Background: The mechanisms underlying U1-70K aggregation in AD are unknown.
Results: AD brain homogenates can induce the aggregation of soluble U1-70K, and disordered low complexity domains are necessary for U1-70K aggregation.
Conclusion: U1-70K in AD may directly sequester normal soluble forms of U1-70K into insoluble aggregates.
Significance: These observations highlight the importance of low complexity domains in RNA-binding protein aggregation in neurodegenerative disease.
Keywords: Neurodegenerative Disease, Protein Aggregation, RNA, Spliceosome, Tau Protein (Tau)
Abstract
Recent evidence indicates that U1-70K and other U1 small nuclear ribonucleoproteins are Sarkosyl-insoluble and associate with Tau neurofibrillary tangles selectively in Alzheimer disease (AD). Currently, the mechanisms underlying the conversion of soluble nuclear U1 small nuclear ribonucleoproteins into insoluble cytoplasmic aggregates remain elusive. Based on the biochemical and subcellular distribution properties of U1-70K in AD, we hypothesized that aggregated U1-70K itself or other biopolymers (e.g. proteins or nucleic acids) interact with and sequester natively folded soluble U1-70K into insoluble aggregates. Here, we demonstrate that total homogenates from AD brain induce soluble U1-70K from control brain or recombinant U1-70K to become Sarkosyl-insoluble. This effect was not dependent on RNA and did not correlate with detergent-insoluble Tau levels as AD homogenates with reduced levels of these components were still capable of inducing U1-70K aggregation. In contrast, proteinase K-treated AD homogenates and Sarkosyl-soluble AD fractions were unable to induce U1-70K aggregation, indicating that aggregated proteins in AD brain are responsible for inducing soluble U1-70K aggregation. It was determined that the C terminus of U1-70K, which harbors two disordered low complexity (LC) domains, is necessary for U1-70K aggregation. Moreover, both LC1 and LC2 domains were sufficient for aggregation. Finally, protein cross-linking and mass spectrometry studies demonstrated that a U1-70K fragment harboring the LC1 domain directly interacts with aggregated U1-70K in AD brain. Our results support a hypothesis that aberrant forms of U1-70K in AD can directly sequester soluble forms of U1-70K into insoluble aggregates.
Introduction
Many neurodegenerative diseases, including Alzheimer disease (AD),3 Parkinson disease, frontotemporal lobar degeneration (FTLD), and prion diseases, are proteinopathies that are associated with the accumulation of detergent-insoluble protein aggregates (1, 2). Neuropathology of AD is defined by the presence of senile plaques and neurofibrillary tangles (NFTs) composed of detergent-insoluble amyloid-β and phosphorylated Tau (pTau), respectively (3). The clinical progression of AD is characterized by an extended prodromal phase in which amyloid-β and Tau pathology accumulate years prior to cognitive symptoms (4). These proteinaceous aggregates are hypothesized to cause both functional deficits as well as gain-of-function toxicity that ultimately lead to synapse loss and cognitive impairment. However, high amyloid-β burden and NFT pathology are present in 30–50% of cognitively normal older individuals at death (5–7), suggesting that these core pathologies are not sufficient to explain the onset of cognitive decline (4).
To identify other aggregated proteins in AD that could contribute to disease pathogenesis, we recently undertook a comprehensive study of the human brain detergent-insoluble proteome in AD (8). These studies revealed substantial enrichment of U1-70K and other core U1 small nuclear ribonucleoproteins (snRNPs) in Sarkosyl-insoluble fractions in AD but not in other neurodegenerative diseases. Additionally, mis-localized U1 snRNP proteins associated with cytoplasmic tangle-like aggregates in AD (8).
snRNPs are RNA-protein complexes that interact with pre-mRNA and auxiliary splicing factors to form a spliceosome (9, 10). Five distinct snRNP complexes (U1, U2, U4, U5, and U6) act in the spliceosome to remove introns from pre-mRNA, which occurs, with rare exceptions (11, 12), in the nucleus of all eukaryotic cells. The U1 snRNP complex consists of a 165-nucleotide U1 small nuclear RNA (snRNA) and 10 distinct proteins, which together initiate splicing by recognizing the 5′ splice site on nearly 98% of all pre-mRNAs (13, 14). Comparison of RNA from AD and control brains revealed significant defects in RNA maturation with accumulation of unspliced RNA species in AD, consistent with a loss of U1 snRNP nuclear function (8). However, the mechanisms underlying the conversion of highly soluble nuclear U1 snRNP proteins into insoluble cytoplasmic aggregates remain elusive.
Recently McKnight and co-workers (15) used a biotinylated isoxazole compound to precipitate hundreds of RNA splicing factors with intrinsically disordered low complexity (LC) domains from cell and tissue extracts. A large proportion of proteins identified were core or associated members of the spliceosome that included U1-70K, serine-arginine (SR) splicing factors, and several heterogeneous nuclear ribonucleoproteins. LC domains are polypeptide sequences that have a tendency to be comprised of just a few of the 20 amino acids and are typically organized in repetitive arrays (16). Interestingly, at high concentrations in vitro, some LC domains undergo a concentration-dependent phase transition to a hydrogel-like state composed of uniformly polymerized amyloid-like aggregates that can sequester normal soluble forms of the same protein or other RNA-binding proteins with similar, but not identical, LC domains (15). U1-70K harbors two C-terminal LC domains that lack native three-dimensional and secondary structure in solution (17). Other notable RNA-binding proteins with LC domains include TAR DNA-binding protein 43 (TDP-43) and fused in sarcoma (FUS), both of which form detergent-insoluble amyloid-like aggregates in amyotrophic lateral sclerosis and FTLD (18). These proteins also share aggregation properties mediated via their LC domains (15, 19–21). Whether LC domains in U1-70K or other intrinsically disordered U1 snRNP proteins drive their detergent insolubility and aggregation in AD remains unclear.
In this study, immunogold electron microscopy was used to demonstrate that cytoplasmic U1-70K associates with NFTs in AD brain. Paradoxically, nearly all detectable U1-70K was found in the Sarkosyl-insoluble fraction of AD brain tissue following biochemical fractionation, despite the fact that most neurons and glia harbor nonfilamentous nuclear U1-70K. Based on these observations, we hypothesized that following tissue homogenization, aggregated U1-70K itself or other biopolymers (i.e. proteins or nucleic acids) sequester natively folded soluble U1-70K into detergent-insoluble aggregates. Herein, we provide evidence that AD brain homogenate can induce the aggregation of endogenous soluble U1-70K from control brain homogenate or recombinant U1-70K and render it Sarkosyl-insoluble. The mechanisms that underlie this process are not influenced by RNA and require the presence of Sarkosyl-insoluble proteins in AD, including U1-70K. Moreover, levels of insoluble U1-70K do not always correlate with levels of aggregated Tau in AD or other tauopathies, suggesting that Tau does not directly influence U1-70K aggregation. By expressing both N- and C-terminal truncations of recombinant U1-70K (rU1-70K) it was determined that the C terminus, which harbors two intrinsically disordered LC domains (LC1 and LC2), is necessary for U1-70K aggregation. Interestingly, rU1-70K fragments harboring either LC1 or LC2 were sufficient alone for aggregation, although rU1-70K fragments with only the LC1 domain showed enhanced aggregation properties. Finally, using protein cross-linking and mass spectrometry, we show that a rU1-70K fragment harboring the LC1 domain directly interacts with aggregated U1-70K in AD brain. These results support the hypothesis that aggregated U1-70K in AD brain can interact with and sequester normal soluble forms of U1-70K through the intrinsically disordered LC1 domain, thus providing important insight into the mechanisms underlying U1-70K protein aggregation events in AD.
EXPERIMENTAL PROCEDURES
Materials
Primary antibodies used in these studies include an in-house antibody raised against a synthetic keyhole limpet hemocyanin-conjugated peptide corresponding to a C-terminal epitope of U1-70K (clone EM439) (8), an anti-Myc-tag (clone 9B11, Cell Signaling), anti-TDP-43 (10782-2 ProteinTech), anti-Tau2 (clone MAB375, Millipore), anti-α-tubulin (clone DM1A, Cell Signaling), and anti-EEA1 (ab2900, Abcam). Secondary antibodies were conjugated to either Alexa Fluor 680 (Invitrogen) or IRDye800 (Rockland) fluorophores. RNase A from bovine pancreas (R4875, Sigma) was obtained as a lyophilized powder. A stock solution (10 mg/ml) was prepared in 20 mm Tris-HCl, pH 7.5, 10 mm NaCl, and 50% glycerol. A stock solution (1 mg/ml) of linear polyethyleneimine (25 kDa, Polysciences, Inc.) was prepared by heating in water and adjusting the pH to 7.0 with HCl. The solution was sterilized by passing through a 0.2-μm filter and stored in 1-ml aliquots at −20 °C.
Human Brain Tissue
Post-mortem frontal cortex tissue from healthy control cases and pathologically confirmed AD cases were selected for comparison from the Emory Alzheimer's Disease Research Center brain bank (n = 16). Post-mortem neuropathological evaluation of amyloid plaque distribution was performed according to the Consortium to Establish a Registry for Alzheimer's Disease semi-quantitative scoring criteria (22), although neurofibrillary tangle pathology was assessed in accordance with the Braak staging system (23). The corticobasal degeneration (CBD) case included in this study underwent extensive neuropathological characterization required for diagnosis based on established criteria (24, 25). All AD cases met NIA-Reagan criteria for the diagnosis of Alzheimer disease (high likelihood). Cases were matched as closely as possible for age at death, gender, and post-mortem interval, as shown in Table 1. Control and AD cases (n = 18) were also obtained from The Johns Hopkins University Brain Resource Center and Baltimore Longitudinal Study of Aging (Table 2), characterized as described previously (5).
TABLE 1.
Emory case demographics
The abbreviations used are as follows: PMI (h), postmortem interval; w, white/Caucasian; b, black/African American; a, Asian; h, Hispanic; NA, not applicable; m, male; and f, female.
| Case no. | BRAAK score | Neocortical neuritic plaque frequency | PMI | Age at death | Race/sex | Electron microscopy | Western blotting | Co-aggregation assays |
|---|---|---|---|---|---|---|---|---|
| Control | ||||||||
| OS03-299 | II | None | 6 | 69 | wm | x | ||
| E06-41 | II | None | 10 | 57 | wm | x | ||
| OS02-35 | I | None | 6 | 75 | wf | x | ||
| OS03-390 | II | None | 7 | 74 | wf | x | ||
| OS01-112 | I | None | 6 | 65 | wf | x | ||
| A93-03 | I | None | 4.5 | 70 | hm | |||
| A87-50 | II | None | 10 | 66 | wm | x | ||
| AD | ||||||||
| E05-04 | VI | High | 4.5 | 64 | wf | x | x | |
| E04-172 | V | High | 6 | 87 | wf | |||
| E07-69 | VI | High | 6 | 58 | wf | x | ||
| E05-56 | VI | High | 11.5 | 62 | wm | x | ||
| E12-24 | VI | High | 5.5 | 54 | wm | x | ||
| OS02-252 | VI | High | 17 | 50 | wf | x | ||
| OS98-15 | VI | High | 5 | 66 | wm | x | ||
| OS01-02 | VI | High | 5.5 | 69 | wf | x | ||
| CBD | ||||||||
| OS00-39 | NA | None | 3 | 61 | hm | x | ||
TABLE 2.
