Background: Intracellular variants of voltage-gated sodium channels are expressed in macrophages.
Results: Human macrophage SCN5A regulates cAMP signaling and ATF2-mediated transcription in response to pharmacological activation or dsRNA, a pathogen-associated molecular pattern.
Conclusion: Human macrophage SCN5A acts as a pathogen sensor and mediates antiviral responses.
Significance: The SCN5A channel in human macrophages regulates a mechanism to help fight off viral infections.
Keywords: Adenylate Cyclase, Double-stranded RNA (dsRNA), Innate Immunity, Interferon, Sodium Channel
Abstract
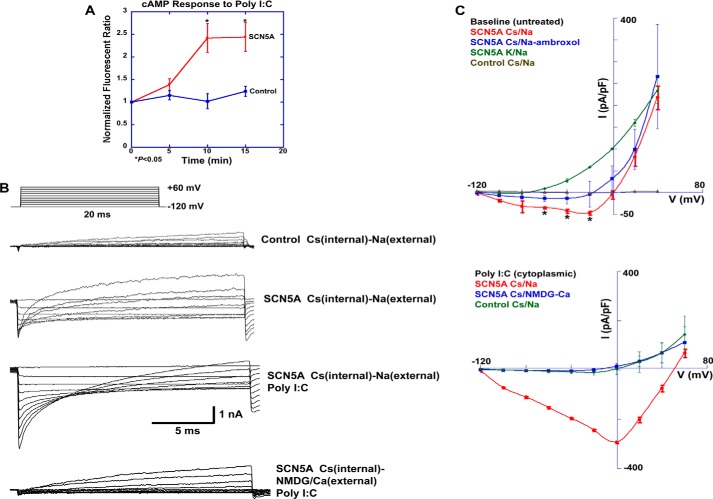
Pattern recognition receptors contain a binding domain for pathogen-associated molecular patterns coupled to a signaling domain that regulates transcription of host immune response genes. Here, a novel mechanism that links pathogen recognition to channel activation and downstream signaling is proposed. We demonstrate that an intracellular sodium channel variant, human macrophage SCN5A, initiates signaling and transcription through a calcium-dependent isoform of adenylate cyclase, ADCY8, and the transcription factor, ATF2. Pharmacological stimulation with a channel agonist or treatment with cytoplasmic poly(I:C), a mimic of viral dsRNA, activates this pathway to regulate expression of SP100-related genes and interferon β. Electrophysiological analysis reveals that the SCN5A variant mediates nonselective outward currents and a small, but detectable, inward current. Intracellular poly(I:C) markedly augments an inward voltage-sensitive sodium current and inhibits the outward nonselective current. These results suggest human macrophage SCN5A initiates signaling in an innate immune pathway relevant to antiviral host defense. It is postulated that SCN5A is a novel pathogen sensor and that this pathway represents a channel activation-dependent mechanism of transcriptional regulation.
Introduction
Characterization of innate immune signal transduction has focused primarily on ligand-receptor interactions between pathogen-associated molecular patterns and pattern recognition receptors (PRR)2 (1, 2). PRR contain a binding domain for pathogen-associated molecular patterns and a signaling domain that initiates downstream activation of transcription factors relevant to host defense. However, alternative biological mechanisms also may initiate signaling in innate immune cells and include those pathways utilized by excitable tissues such as neurons and muscle. These excitable cells express voltage-gated channels that mediate not only ionic flux but also regulate activity-dependent regulation of gene transcription and some long term forms of memory (3).
One candidate molecule is a unique splice variant of the sodium channel gene SCN5A (4–7). Prior studies demonstrated that the SCN5A variant encodes a channel that regulates phagocytosis, endosomal acidification, calcium signaling, and phenotypic differentiation in human macrophages. Unlike voltage-gated sodium channels in excitable tissues, this macrophage variant is expressed on endosomes intracellularly and not at the plasma membrane. Because of an exon deletion in the extracellular selectivity filter, channel activation does not elicit typical action potentials as in excitable tissues but does mediate ionic flux in response to pharmacological stimulation. Because this channel is not expressed in murine macrophages, in vivo characterization has been performed in a knock-in transgenic model (7).
The central questions of this study were to assess how an intracellular voltage-gated channel could mediate innate immune signaling, regulate transcription, and function as a pathogen sensor. In mouse macrophages that express the human SCN5A variant and primary human monocyte-derived macrophages, we demonstrate that channel expression and activation are associated with a signaling pathway that links activation of a calcium-dependent adenylate cyclase isoform, ADCY8, to the transcription factor ATF2. In an ATF2-dependent manner, SCN5A expression and activation in macrophages regulates expression of host antiviral response genes, Sp100 and β-interferon (IFNB), and an innate immune signaling pathway initiated by cytosolic poly(I:C). Channel activation by poly(I:C) markedly increases an inward voltage-sensitive sodium current and decreases a nonspecific outward current.
EXPERIMENTAL PROCEDURES
Mice
Experimental mouse strains (C57BL6cfms-hSCN5A) were bred at our on-campus breeding facility (Biotron Facility), where our transgenic colony is maintained. Experimental mice were transferred to an approved University of Wisconsin animal facility for the performance and monitoring of all in vivo experiments. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals from the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee of the University of Wisconsin, Madison (protocol numbers M024031 and M02544). Genotyping was performed at Mouse Genotype (Escondido, CA).
Cells
Primary mouse bone marrow cells were obtained from femurs and tibias of transgenic and wild type littermate control mice and were differentiated to macrophages in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS), sodium pyruvate, nonessential amino acids, and mCSF (macrophage colony stimulating factor, 20 ng/ml) for 7–10 days. Human CD14+ peripheral blood monocytes were obtained from Lonza and differentiated to macrophages in the same media.
HEK-293F cells were obtained from Invitrogen and maintained as a suspension culture in 293 Freestyle media. Plasmid transfections of empty vector (pcDNA3.1 hygro) or a vector-containing construct (human macrophage SCN5A, accession number KC858891 (7)) were performed using TransIT X2 transfection reagent (Mirus).
Immunoprecipitation
Mouse bone marrow-derived macrophages from transgenic and littermate controls were lysed by passage through a 27-gauge needle in PBS containing 0.1% Triton X-100, protease, and phosphatase inhibitors (Thermo Scientific Pierce). The lysate was cleared by centrifugation and incubated with rabbit anti-NaV1.5 (SCN5A, Alomone Labs) at a 1:100 dilution and anti-rabbit magnetic microbeads (Miltenyi) for 15 min. Beads were then applied to a micro-column, washed, and collected by elution and subsequent centrifugation.
Nano-liquid Chromatography-Mass Spectroscopy/Mass Spectroscopy (Nano LC-MS/MS)
Nano LC-MS/MS was performed at Applied Biomics (Hayward, CA). For sample preparation, proteins were exchanged into 50 mm ammonium bicarbonate buffer. DTT was added to a final concentration of 10 mm and incubated at 60 °C for 30 min, followed by cooling down to room temperature. Iodoacetamide was then added to a final concentration of 10 mm and incubated in the dark for 30 min at room temperature. The proteins were then digested by trypsin (Promega) overnight at 37 °C.
Nano-LC was carried out using a Dionex Ultimate 3000 (Milford, MA). Mobile phase solvents A and B were 0.1% TFA (v/v) in water and 0.1% TFA (v/v) in 80% acetonitrile, respectively. Tryptic peptides were loaded into a μ-Precolumn Cartridge and separated on an acetonitrile gradient (ranging from 5 to 60%) on a Nano LC column. Fractions were collected at 20-s intervals followed by mass spectrometry analysis on AB SCIEX TOF/TOFTM 5800 system (AB SCIEX). Mass spectra were acquired in reflectron positive ion mode. TOF/TOF tandem MS fragmentation spectra were acquired for each ion, averaging 4000 laser shots per fragmentation spectrum (excluding trypsin autolytic peptides and other known background ions).
Both of the resulting peptide mass and the associated fragmentation spectra were submitted to GPS Explorer workstation equipped with MASCOT search engine (Matrix Science, London, UK) to search the database of Swiss-Prot. Searches were performed without constraining protein molecular weight or isoelectric point, with variable carbamidomethylation of cysteine and oxidation of methionine residues, and with one missed cleavage also allowed in the search parameters.
Western Blot
One-dimensional Western blot was performed by Applied Biomics (Hayward, CA). For sample preparation, lysate or immunoprecipitation pellets were exchanged into cell lysis buffer (30 mm Tris-HCl, pH 8.8, containing 7 m urea, 2 m thiourea, and 4% CHAPS). Protein concentration was measured using Bio-Rad protein assay method. For each sample, 30 μg of protein was mixed with 1.0 μl of diluted CyDye and kept in the dark on ice for 10 min. The labeling reaction was stopped by adding 1.0 μl of 10 mm lysine to each sample and incubating in dark on ice for an additional 15 min.
The labeled samples were then mixed with 120 μg of unlabeled material and exchanged into SDS gel loading buffer (100 mm Tris-Cl, pH 6.8, 4% (w/v) SDS, 0.2% (w/v) bromphenol blue, 20% (v/v) glycerol); DTT was added to the final concentration of 200 mm. The sample was heated at 95 °C for 5 min before loading into the wells. The SDS gels were run at 15 °C. Gel images were scanned immediately following SDS-PAGE using Typhoon TRIO (GE Healthcare). The gels were transferred to Immobilon-FL PVDF membranes (EMD Millipore, Billerica, MA), and the resulting images were immediately scanned using Typhoon TRIO.
Membranes were blocked in 2.5% fish gelatin for 2 h with shaking. The membranes were then incubated in primary antibodies (mouse anti-ATF2 and rabbit anti-phospho-ATF2, Santa Cruz Biotechnologies) in 1× TBST buffer with shaking overnight. The membranes were washed four times, 10 min each, with shaking in 1× TBST buffer. The membranes were then incubated using CyDye-conjugated secondary antibodies at a dilution of 1:2000 in 1× TBST buffer with shaking for 2 h. The membranes were then washed four times, 10 min each, with shaking in 1× TBST buffer. The membranes were dried, and the images were scanned using Typhoon TRIO. The scanned images were then analyzed by ImageJ.
