Background: Kalirin-7 is a neuronal Rac1-GEF that regulates spine morphology. X11α is a PDZ scaffolding protein.
Results: X11α binds kalirin-7 and modulates its localization to Golgi outposts, GEF activity, and spine morphology.
Conclusion: X11α is a novel regulator of small GTPase signaling by recruitment to Golgi outposts.
Significance: We identify a novel mechanism for regulation of neuronal GEF localization and function.
Keywords: Dendritic Spine, Neurobiology, Neuron, Neuroscience, Small GTPase, Synapse
Abstract
Pyramidal neurons in the mammalian forebrain receive their synaptic inputs through their dendritic trees, and dendritic spines are the sites of most excitatory synapses. Dendritic spine structure is important for brain development and plasticity. Kalirin-7 is a guanine nucleotide-exchange factor for the small GTPase Rac1 and is a critical regulator of dendritic spine remodeling. The subcellular localization of kalirin-7 is thought to be important for regulating its function in neurons. A yeast two-hybrid screen has identified the adaptor protein X11α as an interacting partner of kalirin-7. Here, we show that kalirin-7 and X11α form a complex in the brain, and this interaction is mediated by the C terminus of kalirin-7. Kalirin-7 and X11α co-localize at excitatory synapses in cultured cortical neurons. Using time-lapse imaging of fluorescence recovery after photobleaching, we show that X11α is present in a mobile fraction of the postsynaptic density. X11α also localizes to Golgi outposts in dendrites, and its overexpression induces the removal of kalirin-7 from spines and accumulation of kalirin-7 in Golgi outposts. In addition, neurons overexpressing X11α displayed thinner spines. These data support a novel mechanism of regulation of kalirin-7 localization and function in dendrites, providing insight into signaling pathways underlying neuronal plasticity. Dissecting the molecular mechanisms of synaptic structural plasticity will improve our understanding of neuropsychiatric and neurodegenerative disorders, as kalirin-7 has been associated with schizophrenia and Alzheimer disease.
Introduction
The architecture of the dendritic arbor and the distribution, morphology, and strength of synapses on dendrites are crucial determinants of neuronal connectivity within brain circuits. Dendritic spines are small mushroom-shaped protrusions that decorate the dendrite and serve as the sites of most excitatory synapses onto neurons in the mammalian forebrain. Dendritic spine remodeling is an important process in activity-independent and -dependent circuit formation and plasticity during development and adulthood (1). Their dysfunction in disease states may have profound effects on connectivity, as altered dendritic and synaptic morphology has been documented in several neurodevelopmental disorders (2).
Dendritic spine morphogenesis relies on the precise regulation of local actin dynamics (3). Although multiple signaling cascades converge onto the actin cytoskeleton, the Rho-like small GTPase, Rac, has been shown to have profound effects on dendritic spine morphology (4). In cortical neurons, the function of Rac is tightly controlled by the brain-specific guanine nucleotide-exchange factor (GEF)4 kalirin-7 (5–7). Kalirin-7 has been shown to be a critical coordinator of activity-independent and -dependent changes in dendritic spine morphology in cortical neurons (5, 7, 8). Functionally, the interaction of kalirin-7 with PDZ domain-containing proteins such as PSD-95 and afadin (also known as AF-6) mediate its localization to the postsynaptic density (9). This localization is thought to be essential for its function, and PDZ proteins may play additional roles in its subcellular localization. However, interactions of kalirin-7 with other PDZ proteins have not been further characterized.
Previously, a yeast two-hybrid screen with the C terminus of kalirin-7 identified a number of interacting PDZ domain-containing proteins, including the X11α protein (10). The X11 family of adaptor/scaffold proteins are mammalian homologs of the Caenorhabditis elegans gene LIN-10 (also referred to collectively as mLIN-10) and consist of three members, X11α, -β, and -γ, encoded by different genes (ABP1, ABP2, and ABP3) (11, 12). X11α (also known as Mint1) contains a Munc-18-interacting region, a CASK-interacting region, a phosphotyrosine-binding domain, and two PDZ domains (Fig. 1A). X11β (also known as Mint2) has a similar domain structure but lacks the CASK-interacting domain, whereas X11γ lacks the Munc18- and CASK-interacting domains (12). X11α and X11β have been shown to be expressed in neurons (12, 13), exhibiting both presynaptic (14) and postsynaptic localization (13, 15, 16). Functionally, LIN-10 and its mammalian homologs act as adaptor proteins involved in the trafficking of ion channels and glutamate receptors. In invertebrates, the Lin-10 protein interacts with the motor protein KIF17; this complex mediates the trafficking of NMDA receptors along microtubules (16) and is required for the delivery of AMPA-like receptors to synapses in C. elegans (17). In vertebrates, X11 proteins interact with the small GTPase Rab6 to control the trafficking of amyloid precursor protein (18–20). In addition, X11 proteins have been shown to directly interact with amyloid precursor protein via their phosphotyrosine-binding domain and are involved in amyloid precursor protein processing and secretion (11, 12). Examination of specific X11 family members have revealed that X11α promotes the presynaptic localization of N-type calcium channels (21). Recently, X11β has been shown to localize to the Golgi apparatus, dendrites, and synapses of hippocampal neurons, where it binds to AMPA receptors in a PDZ-dependent manner and modulates their trafficking (15). However, whether X11α also shows a similar localization and can modulate the trafficking of synaptic proteins is unknown.
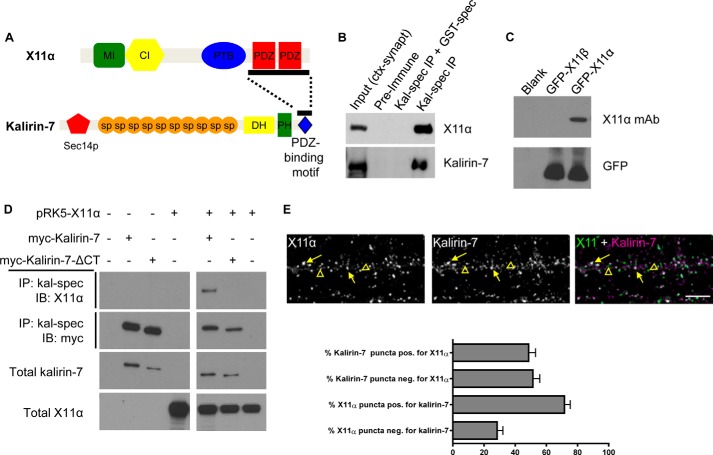
FIGURE 1.
Kalirin-7 and X11α interact and co-localize in dendrites. A, structures of X11α and kalirin-7. Bar along kalirin-7's C terminus indicates the region used as bait in a yeast two-hybrid screen; bar along X11α's sequence indicates sequence contained in a clone that interacted with kalirin-7 bait (10). MI, Munc-18-interacting region; CI, CASK-interacting region; PTB, phosphotyrosine-binding domain; PDZ, PSD-95/Discs large/ZO-1 domain; Sec14p, Sec14p-like domain; sp, spectrin-like domain; DH, Dbl-homology domain; PH, pleckstrin homology domain. B, co-immunoprecipitation (IP) of X11α with kalirin-7 from rat cortical synaptosomes (ctx-synapt). X11α is present in the input lane and in the IP lane, but absent when a GST-tagged spectrin region of kalirin-7 interferes with antigen binding. C, X11α-specific antibody detects GFP-tagged X11α, but not X11β, in HEK293 lysates expressing the indicated constructs. D, co-immunoprecipitation of X11α with Myc-tagged kalirin-7 in transiently transfected HEK293 cells is abrogated by truncation of kalirin-7's C-terminal PDZ-binding domain. E, X11α and kalirin-7 co-localize in dendrites of cultured cortical pyramidal neurons. Yellow arrows indicate puncta of co-localization, and yellow arrowheads indicate X11α puncta that do not localize with kalirin-7. Bar graph indicates quantitative measures of respective co-localization of immunofluorescent puncta.
Here, we show that X11α and kalirin-7 interact via a PDZ-mediated interaction in cortical neurons. We find that X11α localizes to dendrites and spines of cortical neurons, as well as to Golgi outposts, distal specializations of the Golgi complex in dendrites. At synapses, X11α co-localizes with several synaptic proteins; furthermore, FRAP and biochemical analysis reveal that X11α is weakly associated with the postsynaptic density (PSD) and is a mobile protein in spines. Furthermore, overexpression of X11α drives kalirin-7 accumulation in Golgi outposts, attenuates kalirin-7 GEF activity, and reduces dendritic spine size. Taken together, these data provide a novel mechanism of regulation and localization of small GTPase GEFs in dendrites by recruitment to Golgi outposts.