Baltimore Longitudinal Study of Aging case demographics
The abbreviations used are as follows: PMI (h), post-mortem interval; w, white/Caucasian; NA, not available; m, male; and f, female; CERAD, Consortium to Establish a Registry for Alzheimer's Disease.
| Case no. | BRAAK score | CERAD | PMI | Race/sex | Age at death |
|---|---|---|---|---|---|
| Control | |||||
| 1313 | II | 0 | NA | w/f | 92 |
| 1471 | II | 0 | NA | w/m | 87 |
| 1517 | II | A | 16 | w/f | 71 |
| 2027 | IV | 0 | 7 | w/m | 86 |
| 2066 | III | 0 | 17 | w/m | 95 |
| AD | |||||
| 1407 | V | C | 7 | w/m | 81 |
| 1649 | IV | C | 20 | w/m | 98 |
| 1712 | IV | C | 7 | w/m | 92 |
| 1735 | VI | C | 15 | w/m | 86 |
| 1875 | VI | C | 6 | w/f | 82 |
| 1921 | VI | C | 10 | w/f | 72 |
| 1984 | VI | C | 19 | w/f | 92 |
| 2004 | VI | C | 23 | w/m | 82 |
| 2028 | VI | C | 11 | w/f | 83 |
| 2032 | VI | C | 17.5 | w/f | 94 |
| 2157 | VI | C | 18 | w/f | 91 |
| 2184 | VI | C | 14.5 | w/f | 62 |
| 2226 | VI | C | 13 | w/f | 90 |
Plasmids and Cloning
The cDNA of U1-70K containing C-terminal Myc and DDK tags was cloned from pCMV6-Entry vector (Origene) and inserted into the HindIII/BamHI sites in the pcDNA3.1 vector. These U1-70K sequences were also cloned into the EcoRV/XhoI sites in the pLEXM-GST vector (kindly provided by Dr. Richard Kahn, Emory Department of Biochemistry) for the expression of N-terminal GST-tagged proteins. All expression plasmids were confirmed by DNA sequencing.
Tissue Fractionation
Post-mortem frontal cortex tissue was dounce-homogenized in 5 ml/g (20% w/v) of ice-cold homogenization buffer (50 mm HEPES, pH 7.0, 250 mm sucrose, 1 mm EDTA, and 1× HALT (Pierce) protease inhibitor mixture). Following homogenization, Sarkosyl (N-lauroylsarcosine) and NaCl were added to final concentrations of 1% (w/v) and 0.5 m, respectively (Sark-buffer). This fraction was defined as the total brain homogenate and was sonicated (Sonic Dismembrator System, Fisher Scientific) with three 5-s pulses at 30% amplitude (maximum intensity = 40%) using a microtip probe to shear nucleic acids. Protein concentrations were determined using the bicinchoninic acid (BCA) method (Pierce). To generate Sarkosyl-soluble (S1) and -insoluble fractions (P1), total brain homogenates (5 mg of total protein per case) were centrifuged at 180,000 × g for 30 min at 4 °C. The pellet (P1) was resuspended in Sark-buffer and centrifuged at 180,000 × g for an additional 30 min. The final pellet (P2) was solubilized in urea buffer (8 m urea and 2% SDS in 50 mm Tris-HCl, pH 8.5) with brief sonication, affording the Sarkosyl-insoluble fraction.
Recombinant Protein Expression
HEK293T cells (ATCC CRL-3216) were cultured in Dulbecco's modified Eagle's medium (DMEM, high glucose (Invitrogen)) supplemented with 10% (v/v) fetal bovine serum (Invitrogen) and penicillin/streptomycin (Invitrogen). The cells were maintained at 37 °C under a humidified atmosphere of 5% (v/v) CO2 in air. For transient transfection, the cells were grown to 80–90% confluency in 10-cm2 culture dishes and transfected with 10 μg of expression plasmid and 30 μg of linear polyethyleneimine. For co-aggregation experiments, cells were harvested 72 h post-transfection by scraping in ice-cold PBS and lysed in Sark-buffer. Lysates were briefly sonicated and cleared by centrifugation at 180,000 × g for 30 min at 4 °C.
U1-70K Co-aggregation Assays
For each experiment, all post-mortem human brain homogenates were diluted to a protein concentration of 10 mg/ml with Sark-buffer. AD, and control homogenates were combined in six ratios with an increasing proportion of AD homogenate (0.5 ml final volume). The mixtures were incubated 4 h at 4 °C and fractionated into detergent-soluble and detergent-insoluble fractions. For comparison, matching quantities of AD brain homogenate alone were diluted in Sark-buffer (0.5 ml final volume), incubated, and fractionated as before. The Sarkosyl-insoluble fractions were analyzed by quantitative Western blotting for U1-70K as described below. For co-aggregation experiments performed with full-length recombinant U1-70K (rU1-70K), HEK lysate (200 μg) with overexpressed rU1-70K was combined with post-mortem frontal cortex brain homogenates (5 mg of total protein) of control, AD, or CBD cases, incubated at 4 °C for 4 h, and fractionated as outlined above. For experiments involving GST-tagged N- and C-terminal truncation products, the incubation time was extended to 12 h at 4 °C. The Sarkosyl-insoluble fractions were analyzed by Western blotting with an in-house C-terminal U1-70K antibody (EM439) (8), as well as antibodies against the Myc tag (clone 9B11, Cell Signaling).
RNase and Proteinase K Treatment
To assess the effect of RNA on the aggregation of U1-70K, a co-aggregation experiment was performed using HEK lysate and brain homogenate pretreated with RNase A. Briefly, HEK lysates overexpressing rU1-70K and AD brain homogenates were treated with 200 μg/ml RNase A for 20 min at room temperature. RNA degradation was confirmed by resolving on an 8% denaturing urea polyacrylamide gel (urea-PAGE). Similarly, proteinase K (PK) digestion was used to assess the protein dependence of U1-70K aggregation as follows. A 0.5-ml aliquot (5 mg of total protein) of AD brain homogenate was treated with 200 μg/ml PK (Thermo EO0491) at 37 °C for 1 h and quenched with PMSF (5 mm). For comparison, an identical aliquot of AD brain homogenate was prepared without PK and incubated at 37 °C for 1 h. Both PK-treated and untreated AD homogenates were used to seed rU1-70K aggregation.
Immunohistochemistry
A cryostat was used to cut formalin and paraformaldehyde-fixed post-mortem human frontal cortex into 50-μm sections and immuno-stained essentially as described previously (26). Briefly, sections were washed in 0.1 m phosphate buffer (PB), incubated with 3% hydrogen peroxide, washed again with PB, and then incubated with blocking solution (Tris-buffered saline (TBS), 8% goat serum, 0.1% Triton X-100 and Avidin D (10 μg/ml final concentration; Vector Laboratories, A-2000)) for 1 h at 4 °C. Sections were washed with TBS and then incubated with primary antibody solution (primary antibody, TBS, 2% goat serum and biotin (50 μg/ml), Sigma, B0301) for 24–48 h at 4 °C. Sections were washed with TBS and incubated with biotinylated secondary antibody solution (biotinylated secondary antibody, TBS, 2% goat serum) for 1 h at 4 °C. Sections were washed with TBS, incubated with Vectastain Elite ABC amplification reagent (Vector Laboratories, PK-6200) for 1 h at 4 °C, washed again with TBS, incubated with 3,3-diaminobenzidine solution (Sigma, D4418) for 2–10 min at room temperature, and finally washed with TBS. Sections were mounted in standard fashion to glass slides, dried overnight at room temperature, dehydrated through alcohol rinses and Histo-Clear (National Diagnostics, HS-200), and finally coverslipped with DPX mounting medium (VWR, 360294H). All incubation steps were conducted with gentle agitation from a table-top orbital shaker. Images were captured using an Olympus BX51 microscope with Olympus DPS camera (UCMAD3).
Immunogold Transmission Electron Microscopy
A vibratome was utilized to cut 50 μm of free-floating sections from AD frontal cortex. After treatment with primary antibody (see under “Immunohistochemistry”), sections were incubated with secondary antibody conjugated to ultra-small gold particles (Aurion, Wageningen, The Netherlands). After rinsing, sections were fixed in glutaraldehyde for 1 h, enhanced with R-gent SE-EM (Aurion), and postfixed in OsO4. Sections were dehydrated, embedded in Epon, and cut into ultra-thin sections (60–70 nm). Thin sections were counterstained with uranyl acetate, followed by lead citrate, and examined using a Hitachi (Tokyo, Japan) H-7500 electron microscope (27, 28).
Label-free Quantitative Proteomics
For the proteomic analysis of the insoluble fractions from the 18 Baltimore Longitudinal Study of Aging cases, protein samples (50 μg) were reduced with 5 mm dithiothreitol (DTT) for 15 min at 37 °C and then alkylated with 20 mm iodoacetamide for 30 min at 25 °C. The alkylated samples were separated on a 10% SDS gel and stained with Coomassie Blue G-250. Each sample lane was cut into five gel bands corresponding to molecular weight ranges to increase the depth of coverage of the proteome. The gel pieces were then digested overnight in 12.5 μg/ml trypsin at 37 °C. The resulting peptides were analyzed by high resolution LC-MS/MS and quantified by signal intensity using in-house software as described previously (29).
GST Purification of Recombinant U1-70K Proteins
HEK293T cells were grown and transfected as before. For each GST affinity purification, 5–10 × 15-cm2 tissue culture dishes were transiently transfected with 20 μg of plasmid DNA and 60 μg of PEI. The cells were harvested 72 h post-transfection by scraping in ice-cold PBS, and lysates were prepared according to published methods (30), with some minor modifications. Briefly, the cells were resuspended in ∼1.5× packed cell volume of ice-cold lysis buffer (50 mm HEPES, pH 7.4, 200 mm NaCl, 5% glycerol, 1 mm EDTA, 1% (w/v) Sarkosyl, and 1× HALT protease inhibitor mixture). Lysates were sonicated with five 10-s pulses at 30% amplitude (maximum intensity = 40%) using a microtip probe to shear nucleic acids. To render the lysates compatible with GST affinity purification, Triton X-100 was added to a concentration of 1.5% (v/v), and the lysates were cleared by centrifugation at 14,000 × g for 10 min at 4 °C. The resulting supernatants were incubated overnight at 4 °C with 0.5–1 ml of swelled glutathione-agarose resin (Sigma G4510), after which the slurries were loaded onto 10-ml polypropylene columns. The resin was washed with 10 column volumes of GST wash buffer (50 mm HEPES, pH 7.4, 200 mm NaCl, and 1% Triton X-100), and eluted with 4× 1 ml of elution buffer (50 mm Tris-HCl, pH 8.5, 500 mm NaCl, 20 mm reduced l-glutathione (Sigma G4251) and 0.1% Triton X-100). The eluted fractions were concentrated to ∼200 μl using Amicon Ultra-0.5-ml 10,000 molecular weight cutoff centrifugal filter units (EMD Millipore), and dialyzed overnight against 50 mm HEPES, pH 7.4, 200 mm NaCl, and 0.1 mm PMSF using 10,000 molecular weight cutoff Slide-A-Lyzer MINI dialysis units (Thermo). Protein concentrations were determined by running the elution fractions on an SDS-polyacrylamide gel with bovine serum albumin (BSA) standards ranging from 0.2 to 1 μg per lane and staining with Coomassie G-250 (31). Densitometry of the BSA standards was used to calculate the concentration of the purified protein.
