, potentially by modulating the DNA binding activity of FOXP3. Therefore, therapeutic intervention targeting the COT/Tpl2-MEK-ERK pathway might contribute to treating autoimmune diseases and optimize human Treg cell therapy.
Compared with studying FOXP3 functions in cultured primary Treg cells or T cell lines, our FOXP3Luc system in HEK293T cells presents several unique advantages. First, the background FOXP3 expression in FOXP3Luc was negligible because HEK293T cells do not endogenously express FOXP3. Second, FOXP3Luc in HEK293T cells was easy to utilize for large-scale screening experiments. Third, FOXP3Luc reliably translated the DNA binding activity of FOXP3 into luciferase activity. In addition, FOXP3Luc could be useful for evaluating IPEX mutations for clinical diagnosis. Indeed, as shown by analysis of different known FOXP3 alleles, the M370I, F371C, F373A, and R397W mutations linked to severe IPEX presentations caused dramatic reductions in the readout of FOXP3Luc, whereas the mild IPEX syndrome-associated A384T and F324L mutations caused mild or negligible reductions (35). Therefore, FOXP3Luc provides a practical and simple tool to predict the clinical significance of genetic mutations in the FOXP3 gene. Moreover, it is interesting that removal of the N terminus (amino acids 1–181) of FOXP3 increased the DNA binding activity of its FKH domain, as shown by this study and gel shifts findings reported previously (31). These data suggest that an autoinhibitory function of the N-terminal region may be associated with dynamic control of FOXP3 function in vivo. It is possible that Foxp3 requires partners or certain modifications to efficiently bind target gene loci in Treg cells.
Aside from the DNA binding activity of the FKH domain, nonspecific effects caused by the active form of COT and MEK1 may lead to the reduction of FOXP3Luc readout. We set up multiple levels of controls to evaluate such nonspecific effects. First, readouts of FOXP3Luc were normalized to a blank control group to remove the nonspecific effects caused by transfection and the physiological status of the cultured cells. Second, Gal4Luc was used as a mutual control for FOXP3Luc to evaluate VP16 function. Third, regardless of whether VP16-DelN expression was driven by the MSCV or CMV promoter, FOXP3Luc could be inhibited by the active forms of COT and MEK1. Fourth, neither the protein degradation nor the nuclear transport of FOXP3DelN were affected by the active form of COT and MEK1. More importantly, the DNA pulldown assays directly proved that the active form of COT and MEK1 inhibited the DNA binding activity of FOXP3. Moreover, active COT and MEK1 inhibited FOXP3Luc in a dose-dependent manner, implying that the signal strength of the COT-MEK-ERK pathway is important for their roles in Treg cells in vivo. Taken together, we suggest that activation of the COT-MEK-ERK pathway specifically inhibits the DNA binding activity of FOXP3.
It has been reported that an attenuated MEK-ERK pathway favors FOXP3 induction in conventional T cells in vitro (36–38). However, in differentiated Treg cells, which have established Foxp3 expression, the role of the MEK-ERK pathway was not yet clear. In this study, we demonstrated that inhibition of COT and MEK1 activity favors the lineage stabilization of Treg cells in vitro, which was further supported by evidence from a transgenic mouse model with constitutively activated MEK1 in differentiated Treg cells in vivo. Because several target genes of Foxp3, such as Foxp3, CD25, and IL-2, were dysregulated, it was unlikely that the active MEK1 individually disturbed the expression of these genes. The most likely scenario is that the active MEK1 impairs the DNA binding activity of Foxp3, which subsequently elicits downstream alterations of the Foxp3-dependent program in Treg cells. Meanwhile, it cannot be excluded that other functions of Foxp3 or Foxp3-independent programs are altered by the active MEK1. Therefore, we suggest that constitutive activation of the MEK-ERK pathway promotes dysregulation of Treg function and instability of Foxp3 expression in vivo, at least partially through impairing the DNA binding activity of Foxp3. Such defects of Treg cells may aggravate the progression of autoimmune diseases such as EAE.
Birzele et al. (23) combined FoxP3 ChIP-sequencing and mRNA-sequencing techniques to examine the transcriptional differences between primary human CD4+ Th and Treg cells. They found that the expression of the DUSP6 and DUSP4 phosphatases, which selectively inactivate ERK kinases, is up-regulated in Treg cells (23). Consistent with this, CD3/CD28-mediated activation of the MEK-ERK pathway is largely impaired in regulatory T-cell lines from human cord blood (39). Therefore, the activation of ERK caused by MEK1DD transgene expression in Treg cells might be restricted by DUSPs under homeostatic conditions, whereas immune activation could facilitate Treg cells to bypass the threshold on ERK activation, consistent with our observations that Treg cells expressing MEK1DD are more unstable in Peyer's patch or under the EAE induction conditions. Alternatively, it is possible that a properly functioning MEK-ERK pathway favors Treg cell activation and function, whereas an overactive MEK-ERK pathway impairs Treg cell stability and function. This may explain the sharp increase in the clinical score curve in the later phase of EAE induction in TregMEK1 mice.