Immunofluorescence
For immunofluorescence staining, cell monolayers were fixed in 4% paraformaldehyde, washed with PBS, and blocked in PBS containing 5% serum, 0.1% Triton X-100, and 1% BSA. Primary and secondary antibodies were diluted in blocking solution. For primary antibody staining, anti-ATF2 antibodies were from Santa Cruz Biotechnology (goat antiphospho-ATF2-Thr-71; rabbit anti-ATF2 C terminus; 1:50 dilution), and isotype controls were from eBioscience. Alexa dye-labeled secondary antibodies (donkey anti-goat and anti-rabbit) were from Invitrogen. Rhodamine-labeled poly(I:C) was obtained from Invivogen, and staining was performed on live cells at 200 ng/ml for 20 min. Treated cells were washed extensively with PBS prior to fixation and imaging. Fluorescent images were acquired and analyzed using a Zeiss Axiovert 200 fluorescent microscope equipped with an Axiocam-MRm CCD camera (Zeiss) and AxioVision version 4.8 software.
siRNA and Poly(I:C) Transfection
Primary cells were transfected in serum-free OptiMEM media that contained 100 nm retinoic acid using the TransIT X2 transfection reagent (Mirus) and pooled ON-TARGETplus siRNA for each target (Dharmacon). Cells were maintained in media with transfection complexes for 48 h prior to experiments. Knockdown was confirmed by QPCR and was greater than 75%.
For QPCR experiments, cells were treated with veratridine (100 μm; Tocris) or DMSO vehicle in OptiMEM media. For immunoprecipitation and live cell imaging experiments, treatment was performed in HBSS supplemented with 10 mm Hepes. Poly(I:C) (LMW; Invivogen) complexes for acute treatments (2 h) were generated in OptiMEM using 10 μg/ml poly(I:C) and 30 μl of TransIT X2 transfection reagent/ml media (30 min at 25 °C). For transfected and naked poly(I:C), the final concentration during cell treatments was 200 ng/ml.
QPCR
RNA purification, reverse transcription, QPCR, and data analysis were performed as described previously (5). Data were acquired on a Cepheid SmartCycler and analyzed by the ΔCt method with normalization to either mouse GAPDH or human hydroxymethylbilane synthase. The following TaqMan primers were obtained from Applied Biosystems/Invitrogen: Hs00165693_m1 (human SCN5A), Mm99999915_g1 (mouse Gapdh), Mm00449735_m1 (mouse Sp100), Mm04204797_m1 (mouse Gm7609), Hs00162109_m1 (human SP100), Hs01077958_s1 (human IFNB1), AIGJQ71 (HSV thymidine kinase), and AIHSPDQ (HSV glycoprotein B (8)).
Microarray Analysis
Total RNA was prepared from mouse bone marrow-derived macrophages using column purification (Qiagen RNeasy). Labeling, hybridization, and scanning were performed at the Gene Expression Center at the University of Wisconsin Biotechnology Center (Madison, WI). Affymetrix mouse 2.0ST whole transcriptome arrays were utilized. Data were analyzed using NetAffx software (Affymetrix). Fold change (hSCN5A-transgenic macrophages versus wild type cells) and p values were based on three separate RNA preparations for each condition.
Live Cell Microscopy
Primary mouse bone marrow-derived macrophages and human monocyte-derived macrophages were grown on collagen-coated glass coverslips (Mat-Tek). siRNA treatment was performed 48 h prior to use in experiments. For imaging, cells were maintained with HBSS supplemented with 10 mm Hepes and treated with either veratridine (100 μm) or DMSO vehicle. Live cell imaging was performed on a Zeiss Axiovert 200 fluorescent microscope with a ×40/0.75 objective (Neofluar) in an environmental chamber, and time lapse images were acquired with a QuantEM:512SC EMCCD camera. Time-lapse sequences were acquired using Zeiss AxioVision 4.6.3 software and analyzed using the Time Series Analyzer plugin (Balaji) for ImageJ software (rsb.info.nih.gov). Subsequent statistical analysis was performed using KaleidaGraph (Synergy) and MATLAB (MathWorks).
For single cell detection of cAMP, mouse bone marrow-derived macrophages were transfected with a plasmid encoding human GFP-labeled CNGA2 (cyclic nucleotide-gated channel). Two days later, cells were labeled with the red fluorescent dye DiSBAC2(3) (9). Cells were observed in the presence and absence of veratridine, a sodium channel agonist (100 μm at 100 s). Background fluorescent responses over time in DiSBAC2(3) labeled cells were measured in GFP-negative cells. Responses in GFP-positive cells were acquired in the time-lapse mode (every 10 s for 720 s) by AxioVision software, and background responses were subtracted. Further analysis was performed using ImageJ and MATLAB (MathWorks). Normalized fluorescent responses (fluorescent ratio, Ft/F0) versus time were plotted.
For calcium measurements, human monocyte derived macrophages were labeled with 1 μm Fluo 4 NW (Invitrogen) for 30 min. Images were acquired every 10 s for 600 s and then analyzed using ImageJ and MATLAB.
cAMP ELISA
cAMP concentration in lysates from monocyte-derived macrophages was determined by a commercially available ELISA kit (colorimetric; Cell Biolabs, San Diego). Plates were scanned on a Victor3 plate reader (PerkinElmer Life Sciences), and data were analyzed using KaleidaGraph.
Viral Infection
Monocyte-derived macrophages were infected with HSV-1 (KOS strain, ATCC VR-1493) for 2 h at a multiplicity of infection of 1:1 in HBSS supplemented with 10 mm Hepes. Cells were washed and incubated for an additional 22 h in HBSS/Hepes prior to assays.
Whole Cell Patch Clamp
Whole cell patch clamp analysis of transfected HEK-293F cells that were grown as suspension cultures was performed on a planar chip system (Nanion, Munich, Germany). Microchips had an equivalent resistance of 2–5 megohms. Cells were placed on the microchip, and giga-ohm seals (1–4 giga-ohms) were obtained with an automated suction device (port-a-patch). Mean cell capacitance was 6.7 ± 0.3 picofarads (n = 9). Whole cell currents were recorded for 20 millisiemens in 20-mV increments from −120 to +60 mV utilizing pClamp 10.3.1 software. Data were acquired at a sampling frequency of 20 kHz and a low pass Bessel filter setting at 5 kHz. Capacitative transients and series resistance were compensated (∼80%) with amplifier settings (Axopatch 200B) in conjunction with pClamp software. P/4 leak subtraction was performed for all recordings used for analysis. All recordings were performed at room temperature.
Intracellular solutions (pH 7.2, 285 mosm) contained either 50 mm CsCl, 10 mm NaCl, 60 mm cesium fluoride, 20 mm EGTA, and 10 mm Hepes (cesium internal solution) or 50 mm KCl, 10 mm NaCl, 60 mm potassium fluoride, 20 mm EGTA, and 10 mm Hepes (potassium internal solution). For experiments with poly(I:C) treatment, 200 ng/ml LMW poly(I:C) was added directly to the internal solution of the microchip.
Extracellular solutions (pH 7.4, 298 mosm) contained either 140 mm NaCl, 4 mm KCl, 1 mm MgCl2, 2 mm CaCl2, 5 mm d-glucose, and 10 mm Hepes (sodium external solution) or 100 mm NMDG, 30 mm CaCl2, 10 mm d-glucose, and 20 mm Hepes (NMDG-calcium external solution). As indicated, ambroxol (50 μm final concentration), an inhibitor of tetrodotoxin-resistant sodium channels, was added to the external bath solution.
Statistical Analysis
Data were analyzed as indicated using ImageJ, AxioVision, KaleidaGraph (Student's t test and ANOVA), MATLAB, and pClamp.
RESULTS
Protein-Protein Interactions of the Macrophage Variant of Human SCN5A Reveal a Potentially Novel Signaling Network
Although prior work suggested that SCN5A plays a central role in human macrophage regulation, its intracellular signaling mechanisms remained unknown. Our initial goal in this study was to identify potentially novel signaling mechanisms for SCN5A in macrophages by using an unbiased, discovery-based approach.
To identify potential signaling mechanisms, immunoprecipitation of SCN5A from bone marrow-derived macrophages (BMDM) from C57BL6cfms-hSCN5A mice was performed to analyze its interactions by nano-LC-MS/MS. These transgenic mice express the human macrophage splice variant of SCN5A under the control of a macrophage-specific promoter (7). Nano-LC/MS analysis demonstrated known and novel protein-protein interactions with human SCN5A (C.I. >95%). Consistent with prior studies of the cardiac channel variant, the macrophage variant binds to PDZ (PSD95/Dlg/ZO-1 domain) (10) and ankyrin-domain containing proteins (11); these interactions mediate connections to the cytoskeleton (data not shown). In addition, consistent with prior cellular analysis of SCN5A subcellular localization in macrophages, the channel interacts with proteins found in acidic organelles such as late endosomes (4) and lysosomes. These included lysosomal acid phosphatase and a proton transporter. These findings provided an initial validation measure for the specificity of the immunoprecipitation and nano-LC/MS analysis.
In addition to known interactions, a novel interaction was identified (Table 1) between SCN5A and the cAMP-dependent transcription factor ATF2 that is relevant to cellular signaling and transcriptional regulation. To confirm the protein-protein interaction with ATF2, Western blot analysis following immunoprecipitation for macrophage SCN5A was performed (Fig. 1). Co-immunoprecipitation of ATF2 was observed in transgenic BMDM but not in the wild type condition. Although co-immunoprecipitation does not demonstrate direct binding, the results suggested a possible association within a subcellular signaling complex. Based on these results, it was reasoned that macrophage SCN5A could regulate ATF2-dependent transcription.
TABLE 1.
Mass spectroscopy analysis of SCN5A-ATF2 protein-protein interaction
Peaks from liquid chromatography were analyzed by mass spectroscopy. The ion score is based on probability of a peptide match with known database proteins. The total ion score is a sum of the scores for all peptide fragments that match a specific protein. The total ion confidence interval (C.I.) is a normalized probability. PI is isoelectric point.