Protein Labeling with NHS-Diazirine and/or NHS-Biotin
For photocross-linking experiments, purified recombinant U1-70K (rU1-70K) was labeled with NHS-diazirine (Pierce 26167) according to the manufacturer's instructions. All labeling reactions were conducted in an aqueous buffer consisting of 50 mm HEPES, pH 7.4, and 200 mm NaCl. For a general labeling reaction, 40–120 μg of protein was combined with 5 mm NHS-diazirine, from a freshly prepared 100 mm stock in anhydrous DMSO. The reaction was incubated at room temperature for 2 h in the dark, quenched with 50 mm Tris-HCl, pH 8, and desalted using 0.5-ml Zeba Spin 7000 molecular weight cutoff desalting columns (Pierce). Dual labeling with NHS-diazirine and EZ-Link NHS-biotin (Pierce 20217) was performed in a similar manner; however, the purified protein was incubated with a 2:1 molar ratio of NHS-diazirine (5 mm) and NHS-biotin (2.5 mm).
Protein Cross-linking, Affinity Capture, and Mass Spectrometry
For photo-cross-linking experiments, 5 mg of AD total homogenate was fractionated at 180,000 × g as before, and the detergent-insoluble fractions were resuspended in 200 μl of Sark-buffer using the pellet pestle (Sigma Z359971) followed by brief, low intensity sonication (5 s at 20% amplitude). For each experimental condition, 40 μg each of unlabeled or diazirine-labeled rU1-70K fragments were incubated overnight with the insoluble fractions from AD brain. The mixtures were then fractionated at 180,000 × g for 30 min, and the resulting pellets gently resuspended in Sark-buffer using the pellet pestle. Both the labeled and unlabeled co-aggregation mixtures were transferred to a shallow plastic dish (the lid of a 35-mm Petri dish) and irradiated with 365 nm UV light (1 × 8-watt bulb; Pierce 95034) for 15 min on ice, at a distance of roughly 2 cm from the bulb. The mixtures were fractionated as before, and the pellets were resuspended in 8 m urea buffer and analyzed by Western blotting. For streptavidin pulldown experiments, 40 μg of unlabeled or dual-labeled (diazirine and biotin) rU1-70K fragments were UV-cross-linked with the detergent-insoluble fraction of AD brain as outlined above. The resulting pellets (in 8 m urea + 2% SDS buffer) were diluted 10-fold with GST wash buffer and incubated with 1 mg (100 μl) of Dynabeads M280 streptavidin (Invitrogen) for 2 h at room temperature. The beads were washed with 4× 0.5-ml of wash buffer A (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, and 0.1% SDS) and 2× 0.5 ml of wash buffer B (50 mm Tris-HCl pH 7.5, 0.5 m NaCl, and 0.1% Triton X-100). The beads were eluted in 30 μl of SDS sample buffer (without reducing agent) supplemented with 3 mm biotin by incubating at 95 °C for 15 min. The eluted samples were reduced with 5 mm DTT at 95 °C for 5 min and alkylated with 50 mm iodoacetamide at room temperature for 30 min in the dark. Samples were resolved by SDS-PAGE, and regions above 60 kDa were excised and analyzed by LC-MS/MS following in-gel trypsin digestion as described previously (26). A user defined peptide precursor mass tolerance of ±20 ppm was employed for extracted ion chromatogram (XIC) based quantification (32).
Western Blotting
Western blotting was performed according to standard procedures as reported previously (26). Control, CBD, and AD brain homogenates were blotted at 50 μg per lane, HEK lysate with overexpressed rU1-70K at 5 μg per lane, and Sarkosyl-insoluble fractions at 40 μg per lane. Samples in Laemmli sample buffer were resolved by SDS-PAGE before an overnight wet transfer to 0.2-μm nitrocellulose membranes (Bio-Rad). Membranes were blocked with casein blocking buffer (Sigma B6429) and probed with primary antibodies for 2 h at room temperature or overnight at 4 °C. Membranes were incubated with secondary antibodies conjugated to Alexa Fluor 680 (Invitrogen) or IRDye800 (Rockland) fluorophores for 1 h at room temperature. Images were captured using an Odyssey Infrared Imaging System (Li-Cor Biosciences), and band intensities were quantified using Odyssey imaging software. Statistical analysis was performed using Student's t test for independent samples.
RESULTS
Disconnect between the Subcellular Distribution and Biochemical Properties of U1-70K in AD Brain
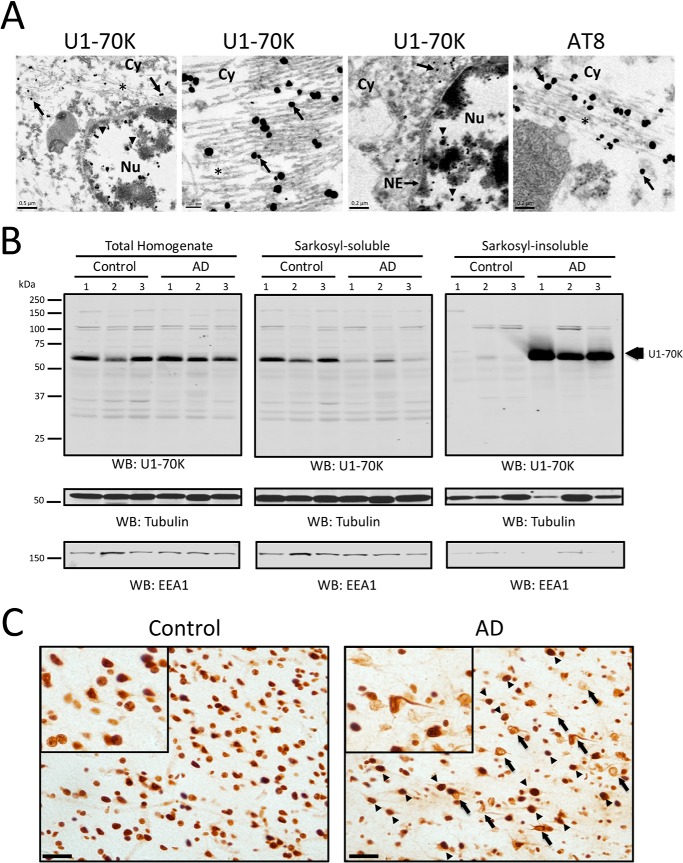
The RNA-binding protein U1-70K and other core U1 snRNP proteins are Sarkosyl-insoluble and associate with Tau in AD cortex (8). To further characterize the precise subcellular localization of U1-70K in AD brain, we used immunogold transmission electron microscopy (EM). Both nuclear and cytoplasmic U1-70K distribution were observed within neurons (Fig. 1A). Notably, clusters of immunogold-labeled U1-70K particles in the cytoplasm were frequently observed associated with filamentous structures resembling AT8-positive NFTs. In contrast, U1-70K in the nucleus was typically associated with dense chromatin structures (Fig. 1A), supporting previous observations that the majority of snRNPs localize in interchromatin granule clusters (33). We did not observe tangle-like U1-70K-positive structures in the nucleus. These observations confirm our previous findings that U1-70K associates with cytoplasmic NFTs in AD brain (8).
FIGURE 1.
Subcellular and biochemical distribution properties for U1-70K in AD. A, immunogold transmission electron microscopy of U1-70K or Tau neurofibrillary tangles (AT8) in AD frontal cortex. Immunogold positive staining for U1-70K (left panel, scale bar, 0.5 μm) is observed in both the nucleus (Nu), denoted by arrowheads, and cytoplasm (Cy), denoted by arrows. The asterisk denotes twisted-ribbon tangle-like structures positive for U1-70K in the cytoplasm (middle panels), resembling AT8-positive NFTs shown at higher magnification (right panel). The nuclear envelope (NE) is also denoted. B, U1-70K displays a near complete solubility shift in AD brain by Western blot (WB). Total brain homogenates prepared from control (n = 3) and AD (n = 3) frontal cortex (left panel) were fractionated into Sarkosyl-soluble (middle panel) and Sarkosyl-insoluble fractions (right panel) and blotted for U1-70K. Upon fractionation U1-70K from AD brain almost exclusively partitions into the Sarkosyl-insoluble fraction. Tubulin served as a relative loading control and early endosomal antigen 1 (EEA1) served as a soluble protein marker. C, immunohistochemisty using antibodies against U1-70K in both control (left panel) and AD brain (right panel) tissue (scale bar, 50 μm; higher magnification shown in the insets). Arrows highlight cytoplasmic U1-70K tangles, and arrowheads highlight U1-70K-positive nuclei.
Using a multistep tissue fractionation approach, we previously showed that U1-70K is highly Sarkosyl-insoluble in AD cases (8). To further characterize the intra-tissue biochemical properties of U1-70K, we used a simplified one-step fractionation protocol to generate Sarkosyl-soluble and -insoluble fractions from three independent control and AD post-mortem cortical homogenates (Fig. 1B). When AD and control brain homogenates with equivalent levels of total U1-70K were fractionated, we observed that U1-70K was almost entirely Sarkosyl-soluble in controls, whereas in AD it was almost exclusively Sarkosyl-insoluble. Interestingly, by immunohistochemistry U1-70K displayed normal nuclear distribution in the majority of cells in AD cortex (Fig. 1C). This suggests that the near complete shift of U1-70K into the Sarkosyl-insoluble fraction in AD brain is not solely attributable to the tangle-like cytoplasmic pool of U1-70K alone (Fig. 1, A and B). Based on these collective observations, we hypothesized that following homogenization of AD brain either aggregated U1-70K itself or other biopolymers (e.g. proteins or nucleic acids) sequester natively folded conformers of soluble U1-70K into insoluble aggregates.