EXPERIMENTAL PROCEDURES
Reagents
Polyclonal antibodies recognizing kalirin or kalirin-7, and plasmids encoding myc-kalirin-7 and myc-kalirin-7-ΔCT, were described previously (10). The following antibodies were purchased: GluA1 rabbit polyclonal (Millipore); X11α mouse monoclonal (sc-136122) and TGN38 rabbit polyclonal (Santa Cruz Biotechnology); giantin rabbit polyclonal (Covance); and PSD-95 (University of California at Davis/National Institutes of Health NeuroMab Facility; clone K28/43). X11α-GFP and pRK5-X11 constructs were the generous gifts from Richard Huganir (Johns Hopkins University). Other constructs used include YFP-GalT (22) and eGFP-N2 and pmCherry-N1 (Clontech).
Cell Culture
Dissociated cultures of primary cortical neurons were prepared from E18 Sprague-Dawley rat embryos and cultured as described (23). Neurons were plated onto coverslips or 60-mm dishes, previously coated with poly-d-lysine (0.2 mg/ml, Sigma), in feeding media (Neurobasal + B27 supplement (Invitrogen) + penicillin/streptomycin + 0.5 mm l-glutamine). Neuron cultures were maintained in the presence of 200 μm dl-amino-phosphonovalerate acid (Ascent Scientific) beginning on DIV 4 to maintain neuron health for long term culturing and to reduce cell death due to excessive Ca2+ cytotoxicity via overactive NMDA receptors (23). Cortical neurons were transfected at DIV 24–28 using Lipofectamine 2000 (Invitrogen) following the manufacturer's recommendations. Transfected constructs were allowed to express for 2 days before being fixed. Human embryonic kidney 293 (HEK293) cells were cultured in DMEM with 10% FBS and penicillin/streptomycin. Cells were plated onto 6-well plates and grown until 40% confluent, when they were transfected using Lipofectamine 2000. Between 2 and 5 μg of DNA was used with 6 μl of Lipofectamine 2000 per well. Transfections proceeded for 48 h before cell lysates were harvested.
Co-immunoprecipitations
Cells (HEK293) were harvested in RIPA buffer (in mm: 150 NaCl, 10 Tris-HCl, pH 7.2, 5 EDTA, 0.1% SDS, 1% Triton X-100, 1% deoxycholate, plus inhibitors). Precleared lysates were incubated with 3–5 μl of antibody for 2–4 h; 60 μl of protein-A-Sepharose was added for 1 h at 4 °C, after which samples were washed with 1 ml of RIPA buffer. Co-immunoprecipitation from cortical neurons or synaptosomal preparations of rat cortical tissue homogenates (6) was carried out essentially as described above. Following treatments, cells were lysed in RIPA buffer and sonicated, and cell debris was removed by centrifugation. Samples were then incubated with antibodies for 3 h before the addition of 60 μl of protein-Sepharose-A, followed by 3–4 washes with 0.5 ml of RIPA buffer. All experiments were performed at least three times from independent cultures.
Rac1 Activation Assays
To examine activation of endogenous Rac1 in neurons, we used the Rac/cdc42 assay kit from Millipore. Neurons were harvested in 0.5 ml of lysis buffer on ice and sonicated. Lysates were cleared by centrifugation at 14,000 × g for 10 min, and supernatants were incubated with 10 μl of PAK-1 p21-binding domain resin for 2 h at 4 °C; positive and negative controls were incubated with 10 mm EDTA and 0.1 mm GTPγS and 0.1 mm GDP, respectively, for 15 min at 30 °C (controls were incubated with the resin for 45 min). The resin pellet was washed three times in 0.5 ml of lysis buffer, loaded on SDS-PAGE, and analyzed by Western blotting with the anti-Rac1 monoclonal antibody. Quantification of bands was performed by measuring the integrated intensity of each band and normalizing it for protein loading using ImageJ.
Immunocytochemistry
Cells were fixed in either 4% formaldehyde, 4% sucrose in PBS or at 4 °C in methanol pre-chilled to −20 °C for 20 min. Fixed neurons were permeabilized and blocked simultaneously in PBS containing 2% normal goat serum and 0.2% Triton X-100 for 1 h at room temperature. Primary antibodies were added in PBS containing 2% normal goat serum overnight at 4 °C, followed by three 10-min washes in PBS. Secondary antibodies were incubated for 1 h at room temperature, also in 2% normal goat serum in PBS. Three further washes (15 min each) were performed before coverslips were mounted using ProLong antifade reagent (Invitrogen).
Quantitative Immunofluorescence
Changes in protein clustering and synaptic localization were quantified using immunofluorescence on fixed neurons and visualized with the indicated antibodies, as described previously (23). Images were taken using a Zeiss LSM5 Pascal confocal microscope and a ×63 objective (N.A. 1.4) or using a Leica SP5 confocal microscope and a ×63 objective (N.A. 1.4). Images of GFP-expressing neurons or endogenous proteins labeled with Alexa 488 were acquired with excitation/detection at 488/505–530 nm, YFP-GalT images with 496/510–555 excitation/detection, Alexa 568 images with 568/570–630 excitation/detection, and Alexa 633 with 647/655–750 excitation/detection. Cultures that were directly compared were stained simultaneously and imaged with the same acquisition parameters. For each condition, 9–15 neurons from three to five separate experiments and 100 μm apical dendrite from each neuron were analyzed. Experiments were performed on sister cultures, and analysis was carried out under blind conditions. Maximum projection images were reconstructed using Metamorph. For fluorescence intensity measurements, the background corresponding to areas without cells were subtracted to generate a “background-subtracted” image. Images were then thresholded equally to include clusters with intensity at least 2-fold above the adjacent dendrite. Regions along dendrites were outlined using the “Perimeters” utility, and the linear density (number/100 μm dendrite length), area, and total gray value (total immunofluorescence intensity) of each AMPA receptor cluster were measured automatically. To determine the degree of co-localization between two channels, each channel was thresholded to select distinct puncta as described above. A 100-μm dendritic region was selected, and puncta counts were made; puncta smaller than 0.08 μm2 were excluded from analysis. Regions representing the measured puncta in one channel were generated using Metamorph and overlaid on the other channel. Puncta were counted as co-localized if the average intensity within the overlaid region exceeded the threshold. Thresholds were set individually for each antibody and held constant across treatment conditions. In the green/purple color scheme, co-localization is indicated in white. Images were taken in the linear range, to allow an accurate representation. Each channel is shown individually in grayscale.
Time-lapse Imaging and FRAP
Neurons on coverslips were transfected with GFP-tagged X11α and 2 days later were preincubated in artificial cerebrospinal fluid (in mm: 125 NaCl, 2.5 KCl, 26.2 NaHCO3, 1 NaH2PO4, 11 glucose, 5 HEPES, 2.5 CaCl2, and 1.25 MgCl2 with 200 μm dl-amino-phosphonovalerate acid), after which they were transferred to a stage chamber and imaged at 37 °C in artificial cerebrospinal fluid. Square regions encompassing dendritic shaft or spines were photobleached using 10 passes (12 s total) of 100% laser power, which was optimized to quench fluorescence in fixed cells. Recovery fluorescence was acquired using 1% laser power, with images taken every 6 s for 144 s. Dendrites were captured through a ×63 objective with 2× averaging. Intensity of recovered fluorescence was measured in the spine region or dendritic shaft region. Focal drift was adjusted for by measuring the intensity of nonbleached area on a different dendrite of the same cell. Healthy neurons with overall pyramidal morphologies expressing GFP-tagged protein were identified and imaged. Kymographs were created using the “polyline kymograph” plugin in ImageJ. Curved processes were straightened using the plugin; resultant kymographs show the process along the x axis and time across the y axis. Following imaging sessions, ×20 images of the neurons were taken to ascertain lack of photodamage. Any neurons exhibiting signs of distress were omitted from quantification. Between 7 and 9 cells from three independent cultures were analyzed.
Tissue Preparation and Subcellular Fractionation
Rat cortex was dissected from adult female Sprague-Dawley rats (Harlan) and homogenized in 10 volumes of 20 mm Tris-HCl, pH 7.5, buffer containing protease inhibitors, as described previously (6). Subcellular fractionations and synaptosomal preparations of rat cortical tissue homogenates were performed as described previously (6).