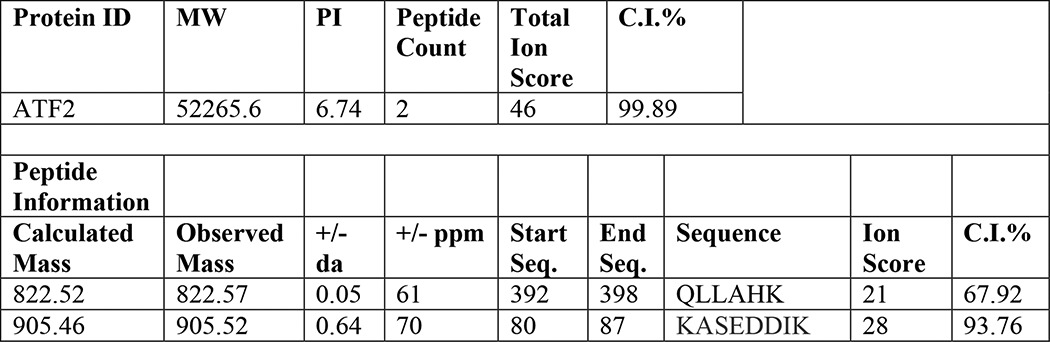
FIGURE 1.
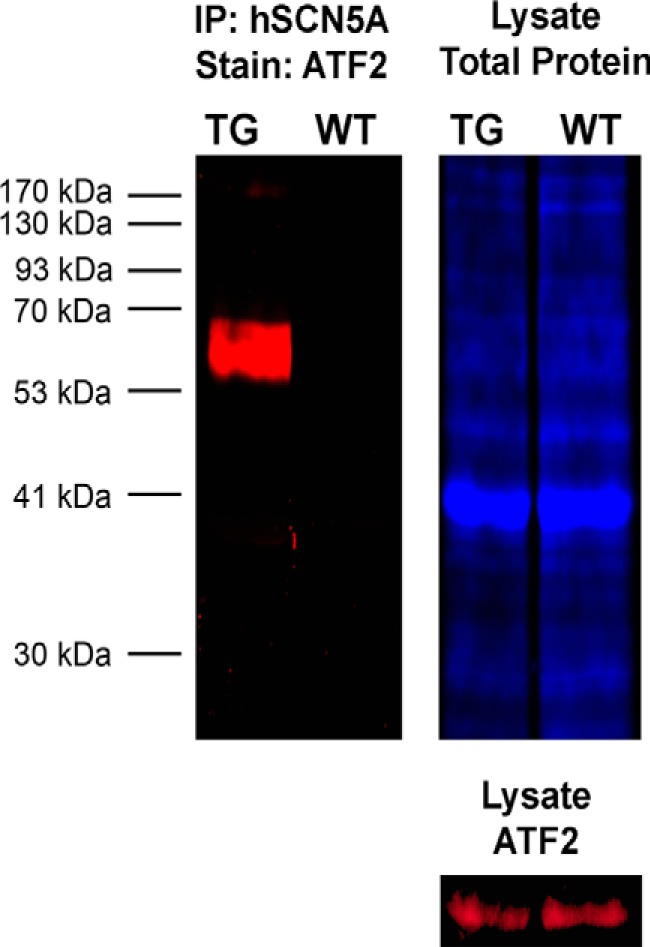
Human macrophage SCN5A splice variant associates with the transcription factor ATF2. To confirm the results from LC/MS (Table 1), immunoprecipitation (IP) of human SCN5A was performed in BMDM from transgenic and wild type conditions. BMDM from the transgenic mice express the human macrophage SCN5A variant under the control of a cfms promoter (24). Co-immunoprecipitation of ATF2 (red) was observed in the transgenic (TG) but not wild type (WT) condition. Difference gel electrophoresis labeling for total protein content in the lysate is shown in blue.
Pharmacological Activation of Macrophage SCN5A Increases Phosphorylation of ATF2 and Nuclear Translocation
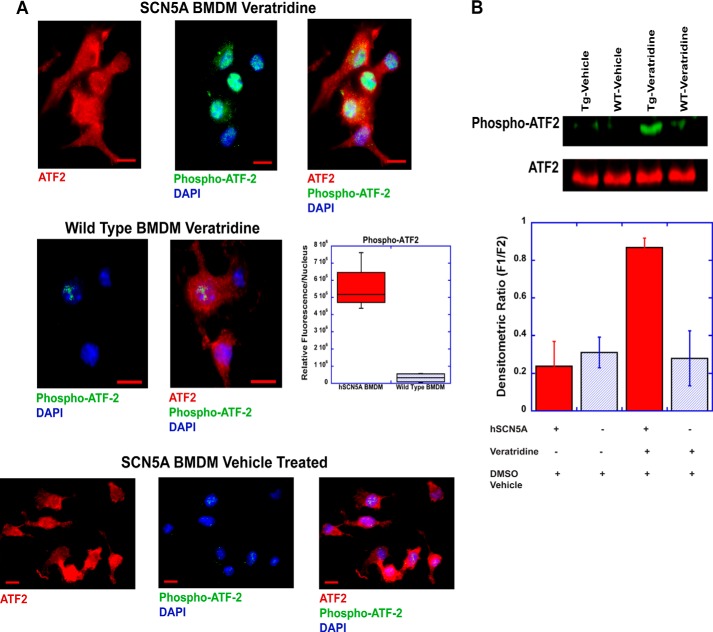
When ATF2 is activated in the cytosol through phosphorylation, it translocates from the cytosol to the nucleus. Treatment of SCN5A+ macrophages with veratridine (100 μm for 15 min), a voltage-gated sodium channel agonist, resulted in a significant increase in phosphorylated ATF2 (Thr-71 phosphorylation site) in the nucleus (Fig. 2A). This translocation and increase in phosphorylated ATF2 was significantly greater than that observed in wild type cells. To confirm these results, Western blot analysis was performed and demonstrated increased levels of phospho-ATF2 in SCN5A+ macrophages that were treated with veratridine but no related increase in wild type cells (Fig. 2B).
FIGURE 2.
Pharmacological activation increases phospho-ATF2 levels. A, human SCN5A+ (hSCN5A) and wild type BMDM were treated with the sodium channel agonist, veratridine (100 μm), for 15 min and then stained for total ATF2 (red), phospho-ATF2 (green), and DNA (DAPI, blue). Scale bar, 10 μm. Quantitative analysis (AxioVision auto-measurement module) revealed a statistically significant difference in nuclear phospho-ATF2 staining between the two conditions (top micrographs). Fluorescent densitometric sum/nucleus was 5.58 × 106 ± 0.71 × 106 in hSCN5A cells versus 0.32 × 106 ± 0.14 × 106 in the wild type condition (p < 0.01, n = 4 cell samples; 46 cells analyzed for the hSCN5A condition and 54 cells for the wild type condition; ± S.E.). Baseline staining in the SCN5A BMDM without treatment is shown on the bottom. Scale bar, 10 μm. B, Western blot analysis was performed to confirm phospho-ATF2 cell staining. Veratridine treatment of mouse BMDM for 15 min led to markedly increased phospho-ATF2 levels in transgenic human SCN5A+ (hSCN5A) macrophages (Tg) but not in the wild type conditions (WT) or following vehicle (DMSO) treatment. Densitometric analysis (ImageJ) revealed relative fluorescent units of 0.24 ± 0.13 for vehicle-treated transgenic BMDM, 0.31 ± 0.08 for vehicle-treated WT, 0.87 ± 0.05 for veratridine-treated transgenic cells (p < 0.01 ANOVA, n = 4 cell preparations, + S.E.), and 0.28 ± 0.15 for veratridine-treated WT. TG, transgenic.
mRNA Expression Profile of SCN5A+ Macrophages Demonstrates Increased Expression of Sp100-related Genes
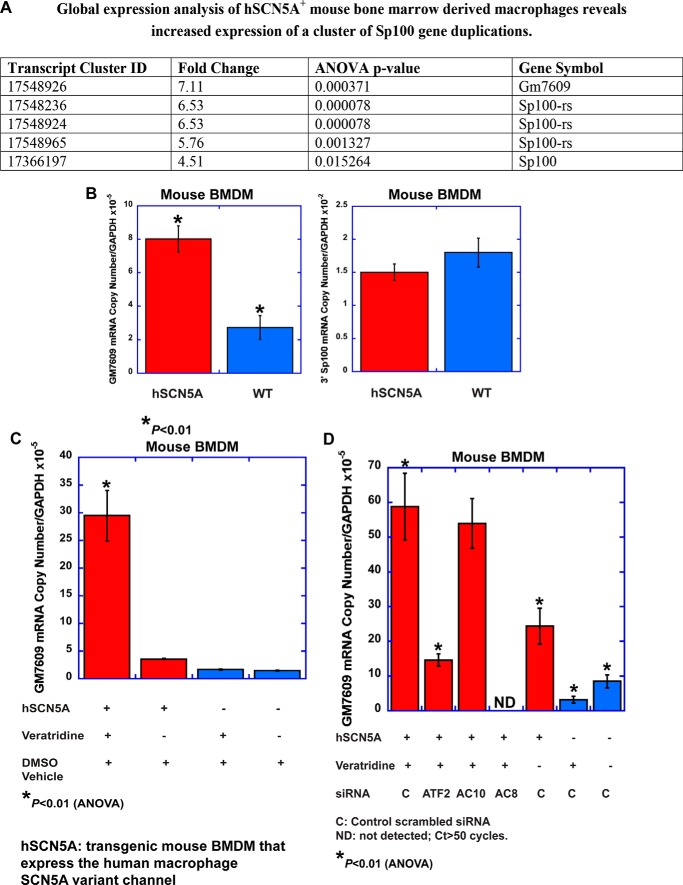
Because cAMP can activate ATF2 (12), this interaction provides a potential link between channel activation, cAMP signaling, and transcriptional pathways. Based on these findings, it was hypothesized that SCN5A regulates cellular phenotype and function through regulation of downstream transcriptional mechanisms. To assess this possible mechanism, global gene expression analysis was performed on BMDM from transgenic and wild type littermate mice (Fig. 3A). In the absence of any exogenous treatment, there was a statistically significant increase in the expression of genes (Gm7609 and Sp100rs) that are duplications or fusion variants of mouse Sp100 (13). Most variants of Sp100 localize to a specialized nuclear structure called the nuclear body; it can function as a transcriptional repressor, mediate antiviral host defense, and regulate mRNA splicing and translation. Many of these variants are splice variants of the Sp100 gene. However, the mouse Sp100rs and Gm7609 variants are gene duplications with distinct promoter regions. Gm7609 is a fusion gene that contains Csprs (component of Sp100rs), the 5′ portion of Sp100, and a predicted G-protein domain.