AD Homogenates Can Induce the Aggregation of Normal Soluble U1-70K
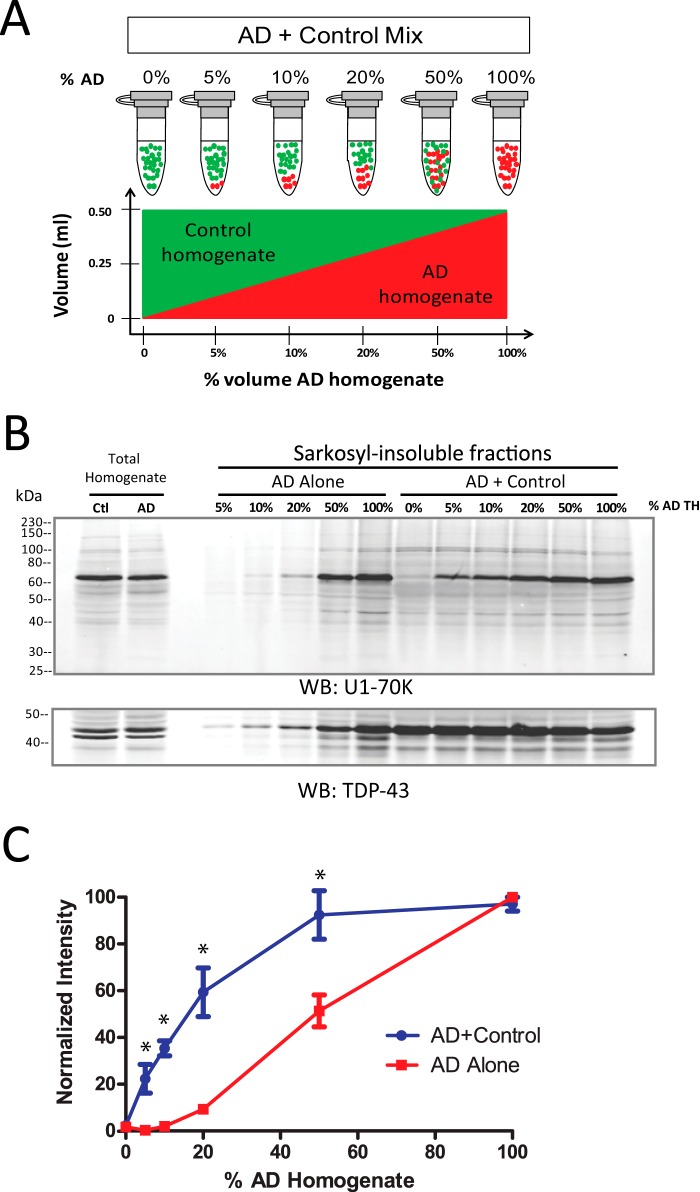
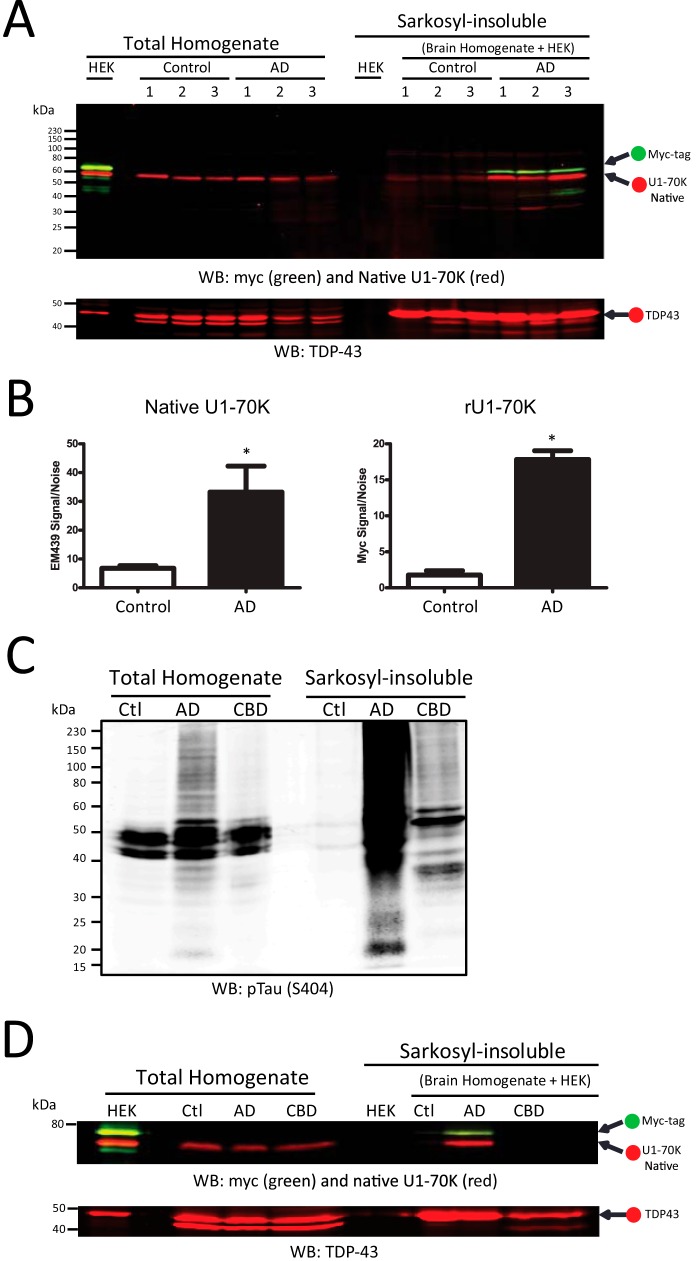
To ascertain whether AD homogenates could induce the aggregation of soluble U1-70K from control tissue, control brain homogenates were mixed with increasing proportions of AD brain homogenate (Fig. 2A). Each AD + control mixture consisted of 5 mg of total protein, with identical volumes (0.5 ml) and protein concentrations (10 mg/ml). For comparison, matching quantities of AD brain homogenate were diluted (AD alone) in buffer to the same final volume. Prior to mixing, we ensured that both AD and control homogenates had equal levels of U1-70K (Fig. 2B, left). After incubation and fractionation, the Sarkosyl-insoluble fractions were analyzed by Western blotting for U1-70K (Fig. 2B). Quantification of Sarkosyl-insoluble U1-70K from AD and control homogenate mixtures revealed a nonlinear increase in levels of insoluble U1-70K, approximating a logarithmic curve (Fig. 2, B and C). In contrast, quantification of Sarkosyl-insoluble U1-70K from AD homogenates alone yielded a signal increase that was generally proportional to the amount of total AD homogenate used in the assay. As expected, U1-70K in the control homogenate alone (0% AD homogenate) was entirely Sarkosyl-soluble in the absence of AD homogenate (Fig. 2B). These results indicate that AD homogenates can sequester soluble control brain-derived U1-70K into a detergent-insoluble aggregated state. TDP-43 was used as a loading control in this assay because Sarkosyl-insoluble levels of TDP-43 were consistent in control and AD fractions (Fig. 2B, lower panel). We extended this assay to assess the ability of AD homogenates to convert soluble recombinant U1-70K (rU1-70K) into an aggregated state. HEK cell lysate with overexpressed Myc-tagged rU1-70K was incubated with AD or control brain homogenates, followed by fractionation and Western blot analysis of the Sarkosyl-insoluble fractions (Fig. 3A). Although rU1-70K incubated at 4 °C for up to 4 h remained Sarkosyl-soluble, incubation with AD brain homogenates under the same conditions induced rU1-70K aggregation. In contrast, rU1-70K incubated with control brain homogenates remained Sarkosyl-soluble.
FIGURE 2.
AD homogenates induce the aggregation of normal soluble U1-70K from control human brain. A, frontal cortex homogenates of AD and control brains were mixed in six ratios with increasing proportions of AD homogenate. The mixtures were incubated 4 h at 4 °C and fractionated. B, representative WB for U1-70K (top panel) of Sarkosyl-insoluble fractions from control and AD (AD + Control) mixtures or from matching quantities of AD homogenates alone (AD alone). TDP-43 insolubility was consistent across control and AD Sarkosyl-insoluble fractions and was therefore used as a loading control (bottom panel). C, quantification of U1-70K in the insoluble fractions from AD+Control mixtures revealed a nonlinear increase in the signal of Sarkosyl-insoluble U1-70K, consistent with the sequestration of soluble U1-70K from control brain into the Sarkosyl-insoluble fraction. In contrast, the AD alone series revealed a roughly proportional relationship between the amount of AD brain homogenate and the Sarkosyl-insoluble U1-70K signal. Analysis of three biological replicates indicates significant enrichment of U1-70K in AD+Control series compared with the AD alone series. Statistical significance (*) was calculated using Student's t test (p < 0.05).
FIGURE 3.
AD homogenates, but not CBD homogenates, induce the aggregation of soluble recombinant U1-70K. A, HEK lysate with overexpressed Myc-tagged rU1-70K (full-length) was incubated with control (n = 3) and AD (n = 3) brain homogenates for 4 h at 4 °C and fractionated. Sarkosyl-insoluble fractions were analyzed by quantitative WB for native U1-70K (red) and Myc-tagged rU1-70K (green). HEK lysate alone did not harbor insoluble rU1-70K, and only AD homogenate was able to induce rU1-70K aggregation (top panel). TDP-43 (red) served as a loading control (bottom panel). B, densitometry analysis revealed a significant increase in both native and rU1-70K in the Sarkosyl-insoluble fractions of the AD + HEK mixtures compared with the control + HEK mixtures. Statistical significance (*) was calculated using Student's t test (p < 0.05). C, both CBD and to a greater extent AD homogenate had increased levels of Sarkosyl-insoluble phosphorylated Tau (Ser(P)-404) levels compared with control brain homogenate. D, compared with AD homogenate, a CBD homogenate was unable to induce rU1-70K into an aggregated state. TDP-43 served as a loading control (bottom panel).
To test the disease specificity of the induced aggregation of rU1-70K, we performed a co-aggregation assay with rU1-70K and brain homogenates generated from an individual with CBD, which is pathologically characterized by the accumulation of hyperphosphorylated detergent-insoluble Tau (Fig. 3C) and has only soluble U1-70K (8, 34). In contrast to AD homogenate, CBD homogenate was ineffective at sequestering rU1-70K into the Sarkosyl-insoluble fraction (Fig. 3D). This suggests that U1-70K aggregation is not mediated by factors present in the CBD brain, which is consistent with our previous findings that U1-70K aggregation is specific to AD and not observed in other neurodegenerative diseases, including tauopathies (8). Our findings indicate that AD homogenates are uniquely effective at inducing the aggregation of both soluble U1-70K from control brain and rU1-70K expressed in cells.