Statistics
Bars represent means, and error bars are standard errors. To identify differences among conditional means, statistical analyses (Student's unpaired t test, analysis of variance) were performed in GraphPad Prism 5. Tukey-b post hoc analysis was used for multiple comparisons.
RESULTS
X11α and Kalirin-7 Interact and Co-localize in Neurons
In a previous study, the C terminus of kalirin-7 was shown to interact with a fragment containing the two PDZ domains of X11α in a yeast two-hybrid screen (Fig. 1A) (10); however, this interaction has not been shown in neurons. To validate this interaction, we performed co-immunoprecipitation experiments from rat brain and transfected heterologous cells. We first used an antibody recognizing the spectrin region of kalirin (kal-spect) to immunoprecipitate kalirin proteins from rat cerebral cortex synaptosomal preparations (Fig. 1B), and we probed them using an antibody that specifically detected X11α (Fig. 1C). In these experiments, X11α co-immunoprecipitated with kalirin-7, indicating that these two proteins form a protein complex. Notably, X11α and kalirin-7 did not co-precipitate in the presence of kalirin-7 preimmune serum or when the kal-spec antibody was preadsorbed to its antigen (GST-spectrin domains) (Fig. 1B). To determine whether this interaction was dependent on the C terminus of kalirin-7, we performed co-immunoprecipitation experiments from HEK293 cells transfected with Myc-tagged kalirin-7 or its C-terminally truncated mutant that lacks the C-terminal PDZ-binding domain (kal7-ΔCT). Consistent with our data from rat cerebral cortex, X11α co-immunoprecipitated with kalirin-7 (Fig. 1D). However, X11α did not co-immunoprecipitate with myc-kalirin-7-ΔCT, indicating that the C-terminal portion of kalirin-7 was critical for its interaction with X11α (Fig. 1D). These data show that kalirin-7 interacts with X11α in the cerebral cortex, and this is likely mediated by the interaction of the kalirin-7 C-terminal tail with the PDZ domains of X11α.
As kalirin-7 and X11α co-immunoprecipitated together, we sought to determine whether these proteins co-localized along dendrites and dendritic spines of cortical neurons. Endogenous kalirin-7 and X11α were detected by immunofluorescence using specific antibodies. As described previously, kalirin-7 was present along dendrites and at synapses at 24 days in vitro (DIV) (Fig. 1E) (7). Consistent with previous suggestions (11, 12), X11α was also present along dendrites and at a subset of spine-like structures (Fig. 1E). Moreover, X11α and kalirin-7 were found to co-localize along the dendrite and in a subset of spine-like structures along dendrites of cortical pyramidal neurons (Fig. 1E). Quantification of co-localization revealed that ∼70% of X11α was positive for kalirin-7, whereas only 50% of kalirin-7 puncta was positive for X11α (Fig. 1E). Taken together, these data indicate that kalirin-7 interacts with X11α in the cerebral cortex, via its C-terminal tail. Furthermore, X11α and kalirin-7 co-localize in a subset of punctate structures along the dendrites of cultured cortical pyramidal neurons.
X11α Is Present in Excitatory Synapses and Spines
Although X11α has been suggested to be present along dendrites and at the PSD (11, 12, 14), a detailed analysis of X11α's subcellular distribution has not been performed. Interestingly, a previous study using a pan-X11 antibody, which detects both α and β isoforms, observed X11/mLIN-10 within synaptosomes prepared from cultured hippocampal neurons (15). This suggests that X11α is enriched in synapses, consistent with our observations that X11α co-localizes with kalirin-7. Thus, we investigated the localization of X11α in dendrites of mature cultured cortical pyramidal neurons (DIV 24) using immunofluorescence microscopy and an X11α-specific antibody (Fig. 1C). X11α localized to punctate structures along dendrites and in dendritic shafts where it partially co-localized with the postsynaptic protein PSD-95, a marker for excitatory synapses (Fig. 2A). Moreover, X11α also partially co-localized with the AMPA receptor subunit GluA1 (Fig. 2B). Quantification of co-localization revealed that approximately 55% of X11α puncta was positive for PSD and 50% of PSD-95 puncta was positive for X11α. Conversely, ∼60% of X11α puncta was positive for GluA1, and ∼65% of GluA1 puncta was positive for X11α (Fig. 2C). These data indicate that a subpopulation of X11α is enriched at the postsynaptic density of excitatory synapses.
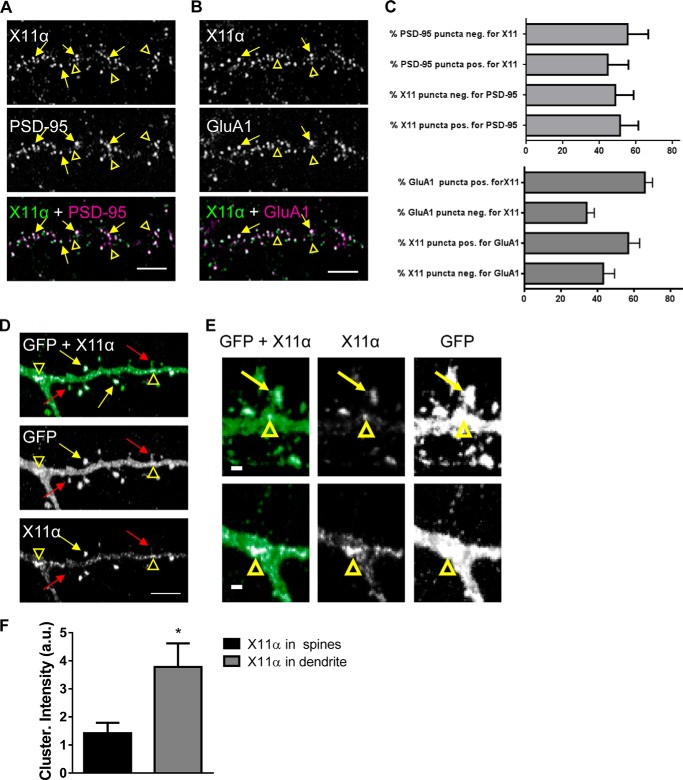
FIGURE 2.
X11α is enriched in cortical synapses and localizes to dendritic spines. A, X11α partially co-localizes with PSD-95 in cultured rat cortical pyramidal neurons (DIV 24). Yellow arrows indicate sites of co-localization, and yellow arrowheads indicate X11α puncta that do not localize with PSD-95 (overlay of green and magenta = white). B, X11α partially co-localizes with AMPA receptor subunit GluA1 in cultured rat cortical pyramidal neurons. Yellow arrows indicate sites of co-localization, and yellow arrowheads indicate X11α puncta that do not localize with GluA1. C, bar graphs are quantitative measures of respective co-localization of immunofluorescent puncta (PSD-95 and X11α; GluA1 and X11α). D and E, representative confocal images of cortical neurons (DIV 24) expressing GFP and pRK5-X11α. Overexpressed pRK5-X11α localizes to dendritic spine heads (yellow arrows) and dendritic shaft (yellow arrowheads). Red arrows indicate dendritic spines containing little or no X11α signal (D). X11α accumulated at the base of spines and at dendritic branching points (yellow arrowheads) (E). F, mean fluorescence intensity of X11α signal in dendritic spines is increased compared with X11α signal in dendritic shaft (*, p < 0.05, Student's unpaired t test). Scale bars, 5 μm (A, B, and D) and 1 μm (E).
To further characterize the localization of X11α at synapses, we immunostained GFP-expressing neurons for the endogenous protein (Fig. 2D). Consistent with its co-localization with PSD-95 and GluA1, X11α was observed in a subset of dendritic spines (Fig. 2, D and E, yellow arrows). In addition, X11α could also be seen in large clusters along the dendrites (Fig. 2D, open arrowheads); X11α clusters were especially prominent at the dendrite branch points and were observed at the base of spines (Fig. 2E). To analyze the distribution of X11α further, we examined X11α clusters within spines or along the dendritic shaft. X11α clusters along the dendritic shaft exhibited increased fluorescence intensity compared with clusters within dendritic spines (Fig. 2F). Collectively, these data demonstrate that X11α is present at a subset of excitatory synapses, within dendritic spines and along the dendritic shaft of cortical neurons.