FIGURE 3.
Increased expression of an Sp100 variant, Gm7609, is associated with human SCN5A expression and pharmacological activation. A, Affymetrix mouse 2.0ST whole transcriptome arrays were utilized to examine global gene expression in mouse BMDM that express the human macrophage SCN5A transgene and in control wild type cells. Data were analyzed using NetAffx software (Affymetrix). Fold change (transgenic versus wild type) and p values are based on three separate RNA preparations for each condition (error bars, ± S.E.). B, QPCR analysis confirmed increased expression of Gm7609 in transgenic cells but not the 3′ prime segment of Sp100 (Table 2, statistical analysis). C, pharmacological activation with the sodium channel agonist veratridine (100 μm for 2 h) of transgenic BMDM led to a further increase in Gm7609 mRNA that was not observed in wild type cells (Table 3). D, siRNA knockdown of ATF2 and Adcy8, a calcium-dependent isoform of adenylate cyclase, but not Adcy10, inhibited the veratridine-induced response (Table 4). The transgenic condition in the graphs is shown in red and the wild type condition in blue.
To validate the microarray analysis, QPCR analysis was performed to analyze mRNA expression of mouse Gm7609 and the 3′ region of Sp100. As described above, Gm7609 contains unique domains and the 5′ region of Sp100. Consistent with the microarray data, significantly increased expression of Gm7609 mRNA was observed in SCN5A+ macrophages as compared with wild type cells but not in transcripts that contain the 3′ region of Sp100 (Fig. 3B and Table 2). These results suggested that expression of human macrophage SCN5A in mouse BMDM increases expression of an ATF2-regulated gene.
TABLE 2.
QPCR analysis of hSCN5A+ bone marrow-derived macrophages demonstrates increased expression of mouse Gm7609 mRNA but not Sp100 transcripts that contain the 3′ gene sequences
| Condition | Sp100-related Gm7609 mRNA copies/Gapdh × 10−5 | Sp100 3′ region copies/Gapdh × 10−2 |
|---|---|---|
| hSCN5A-untreated | 8.0 ± 0.8a | 1.5 ± 0.1 |
| Wild type-untreated | 2.7 ± 0.7a | 1.8 ± 0.2 |
a p < 0.01 (Student t test, n = 5).
Pharmacological Activation of Macrophage SCN5A Increases Expression of an ATF2-regulated Gene
The next goal was to determine whether channel activation regulates this pathway. Activity-dependent regulation of gene transcription occurs in neurons to regulate cellular plasticity and memory. Based on this mechanism, it was reasoned that channel activation in macrophages also could be associated with transcriptional regulation. To test this hypothesis, expression of Gm7609 transcripts was examined in the presence and absence of pharmacological activation. Treatment with the channel agonist veratridine (100 μm for 2 h) induced an early transcriptional response in transgenic cells but not wild type (Fig. 3C and Table 3). This increase in Gm7609 transcripts was ∼3–4-fold greater than that observed in the untreated condition. There was no effect on the expression of transcripts that contain the 3′ region of Sp100.
TABLE 3.
Veratridine treatment increases Gm7609 transcription in hSCN5A+ BMDM
| Condition | Sp100-related Gm7609 mRNA copies/Gapdh × 10−5 |
|---|---|
| hSCN5A-veratridine | 29.5 ± 4.5a |
| hSCN5A-DMSO vehicle | 3.6 ± 0.1a |
| Wild type-veratridine | 1.6 ± 0.1a |
| Wild type-DMSO vehicle | 1.5 ± 0.1a |
a p < 0.01 for hSCN5A-veratridine treated versus all conditions (ANOVA, n = 5).
To analyze the relevance of ATF2 to transcriptional regulation by SCN5A, the promoter sequence of the mouse Gm7609 and Sp100 genes was examined (Genomatix MatInspector). The promoter region of mouse Gm7609 contains a high affinity ATF2-binding site (TGACAACA sequence at positions 557–564 on the +strand, EMBL sequence (14)), but the mouse Sp100 gene does not. Therefore, the increased expression of mouse Gm7609 in BMDM that expresses the human macrophage variant SCN5A can be used as a reporter gene to examine ATF2-dependent transcription.
In addition, in contrast to the mouse promoter, human Sp100 contains potential ATF2-binding sites near the transcription initiation site (Genomatix software analysis). The relevance of increased expression of mouse Gm7609 in cells that express the macrophage variant human SCN5A is that this mechanism can potentially mimic human Sp100 gene regulation.
To assess the role of ATF2 in transcriptional control of Gm7609, siRNA knockdown of ATF2 was performed in SCN5A macrophages. Knockdown resulted in an ∼3-fold reduction in Gm7609 mRNA levels as compared with the control siRNA condition (Fig. 3D and Table 4). These results suggested that SCN5A expression and activation regulate Gm7609 transcription in transgenic mouse BMDM through an ATF2-dependent mechanism.
TABLE 4.
The transcriptional response to veratridine in hSCN5A+ bone marrow-derived macrophages is dependent on Adcy8 and ATF2 expression
| Condition/treatment | siRNA | Sp100-related Gm7609 mRNA copies/Gapdh × 10−5 |
|---|---|---|
| hSCN5A-veratridine | Scrambled control | 58.8 ± 9.6a |
| hSCN5A-veratridine | ATF2 | 14.6 ± 1.8a |
| hSCN5A-veratridine | Adcy10 | 53.9 ± 7.2 |
| hSCN5A-veratridine | Adcy8 | NDb |
| hSCN5A-DMSO vehicle | Scrambled control | 24.4 ± 5.1a |
| Wild type-veratridine | Scrambled control | 3.1 ± 1.0a |
| Wild type-DMSO vehicle | Scrambled control | 8.5 ± 1.9a |
a p < 0.01 for hSCN5A-veratridine control versus all conditions except Adcy10 (ANOVA, n = 7).
b ND means not detected (Ct >50).
Calcium-dependent Isoform of Adenylate Cyclase, Adcy8, Regulates the SCN5A Pathway
The role of two calcium-dependent adenylate cyclase genes on this pathway, the soluble adenylate cyclase, Adcy10, and Adcy8, also was assessed (Fig. 3D and Table 4). Knockdown of Adcy10 appeared to regulate baseline expression of Gm7609 in transgenic cells, but reduced expression did not inhibit activation-induced transcription. In contrast, knockdown of an alternative calcium-dependent adenylate cyclase, Adcy8, did inhibit veratridine-induced expression of Gm7609 in transgenic cells that express SCN5A. These results suggested that activation-dependent regulation of transcription by SCN5A requires signaling through adenylate cyclase and ATF2.
Activation of SCN5A in Macrophages Increases cAMP Levels
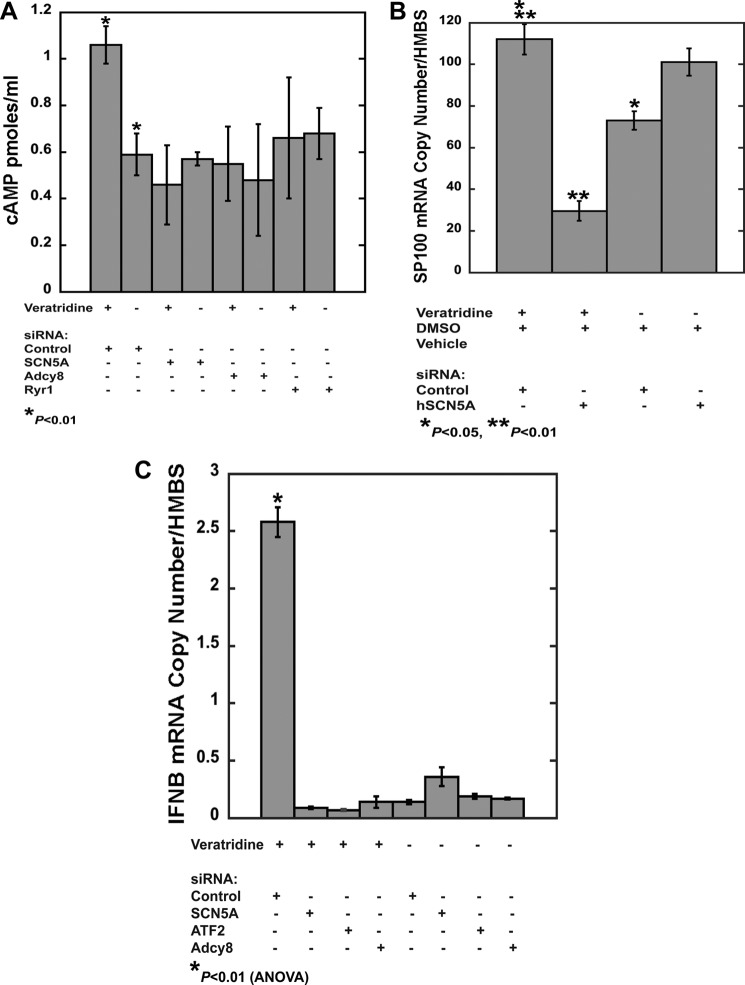
Based on these results, it was hypothesized that channel activation directly links localized calcium ion flux to cAMP signaling, ATF2, and downstream transcription. To examine whether channel activation regulates cAMP, we first measured cAMP levels at the single cell level (Fig. 4) (9). When treated with the sodium channel agonist veratridine, the magnitude and peak of the cAMP response were significantly greater in BMDM from SCN5A than in wild type cells.
FIGURE 4.