Induced Aggregation of rU1-70K by AD Homogenate Is Protein-dependent
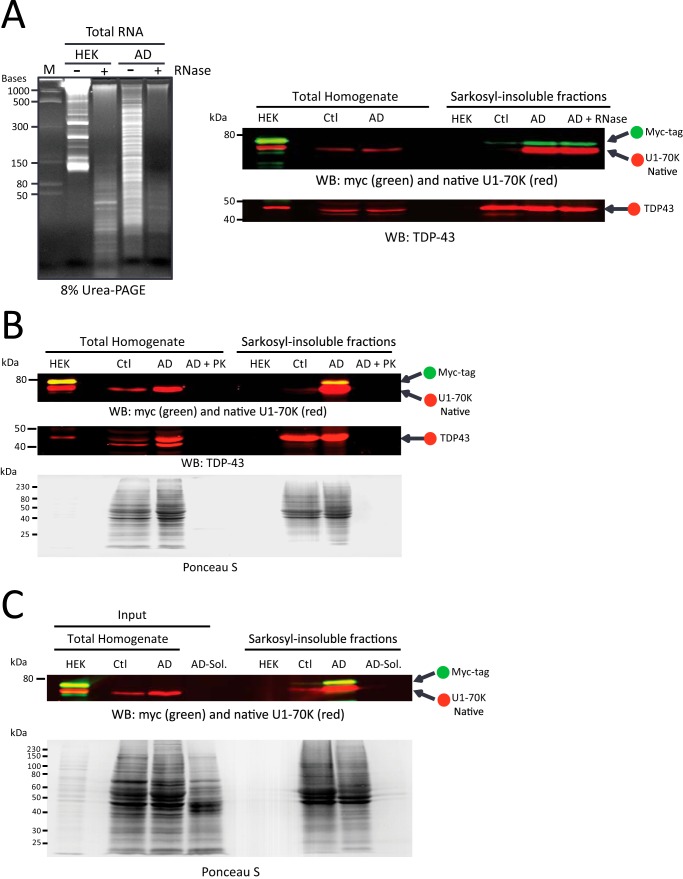
We previously reported that U1 snRNA co-aggregates with U1-70K in AD brain (8, 35) suggesting that RNA may play an important role in rU1-70K aggregation in AD homogenates. To determine whether RNA is required for the induced aggregation of rU1-70K by AD homogenate, we pretreated both HEK lysate and AD brain homogenates with RNase A prior to co-aggregation assays (Fig. 4A). Urea-PAGE was performed on RNA extracts of pre- and post-digested aliquots to confirm near complete degradation of intact RNA in both AD homogenates and HEK lysates with overexpressed rU1-70K. Both RNase-treated and -untreated AD brain homogenates were able to induce the aggregation of proportional levels of rU1-70K. Native levels of Sarkosyl-insoluble U1-70K in AD were similarly unaffected by RNase treatment, suggesting that U1-70K Sarkosyl insolubility is independent of RNA. To ascertain whether rU1-70K aggregation is protein-dependent, we pretreated AD homogenate with PK followed by inhibition with PMSF. PK inhibition was confirmed by incubating the PK-treated AD homogenates with rU1-70K and analyzing aliquots taken at different time points by Western blot (data not shown). PK inhibition was only considered complete if proteolytic degradation of rU1-70K was not observed. We then assayed the ability of PK-treated homogenates to aggregate rU1-70K (Fig. 4B). Ponceau S staining of the subsequent transfer membrane confirmed near complete protein degradation in AD homogenate following PK treatment. Compared with untreated AD homogenate, the PK-digested homogenates were ineffective at inducing the aggregation of rU1-70K. Finally, to assess whether proteins harbored within the detergent-soluble protein pool could induce rU1-70K aggregation, we performed co-aggregation assays with Sarkosyl-soluble fractions of AD brain (Fig. 4C). Unlike AD total homogenates, the Sarkosyl-soluble fractions were unable to induce the aggregation of rU1-70K. Together, these data indicate that rU1-70K aggregation is induced by Sarkosyl-insoluble proteins, which includes U1-70K and Tau.
FIGURE 4.
U1-70K aggregation is not dependent on RNA and requires Sarkosyl-insoluble proteins in AD homogenate. A, AD brain homogenate and HEK lysate overexpressing rU1-70K were pretreated with RNase A prior to performing co-aggregation experiments. Total RNA extracts were analyzed by urea-PAGE to confirm RNA degradation (left panel). HEK lysate with overexpressed rU1-70K (± RNase) was incubated alone or with control and AD homogenates (± RNase) for 4 h at 4 °C and fractionated. WB analysis of Sarkosyl-insoluble fractions revealed equivalent levels of native U1-70K (red) and rU1-70K (green) in both RNase-treated and -untreated samples (right panel). TDP-43 served as a loading control (bottom panel). B, AD homogenate was pretreated with PK for 1 h at 37 °C and then inhibited with the addition of excess PMSF. The PK-treated AD homogenate was incubated with rU1-70K lysate for 4 h at 4 °C and fractionated. Ponceau S staining of the transfer membrane indicates near complete digestion of protein in the PK-treated samples (bottom panel). PK digestion abolished the ability of AD homogenate to seed rU1-70K. C, rU1-70K HEK lysate was incubated with control and AD brain homogenates as well as the Sarkosyl-soluble fraction of AD brain (AD sol.) as before. The resulting Sarkosyl-insoluble fractions were subjected to Western blot analysis for native U1-70K (red) and rU1-70K (green), suggesting that the Sarkosyl-insoluble protein fraction is responsible for inducing rU1-70K aggregation (top panel). Ponceau S staining of the transfer membrane shows equal protein loading across total control homogenate, AD homogenate, and Sarkosyl-soluble AD fractions (bottom panel).
U1-70K and Tau Do Not Always Co-aggregate in AD
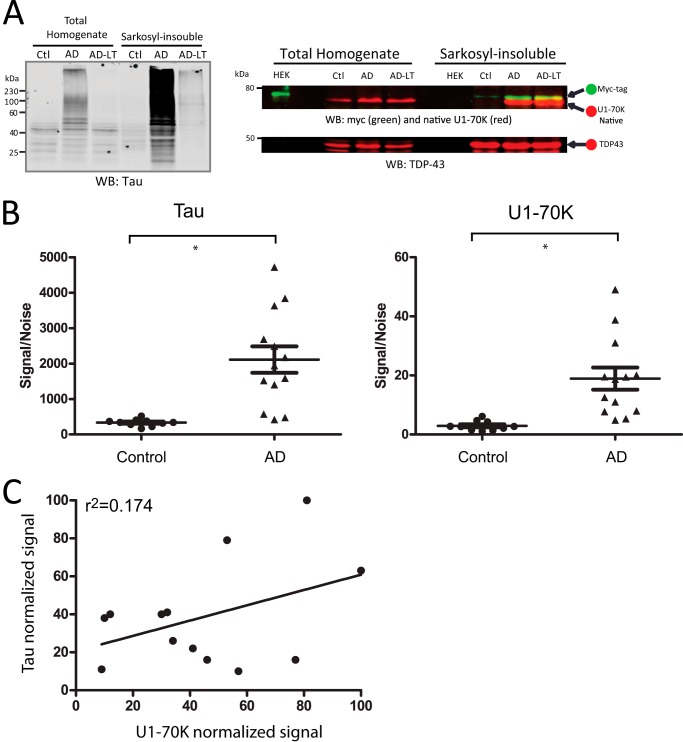
Both Tau and U1-70K proteins are highly Sarkosyl-insoluble in AD homogenates, and both associate with NFTs in AD frontal cortex. However, our previous immunohistochemical studies showed discordant distributions of U1-70K and Tau aggregates in certain brain regions, including the hippocampus (8). These data suggest that U1-70K and Tau do not always co-aggregate in AD brain. Thus, we sought to perform a co-aggregation assay with an AD case that harbored markedly reduced levels of insoluble Tau relative to other AD cases. Using quantitative mass spectrometry, we identified an AD case with significantly lower levels of phosphorylated Tau (36). More specifically, AD case E04-172 (Table 1) had on average 14-fold less pTau based on Tau phosphopeptide signal intensity in the frontal cortex compared with the average of other AD cases we analyzed (36). Interestingly, this “low pTau” AD case had comparable levels of native Sarkosyl-insoluble U1-70K and was equally effective at inducing rU1-70K aggregation, as compared with an AD case (case E05-04) with significantly higher levels of Sarkosyl-insoluble pTau (Fig. 5A). This result suggests that U1-70K aggregation in AD does not correlate with aggregated Tau levels. In further support of this hypothesis, we directly quantified Tau and U1-70K levels in the Sarkosyl-insoluble brain proteome across 18 individual cases representing 5 controls (analyzed in technical replicate) and 13 AD cases (Fig. 5B). All cortical tissues were collected from the Baltimore Longitudinal Study of Aging (5). The protein abundance derived from peptide signal intensities by mass spectrometry for both U1-70K and Tau were measured as described previously (29). As expected, both U1-70K and Tau levels were significantly enriched in AD cases compared with controls (Fig. 5B). The coefficient of determination (R2 = 0.174) revealed weak correlation between levels of Sarkosyl-insoluble Tau and U1-70K within the 13 individual AD cases (Fig. 5C). Therefore, many AD cases had significant levels of either Sarkosyl-insoluble U1-70K or Tau but not both proteins. Together these data suggest that U1-70K does not exclusively co-aggregate with Tau in AD.
FIGURE 5.
U1-70K and Tau do not always co-aggregate in AD. A, AD case with low levels of Sarkosyl-insoluble Tau (AD-LT) was subjected to WB analysis for Tau, along with a typical “high Tau” AD case (AD) and a control case (left panel). The control (Ctl), AD, and AD-LT cases were incubated with rU1-70K lysate for a co-aggregation assay. The resulting Sarkosyl-insoluble fractions were subjected to WB analysis for native U1-70K (red) and rU1-70K (green) (top right panel). Both the AD and AD-LT homogenates were equally capable of inducing rU1-70K aggregation, in contrast to the control (Ctl) homogenate. The rU1-70K lysate alone (HEK) did not harbor any Sarkosyl-insoluble rU1-70K. TDP-43 served as the loading control (bottom right panel). B, U1-70K and Tau levels in the Sarkosyl-insoluble proteome were quantified by peptide ion intensities across 18 individual cases representing five controls (each run in technical replicate) and 13 AD cases. Both U1-70K and Tau levels were significantly enriched in the Sarkosyl-insoluble fractions of AD cases compared with control cases (*, p value < 0.01, Student's t test). C, coefficient of determination across AD (n = 13) reveals a weak correlation between Tau and U1-70K levels in the insoluble fractions. Protein signal intensities were normalized to the individual AD cases with the maximum signal of U1-70K and Tau, respectively.