X11α Is a Mobile Protein in Dendrites and at Synapses
Based on our localization data, X11α localizes to spines and dendritic shaft. To examine the possibility that X11α is a mobile protein, we examined the mobility and persistence of X11α at synapses and within the dendritic shaft in live cells. We monitored GFP-tagged X11α in live mature cortical neurons (DIV 24) also expressing mCherry, allowing visualization of neuronal morphology. Similar to the endogenous protein, GFP-X11α localized to both dendrites and dendritic spines (Fig. 3A). To monitor the behavior of GFP-X11α puncta, we used kymograph analysis to plot to the fluorescence intensity of X11α puncta in the dendrite. Within this region, motile and stationary X11α puncta could be visualized (Fig. 3, B and C); stationary puncta were seen as vertical bands, and mobile puncta are indicated by diagonal bands within the kymograph. The percentage of X11α puncta that were moving, assessed over a period of 3 min, was ∼45% (data not shown), indicating that a subpopulation of X11α is a mobile protein within the shaft (Fig. 3C). Interestingly, kymograph analysis of X11α puncta within dendritic spines also demonstrated that GFP-X11α was being trafficked in spines (Fig. 3D). Occasionally, GFP-X11α puncta could be observed moving to or from the dendrite into a spine (Fig. 3D), strongly suggesting that X11α can traffic to and from synapses.
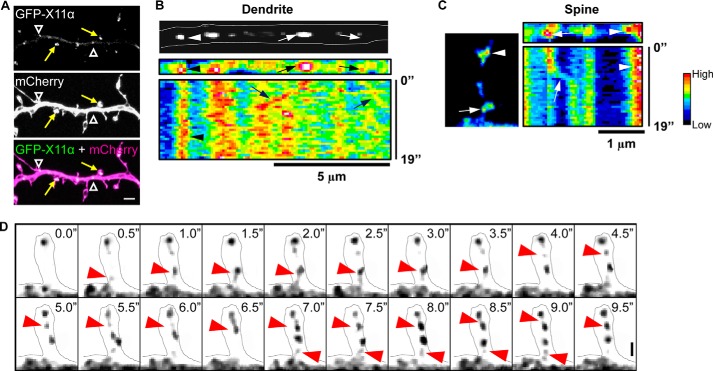
FIGURE 3.
Trafficking of X11α in cortical neurons. A, GFP-tagged X11α localizes to dendrites (white arrowheads) and dendritic spines (yellow arrows) in mCherry-expressing cortical neurons. B, top, image of a segment of dendrite showing mobile (arrows) and stationary (arrowheads) GFP-X11α puncta. Bottom, kymograph (pseudocolored) of GFP-X11α in dendrite segment. Vertical bands of high fluorescence intensity indicate stable fluorescent puncta (arrowheads), although diagonal bands (arrows) indicate mobile puncta. Multiple X11α puncta displayed movement within the dendritic shaft. C, representative kymograph analysis (19 s) of GFP-X11α puncta within the dendritic spine (left). Both mobile (arrows) and stationary (arrowhead) GFP-X11α puncta can be seen in spines. D, time-lapse imaging of GFP-X11α immunofluorescence in dendritic spine (outlined). Red arrows indicate the movement of X11α puncta into and out of a spine during a 10-s imaging session. Scale bars, 5 μm (A), 5 μm (B), and 1 μm (C and D).
To quantitatively examine the mobility of X11α puncta in the dendritic shaft and within spines, we performed FRAP experiments. A lack of fluorescence recovery in FRAP experiments indicated an immobile protein, although a recovery of fluorescence indicated the exchange of the bleached molecule in the spine or dendrite with unbleached molecules from other pools of protein. Cultured cortical neurons (DIV 24) were transfected with GFP-tagged X11α and imaged in artificial cerebrospinal fluid, and regions encompassing individual spines or dendritic shaft were photobleached (Fig. 4, A and B). Intensity of recovered fluorescence was measured in the spine region or dendritic shaft region. We found that GFP-X11α fluorescence signal was effectively depleted by bleaching and partially recovered after photobleaching in both dendritic shaft and spine, as would be expected for a mobile protein (Fig. 4, A–C). Indeed, over 60% of GFP-X11α recovered within 150 s, indicating that a large portion of X11α is mobile. Interestingly, the absolute mobile fraction was similar in the spine and dendritic shaft (% mobile fraction: spine, 62.7 ± 8.2; shaft, 60.4 ± 7.0; p = 0.84; Fig. 4D). However, the fluorescence recovery pattern was not the same; the recovery of GFP-X11α fluorescence within the dendritic shaft occurred more rapidly within the first 30 s as compared with the recovery within spines (Fig. 4C). Consistent with this, the time at which 50% of equilibrium fluorescence level is recovered (τ½) from GFP-X11α in the dendritic shaft was significantly lower than in the dendritic spines (τ½ (seconds): spine, 67.3 ± 8.2; shaft, 29.4 ± 5.2; p < 0.001; Fig. 4E). This difference in mobility dynamics for X11α in each compartment suggests that X11α turnover in spines is slower than in the dendritic shaft, potentially due to increased stability of X11α within spines. Overall, these data demonstrate that X11α is a mobile protein, with a high degree of mobility within both spines and shaft, but it is partly stabilized in spines as compared with the shaft.
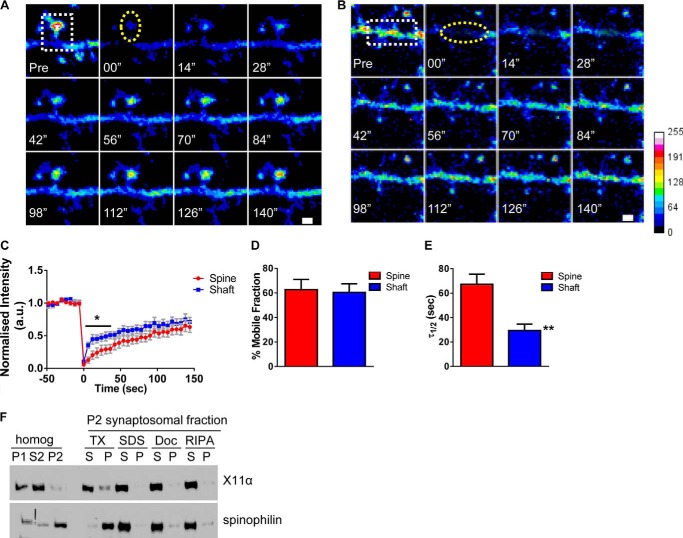
FIGURE 4.
X11α is a mobile protein in dendrites and synapses. A and B, time-lapse FRAP imaging of GFP-X11α in dendritic spine (A) and in dendrites (B). Square regions encompassing spines or dendritic shaft were photobleached (white dotted square) using 10 passes (12 s total) of 100% laser power, which was optimized to quench fluorescence in fixed cells. Recovery fluorescence was acquired using 1% laser power, with images taken every 6 s for 144 s. Intensity of recovered fluorescence was measured in spine region or dendritic shaft region (yellow dashed ovals). Focal drift was adjusted for by measuring intensity of nonbleached area on a different dendrite of the same cell. C, normalized intensity of GFP-X11α signal during fluorescence recovery after photobleaching. D, percentage of GFP-X11α that is mobile in spines and dendritic shaft, as measured by percentage of fluorescence that recovers after photobleaching. E, τ½ (time at which 50% of equilibrium fluorescence level is recovered) for regions of interest in spines and dendritic shafts. τ½ of GFP-X11α in dendritic shaft is significantly shorter than dendritic spines. F, relative abundance of X11α and spinophilin in rat cortex PSD fractions. Synaptosomes were prepared from rat cortical tissue using a HEPES/sucrose gradient, separated by SDS-PAGE, and analyzed by Western blotting using X11α (upper) and spinophilin (lower) antibodies. Fractions indicated are as follows: P1, nuclear pellet; S2, supernatant from crude synaptosomal fraction; P2, crude synaptosomal pellet. The P2 fraction was further solubilized in buffers with detergents of different strengths, and soluble (S) or insoluble and particulate (P) fractions were separated by centrifugation. Detergents used were as follows: TX, Triton X-100; SDS, sodium dodecyl sulfate; Doc, deoxycholate. X11α was present in P1, S2, and P2 fractions and was extracted from synaptosome fractions by all detergents tested, suggesting a weaker association with the PSD. **, p < 0.001. Scale bar, 1 μm.