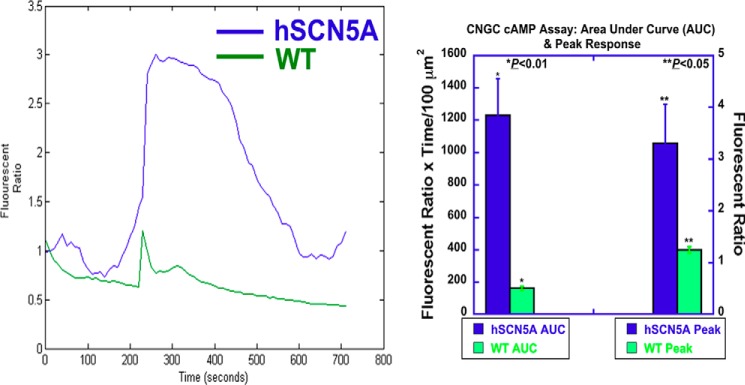
Cyclic nucleotide-gated channel cAMP assay demonstrates markedly enhanced responses in hSCN5A bone marrow-derived macrophages. Cells were transfected with a plasmid encoding human GFP-labeled CNGA2. Two days later, cells were labeled with the red fluorescent dye DiSBAC2(3). Cells were observed in the presence and absence of veratridine, a sodium channel agonist (100 μm at 100 s). Background fluorescent responses over time in DiSBAC2(3)-labeled cells were measured in GFP-negative cells. Responses in GFP-positive cells were acquired in the time-lapse mode (every 10 s for 720 s) by AxioVision software, and background responses were subtracted. Further analysis was performed using MATLAB (MathWorks). Normalized fluorescent responses (fluorescent ratio: Ft/F0) versus time were plotted and are shown on the left. hSCN5A BMDM (blue) demonstrated increased magnitude (area under the curve; AUC) and peak of response than WT (green). AUC was 1229 ± 229 in the hSCN5A cells (n = 10 regions of interest, error bars, ± S.E.) versus 163 ± 13 fluorescent ratio × time/μm2 in WT (n = 8; p < 0.01)). Peak responses were 3.30 ± 0.76 in the hSCN5A cells versus 1.25 ±0.15 ratiometric fluorescent units in WT (p < 0.05).
Veratridine-induced Increases in Cytosolic Calcium in Human Monocyte-derived Macrophages Demonstrate an SCN5A and RYR1-dependent Component
During bacterial phagocytosis in human macrophages, SCN5A regulates mitochondrial sodium-calcium exchange within polarized subcellular regions that contain clusters of endosomes, phagosomes, and mitochondria (5). Although a similar mechanism links activation of a different sodium channel, SCN8A, in mouse BMDM (6), the role of SCN5A in calcium regulation in human macrophages under alternative conditions was not known.
Calcium and cAMP responses were further analyzed in primary human monocyte-derived macrophages (MDM). Analysis of calcium responses following veratridine treatment of human MDM revealed SCN5A-dependent and -independent features (Fig. 5). Knockdown of either SCN5A or a ryanodine channel, RYR1, reduced the initial peak response and prevented a more persistent elevation of cytosolic calcium.
FIGURE 5.
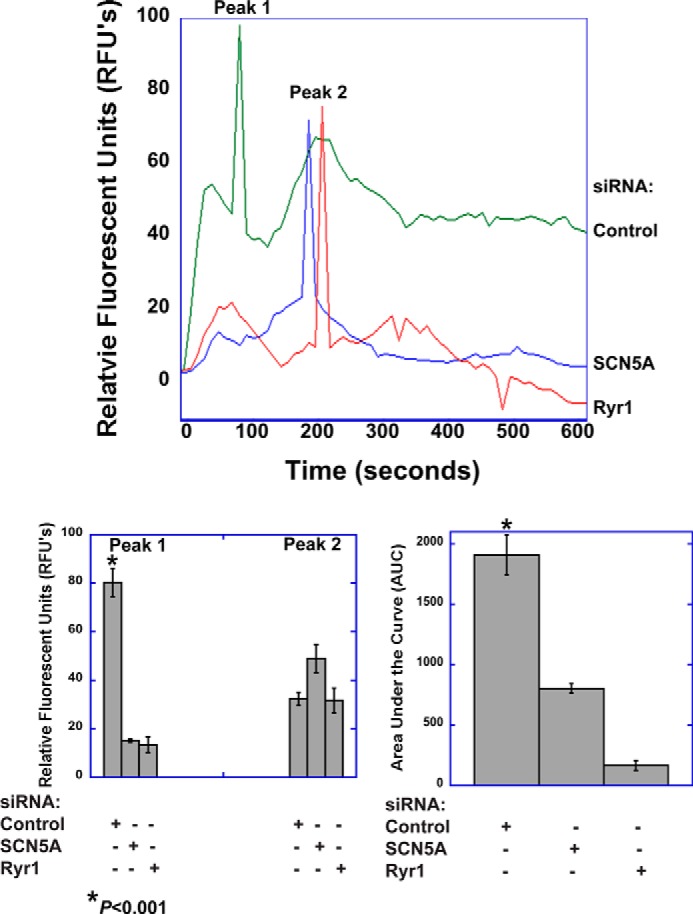
SCN5A mediates a veratridine-induced calcium response in human monocyte-derived macrophages. Human MDM were labeled with the calcium indicator Fluo4 and imaged by time-lapse live cell microscopy. Stimulation of human MDM with veratridine (100 μm) elicited a biphasic peak response and a more prolonged elevation in cytosolic calcium. The initial peak response (Peak 1) and the sustained response, but not Peak 2, were dependent on expression of SCN5A and RYR1. Images were acquired at 10-s intervals for 10 min and subsequently analyzed in ImageJ and MATLAB. The top graph shows representative tracings for each condition. The bar graphs show quantitative analysis of peak responses and the cumulative response (AUC, area under the curve). For peak 1, the mean responses were as follows: 80.2 ± 5.8 RFUs for control siRNA, 15.1 ± 4.9 for SCN5A knockdown, and 13.4 ±3.2 for RYR1 knockdown (p < 0.001 for control condition, ANOVA, n = 10 regions of interest, error bars, ±S.E.). For peak 2, 32.3 ± 2.7 for control siRNA, 48.9 ±5.8 for SCN5A knockdown, and 31.7 ± 5.1 for RYR1 knockdown (not significant, ANOVA, n = 10). The AUC was 1909.0 ± 163.1 for the siRNA control condition, 803.3 ± 192.5 for SCN5A knockdown, and 164.6 ± 40.3 for RYR1 knockdown (p < 0.001 for control condition, ANOVA, n = 10).
To confirm the cAMP results from transgenic mouse macrophages, veratridine-induced activation of cAMP was analyzed in human MDM in the presence and absence of siRNA-mediated knockdown of SCN5A, RYR1, or ADCY8. Increased cAMP levels in response to veratridine were only observed in control siRNA-transfected cells (Fig. 6A and Table 5). These results suggested that the human macrophage splice variant of SCN5A and RYR1 regulate calcium-dependent cAMP signaling in its native cell type.
FIGURE 6.
Pharmacological activation of human monocyte-derived macrophages regulates signal transduction and transcription that is dependent on SCN5A expression. A, veratridine treatment (100 μm for 15 min) increased cAMP levels in human MDM in control siRNA conditions but not following knockdown of SCN5A, ADCY8, or RYR1 (Table 5). B, veratridine treatment (100 μm for 2 h) increased Sp100 mRNA levels in control siRNA-treated MDM but not following SCN5A knockdown (Table 6). C, veratridine treatment (100 μm for 2 h) markedly increased IFNB transcription in MDM treated with control siRNA. Knockdown of SCN5A, ADCY8, or ATF2 inhibited this response (Table 7). Error bars, ± S.E., from pooled replicates from a representative donor. Similar results were obtained from a second donor.
TABLE 5.
Veratridine-induced increases in cAMP in human monocyte-derived macrophages is dependent on expression of SCN5A, ADCY8, and RYR1
| Treatment | siRNA | cAMP |
|---|---|---|
| pmol/ml | ||
| Veratridine | Scrambled control | 1.06 ± 0.08a |
| DMSO vehicle | Scrambled control | 0.59 ± 0.09a |
| Veratridine | SCN5A | 0.46 ± 0.17 |
| DMSO vehicle | SCN5A | 0.57 ± 0.28 |
| Veratridine | ADCY8 | 0.55 ± 0.16 |
| DMSO vehicle | ADCY8 | 0.48 ± 0.24 |
| Veratridine | RYR1 | 0.66 ± 0.26 |
| DMSO vehicle | RYR1 | 0.68 ± 0.11 |
a p < 0.01 for hSCN5A-veratridine control versus DMSO vehicle (Student's t test, n = 4).
The combined results in mouse and human cells were consistent with our hypothesis that channel activation is associated with calcium-dependent adenylate cyclase activation and cAMP production. These interactions provide a mechanism to link human macrophage SCN5A function to downstream signaling and regulation of transcription.
Activation-dependent Increases in IFNB Transcription in Human Monocyte-derived Macrophages Is Dependent on SCN5A, ATF2, and ADCY8 Expression
To assess transcriptional regulation in human macrophages, veratridine-induced gene expression was analyzed in primary MDM in the presence and absence of SCN5A expression. Cells treated with control siRNA exhibited an increase in SP100 expression with veratridine treatment (100 μm for 2 h), whereas SCN5A knockdown cells showed decreased levels of expression relative to the vehicle-treated condition (Fig. 6B and Table 6).
TABLE 6.
Veratridine treatment increases transcription of SP100 in human monocyte-derived macrophages and is dependent on expression of SCN5A
HMBS is hydroxymethylbilane synthase.
| Treatment | siRNA | SP100 mRNA copies/HMBS |
|---|---|---|
| Veratridine | Scrambled control | 112.13 ± 7.34 |
| DMSO vehicle | Scrambled control | 73.13 ± 4.37a |
| Veratridine | SCN5A | 29.55 ± 4.80b |
| DMSO vehicle | SCN5A | 101.1 ± 6.54 |
a p < 0.05 for veratridine control versus DMSO vehicle control (Student's t test, n = 4).
b p < 0.01 for veratridine control versus veratridine SCN5A knockdown (Student's t test, n = 4).
In human cells, prior studies demonstrated that ATF2 is a regulator of IFNB transcription (15). Therefore, the effects of SCN5A expression and activation in human MDM were examined. Veratridine treatment in control-transfected human MDM markedly increased IFNB expression but did not in the SCN5A knockdown condition. Knockdown of ATF2 or ADCY8 also inhibited this response (Fig. 6C and Table 7).