The MEK-ERK pathway is an important downstream branch of TCR signaling (40). To a large extent, constitutive activation of MEK1 mirrors strong TCR stimulation both in intensity and duration. It is known that the quality and quantity of TCR signaling usually determines the fate of T cells. For example, a TCR signal that is too strong induces apoptosis of thymocytes during the negative selection process (41). In addition, strong TCR signaling enhances Th1 differentiation, whereas weak TCR signaling favors Th2 polarization (42). Consistent with this scenario, we found previously that approximately half of Vβ4-positive Treg cells lose Foxp3 expression in the pancreatic lymph nodes of NOD BDC2.5 TCR-transgenic mice, which harbor a transgenic TCR specific for pancreatic antigen (13). More recently, another study reported that unstable Foxp3 expression occurs selectively in MOG38–49-specific Treg cells rather than polyclonal Treg cells from the same inflamed sites in an EAE induction model (15). This evidence suggests that TCR overstimulation by autoantigens promotes Treg cells instability in vivo. Our findings support this hypothesis and highlight the MEK-ERK pathway as an important transmitter/effector for the overly strong TCR signal in the induction of Treg cell instability. Our study also provides a model showing that a fraction of Treg cells in situ is subjected to too much engagement with antigens in autoimmune and inflammatory settings, resulting in strong activation of the MEK-ERK pathway, which promotes instability of Foxp3 expression, partially through breaking the CNS2-Cbfβ-Runx1-Foxp3 feedback loop.
Aside from the TCR signal, other inflammatory signals that lead to activation of the MEK-ERK pathway potentially modulate Treg cell stability, such as LPS, which may activate the MEK-ERK pathway though the TLR4/COT cascade if this signaling pathway is functional in Treg cells as it is in macrophages (43). Together, our results support the notion that outside inflammatory signals act through the Cot/Tpl2-MEK-ERK signaling pathway to destabilize the Treg lineage. Further studies are needed to elucidate the mechanisms of the down-regulation of Foxp3 DNA binding activity by the MEK-ERK pathway in Treg cells, which will provide new insights into the generation of Treg cell instability.
Acknowledgments
We thank Dr. Baidong Hou, Dr. Zhaolin Hua, and Dr. Fuping Zhang for critical and constructive comments regarding the manuscript. We also thank Prof. William C. Hahn (Dana Farber Cancer Institute), for the myristoylated kinase library, Prof. Igor Astsaturov (Fox Chase Cancer Center), for the pLEX-DUSP6 plasmid, and Prof. Klaus Rajewsky (Immune Disease Institute, Harvard Medical School) for the C57BL/6-Gt (ROSA) 26Sortm8 (Map2k1*, EGFP) Rsky/J mice.
This work was supported by National Key Basic Research and Development 973 Program of China Grant 2012CB917102, by National Natural Science Foundation of China Grant 31070804, and by China Postdoctoral Science Foundation Grant 2011M500422.
- Treg cell
- regulatory T cell
- FKH
- forkhead/winged helix
- IPEX
- immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome
- Luc
- luciferase
- mLN
- mesenteric lymph node(s)
- pLN
- axillary and inguinal lymph node(s)
- EAE
- experimental autoimmune encephalitis
- TCR
- T cell receptor.