As our FRAP data demonstrate that X11α is a mobile protein at synapses, we examined the strength of association of X11α with the PSD using subcellular fractionation and detergent solubilization experiments. We first prepared synaptosomes using a HEPES/sucrose gradient and PSD fractions by solubilization of synaptosomes using Triton X-100, SDS, or deoxycholate from rat cortical tissue. X11α was present in P1, S2, and P2 homogenates indicating that this protein is abundant in synaptosomes (Fig. 4F), similar to other proteins, including kalirin-7, localized to spines (6, 24). Interestingly, X11α could be extracted by all detergents tested, even weaker detergents like Triton X-100 or deoxycholate (Fig. 4F). In comparison, spinophilin, another PDZ domain-containing protein that is tightly associated with the PSD, was not readily extracted by Triton X-100 (Fig. 4F), consistent with previous reports (25). Thus, based on our biochemical data, X11α appears to associate weakly with the PSD, which would be consistent with a mobile protein capable of trafficking to and from synapses.
X11α Localizes to Golgi Outposts in Dendrites
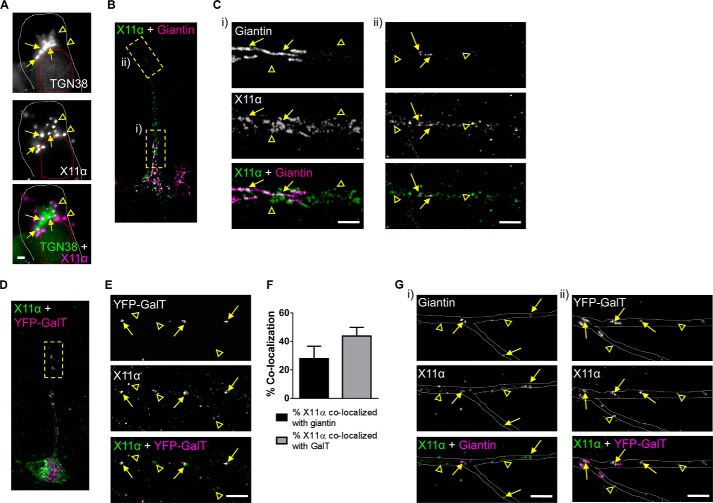
It is now generally accepted that many neurons contain both somatic and discrete discontinuous Golgi complexes (also known as Golgi outposts) located in dendrites. These satellite Golgi complexes are thought to play an important role in controlling the synthesis, sorting, processing, and trafficking of synaptic proteins (26, 27). In neuronal cells, both X11α and -β have been suggested to associate with the Golgi complex (12, 15), although whether X11α is present in Golgi outposts along dendrites is not known. We thus examined in detail the localization of X11α to Golgi complexes and related structures. In agreement with previous studies (15), X11α localized in the vicinity of the Golgi complex (Fig. 5A), as indicated by proximity to TGN38, a marker of trans-Golgi complexes. To further investigate the relationship between X11α and the Golgi network throughout the neuron, we examined the co-localization of X11α with giantin, a marker for cis-Golgi and medial Golgi complexes located in the soma and in dendrites (28). Consistent with co-localization with TGN38, X11α immunoreactive puncta partially overlapped with giantin in the soma (Fig. 5, B and C, panel i). In the soma and proximal regions of the primary dendrite, giantin was enriched in tubulovesicular structures in the soma and proximal regions of the primary dendrite (Fig. 5C, arrows), and X11α was more abundant in vesicular structures (Fig. 5C, panel i, open arrowheads). Consistent with previous studies (28), giantin was present in vesicular structures in distal portions of the dendrite, consistent with the presence of Golgi outposts (Fig. 5, B and C, panel ii) (26, 29). Remarkably, a subset of X11α puncta co-localized with giantin in these structures along dendrites (Fig. 5, B and C, panel ii, yellow arrows). Recently, it has been suggested that the medial, cis-, and trans-compartments of Golgi complexes are disconnected the dendrites of Drosophila neurons (30). Therefore, to assess whether X11α preferentially localized to different Golgi compartments, we next examined the localization of X11α with the trans-Golgi compartment. Thus, we labeled Golgi outposts with fusion proteins consisting of the N-terminal 81 amino acids of the human β-1,4-galactosyltransferase and YFP (YFP-GalT) (31). Once more, we found that X11α and YFP-GalT co-localized in punctate structures in distal dendrites (Fig. 5, D and E, arrows). Interestingly, only 27.9 ± 8.74% of X11α co-localizes with giantin and 43.8 ± 6.11% with YFP-GalT (Fig. 5F), suggesting that is predominantly located in trans-Golgi compartments. Interestingly, X11α was found to co-localize with both giantin and YFP-GalT at dendritic branch points (Fig. 5G), a region where Golgi complexes consisting of medial, cis-, and trans-Golgi compartments (30). Taken together, these data indicate that X11α is preferentially associated with a vesicle pool in proximity of the Golgi apparatus, as well as with dendritic Golgi outposts.
FIGURE 5.
X11α localizes to Golgi outposts in dendrites. A, relative co-localization of X11α with TGN38, a marker of the trans-Golgi network, in the soma of rat cultured cortical neurons (DIV 24). Arrows indicate co-localization, and arrowheads indicate X11α puncta that do not co-localize with TGN38. White dashed line denotes cell soma; red dashed line denotes cell nucleus. B, relative localization of X11α with giantin, a general marker for the Golgi complex. Dashed boxes indicate regions magnified in C. C, insets from B from proximal (panel i) and distal (panel ii) regions of the apical dendrite. Arrows indicate co-localization, and arrowheads indicate X11α puncta that do not co-localize with giantin. D, relative localization of endogenous X11α with overexpressed YFP-GalT in Golgi outposts in dendritic shaft. Dashed box indicates dendrite region magnified in E. E, magnified dendrite region from D. Arrows indicate co-localized puncta between YFP-GalT and X11α (overlay in white). Arrowheads indicate X11α puncta that do not co-localize with YFP-GalT. F, quantification of X11α co-localization with either giantin or YFP-GalT in dendrites. G, co-localization of X11α with giantin and YFP-GalT at dendrite branch points. Gray dashed lines outline the dendritic shaft. Scale bars, 1 μm (A); 5 μm (C, E, and G).
X11α Modulates Kalirin-7 Localization in Dendrites and Recruits It to Golgi Outposts
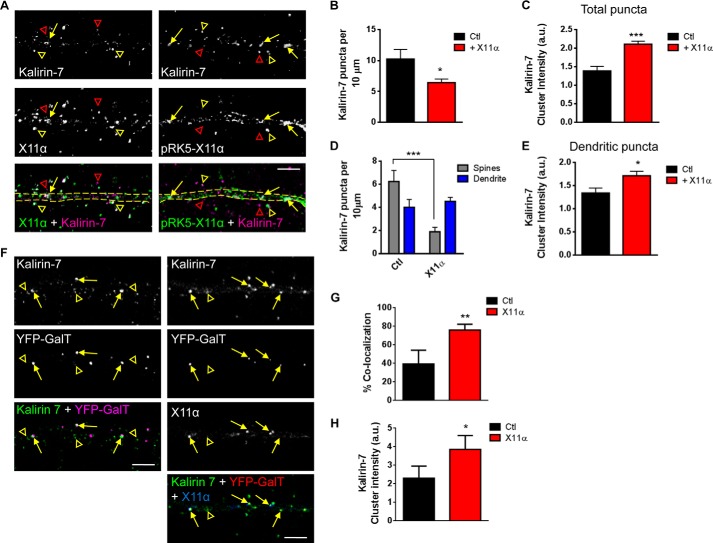
Because X11α appeared to be a highly motile dendritic protein, we hypothesized that it may mediate the rapid translocation of specific proteins between the spines and shaft. Interestingly, m-Lin-10/X11 and X11β proteins interact with and regulate the trafficking of both NMDA and AMPA receptors (15–17), but whether X11α also regulates the trafficking of synaptic proteins is unknown. Thus, we reasoned that X11α may be well suited to regulate the trafficking of interacting partners such as kalirin-7 within dendrites. We first examined whether X11α modulated the synaptic localization of kalirin-7. Similar to the endogenous proteins, kalirin-7 co-localized with overexpressed X11α (pRK5-X11α) within the dendritic shaft (yellow arrows) and in spine-like structures adjacent to the dendrite (yellow arrowheads) (Fig. 6A). When we examined the distribution of kalirin-7, we found that overexpression of X11α resulted in a reduction in linear density of kalirin-7 puncta (kalirin-7 linear density (per 10 μm): Ctl, 10.2 ± 1.5; pRK5-X11α, 6.4 ± 0.59; p < 0.05; Fig. 6B). Interestingly, the average fluorescence intensity of kalirin-7 puncta was significantly increased in the presence of exogenous X11α (kalirin-7 cluster intensity (a.u.): Ctl, 1.4 ± 0.12; pRK5-X11α, 2.1 ± 0.08; p < 0.001; Fig. 6C), suggesting that this loss of kalirin-7 linear density could be due to the accumulation of the protein into distinct pools. To determine whether overexpression of X11α could alter the distribution of kalirin-7 in the dendrite shaft and spines, we measured kalirin-7 puncta in the dendrite and spines. This revealed that in the presence of exogenous X11α, kalirin-7 levels were reduced in spines, whereas the kalirin-7 linear density in dendrites remained unchanged (kalirin-7 linear density (per 10 μm): Ctl spines, 6.2 ± 0.95 versus pRK5-X11α spines, 1.9 ± 0.38; Ctl dendrite, 4.0 ± 0.67 versus pRK5-X11α dendrite, 4.5 ± 0.34; p < 0.001, Fig. 6D). In addition, kalirin-7 dendritic puncta were larger when X11α was overexpressed (kalirin-7 cluster intensity (a.u.): Ctl dendrite, 1.3 ± 0.11 versus pRK5-X11α dendrite, 1.7 ± 0.09; p < 0.05, Fig. 6E), further indicating that synaptic kalirin-7 was being redistributed into distinct dendritic pools.