TABLE 7.
Veratridine-induced increases in IFNB transcripts in human monocyte-derived macrophages is dependent on expression of SCN5A, ATF2, and ADCY8
HMBS is hydroxymethylbilane synthase.
| Treatment | siRNA | IFNB mRNA copies/HMBS |
|---|---|---|
| Veratridine | Scrambled control | 2.58 ± 0.13a |
| DMSO vehicle | Scrambled control | 0.14 ± 0.02 |
| Veratridine | SCN5A | 0.09 ± 0.01 |
| DMSO vehicle | SCN5A | 0.36 ± 0.08 |
| Veratridine | ATF2 | 0.07 ± 0.01 |
| DMSO vehicle | ATF2 | 0.19 ± 0.02 |
| Veratridine | ADCY8 | 0.14 ± 0.05 |
| DMSO vehicle | ADCY8 | 0.17 ± 0.01 |
a p < 0.01 for veratridine control (ANOVA, n = 4).
SCN5A Expression Enhances Cytosolic Poly(I:C) Responses, and Knockdown Is Associated with Increased Viral Transcription in HSV-infected Human Monocyte-derived Macrophages
The regulation of IFNB and SP100 in human macrophages suggested a role in host antiviral defense. The effects of a dsRNA mimic, poly(I:C) (16), were examined in BMDM from transgenic mice that express the human SCN5A transgene. To assess relative uptake of poly(I:C) transfection complexes in the presence and absence of SCN5A, we first challenged macrophages with fluorescently labeled poly(I:C) (LMW, rhodamine labeled, 200 ng/ml). Channel expression did not affect the efficiency of transfection of poly(I:C) into the cytosol (Fig. 7).
FIGURE 7.
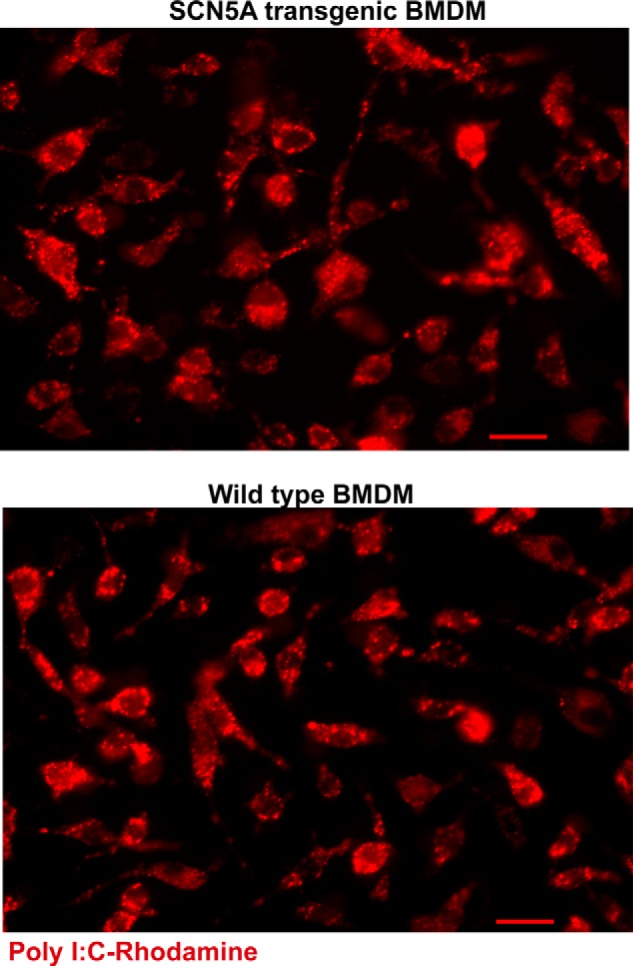
Transfection of poly(I:C) complexes is similar in the presence or absence of macrophage SCN5A. The indicated cell types were challenged with rhodamine-labeled poly(I:C) (LMW, 200 nm for 20 min) and subsequently washed and fixed for microscopic analysis (scale bars, 20 μm).
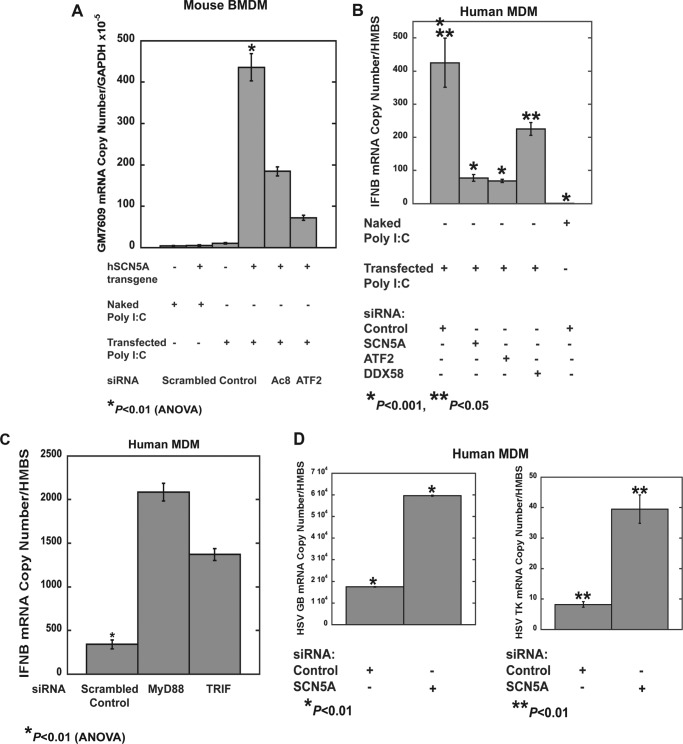
Treatment of mouse BMDM with transfected poly(I:C) (200 ng/ml for 2 h), but not the naked form, resulted in an increase in Gm7609 expression that was not observed in wild type cells. Knockdown of ATF2 or Adcy8 in SCN5A+ cells inhibited the increase in Gm7609 expression (Fig. 8A and Table 8). These results suggested that cytosolic poly(I:C), but not endosomal, activates the SCN5A signaling pathway.
FIGURE 8.
Poly(I:C) treatment initiates SCN5A-dependent transcription. A, in mouse BMDM that express the human macrophage SCN5A variant, transfection of poly(I:C) (low molecular weight, 200 ng/ml for 2 h), but not treatment with naked poly(I:C) (200 ng/ml for 2 h), increased expression of Gm7609 transcripts. This increase was not observed in wild type, SCN5A-negative cells, or in transgenic cells following siRNA knockdown of Adcy8 or ATF2 (Table 8) (error bars, ± S.E. from pooled replicates of multiple RNA preparations). B, in human MDM treated with control siRNA, increased transcription of IFNB was observed following treatment with transfected poly(I:C) (200 ng/ml for 2 h). This increase was inhibited by siRNA knockdown of SCN5A or ATF2; knockdown of DDX58 (RIG-1) also reduced the response but to a lesser degree (Table 9). Error bars, ± S.E., from pooled replicates from a representative donor. Similar results were obtained from a second donor. C, knockdown of MyD88 or TRIF (TICAM-1) increased expression of IFNB in response to cytosolic poly(I:C) in human MDM (Table 10). D, human MDM were infected with HSV-1 for 24 h, and transcription of viral transcripts glycoprotein B (GB) and thymidine kinase (TK) was analyzed. Knockdown of SCN5A, but not treatment with control siRNA, increased transcription of glycoprotein B and thymidine kinase (Table 11).
TABLE 8.
Cytosolic poly(I:C)-induced increases in mouse Gm7609 transcripts in hSCN5A+ bone marrow-derived macrophages is dependent on expression of SCN5A, ATF2, and ADCY8
| Treatment/condition | siRNA | Sp100-related Gm7609 mRNA copies/Gapdh × 10−5 |
|---|---|---|
| Transfected poly(I:C)/hSCN5A | Scrambled control | 436.0 ± 33.0a |
| Naked poly(I:C)/hSCN5A | Scrambled control | 5.2 ± 2.0 |
| Transfected poly(I:C)/hSCN5A | Adcy8 | 184.5 ± 11.0 |
| Transfected poly(I:C)/hSCN5A | ATF2 | 72.0 ± 6.3 |
| Transfected poly(I:C)/wild type | Scrambled control | 10.3 ± 1.6 |
| Naked poly(I:C)/wild type | Scrambled control | 4.2 ± 1.1 |
a p < 0.01 for hSCN5A-transfected poly(I:C) control (ANOVA, n = 4).
We next examined the effects of cytosolic poly(I:C) on primary human cells. Knockdown of either SCN5A or ATF2 in human MDM reduced the increases in IFNB transcription in response to cytosolic poly(I:C) (Fig. 8B and Table 9). Knockdown of a known cytoplasmic PRR, DDX58 (also known as RIG-1), decreased IFNB transcription but to a lesser extent.
TABLE 9.
Cytosolic poly(I:C)-induced increases in IFNB transcripts in human monocyte-derived macrophages is dependent on expression of SCN5A and ATF2 but less so on DDX58
HMBS is hydroxymethylbilane synthase.
| Treatment | siRNA | IFNB mRNA copies/HMBS |
|---|---|---|
| Transfected poly(I:C) | Scrambled control | 425.4 ± 74.3a,b |
| Transfected poly(I:C) | SCN5A | 77.7 ± 10.2a |
| Transfected poly(I:C) | ATF2 | 68.7 ± 4.8a |
| Transfected poly(I:C) | DDX58 (RIG-1) | 225.3 ± 19.1b |
| Naked poly(I:C) | Scrambled control | 0.9 ± 0.3a |
a p < 0.01 for transfected poly(I:C) control (ANOVA, n = 4).
b p < 0.05 for transfected poly(I:C) control versus DDX58 knockdown (ANOVA, n = 4).
For these experiments in human MDM, primary monocytes were differentiated with mCSF and subsequently transfected with siRNA in serum-free media. With these conditions, there was no response to treatment with naked poly(I:C), which can trigger a toll-like receptor response in cells polarized to an inflammatory phenotype (Fig. 8B). Knockdown of components of the toll pathway, MyD88 and TRIF (also known as TICAM-1), increased rather than decreased the transcriptional response to cytosolic poly(I:C) (Fig. 8C and Table 10). Taken together, these results suggest, in human MDM, the SCN5A pathway of cytosolic dsRNA recognition mediates a more robust effect on interferon transcription as compared with the RIG pathway and that the TLR pathway may regulate an inhibitory effect in some individuals.