REFERENCES
- 1. Mocci S., Lafferty K., Howard M. (2000) The role of autoantigens in autoimmune disease. Curr. Opin. Immunol. 12, 725–730 [DOI] [PubMed] [Google Scholar]
- 2. Miyara M., Sakaguchi S. (2007) Natural regulatory T cells: mechanisms of suppression. Trends Mol. Med. 13, 108–116 [DOI] [PubMed] [Google Scholar]
- 3. Josefowicz S. Z., Lu L. F., Rudensky A. Y. (2012) Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brunstein C. G., Miller J. S., Cao Q., McKenna D. H., Hippen K. L., Curtsinger J., Defor T., Levine B. L., June C. H., Rubinstein P., McGlave P. B., Blazar B. R., Wagner J. E. (2011) Infusion of ex vivo expanded T regulatory cells in adults transplanted with umbilical cord blood: safety profile and detection kinetics. Blood 117, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tang Q., Bluestone J. A. (2013) Regulatory T-cell therapy in transplantation: moving to the clinic. Cold Spring Harb. Perspect. Med. 10.1101/cshperspect.a015552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fontenot J. D., Rasmussen J. P., Williams L. M., Dooley J. L., Farr A. G., Rudensky A. Y. (2005) Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity 22, 329–341 [DOI] [PubMed] [Google Scholar]
- 7. Williams L. M., Rudensky A. Y. (2007) Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 8, 277–284 [DOI] [PubMed] [Google Scholar]
- 8. Fontenot J. D., Gavin M. A., Rudensky A. Y. (2003) Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 [DOI] [PubMed] [Google Scholar]
- 9. Brunkow M. E., Jeffery E. W., Hjerrild K. A., Paeper B., Clark L. B., Yasayko S. A., Wilkinson J. E., Galas D., Ziegler S. F., Ramsdell F. (2001) Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27, 68–73 [DOI] [PubMed] [Google Scholar]
- 10. Wildin R. S., Freitas A. (2005) IPEX and FOXP3: clinical and research perspectives. J. Autoimmun. 25, 56–62 [DOI] [PubMed] [Google Scholar]
- 11. Hori S., Nomura T., Sakaguchi S. (2003) Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 [DOI] [PubMed] [Google Scholar]
- 12. Tsuji M., Komatsu N., Kawamoto S., Suzuki K., Kanagawa O., Honjo T., Hori S., Fagarasan S. (2009) Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer's patches. Science 323, 1488–1492 [DOI] [PubMed] [Google Scholar]
- 13. Zhou X., Bailey-Bucktrout S. L., Jeker L. T., Penaranda C., Martínez-Llordella M., Ashby M., Nakayama M., Rosenthal W., Bluestone J. A. (2009) Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat. Immunol. 10, 1000–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Komatsu N., Mariotti-Ferrandiz M. E., Wang Y., Malissen B., Waldmann H., Hori S. (2009) Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc. Natl. Acad. Sci. U.S.A. 106, 1903–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bailey-Bucktrout S. L., Martinez-Llordella M., Zhou X., Anthony B., Rosenthal W., Luche H., Fehling H. J., Bluestone J. A. (2013) Self-antigen-driven activation induces instability of regulatory T cells during an inflammatory autoimmune response. Immunity 39, 949–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Komatsu N., Okamoto K., Sawa S., Nakashima T., Oh-hora M., Kodama T., Tanaka S., Bluestone J. A., Takayanagi H. (2014) Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat. Med. 20, 62–68 [DOI] [PubMed] [Google Scholar]
- 17. Yang X. O., Nurieva R., Martinez G. J., Kang H. S., Chung Y., Pappu B. P., Shah B., Chang S. H., Schluns K. S., Watowich S. S., Feng X. H., Jetten A. M., Dong C. (2008) Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity 29, 44–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lal G., Zhang N., van der Touw W., Ding Y., Ju W., Bottinger E. P., Reid S. P., Levy D. E., Bromberg J. S. (2009) Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J. Immunol. 182, 259–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Z., Barbi J., Bu S., Yang H. Y., Li Z., Gao Y., Jinasena D., Fu J., Lin F., Chen C., Zhang J., Yu N., Li X., Shan Z., Nie J., Gao Z., Tian H., Li Y., Yao Z., Zheng Y., Park B. V., Pan Z., Zhang J., Dang E., Li Z., Wang H., Luo W., Li L., Semenza G. L., Zheng S. G., Loser K., Tsun A., Greene M. I., Pardoll D. M., Pan F., Li B. (2013) The ubiquitin ligase Stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor Foxp3. Immunity 39, 272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou X., Jeker L. T., Fife B. T., Zhu S., Anderson M. S., McManus M. T., Bluestone J. A. (2008) Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J. Exp. Med. 205, 1983–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rudra D., Egawa T., Chong M. M., Treuting P., Littman D. R., Rudensky A. Y. (2009) Runx-CBFβ complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 10, 1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng Y., Josefowicz S., Chaudhry A., Peng X. P., Forbush K., Rudensky A. Y. (2010) Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Birzele F., Fauti T., Stahl H., Lenter M. C., Simon E., Knebel D., Weith A., Hildebrandt T., Mennerich D. (2011) Next-generation insights into regulatory T cells: expression profiling and FoxP3 occupancy in human. Nucleic Acids Res. 39, 7946–7960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ono M., Yaguchi H., Ohkura N., Kitabayashi I., Nagamura Y., Nomura T., Miyachi Y., Tsukada T., Sakaguchi S. (2007) Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 446, 685–689 [DOI] [PubMed] [Google Scholar]