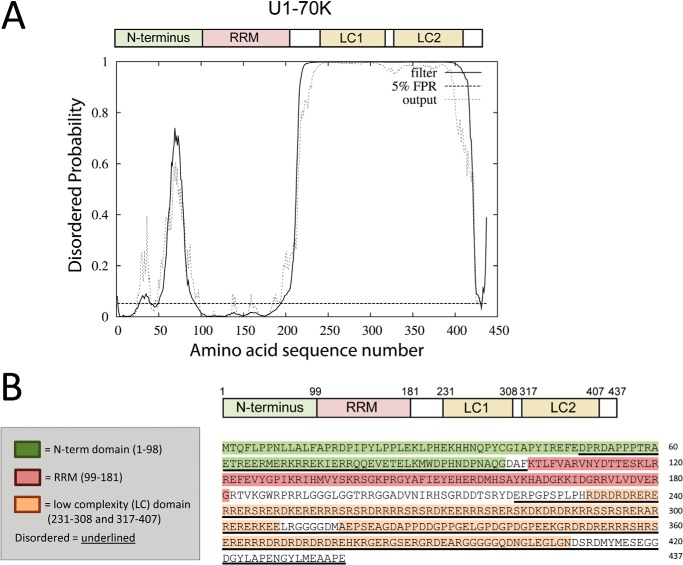
Disordered C Terminus Is Necessary for rU1-70K Co-aggregation
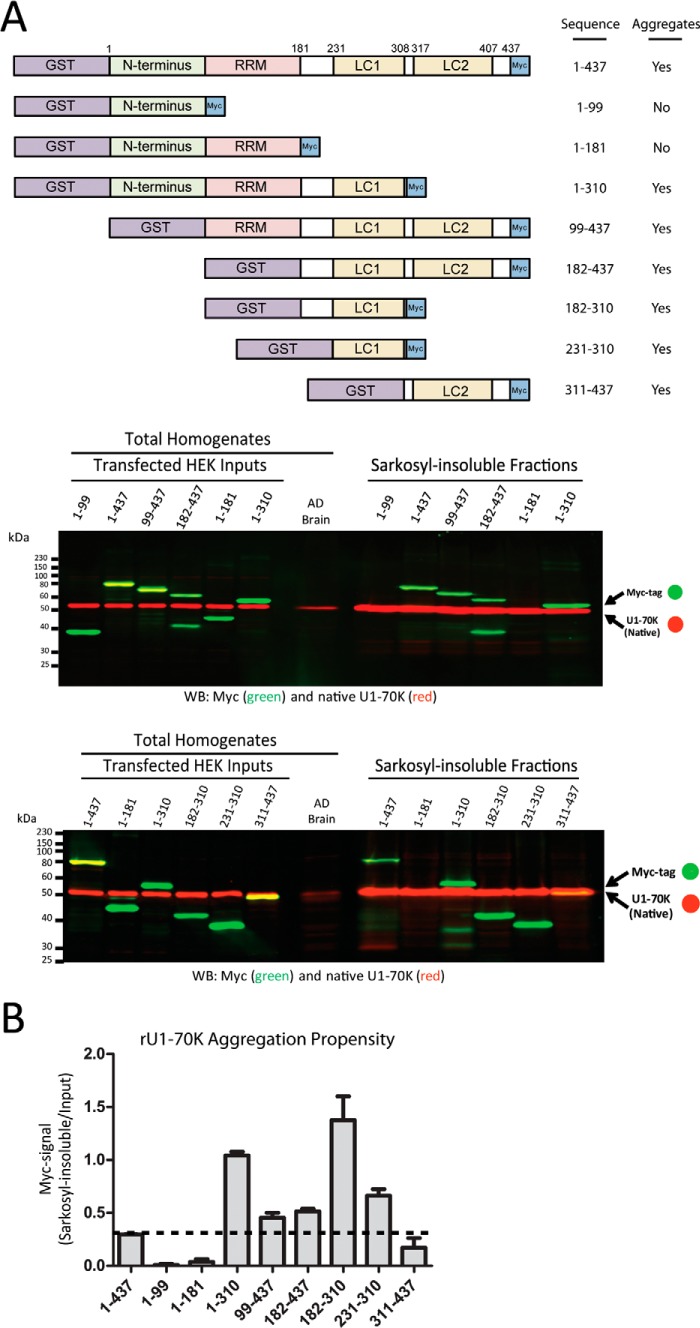
Several RNA-binding proteins, including TDP-43, FUS, hnRNPA1, and ataxinA2 harbor low complexity (LC) domains that are proposed to promote the formation of detergent-insoluble amyloid-like aggregates in neurodegenerative disease (15). U1-70K harbors two such domains, LC1 (amino acids 231–308) and LC2 (amino acids 317–407), which together comprise a majority of the C-terminal primary sequence. The DISOPRED algorithm (37) predicts intrinsically disordered regions within the N-terminal (amino acids 50–100) and C-terminal (amino acids 220–437) regions of U1-70K (Fig. 6). The C-terminal region is predicted to be almost completely disordered as a consequence of the low amino acid complexity of the LC1 and LC2 domains. To determine whether LC region(s) or other domains of U1-70K are required for rU1-70K co-aggregation, we assessed the ability of AD brain homogenate to sequester GST-tagged full-length rU1-70K as well as N- and C-terminal truncated fragments (Fig. 7A). Co-aggregation experiments were performed in which rU1-70K lysates were incubated with AD total homogenates and fractionated. Western blot analysis of the resulting Sarkosyl-insoluble fractions demonstrated that N-terminal fragments (1–99 and 1–181) were not competent for aggregation compared with full-length or C-terminal fragments containing the LC domains (Fig. 7A). Moreover, rU1-70K fragments harboring LC1 or LC2 were sufficient for aggregation, and fragments that contained only the LC1 domain were more aggregate-prone as measured by the ratio of Sarkosyl-insoluble rU1-70K to rU1-70K levels in the input (Fig. 7B). For example, rU1-70K fragments 1–310, 182–310, and 231–310 each had a higher proportion of protein in the AD detergent-insoluble homogenate compared with full-length rU1-70K or other C-terminal fragments harboring both LC1 and LC2. Overall, these results suggest that the induced aggregation of U1-70K is mediated via the LC domains of the intrinsically disordered C terminus.
FIGURE 6.
U1-70K is highly disordered and has C-terminal low complexity domains. A, DISOPRED algorithm predicts intrinsically disordered regions within the N-terminal (amino acids 50–100) and C-terminal (amino acids 220–437) portion of U1-70K, which harbors two distinct low complexity domains, LC1 (amino acids 231–308) and LC2 (amino acids 317–407). B, primary amino acid sequence of U1-70K with the N-terminal (green), RNA recognition motif (RRM) domain (red), and two LC domains (orange) are highlighted. Regions predicted to be intrinsically disordered are underlined.
FIGURE 7.
Intrinsically disordered C terminus is necessary and sufficient for U1-70K aggregation. A, to determine which region(s) or amino acid sequences of U1-70K are required for aggregation, we assessed the ability of AD brain homogenate to induce the aggregation of full-length rU1-70K and a number of GST-tagged N- and C-terminal truncations (top panel). Two representative Western blots are shown. Each rU1-70K fragment was expressed in HEK cells, and individual lysates harbored equivalent amounts of recombinant protein (inputs). Each rU1-70K lysate was incubated with AD brain homogenate (bottom panels, 7th lanes) for 12 h at 4 °C prior to fractionation. The Sarkosyl-insoluble fractions were subjected to WB analysis for native U1-70K (red) and rU1-70K (green). rU1-70K residues 182–437 also displays a C-terminal fragment (∼37 kDa) caused by in vivo proteolysis. B, aggregation propensity of the full-length rU1-70K and various N- and C-terminal truncations was determined by quantifying the recombinant (green) signal ratio of Sarkosyl-insoluble/input fractions. The rU1-70K fragment 182–310 demonstrated the highest degree of aggregation. The dashed line represents the baseline level for full-length rU1-70K (residues 1–437) aggregation. Co-aggregation assays were performed in duplicate for each rU1-70K fragment (± S.E.).
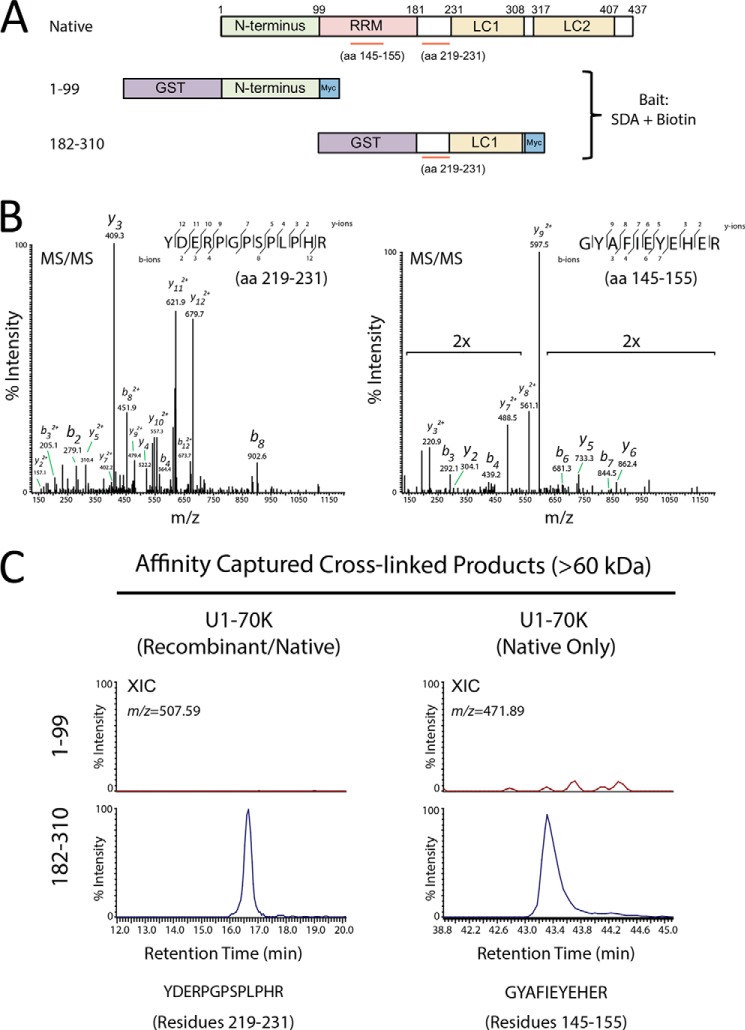
U1-70K in AD Brain Directly Interacts with rU1-70K Fragments Harboring the LC1 Domain
Our data suggest that rU1-70K aggregation is induced by Sarkosyl-insoluble proteins in AD brain, which includes U1-70K. However, it is unclear whether this process involves the direct interaction of aggregated U1-70K in AD brain with soluble rU1-70K. To address this question, we employed a protein cross-linking approach (Fig. 8). Following GST purification, aggregation-prone (182–310) and nonaggregation-prone (1–99) rU1-70K fragments were derivatized with the amine-reactive photoactivatable cross-linker NHS-diazirine (succinimidyl 4,4-azipentanoate (SDA)). Upon photoactivation at 365 nm, SDA will form covalent bonds with any protein within roughly 3.9 Å (38). Based on this principle, we performed a co-aggregation experiment with unlabeled or SDA-labeled purified rU1-70K fragments (182–310 or 1–99) and the Sarkosyl-insoluble fraction of AD homogenate. Following UV cross-linking, the insoluble fractions were analyzed by Western blotting for both rU1-70K and native U1-70K, in which a clear shift to higher molecular weight cross-linked species was only observed in the SDA-labeled C-terminal 182–310-residue sample (Fig. 8B). As expected, no cross-linking was observed in the 1–99 N-terminal samples. Notably, in the samples cross-linked with the SDA-labeled 182–310-residue fragment there was an approximate 60% reduction in the signal of native monomeric U1-70K, indicating that over half of the detergent-insoluble U1-70K in AD brain directly interacted with the 182–310-residue fragment (Fig. 8C). In contrast, TDP-43 levels in the same samples were reduced by ∼14% compared with the control (no SDA), indicative of the selectivity of the diazirine photo-cross-linker. To further demonstrate that rU1-70K interacts with aggregated U1-70K in AD brain, we performed a similar co-aggregation experiment with double-labeled (biotin + SDA) 182–310- and 1–99-residue rU1-70K fragments (Fig. 9). Following UV cross-linking and fractionation, the cross-linked products were affinity-captured with streptavidin and resolved by SDS-PAGE (data not shown). The high molecular mass products (>60 kDa) were subsequently analyzed by LC-MS/MS. Two U1-70K peptides were identified mapping to residues 219–231 (YDERPGPSPLPHR) or 145–155 (GYAFIEYEHER). The first peptide (amino acids 219–231) could be derived from either the rU1-70K fragment 182–310 or native full-length U1-70K from AD brain, whereas the second peptide (amino acids 145–155) could only be derived from native full-length U1-70K (Fig. 9, A and B). Relative quantification for both the shared (219–231 residues) and AD-derived (145–155 residues) U1-70K peptides was measured by extracted ion intensity across the 1–99- and 182–310-residue rU1-70K cross-linked samples (Fig. 9C). Notably, the peptide derived from native U1-70K (145–155 residues) was only detected in the 182–310-residue rU1-70K cross-linked sample, consistent with a direct interaction with aggregated U1-70K in AD brain. These quantitative mass spectrometry results are consistent with Western blot analysis, which show high molecular mass cross-linked U1-70K species only in the 182–310-residue rU1-70K cross-linked samples (Fig. 8B). Together, our cross-linking studies demonstrate that aggregated U1-70K in AD brain directly interacts with a rU1-70K fragment that harbors the LC1 domain.