TABLE 10.
Increased expression of IFNB transcripts in human monocyte-derived macrophages in response to cytosolic poly(I:C) response is not dependent on the toll-like receptor pathway
HMBS is hydroxymethylbilane synthase.
| Treatment | siRNA | IFNB mRNA copies/HMBS |
|---|---|---|
| Transfected poly(I:C) | Scrambled control | 341.1 ± 49.5a |
| Transfected poly(I:C) | MyD88 | 2083.7 ± 101.1 |
| Transfected poly(I:C) | TRIF (TICAM-1) | 1370.2 ± 160.8 |
a p < 0.01 (ANOVA, n = 3).
In addition, knockdown of SCN5A resulted in an increase in transcription of herpes simplex virus (HSV-1) genes in infected human MDM (Fig. 8D and Table 11). These results provided further evidence that SCN5A enhances antiviral innate immune responses.
TABLE 11.
SCN5A knockdown in human monocyte-derived macrophages increases viral transcripts in HSV-1 infected cells
HMBS is hydroxymethylbilane synthase.
| Transcript | siRNA | mRNA copies/HMBS |
|---|---|---|
| Glycoprotein B | Scrambled control | 17,508 ± 132a |
| Glycoprotein B | SCN5A | 59,661 ± 244a |
| Thymidine kinase | Scrambled control | 8.20 ± 0.95b |
| Thymidine kinase | SCN5A | 39.5 ± 4.7b |
a p < 0.01 (Student's t test, n = 4).
b p < 0.01 (Student's t test, n = 4).
Poly(I:C) Increases a Voltage-sensitive Sodium Current Mediated by Human Macrophage SCN5A
To characterize a more direct interaction between poly(I:C) treatment and receptor activation, the channel was heterologously expressed in HEK-293 cells. Consistent with our data in human and mouse transgenic macrophages, cytosolic poly(I:C) treatment (200 ng/ml for 15 min) increased cytosolic cAMP levels in a time-dependent manner in SCN5A-expressing cells but not vector control transfected cells (Fig. 9A).
FIGURE 9.
Poly(I:C) increases cAMP levels and currents in HEK-293 cells that heterologously express human macrophage SCN5A. A, cAMP levels were monitored by live cell imaging using the cyclic nucleotide-gated channel assay (Fig. 4). Poly(I:C) treatment (low molecular weight, 200 ng/ml) increased cAMP levels 10 and 15 min post-challenge in SCN5A-transfected cells but not mock-transfected HEK-293 cells. Normalized fluorescent ratios were 1.39 ± 0.13 (5 min), 2.42 ± 0.32 (10 min), and 2.44 ± 0.32 (15 min) for the SCN5A condition and 1.15 ± 0.10 (5 min), 1.02 ± 0.17 (10 min), and 1.24 ± 0.11 (15 min) for the control condition (error bars, ± S.E., n = 10 regions of interest for each condition, p < 0.05 for 10 and 15 min conditions). B, whole cell patch clamp analysis revealed outward and inward currents in SCN5A-expressing cells with cesium (Cs) as the predominant intracellular cation and sodium in the extracellular solution (second recording from top). Outward currents were reduced in control transfected cells (top current recording), and an inward current was not seen. Poly(I:C) in the intracellular solution (200 ng/ml; third from top) markedly increased a voltage-dependent inward current in SCN5A-positive cells and reduced the outward current. Replacement of sodium (Na) with NMDG in the extracellular solution (bottom current recording) prevented the poly(I:C) induced inward current. C, I-V curve analysis of the experiments shown in B. Ambroxol (50 μm), an inhibitor of tetrodotoxin-resistant sodium channels, reduced inward but not outward currents in SCN5A cells. Data in Tables 12–15 show statistical analysis of currents at +40 mV and −20 mV in the absence and presence of intracellular poly(I:C). Data are pooled, and normalized (pA/picofarad (pF)) current recordings and are expressed as ± S.D.
Our next goal was to perform electrophysiological analysis of human macrophage SCN5A in the presence and absence of intracellular poly(I:C). Because of its intracellular localization (4) and the deletion in the extracellular selectivity filter and a portion of the outer pore (7), the prediction was that the channel would not initiate typical action potentials and not be ion-selective. This deletion renders a related, but different, channel variant “nonfunctional” as a voltage-gated and sodium-selective channel (17).
Consistent with this prediction, channel-expressing HEK-293 cells demonstrated a predominant outward current that was observed with either cesium or potassium as the intracellular cation (Fig. 9, B and C). However, with cesium as the predominant intracellular cation, current recordings demonstrated a small but detectable inward current in addition to the larger outward current (Fig. 9, B and C). In control transfected cells, the outward current was greatly diminished, and the inward current was not observed. Addition of ambroxol (50 μm), an inhibitor of tetrodotoxin-resistant sodium channels, inhibited the inward current but not the outward current (Fig. 9C and Tables 12–15) (18). These results suggested that, in the absence of ligand-mediated regulation, human macrophage SCN5A is a nonselective cation channel. Despite the absence of the selectivity filter in this variant, the results also suggested the inward component may be voltage-dependent.
TABLE 12.
I-V analysis
Current measurements at +40 mV in the untreated condition. pF is picofarads.
| Cell type | Ionic conditions (Internal/External) | Treatment | Normalized current (pA/pF ± S.D.) |
|---|---|---|---|
| HEK-293F SCN5A | Cesium/sodium | None | 232.7 ± 107.2 |
| HEK-293F SCN5A | Cesium/sodium | Ambroxol (external) | 224.0 ± 125.2 |
| HEK-293F SCN5A | Potassium/sodium | None | 214.0 ± 68.9 |
| HEK-293F Control | Cesium/sodium | None | 3.0 ± 5.3a |
a p < 0.001 (ANOVA, n = pooled recordings from six cells).
TABLE 13.
I-V analysis
Current measurements at −20 mV in the untreated condition. pF is picofarads.
| Cell type | Ionic conditions (internal/external) | Treatment | Normalized current (pA/pF ± S.D.) |
|---|---|---|---|
| HEK-293F SCN5A | Cesium/sodium | None | −31.5 ± 19.0a |
| HEK-293F SCN5A | Cesium/sodium | Ambroxol (external) | 2.0 ± 33.8 |
| HEK-293F SCN5A | Potassium/sodium | None | 42.2 ± 22.7 |
| HEK-293F Control | Cesium/sodium | None | 0.1 ± 2.3 |
a p < 0.005 (ANOVA, n = 6).
TABLE 14.
I-V analysis
Current measurements at +40 mV in the poly(I:C) condition. pF is picofarads.
| Cell type | Ionic conditions (internal/external) | Treatment | Normalized current (pA/pF ± S.D.) |
|---|---|---|---|
| HEK-293F SCN5A | Cesium/sodium | Poly I:C (internal) | −10.7 ± 13.4a |
| HEK-293F SCN5A | Cesium/NMDG-calcium | Poly(I:C) (internal) | 71.9 ± 39.9 |
| HEK-293F SCN5A | Cesium/sodium | Poly(I:C) (internal) Ambroxol (external) | 619.8 ± 97.5b |
| HEK-293F Control | Cesium/sodium | Poly(I:C) (internal) | 69.2 ± 17.3b |
a p < 0.005 (ANOVA, n = pooled recordings from 10 cells).
b p < 0.001 (ANOVA, n = 10).
TABLE 15.
I-V analysis
Current measurements at −20 mV in the poly(I:C) condition. pF is picofarads.
| Cell type | Ionic conditions (internal/external) | Treatment | Normalized current (pA/pF ± S.D.) |
|---|---|---|---|
| HEK-293F SCN5A | Cesium/sodium | Poly(I:C) (internal) | −180.6 ± 4.1a |
| HEK-293F SCN5A | Cesium/NMDG-calcium | Poly(I:C) (internal) | 3.0 ± 6.8 |
| HEK-293F SCN5A | Cesium/sodium | Poly(I:C) (internal) Ambroxol (external) | −18.7 ± 10.6 |
| HEK-293F Control | Cesium/sodium | Poly(I:C) (internal) | −10.8 ± 10.2 |
a p < 0.001 (ANOVA, n = 10).
Addition of poly(I:C) (200 ng/ml) to the intracellular solution markedly increased the magnitude of the inward current, which demonstrated voltage-dependent characteristics (Fig. 9C and Tables 12–15). Poly(I:C) also decreased the magnitude of the outward current. In contrast, ambroxol inhibited the increase in the inward current and the decrease in the outward current (Tables 12–15). Replacement of extracellular sodium with NMDG (100 mm) and calcium (30 mm) also blocked the inward current in response to poly(I:C) but had no effect on the decrease in the outward current. There was no evidence of an inward calcium current under these conditions.
These data suggested that human macrophage SCN5A functions as a nonspecific cation channel in the absence of ligand stimulation, but cytoplasmic poly(I:C) activates and partially restores a voltage-sensitive sodium current.
DISCUSSION
Our work demonstrates that human SCN5A channel expression and activation in macrophages are associated with a cyclic AMP-dependent pathway that regulates transcription. The results suggest that SCN5A expression and activation in human macrophages is a central regulator of a pathogen recognition and signaling pathway associated with antiviral host defense. This pathway integrates a novel combination of signaling molecules to mediate a channel activation-dependent mechanism in a cell type that lacks excitable membranes.
These findings were consistent with the hypothesis that agonist and pathogen-associated molecular pattern-mediated channel activation is associated with a calcium-dependent adenylate cyclase and cAMP production. These interactions directly link SCN5A function to downstream signaling and regulation of transcription through ATF2. Our sequential model is that either pharmacological treatment or the presence of cytosolic dsRNA increase intracellular channel activity. Although the channel does not directly regulate calcium flux, its activation is associated with localized fluctuations in cytosolic calcium that are then amplified by calcium-induced calcium release through a ryanodine channel (RYR1) (19). This localized increase in cytosolic calcium activates ADCY8 (20), stimulates cAMP production, and leads to phosphorylation and nuclear translocation of the transcription factor ATF2 (21). Related signaling networks in neurons that link channel activation, calcium and cAMP signaling, and transcriptional regulation have immediate and long term effects on cellular function (3). We hypothesize that SCN5A utilizes similar mechanisms to regulate macrophage function and phenotype and could potentially regulate a primitive form of cellular memory.