FIGURE 6.
X11α modulates kalirin-7 localization in dendrites and recruitment to Golgi outposts. A, representative confocal images of kalirin-7 with endogenous X11α (left panel) or exogenous X11α (pRK5-X11α; right panel). Yellow arrows and open arrowheads indicate kalirin-7 and X11α co-localized puncta in dendritic shaft and dendritic spines, respectively; open red arrowheads indicate kalirin-7 puncta not localized with X11α. Dashed yellow lines indicate dendritic shaft. B and C, overexpression of X11α induced the clustering of kalirin-7 in dendrites of cortical pyramidal neurons. Kalirin-7 puncta linear density is significantly reduced in neurons overexpressing X11α compared with control cells (B). Furthermore, exogenous expression of X11α induces an overall increase in kalirin-7 puncta clustering (C). D, comparison of kalirin-7 linear density in dendritic shaft (dendrites) and dendritic spines (spines) reveals that neurons expressing pRK5-X11α have decreased kalirin-7 puncta in dendritic spines, but no change in puncta linear density in dendrites. E, quantification of dendritic kalirin-7 cluster intensity in the presence or absence of exogenous X11α; X11α overexpression promotes the formation of larger kalirin-7 dendritic puncta. F, representative confocal images of kalirin-7 (stained with Alexa 568) localization to Golgi outposts (marked by YFP-GalT) in neurons overexpressing, or not, pRK5-X11α (stained with Alexa 633). Yellow arrows indicate co-localization of kalirin-7 with YFP-GalT, and yellow open arrowheads indicate kalirin-7 not localized to Golgi outposts. F and G, quantification of co-localization between kalirin-7 and YFP-GalT (F) and mean cluster intensity of kalirin-7 signal in YFP-GalT puncta (G). *, p < 0.05; **, p < 0.01; ***, p < 0.001. Scale bars, 5 μm.
Because in the presence of overexpressed X11α kalirin-7 accumulated in larger structures in the dendritic shaft, we hypothesized that X11α might localize kalirin-7 to Golgi outposts. Thus, we examined the effect of X11α overexpression on kalirin-7 localization in Golgi outposts. Under basal (Ctl) conditions, kalirin-7 only partially co-localized with YFP-GalT (Fig. 6F, left panels, yellow arrows), indicating that only a small amount of kalirin-7 resides within these secretory structures. However, in the presence of exogenous X11α, a greater number of YFP-GalT puncta were positive for kalirin-7 (% co-localization: Ctl, 39.4 ± 8.6; pRK5-X11α, 76.0 ± 2.8; p < 0.01; Fig. 6, F and G). Furthermore, kalirin-7 puncta were larger in Golgi outposts in neurons expressing pRK5-X11α (kalirin-7 cluster intensity in Golgi outposts: Ctl, 2.3 ± 0.37; pRK5-X11α, 3.9 ± 0.34; p < 0.05; Fig. 6G), suggesting that X11α overexpression enhanced the localization of endogenous kalirin-7 to Golgi outposts in primary dendrites. Taken together, these data suggest X11α modulates the synaptic localization of kalirin-7 by recruiting kalirin-7 to Golgi outposts.
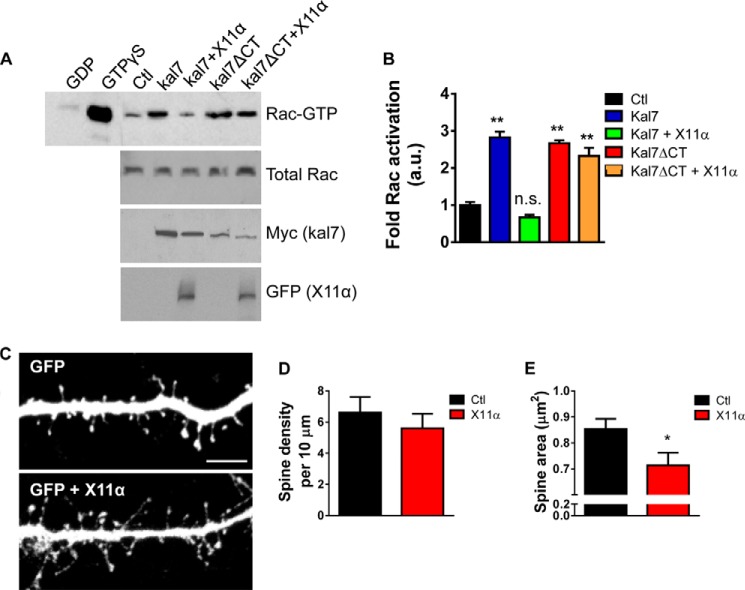
X11α Reduces Kalirin-7-dependent Rac1 Activation and Alters Dendritic Spine Morphology
We hypothesized that interaction of kalirin-7 with X11α might modulate kalirin-7's function, in addition to its localization. As seen previously (7), overexpression of kalirin-7 increased active Rac levels in HEK293 cells (Fig. 7A). When both proteins were co-expressed in HEK293 cells, we found that X11α significantly reduced kalirin-7-dependent Rac1 activation (Fig. 7A). This inhibition of kalirin-7-dependent Rac activation by X11α required the C terminus of kalirin-7, as it did not occur in the absence of kalirin-7's C-terminal PDZ-binding motif (Fig. 7B). Previously, we have shown that disrupting kalirin-7's PDZ binding domain with an interfering peptide results in the loss of synaptic kalirin-7, reducing its GEF activity toward Rac and resulting in the emergence of spines with a thin morphology (7). Therefore, we reasoned that the X11α-dependent recruitment of kalirin-7 to Golgi outposts and the reduction of its GEF activity would impact dendritic spine morphology. We imaged cortical neurons (DIV 24) fixed after overexpression of GFP alone or GFP and pRK5-X11α, and we quantified spine morphology parameters (Fig. 7, C–E). Exogenous X11α did not alter the dendritic spine linear density (spine linear density (per 10 μm): Ctl, 6.6 ± 0.99; pRK5-X11α, 5.6 ± 0.92; p = 0.47; Fig. 7D). However, dendritic spine area was significantly reduced by exogenous expression of X11α (spine area (μm2): Ctl, 0.85 ± 0.039; pRK5-X11α, 0.71 ± 0.048; p < 0.05; Fig. 7E). Taken together, these data suggest that in addition to recruiting kalirin-7 to Golgi outposts, overexpression of X11α inhibits kalirin-7 GEF function at the synapse, thereby inducing a reduction of dendritic spine size.
FIGURE 7.