- 25. Barzaghi F., Passerini L., Bacchetta R. (2012) Immune dysregulation, polyendocrinopathy, enteropathy, x-linked syndrome: a paradigm of immunodeficiency with autoimmunity. Front. Immunol. 3, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boehm J. S., Zhao J. J., Yao J., Kim S. Y., Firestein R., Dunn I. F., Sjostrom S. K., Garraway L. A., Weremowicz S., Richardson A. L., Greulich H., Stewart C. J., Mulvey L. A., Shen R. R., Ambrogio L., Hirozane-Kishikawa T., Hill D. E., Vidal M., Meyerson M., Grenier J. K., Hinkle G., Root D. E., Roberts T. M., Lander E. S., Polyak K., Hahn W. C. (2007) Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 129, 1065–1079 [DOI] [PubMed] [Google Scholar]
- 27. Bagnyukova T. V., Restifo D., Beeharry N., Gabitova L., Li T., Serebriiskii I. G., Golemis E. A., Astsaturov I. (2013) DUSP6 regulates drug sensitivity by modulating DNA damage response. Br. J. Cancer 109, 1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Srinivasan L., Sasaki Y., Calado D. P., Zhang B., Paik J. H., DePinho R. A., Kutok J. L., Kearney J. F., Otipoby K. L., Rajewsky K. (2009) PI3 kinase signals BCR-dependent mature B cell survival. Cell 139, 573–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ivanov I. I., McKenzie B. S., Zhou L., Tadokoro C. E., Lepelley A., Lafaille J. J., Cua D. J., Littman D. R. (2006) The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 [DOI] [PubMed] [Google Scholar]
- 30. Sadowski I., Ma J., Triezenberg S., Ptashne M. (1988) GAL4-VP16 is an unusually potent transcriptional activator. Nature 335, 563–564 [DOI] [PubMed] [Google Scholar]
- 31. Koh K. P., Sundrud M. S., Rao A. (2009) Domain requirements and sequence specificity of DNA binding for the forkhead transcription factor FOXP3. PLoS ONE 4, e8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johannessen C. M., Boehm J. S., Kim S. Y., Thomas S. R., Wardwell L., Johnson L. A., Emery C. M., Stransky N., Cogdill A. P., Barretina J., Caponigro G., Hieronymus H., Murray R. R., Salehi-Ashtiani K., Hill D. E., Vidal M., Zhao J. J., Yang X., Alkan O., Kim S., Harris J. L., Wilson C. J., Myer V. E., Finan P. M., Root D. E., Roberts T. M., Golub T., Flaherty K. T., Dummer R., Weber B. L., Sellers W. R., Schlegel R., Wargo J. A., Hahn W. C., Garraway L. A. (2010) COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468, 968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feng Y., Arvey A., Chinen T., van der Veeken J., Gasteiger G., Rudensky A. Y. (2014) Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 158, 749–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li X., Liang Y., LeBlanc M., Benner C., Zheng Y. (2014) Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell 158, 734–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. d'Hennezel E., Bin Dhuban K., Torgerson T., Piccirillo C. A. (2012) The immunogenetics of immune dysregulation, polyendocrinopathy, enteropathy, X linked (IPEX) syndrome. J. Med. Genet. 49, 291–302 [DOI] [PubMed] [Google Scholar]
- 36. Mor A., Keren G., Kloog Y., George J. (2008) N-Ras or K-Ras inhibition increases the number and enhances the function of Foxp3 regulatory T cells. Eur. J. Immunol. 38, 1493–1502 [DOI] [PubMed] [Google Scholar]
- 37. Chang C. F., D'Souza W. N., Ch'en I. L., Pages G., Pouyssegur J., Hedrick S. M. (2012) Polar opposites: Erk direction of CD4 T cell subsets. J. Immunol. 189, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luo X., Zhang Q., Liu V., Xia Z., Pothoven K. L., Lee C. (2008) Cutting edge: TGF-β-induced expression of Foxp3 in T cells is mediated through inactivation of ERK. J. Immunol. 180, 2757–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li L., Godfrey W. R., Porter S. B., Ge Y., June C. H., Blazar B. R., Boussiotis V. A. (2005) CD4+CD25+ regulatory T-cell lines from human cord blood have functional and molecular properties of T-cell anergy. Blood 106, 3068–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin J., Weiss A. (2001) T cell receptor signalling. J. Cell Sci. 114, 243–244 [DOI] [PubMed] [Google Scholar]
- 41. Starr T. K., Jameson S. C., Hogquist K. A. (2003) Positive and negative selection of T cells. Annu. Rev. Immunol. 21, 139–176 [DOI] [PubMed] [Google Scholar]
- 42. Constant S., Pfeiffer C., Woodard A., Pasqualini T., Bottomly K. (1995) Extent of T cell receptor ligation can determine the functional differentiation of naive CD4+ T cells. J. Exp. Med. 182, 1591–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dumitru C. D., Ceci J. D., Tsatsanis C., Kontoyiannis D., Stamatakis K., Lin J. H., Patriotis C., Jenkins N. A., Copeland N. G., Kollias G., Tsichlis P. N. (2000) TNF-α induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell 103, 1071–1083 [DOI] [PubMed] [Google Scholar]