FIGURE 8.

SDA-labeled rU1-70K fragments harboring the LC1 domain directly interact with aggregated U1-70K in AD brain. A, GST-tagged N- or C-terminal rU1-70K fragments (residues 1–99 and 182–310) were expressed in HEK cells, affinity-purified with glutathione-agarose, and assessed for purity by SDS-PAGE and Coomassie Blue staining (left panel). Purified fragments were derivatized with the amine-reactive photo-activatable cross-linker NHS-diazirine (SDA) (right panel). B, co-aggregation assays using Sarkosyl-insoluble fractions from AD brain were performed with unlabeled or SDA-labeled purified rU1-70K fragments (residues 1–99 or 182–310). Following UV treatment and fractionation, the cross-linked products were analyzed by Western blotting for both rU1-70K (green) and native U1-70K (red) (top panel). Higher molecular weight cross-linked species were only observed in co-aggregation experiments with SDA-labeled 182–310-residue rU1-70K fragment. Notably, lower molecular weight C-terminal fragments in the 182–310-residue rU1-70K co-aggregation assays were also observed to form cross-linked products. TDP-43 was used as a loading control (bottom panel). C, approximately 60% of native monomeric U1-70K from AD brain was cross-linked in co-aggregation assays using SDA-labeled 182–310-residue rU1-70K fragments compared with the control unlabeled fragments (no SDA). In contrast, ∼14% of native TDP-43 was cross-linked in these same experiments, which were performed in technical quadruplicate (n = 4). Student's t test was performed for significance (p < 0.05).
FIGURE 9.
Mass spectrometry analysis of affinity-captured cross-linked products confirm a direct interaction between rU1-70K and aggregated U1-70K in AD brain. A, co-aggregation assays using Sarkosyl-insoluble fractions from AD brain were performed with dual-labeled (SDA+biotin) purified rU1-70K fragments (amino acids (aa) 1–99 or 182–310). Following UV cross-linking and fractionation, the cross-linked products were affinity-captured with streptavidin magnetic beads. The eluted products were resolved by SDS-PAGE, and the gel regions of >60 kDa were excised, subjected to in-gel trypsin digestion, and analyzed by LC-MS/MS. Both rU1-70K and native U1-70K peptides from AD brain were identified by MS/MS and quantified by XIC. B, representative MS/MS spectra for triply charged U1-70K peptides mapping to amino acids 219–231 (YDERPGPSPLPHR) or 145–155 (GYAFIEYEHER). C, XIC for U1-70K peptide sequence 219–231 (m/z = 507.59) and 145–155 (m/z = 471.89) from high molecular weight species cross-linked with rU1-70K fragments 1–99 (negative control) and 182–310. Notably, the peptide derived from native U1-70K (residues 145–155) was only detected in the 182–310-residue rU1-70K cross-linked sample, consistent with a direct interaction with aggregated U1-70K in AD brain. Peptide signal intensities were normalized to levels in the 182–310-residue samples (100% maximum intensity).
DISCUSSION
In this study, we show that AD brain homogenate can induce the aggregation and insolubility of normal soluble U1-70K from control brain homogenate or rU1-70K. We determined that the C terminus of U1-70K, which harbors two disordered LC domains, is necessary for U1-70K aggregation and that fragments containing only the LC1 domain were highly aggregation-prone. Finally, protein cross-linking and mass spectrometry studies demonstrated that a rU1-70K fragment harboring the LC1 domain directly interacts with aggregated U1-70K in AD brain. These data suggest that aggregated forms of U1-70K in AD directly bind and sequester normal soluble forms of U1-70K into detergent-insoluble aggregated species.
Recently, a number of RNA-binding proteins were shown to undergo concentration-dependent phase transition to a hydrogel-like state composed of uniformly polymerized amyloid-like fibers in vitro (15). Subsequent investigation identified LC domains within these RNA-binding proteins as necessary for polymerization and hydrogel formation. Once transformed to a hydrogel-like state, these proteins can sequester soluble proteins of the same primary sequence (homotypic trapping) or other proteins with similar, but not identical, LC domains (heterotypic trapping). These results were proposed as a mechanism for understanding how higher order cellular structures such as RNA granules assemble in the absence of lipid membranes. To this end, the spliceosome is also a higher order cellular structure comprised of RNA and proteins, a majority of which have intrinsically disordered LC sequences (17). Therefore, we propose that the mechanisms underlying spliceosome assembly could parallel those of RNA granule assembly. Although hydrogel formation is thought to be dynamic and reversible (15), it has been hypothesized that this state constitutes an intermediate stage in the development of irreversible protein aggregates (39, 40). Thus, one hypothesis is that U1-70K and other U1 snRNP proteins containing similar disordered LC domains collectively bind and co-aggregate in the AD brain. It should be noted that AD homogenates did not induce the aggregation of TDP-43 (Fig. 2). Conversely, despite both proteins harboring LC domains, U1-70K does not aggregate in FTLD cases with TDP-43 pathology (8). This lack of association may be due to differences in the amino acid composition of their respective LC domains.
A majority of known yeast prion proteins harbor unique “prion domains” enriched in glutamine/asparagine (Q/N-rich) and to a lesser degree tyrosine and glycine (41–43). These domains are usually at least 60 amino acids in length and are predicted to be intrinsically disordered. Of the 210 RNA recognition motif-bearing proteins in the human genome, 29 have a putative prion-like Q/N-rich domain (43). Nearly half of these 29 proteins were identified as harboring low complexity domains (15). Included in this list were the RNA-binding proteins TAF15, TDP-43, FUS, and hnRNPA1, all of which aggregate in neurodegenerative disease (44–46). In contrast, the 78-amino acid LC1 domain in U1-70K (Fig. 6B) (amino acids 231–308), which we have shown is sufficient for aggregation, is comprised almost exclusively (∼99%) of five amino acids (Arg, Glu, Ser, Asp, and Lys). Of these five amino acids, four are organized in highly repetitive tandem arrays of a basic amino acid (R/K) followed by an acidic amino acid (D/E). Therefore this domain does not conform to the canonical Q/N-rich LC sequences observed in yeast prion proteins or the prion-like domains of other RNA-binding proteins known to aggregate in neurodegenerative disease. It remains unknown how the amino acid composition of the C-terminal low complexity domains in U1-70K contributes to its aggregation. Future studies analyzing the functional roles and binding properties of the LC domains in U1-70K as well as similar LC domains found in other aggregate-prone U1 snRNP proteins in AD is warranted.
Interestingly, we also show that the absence of RNA does not affect biochemical insolubility of U1-70K in AD brain homogenates. However, it is plausible that the U1 snRNA serves as an initial scaffold for aberrant U1-70K aggregation, which is supported by the presence U1 snRNA tangle-like pathology in AD brain (35). This underscores the importance of defining the causes underlying U1-70K cytoplasmic mis-localization and aggregation selectively in AD. These causes remain elusive as U1-70K aggregation does not correlate with Tau aggregation in AD brain, and U1-70K insolubility is not observed in other tauopathies, including CBD and familial forms of FTLD with Tau mutations (8). Thus, it is unlikely that U1-70K aggregation in AD brain is solely due to interactions with Tau. However, this does not preclude the possibility that U1-70K or other intrinsically disordered RNA-binding proteins can induce Tau aggregation in neurodegenerative disease. In support of this hypothesis, recent evidence suggests that RNA-binding proteins with LC domains can induce formation of, and co-aggregate with, Tau pathology in animal and cell models of tauopathies (47). Defining the exact relationship between U1 snRNP abnormalities and other pathologies, including Tau and β-amyloid deposition, will be important for understanding the role of U1-70K aggregation in AD.
In sum, these studies provide insight into the biochemical mechanisms underlying U1-70K protein aggregation events in AD. Our evidence suggests that this is a protein-directed process mediated by the intrinsically disordered low complexity C terminus of U1-70K. Interestingly, prion-like transmission of pathogenic protein aggregates has been shown to occur in neurodegenerative disease and is thought to play a central role in disease progression (48, 49). Based on the evidence provided in this study, we propose the hypothesis that self-directed propagation of intracellular U1-70K aggregates may contribute to the pathogenesis of AD.
Acknowledgments
We are grateful to participants in the Baltimore Longitudinal Study of Aging for their invaluable contribution. We thank Drs. Richard Kahn, Daniel Reines, and Eric Dammer for comments and critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant P50AG025688 (to the Emory Alzheimer's Disease Center) and Grant P30NS055077 (to the Emory Neuroscience NINDS Core Facilities), the National Institutes of Health Intramural Research Program, and the National Institute on Aging. This work was also supported by Alzheimer's Association New Investigator Research Award NIRG-12-242297 (to N. T. S.).
- AD
- Alzheimer disease
- snRNP
- small nuclear ribonucleoprotein
- LC
- low complexity
- FUS
- fused in sarcoma
- FTLD
- frontotemporal lobar degeneration
- NFT
- neurofibrillary tangle
- CBD
- corticobasal degeneration
- NHS
- N-hydroxysuccinimide
- SDA
- succinimidyl 4,4-azipentanoate
- XIC
- extracted ion chromatogram
- PK
- proteinase K
- WB
- Western blot.