Our electrophysiological analysis suggests that cytoplasmic poly(I:C) directly increases human macrophage SCN5A activity and that the channel acts as a pathogen sensor rather than a regulator of other pattern recognition pathways. As predicted by the exon deletion in the selectivity filter, the human macrophage variant functions as a nonselective channel. However, the presence of cytoplasmic poly(I:C) increases an inward voltage-dependent sodium current and decreases the nonselective outward current. Ligand activation appears to restore the properties of a more typical voltage-gated sodium channel.
Under the conditions used here, we did not detect an inward calcium current, and, as suggested by prior work, the increases in cytosolic calcium in response to channel activation likely occur by more indirect mechanisms. These mechanisms include not only amplification through ryanodine receptors as shown here but also by mitochondrial sodium calcium exchange (5, 6).
Expression of the SCN5A splice variant in human, but not mouse, macrophages suggests that this pathway is a more restricted evolutionary adaptation to innate immune antiviral host defense. It is hypothesized that this macrophage variant of the SCN5A channel has particular relevance to the pathogenesis of infectious and inflammatory diseases in humans. Although this pathway does not demonstrate the same level of evolutionary conservation as other innate immune pathways, its specific relevance may be the development of host defense against viruses that replicate in macrophages and are particularly virulent in humans. These would include vector-borne flaviviruses such as West Nile, yellow fever, and dengue viruses (22). These single-stranded RNA viruses generate dsRNA during their intracellular replication and productively infect a limited range of insect and mammalian hosts.
Because expression of this human splice variant in mouse macrophages polarizes the cell to an anti-inflammatory phenotype (7), this pathway also may preserve antiviral host defense in alternatively activated cells. This mechanism could provide for simultaneous wound healing and control of viral infection. Recent work from another group suggest that skewing of macrophage phenotype during helminth infection leads to reactivation of herpes viruses and has detrimental effects on the host (23). Utilization of the SCN5A pathway in human macrophages may prevent viral reactivation in inflammatory microenvironments that contain large numbers of alternatively activated cells. In this setting, other PRR pathways such as DDX58 (RIG-1) may not provide as robust a response to cytosolic viral pathogen as does the SCN5A pathway. Our data also suggest the possibility that the toll pathway may not regulate antiviral responses in SCN5A-positive macrophages. In this context, cytoplasmic pathogen recognition mediated by complementary signaling pathways could be the primary mechanism of innate antiviral immunity.
Acknowledgments
We acknowledge the technical assistance of Dr. Wayne Davis at the University of Wisconsin Biotechnology Facility for probe labeling, microarray hybridization, and slide scanning; Franklin Chin and Hongjin Huang at Applied Biomics (Hayward, CA) for proteomic analysis; Ivan Delgado at Mouse Genotype; and Jody Peter at the University of Wisconsin Biotron Facility for mouse breeding and care. We are grateful for the helpful suggestions of Rodolfo Haedo (Nanion, Munich, Germany).
This work was supported by a Merit Award from the Department of Veterans Affairs and the University of Wisconsin at Madison.
- PRR
- pattern recognition receptor
- Adcy
- adenylate cyclase
- ATF2
- activating transcription factor 2
- BMDM
- bone marrow-derived macrophage
- MDM
- monocyte-derived macrophage
- NMDG
- N-methyl-d-glucamine
- poly(I:C)
- polyinosinic:polycytidylic acid
- RYR
- ryanodine receptor
- SCN
- sodium channel
- ANOVA
- analysis of variance
- AUC
- area under the curve
- HBSS
- Hanks' buffered saline solution
- QPCR
- quantitative PCR
- IFNB
- interferon β
- LMW
- low molecular weight
- DiSBAC2(3)
- bis-(1,3-diethylthiobarbituric acid) trimethine oxonol.
REFERENCES
- 1. Janeway C. A., Jr. (2001) How the immune system works to protect the host from infection: a personal view. Proc. Natl. Acad. Sci. U.S.A. 98, 7461–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takeuchi O., Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 [DOI] [PubMed] [Google Scholar]
- 3. Lynch M. A. (2004) Long-term potentiation and memory. Physiol. Rev. 84, 87–136 [DOI] [PubMed] [Google Scholar]
- 4. Carrithers M. D., Dib-Hajj S., Carrithers L. M., Tokmoulina G., Pypaert M., Jonas E. A., Waxman S. G. (2007) Expression of the voltage-gated sodium channel NaV1.5 in the macrophage late endosome regulates endosomal acidification. J. Immunol. 178, 7822–7832 [DOI] [PubMed] [Google Scholar]
- 5. Carrithers L. M., Hulseberg P., Sandor M., Carrithers M. D. (2011) The human macrophage sodium channel NaV1.5 regulates mycobacteria processing through organelle polarization and localized calcium oscillations. FEMS Immunol. Med. Microbiol. 63, 319–327 [DOI] [PubMed] [Google Scholar]
- 6. Carrithers M. D., Chatterjee G., Carrithers L. M., Offoha R., Iheagwara U., Rahner C., Graham M., Waxman S. G. (2009) Regulation of podosome formation in macrophages by a splice variant of the sodium channel SCN8A. J. Biol. Chem. 284, 8114–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rahgozar K., Wright E., Carrithers L. M., Carrithers M. D. (2013) Mediation of protection and recovery from experimental autoimmune encephalomyelitis by macrophages expressing the human voltage-gated sodium channel NaV1.5. J. Neuropathol. Exp. Neurol. 72, 489–504 [DOI] [PubMed] [Google Scholar]
- 8. Mott K. R., Underhill D., Wechsler S. L., Town T., Ghiasi H. (2009) A role for the JAK-STAT1 pathway in blocking replication of HSV-1 in dendritic cells and macrophages. Virol. J. 6, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Willoughby D., Cooper D. M. (2008) Live-cell imaging of cAMP dynamics. Nat. Methods 5, 29–36 [DOI] [PubMed] [Google Scholar]
- 10. Ou Y., Strege P., Miller S. M., Makielski J., Ackerman M., Gibbons S. J., Farrugia G. (2003) Syntrophin γ2 regulates SCN5A gating by a PDZ domain-mediated interaction. J. Biol. Chem. 278, 1915–1923 [DOI] [PubMed] [Google Scholar]
- 11. Mohler P. J., Rivolta I., Napolitano C., LeMaillet G., Lambert S., Priori S. G., Bennett V. (2004) Nav1.5 E1053K mutation causing Brugada syndrome blocks binding to ankyrin-G and expression of Nav1.5 on the surface of cardiomyocytes. Proc. Natl. Acad. Sci. U.S.A. 101, 17533–17538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liao H., Hyman M. C., Baek A. E., Fukase K., Pinsky D. J. (2010) cAMP/CREB-mediated transcriptional regulation of ectonucleoside triphosphate diphosphohydrolase 1 (CD39) expression. J. Biol. Chem. 285, 14791–14805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guldner H. H., Szostecki C., Schröder P., Matschl U., Jensen K., Lüders C., Will H., Sternsdorf T. (1999) Splice variants of the nuclear dot-associated Sp100 protein contain homologies to HMG-1 and a human nuclear phosphoprotein-box motif. J. Cell Sci. 112, 733–747 [DOI] [PubMed] [Google Scholar]
- 14. Ronai Z., Yang Y. M., Fuchs S. Y., Adler V., Sardana M., Herlyn M. (1998) ATF2 confers radiation resistance to human melanoma cells. Oncogene 16, 523–531 [DOI] [PubMed] [Google Scholar]
- 15. Du W., Maniatis T. (1992) An ATF/CREB-binding site is required for virus induction of the human interferon beta gene (corrected). Proc. Natl. Acad. Sci. U.S.A. 89, 2150–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 17. Schroeter A., Walzik S., Blechschmidt S., Haufe V., Benndorf K., Zimmer T. (2010) Structure and function of splice variants of the cardiac voltage-gated sodium channel Na(v) 1.5. J. Mol. Cell. Cardiol. 49, 16–24 [DOI] [PubMed] [Google Scholar]
- 18. Weiser T., Wilson N. (2002) Inhibition of tetrodotoxin (TTX)-resistant and TTX-sensitive neuronal Na+ channels by the secretolytic ambroxol. Mol. Pharmacol. 62, 433–438 [DOI] [PubMed] [Google Scholar]
- 19. Nikolaeva M. A., Mukherjee B., Stys P. K. (2005) Na+-dependent sources of intra-axonal Ca2+ release in rat optic nerve during in vitro chemical ischemia. J. Neurosci. 25, 9960–9967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cali J. J., Zwaagstra J. C., Mons N., Cooper D. M., Krupinski J. (1994) Type VIII adenylyl cyclase. A Ca2+/calmodulin-stimulated enzyme expressed in discrete regions of rat brain. J. Biol. Chem. 269, 12190–12195 [PubMed] [Google Scholar]
- 21. Lau E., Ronai Z. A. (2012) ATF2-at the crossroad of nuclear and cytosolic functions. J. Cell Sci. 125, 2815–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gould E. A., Solomon T. (2008) Pathogenic flaviviruses. Lancet 371, 500–509 [DOI] [PubMed] [Google Scholar]
- 23. Reese T. A., Wakeman B. S., Choi H. S., Hufford M. M., Huang S. C., Zhang X., Buck M. D., Jezewski A., Kambal A., Liu C. Y., Goel G., Murray P. J., Xavier R. J., Kaplan M. H., Renne R., Speck S. H., Artyomov M. N., Pearce E. J., Virgin H. W. (2014) Coinfection. Helminth infection reactivates latent γ-herpesvirus via cytokine competition at a viral promoter. Science 345, 573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sasmono R. T., Oceandy D., Pollard J. W., Tong W., Pavli P., Wainwright B. J., Ostrowski M. C., Himes S. R., Hume D. A. (2003) A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood 101, 1155–1163 [DOI] [PubMed] [Google Scholar]