X11α overexpression reduces dendritic spine size in cultured cortical neurons. A, co-expression of X11α reduces kalirin-7-dependent Rac1 activation in HEK293 cells. HEK293 cells were transfected with plasmids expressing kalirin-7 kalirin-7-ΔCT, and X11α alone or together; Rac1-GTP was measured using an affinity binding assay. Expression of kalirin-7 or kalirin-7-ΔCT alone resulted in an increase in active Rac1 levels. However, in the presence of X11α, kalirin-7 was no longer able to increase Rac1-GTP levels. Inhibition of kalirin-7-dependent Rac activation by X11α requires the C terminus of kalirin-7 as kalirin-7-ΔCT-induced Rac1 activation was not altered by expression of X11α. B, quantification of Rac1-GTP levels from A. C, representative confocal image of cortical neuron (DIV 24) co-expressing GFP with or without pRK5-X11α. X11α overexpression reduces dendritic spine size in cortical neurons. D and E, quantification of dendritic spine linear density (D) and mean dendritic spine area (E) in neurons expressing GFP or GFP + X11α. *, p < 0.05. Scale bar, 5 μm.
DISCUSSION
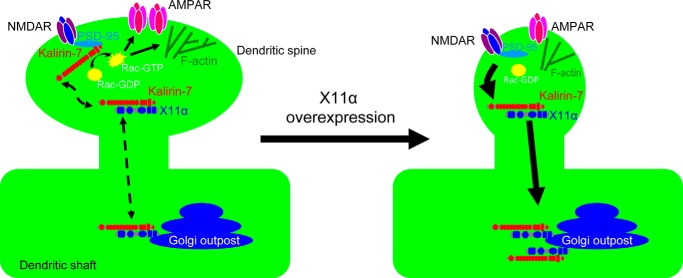
Based on our data, we propose the following model of X11α/kalirin-7 interaction (Fig. 8). Kalirin-7 is targeted to dendritic spines through its interactions with several PDZ domain-containing proteins, including PSD-95 and afadin. In dendritic spines, kalirin-7 enhances Rac1 activation, leading to actin cytoskeletal rearrangements, spine growth, and recruitment of AMPA receptors. In addition to PSD-95, kalirin-7 also interacts with X11α in spines. This interaction with X11α may inhibit kalirin-7's Rac1-GEF activity, similarly to what has been observed with PSD-95 (10). At the same time, X11α promotes the removal of kalirin-7 from spines and the sequestration to Golgi outposts present in the dendrite. This leads to a reduction of kalirin-7 activity in spines and its enrichment in Golgi outposts, where it could potentially participate in compartment-specific signaling pathways.
FIGURE 8.
Model of X11α/kalirin-7 interaction. Kalirin-7 enhances Rac1 activation in dendritic spines, leading to actin cytoskeletal rearrangements, spine growth, and recruitment of AMPA receptors. X11α binding to kalirin-7 inhibits its GEF activity, recruits it to Golgi outposts in the dendrite, and induces spine shrinkage.
We have previously shown that X11α interacts with the C terminus of kalirin-7 in a yeast two-hybrid assay, but whether this occurred in physiological preparations was not clear. Here, we show that X11α and kalirin-7 interact in native brain tissue and in cortical neurons and,, moreover, that this interaction occurs in a PDZ-dependent manner. Kalirin-7 interacts with a number of distinct classes of PDZ domain-containing proteins, and many such interactions appear to fulfill different functions and mediate the association of kalirin-7 with different protein complexes (32). For example, PSD-95 mediates the association of kalirin-7 with glutamate receptors, ErbB4, and potentially serotonin receptors (7, 8, 10, 33). Afadin mediates the association of kalirin-7 with N-cadherin complexes and EphB receptors (5, 34). Interestingly, several of the proteins found to interact with the C terminus of kalirin-7 in a yeast two-hybrid screen, including PSD-95 and afadin, are involved in membrane trafficking, suggesting roles for kalirin signaling in trafficking (10).
X11α and its homolog X11β are mLin-10 family members encoded by different genes, both present in neurons. Although these proteins play important roles in axons and the presynaptic terminal (35, 36), several laboratories have investigated their dendritic roles. Stricker and Huganir (15) have examined the functions of X11/mLin-10 in neuronal dendrites and synapses, with special emphasis on X11β. They have shown that X11β is highly enriched in the trans-Golgi complex of neurons and is present in punctate structures in dendrites. However, X11β was only occasionally present in spines and synapses. Our data indicate that X11α is enriched in dendrites and spines, as well as Golgi outposts and the Golgi complex. Interestingly, Stricker and Huganir (15) show that X11β associates with GluA1 and GluA2 subunits of the AMPA receptors, and it affects GluA1 surface expression. In addition, Setou et al. (16) have shown that mLin-10 interacts with KIF17 to mediate the trafficking of the NR2B subunit of NMDA receptors in dendrites. Here, we show that in addition to structures in dendrites, X11α is present in synapses, and it co-localizes with the postsynaptic and excitatory synapse proteins, PSD-95, GluA1, and kalirin-7.
Our live-cell imaging data also demonstrate that X11α is a moderately mobile protein. Comparisons of the half-recovery time (τ½) for GFP-X11α with that previously published for SAP102 and PSD-95 (37) reveal that X11α is less mobile than these synaptic proteins in spines: τ½/2 for both SAP102 and PSD-95 was around 45 s (37), although our data indicate that X11α has a τ½of ∼67 s. Importantly, both SAP102 and PSD-95 are considered to be highly mobile proteins at synapses (37); therefore, our data suggest that X11α is a moderately mobile protein in the spine. Moreover, the increased stability of X11α in comparison with SAP102 or PSD-95 further supports the suggestion that X11α weakly or transiently interacts with other proteins in the PSD, such as kalirin-7. Interestingly, GFP-X11α displayed differential mobility dynamics in dendritic spines versus the dendritic shaft. This difference in mobility dynamics is likely due to an increased residency of X11α within spines. However, differences in mobility may also be due to the mechanism(s) that regulate X11α trafficking within these two cellular compartments. Moreover, the diffusion coefficient of GFP-X11α in both compartments was not significantly different (diffusion coefficient (μm2/s)): spine, 0.0025 ± 0.00057, and shaft, 0.0024 ± 0.00066; p = 0.96, graph not shown), suggesting that the observed difference in mobility was not due to a differential ability of GFP-X11α to diffuse into an area of similar size in either structure. This further supports the idea that the difference in mobility dynamics of this protein in each compartment is due to either an increase in stability of X11α in spines or differential trafficking mechanisms. However, we need to note that the spine neck may also play a part in increasing the way that X11α is trafficked, as well as its residency within the spine. Indeed, to assess how membrane topology influences the mobility and diffusion of X11α, it would be necessary to take into account the morphology of the subcellular compartment being investigated (38). Nevertheless, these data suggest that X11α may fulfill multiple functions in these locations, likely modulating the localization and function of dendritic and synaptic proteins. We also report that X11α is present at synapses where it weakly associates with the PSD. Furthermore, the weak presence of X11α in detergent fractions is consistent with the subcellular localization of kalirin-7 (6, 10), strengthening the suggestion that these proteins are capable of interacting in vivo.
An interesting observation in this study was that X11α associates with Golgi outposts in dendrites and modulates the localization of kalirin-7 to these structures. Although the Golgi apparatus localizes in a perinuclear region of the neuronal soma, cellular compartments called “Golgi outposts” have also been identified in dendrites (28, 31). Golgi outposts are often localized near synapses or at dendritic branch points (29). Recently, it has been shown that not all Golgi outposts contain the biochemcially distinct medial, cis-, and trans-Golgi compartments (30). Indeed, although we observed co-localization of X11α with giantin, a resident protein found in the medial and cis-Golgi compartment protein, a larger portion of the protein was found to co-localize with the trans-Golgi compartment marker, YFP-GalT. Importantly, it should be noted that it is not possible to determine whether X11α's co-localization with giantin or YFP-GlaT indicates that it present in only single compartment Golgi or whether a subset are multicompartment Golgi. Moreover, the implications of X11α's apparent predominance in trans-Golgi compartment is currently unknown, as the functional significance of single or multicompartment Golgi in dendrites is currently unknown (30). However, X11α is present at branch points where it co-localizes with both giantin or YFP-GalT, indicating that X11α is present in Golgi complexes consisting of medial, cis-, and trans-Golgi compartments. Golgi outposts play central roles in local post-endoplasmic reticulum trafficking of proteins used in dendrite growth and synaptic plasticity (27). These structures may function as a passive site for docking inactivated kalirin-7, to be released when needed. However, kalirin-7 may fulfill specific functions in Golgi outposts, possibly associated with protein trafficking, as kalirin has been linked to the trafficking of transmembrane proteins, such as peptidylglycine α-amidating monooxygenase (39). Future studies will need to explore the functional impact of the targeting of kalirin-7 and X11α to Golgi outposts. One potential role may be in actin remodeling in Golgi outposts. Actin remodeling by small GTPases plays an important role in the morphology and function of the Golgi apparatus (27), and kalirin-7 may play such a regulatory role in Golgi outposts.