REFERENCES
- 1. Taylor J. P., Hardy J., Fischbeck K. H. (2002) Toxic proteins in neurodegenerative disease. Science 296, 1991–1995 [DOI] [PubMed] [Google Scholar]
- 2. Ross C. A., Poirier M. A. (2004) Protein aggregation and neurodegenerative disease. Nat. Med. 10, S10–S17 [DOI] [PubMed] [Google Scholar]
- 3. Querfurth H. W., LaFerla F. M. (2010) Alzheimer's disease. New Engl. J. Med. 362, 329–344 [DOI] [PubMed] [Google Scholar]
- 4. Sperling R. A., Aisen P. S., Beckett L. A., Bennett D. A., Craft S., Fagan A. M., Iwatsubo T., Jack C. R., Jr., Kaye J., Montine T. J., Park D. C., Reiman E. M., Rowe C. C., Siemers E., Stern Y., Yaffe K., Carrillo M. C., Thies B., Morrison-Bogorad M., Wagster M. V., Phelps C. H. (2011) Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dementia 7, 280–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O'Brien R. J., Resnick S. M., Zonderman A. B., Ferrucci L., Crain B. J., Pletnikova O., Rudow G., Iacono D., Riudavets M. A., Driscoll I., Price D. L., Martin L. J., Troncoso J. C. (2009) Neuropathologic Studies of the Baltimore Longitudinal Study of Aging (BLSA). J. Alzheimer's Dis. 18, 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Price J. L., McKeel D. W., Jr., Buckles V. D., Roe C. M., Xiong C., Grundman M., Hansen L. A., Petersen R. C., Parisi J. E., Dickson D. W., Smith C. D., Davis D. G., Schmitt F. A., Markesbery W. R., Kaye J., Kurlan R., Hulette C., Kurland B. F., Higdon R., Kukull W., Morris J. C. (2009) Neuropathology of nondemented aging: Presumptive evidence for preclinical Alzheimer disease. Neurobiol. Aging 30, 1026–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Driscoll I., Troncoso J. (2011) Asymptomatic Alzheimer's disease: a prodrome or a state of resilience? Curr. Alzheimer Res. 8, 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bai B., Hales C. M., Chen P. C., Gozal Y., Dammer E. B., Fritz J. J., Wang X., Xia Q., Duong D. M., Street C., Cantero G., Cheng D., Jones D. R., Wu Z., Li Y., Diner I., Heilman C. J., Rees H. D., Wu H., Lin L., Szulwach K. E., Gearing M., Mufson E. J., Bennett D. A., Montine T. J., Seyfried N. T., Wingo T. S., Sun Y. E., Jin P., Hanfelt J., Willcock D. M., Levey A., Lah J. J., Peng J. (2013) U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 110, 16562–16567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Will C. L., Lührmann R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13, 290–301 [DOI] [PubMed] [Google Scholar]
- 10. Nilsen T. W. (2003) The spliceosome: the most complex macromolecular machine in the cell? BioEssays 25, 1147–1149 [DOI] [PubMed] [Google Scholar]
- 11. Denis M. M., Tolley N. D., Bunting M., Schwertz H., Jiang H., Lindemann S., Yost C. C., Rubner F. J., Albertine K. H., Swoboda K. J., Fratto C. M., Tolley E., Kraiss L. W., McIntyre T. M., Zimmerman G. A., Weyrich A. S. (2005) Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell 122, 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glanzer J., Miyashiro K. Y., Sul J.-Y., Barrett L., Belt B., Haydon P., Eberwine J. (2005) RNA splicing capability of live neuronal dendrites. Proc. Natl. Acad. Sci. U.S.A. 102, 16859–16864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rogers J., Wall R. (1980) A mechanism for RNA splicing. Proc. Natl. Acad. Sci. U.S.A. 77, 1877–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sheth N., Roca X., Hastings M. L., Roeder T., Krainer A. R., Sachidanandam R. (2006) Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 34, 3955–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J., Grishin N. V., Frantz D. E., Schneider J. W., Chen S., Li L., Sawaya M. R., Eisenberg D., Tycko R., McKnight S. L. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J. L., McKnight S. L. (2013) Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Korneta I., Bujnicki J. M. (2012) Intrinsic disorder in the human spliceosomal proteome. PLoS Comput. Biol. 8, e1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lagier-Tourenne C., Polymenidou M., Cleveland D. W. (2010) TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 19, R46–R64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nonaka T., Masuda-Suzukake M., Arai T., Hasegawa Y., Akatsu H., Obi T., Yoshida M., Murayama S., Mann D. M., Akiyama H., Hasegawa M. (2013) Prion-like properties of pathological TDP-43 aggregates from diseased brains. Cell Rep. 4, 124–134 [DOI] [PubMed] [Google Scholar]
- 20. Guo W., Chen Y., Zhou X., Kar A., Ray P., Chen X., Rao E. J., Yang M., Ye H., Zhu L., Liu J., Xu M., Yang Y., Wang C., Zhang D., Bigio E. H., Mesulam M., Shen Y., Xu Q., Fushimi K., Wu J. Y. (2011) An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat. Struct. Mol. Biol. 18, 822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang I. F., Chang H.-Y., Hou S.-C., Liou G.-G., Way T.-D., James Shen C. K. (2012) The self-interaction of native TDP-43 C terminus inhibits its degradation and contributes to early proteinopathies. Nat. Commun. 3, 766. [DOI] [PubMed] [Google Scholar]
- 22. Mirra S. S., Heyman A., McKeel D., Sumi S. M., Crain B. J., Brownlee L. M., Vogel F. S., Hughes J. P., van Belle G., Berg L. (1991) The consortium to establish a registry for Alzheimer's disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology 41, 479–486 [DOI] [PubMed] [Google Scholar]
- 23. Braak H., Braak E. (1991) Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 [DOI] [PubMed] [Google Scholar]
- 24. Dickson D. W., Bergeron C., Chin S. S., Duyckaerts C., Horoupian D., Ikeda K., Jellinger K., Lantos P. L., Lippa C. F., Mirra S. S., Tabaton M., Vonsattel J. P., Wakabayashi K., Litvan I. (2002) Office of rare diseases neuropathologic criteria for corticobasal degeneration. J. Neuropathol. Exp. Neurol. 61, 935–946 [DOI] [PubMed] [Google Scholar]
- 25. Cairns N. J., Bigio E. H., Mackenzie I. R., Neumann M., Lee V. M., Hatanpaa K. J., White C. L., 3rd, Schneider J. A., Grinberg L. T., Halliday G., Duyckaerts C., Lowe J. S., Holm I. E., Tolnay M., Okamoto K., Yokoo H., Murayama S., Woulfe J., Munoz D. G., Dickson D. W., Ince P. G., Trojanowski J. Q., Mann D. M. (2007) Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol. 114, 5–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seyfried N. T., Gozal Y. M., Donovan L. E., Herskowitz J. H., Dammer E. B., Xia Q., Ku L., Chang J., Duong D. M., Rees H. D., Cooper D. S., Glass J. D., Gearing M., Tansey M. G., Lah J. J., Feng Y., Levey A. I., Peng J. (2012) Quantitative analysis of the detergent-insoluble brain proteome in frontotemporal lobar degeneration using SILAC internal standards. J. Proteome Res. 11, 2721–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ferguson S. M., Savchenko V., Apparsundaram S., Zwick M., Wright J., Heilman C. J., Yi H., Levey A. I., Blakely R. D. (2003) Vesicular localization and activity-dependent trafficking of presynaptic choline transporters. J. Neurosci. 23, 9697–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yi H., Leunissen J., Shi G., Gutekunst C., Hersch S. (2001) A novel procedure for pre-embedding double immunogold–silver labeling at the ultrastructural level. J. Histochem. Cytochem. 49, 279–284 [DOI] [PubMed] [Google Scholar]
- 29. Dammer E. B., Duong D. M., Diner I., Gearing M., Feng Y., Lah J. J., Levey A. I., Seyfried N. T. (2013) Neuron enriched nuclear proteome isolated from human brain. J. Proteome Res. 12, 3193–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frangioni J. V., Neel B. G. (1993) Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210, 179–187 [DOI] [PubMed] [Google Scholar]
- 31. Seyfried N. T., Xu P., Duong D. M., Cheng D., Hanfelt J., Peng J. (2008) Systematic approach for validating the ubiquitinated proteome. Anal. Chem. 80, 4161–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Herskowitz J. H., Seyfried N. T., Duong D. M., Xia Q., Rees H. D., Gearing M., Peng J., Lah J. J., Levey A. I. (2010) Phosphoproteomic analysis reveals site-specific changes in GFAP and NDRG2 phosphorylation in frontotemporal lobar degeneration. J. Proteome Res. 9, 6368–6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bridge E., Carmo-Fonseca M., Lamond A., Pettersson U. (1993) Nuclear organization of splicing small nuclear ribonucleoproteins in adenovirus-infected cells. J. Virol. 67, 5792–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mahapatra R. K., Edwards M. J., Schott J. M., Bhatia K. P. (2004) Corticobasal degeneration. Lancet Neurol. 3, 736–743 [DOI] [PubMed] [Google Scholar]
- 35. Hales C. M., Dammer E. B., Diner I., Yi H., Seyfried N. T., Gearing M., Glass J. D., Montine T. J., Levey A. I., Lah J. J. (2014) Aggregates of small nuclear ribonucleic acids (snRNAs) in Alzheimer's disease. Brain Pathol. 24, 344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dammer E. B., Lee A. K., Duong D. M., Gearing M., Lah J. J., Levey A., Seyfried N. T. (2014) Quantitative phosphoproteomics of Alzheimer's disease reveals crosstalk between kinases and small heat shock proteins. Proteomics 10.1002/pmic.201400189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ward J. J., McGuffin L. J., Bryson K., Buxton B. F., Jones D. T. (2004) The DISOPRED server for the prediction of protein disorder. Bioinformatics 20, 2138–2139 [DOI] [PubMed] [Google Scholar]
- 38. Sinz A. (2007) Isotope-labeled photoaffinity reagents and mass spectrometry to identify protein-ligand interactions. Angew. Chem. Int. Ed. Engl. 46, 660–662 [DOI] [PubMed] [Google Scholar]
- 39. Wolozin B. (2012) Regulated protein aggregation: stress granules and neurodegeneration. Mol. Neurodegener. 7, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han T. W., Kato M., Xie S., Wu L. C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., McKnight S. L. (2012) Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 [DOI] [PubMed] [Google Scholar]
- 41. Alberti S., Halfmann R., King O., Kapila A., Lindquist S. (2009) A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Toombs J. A., McCarty B. R., Ross E. D. (2010) Compositional determinants of prion formation in yeast. Mol. Cell. Biol. 30, 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. King O. D., Gitler A. D., Shorter J. (2012) The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 1462, 61–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Neumann M., Bentmann E., Dormann D., Jawaid A., DeJesus-Hernandez M., Ansorge O., Roeber S., Kretzschmar H. A., Munoz D. G., Kusaka H., Yokota O., Ang L.-C., Bilbao J., Rademakers R., Haass C., Mackenzie I. R. (2011) FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain 134, 2595–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim H. J., Kim N. C., Wang Y.-D., Scarborough E. A., Moore J., Diaz Z., MacLea K. S., Freibaum B., Li S., Molliex A., Kanagaraj A. P., Carter R., Boylan K. B., Wojtas A. M., Rademakers R., Pinkus J. L., Greenberg S. A., Trojanowski J. Q., Traynor B. J., Smith B. N., Topp S., Gkazi A.-S., Miller J., Shaw C. E., Kottlors M., Kirschner J., Pestronk A., Li Y. R., Ford A. F., Gitler A. D., Benatar M., King O. D., Kimonis V. E., Ross E. D., Weihl C. C., Shorter J., Taylor J. P. (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 47. Vanderweyde T., Yu H., Varnum M., Liu-Yesucevitz L., Citro A., Ikezu T., Duff K., Wolozin B. (2012) Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J. Neurosci. 32, 8270–8283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jucker M., Walker L. C. (2013) Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501, 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guo J. L., Lee V. M. (2014) Cell-to-cell transmission of pathogenic proteins in neurodegenerative diseases. Nat. Med. 20, 130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]