We have previously reported that disrupting the interaction of PSD-95 with kalirin-7 removes it from synapses and inhibits its GEF activity (7, 10). Our data suggest hat X11α is capable of modulating kalirin-7's presence at synapses by recruiting it to Golgi outposts, and overexpression of X11α results in a reduction in dendritic spine size, a consequence of inhibiting kalirin's GEF activity and removing it from synapses (7, 10). Therefore, it possible that by recruiting kalirin-7 to Golgi outposts, one outcome would be to reduce its GEF activity, thus resulting in a reduction in spine size. Such inhibitory interactions might protect spines from overactivation of Rac1, which has detrimental effects. Inhibition of kalirin-7 GEF activity by another protein, DISC1, has been shown to protect spines from the detrimental effects of Rac1 overactivation that might lead to disease (40). Interestingly, X11α modulation of kalirin-7 localization opposes that of PSD-95; PSD-95 recruits kalirin-7 to spines, whereas X11α seems to remove kalirin-7 from spines. However, both proteins may reduce kalirin-7's GEF activity (10). Thus, X11α-mediated regulation of kalirin-7 localization thus might be a more general mechanism of repressing kalirin-7 GEF activity in spines.
Proper regulation of dendritic spine morphology is a critical component of healthy brain function. Conversely, disruptions in the mechanisms and proteins that control spine morphology have consistently been associated with neurodevelopmental, psychiatric, and neurodegenerative diseases (2). Abnormal kalirin-7 signaling has also been linked with several disorders, including schizophrenia, Alzheimer disease, and Alzheimer disease with psychosis (32). X11/mLIN-10 proteins have been implicated in amyloid precursor protein processing (41) and apoE receptor trafficking (42), and their deletion decreases amyloid production in Alzheimer mouse models (41). Future investigations of kalirin-7/X11α interaction might provide insight into the pathogenesis of neurological disorders.
This work was supported, in whole or in part, by National Institutes of Health Grants MH071316 and MH097216 from NIMH (to P. P.) and NRSA Ruth L. Kirschstein Award F31MH085362 (to K. A. J.). This work was also supported by grants from the Royal Society United Kingdom, Brain and Behavior Foundation (formally National Alliance for Research on Schizophrenia and Depression (NARSAD)), Psychiatric Research Trust, American Heart Association (to D. P. S.), and National Alliance for Research on Schizophrenia and Depression, Brain Research Foundation.
- GEF
- guanine nucleotide-exchange factor
- DIV
- days in vitro
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- FRAP
- fluorescence recovery after photobleaching
- PSD
- postsynaptic density
- Ctl
- control.
REFERENCES
- 1. Holtmaat A., Svoboda K. (2009) Experience-dependent structural synaptic plasticity in the mammalian brain. Nat. Rev. Neurosci. 10, 647–658 [DOI] [PubMed] [Google Scholar]
- 2. Penzes P., Cahill M. E., Jones K. A., VanLeeuwen J. E., Woolfrey K. M. (2011) Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 14, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Penzes P., Cahill M. E. (2012) Deconstructing signal transduction pathways that regulate the actin cytoskeleton in dendritic spines. Cytoskeleton 69, 426–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Penzes P., Cahill M. E., Jones K. A., Srivastava D. P. (2008) Convergent CaMK and RacGEF signals control dendritic structure and function. Trends Cell Biol. 18, 405–413 [DOI] [PubMed] [Google Scholar]
- 5. Penzes P., Beeser A., Chernoff J., Schiller M. R., Eipper B. A., Mains R. E., Huganir R. L. (2003) Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron 37, 263–274 [DOI] [PubMed] [Google Scholar]
- 6. Penzes P., Johnson R. C., Alam M. R., Kambampati V., Mains R. E., Eipper B. A. (2000) An isoform of kalirin, a brain-specific GDP/GTP exchange factor, is enriched in the postsynaptic density fraction. J. Biol. Chem. 275, 6395–6403 [DOI] [PubMed] [Google Scholar]
- 7. Xie Z., Srivastava D. P., Photowala H., Kai L., Cahill M. E., Woolfrey K. M., Shum C. Y., Surmeier D. J., Penzes P. (2007) Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron 56, 640–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jones K. A., Srivastava D. P., Allen J. A., Strachan R. T., Roth B. L., Penzes P. (2009) Rapid modulation of spine morphology by the 5-HT2A serotonin receptor through kalirin-7 signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 19575–19580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Penzes P., Jones K. A. (2008) Dendritic spine dynamics–a key role for kalirin-7. Trends Neurosci. 31, 419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Penzes P., Johnson R. C., Sattler R., Zhang X., Huganir R. L., Kambampati V., Mains R. E., Eipper B. A. (2001) The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron 29, 229–242 [DOI] [PubMed] [Google Scholar]
- 11. Biederer T., Cao X., Südhof T. C., Liu X. (2002) Regulation of APP-dependent transcription complexes by Mint/X11s: differential functions of Mint isoforms. J. Neurosci. 22, 7340–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rogelj B., Mitchell J. C., Miller C. C., McLoughlin D. M. (2006) The X11/Mint family of adaptor proteins. Brain Res. Rev. 52, 305–315 [DOI] [PubMed] [Google Scholar]
- 13. Nakajima Y., Okamoto M., Nishimura H., Obata K., Kitano H., Sugita M., Matsuyama T. (2001) Neuronal expression of mint1 and mint2, novel multimodular proteins, in adult murine brain. Brain Res. Mol. Brain Res. 92, 27–42 [DOI] [PubMed] [Google Scholar]
- 14. Okamoto M., Matsuyama T., Sugita M. (2000) Ultrastructural localization of mint1 at synapses in mouse hippocampus. Eur. J. Neurosci. 12, 3067–3072 [DOI] [PubMed] [Google Scholar]
- 15. Stricker N. L., Huganir R. L. (2003) The PDZ domains of mLin-10 regulate its trans-Golgi network targeting and the surface expression of AMPA receptors. Neuropharmacology 45, 837–848 [DOI] [PubMed] [Google Scholar]
- 16. Setou M., Nakagawa T., Seog D. H., Hirokawa N. (2000) Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science 288, 1796–1802 [DOI] [PubMed] [Google Scholar]
- 17. Rongo C., Whitfield C. W., Rodal A., Kim S. K., Kaplan J. M. (1998) LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell 94, 751–759 [DOI] [PubMed] [Google Scholar]
- 18. King G. D., Cherian K., Turner R. S. (2004) X11α impairs γ- but not β-cleavage of amyloid precursor protein. J. Neurochem. 88, 971–982 [DOI] [PubMed] [Google Scholar]
- 19. King G. D., Perez R. G., Steinhilb M. L., Gaut J. R., Turner R. S. (2003) X11α modulates secretory and endocytic trafficking and metabolism of amyloid precursor protein: mutational analysis of the YENPTY sequence. Neuroscience 120, 143–154 [DOI] [PubMed] [Google Scholar]
- 20. Teber I., Nagano F., Kremerskothen J., Bilbilis K., Goud B., Barnekow A. (2005) Rab6 interacts with the mint3 adaptor protein. Biol. Chem. 386, 671–677 [DOI] [PubMed] [Google Scholar]
- 21. Maximov A., Bezprozvanny I. (2002) Synaptic targeting of N-type calcium channels in hippocampal neurons. J. Neurosci. 22, 6939–6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ye B., Zhang Y., Song W., Younger S. H., Jan L. Y., Jan Y. N. (2007) Growing dendrites and axons differ in their reliance on the secretory pathway. Cell 130, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Srivastava D. P., Woolfrey K. M., Penzes P. (2011) Analysis of dendritic spine morphology in cultured CNS neurons. J. Vis. Exp. 53, e2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Penzes P., Johnson R. C., Kambampati V., Mains R. E., Eipper B. A. (2001) Distinct roles for the two Rho GDP/GTP exchange factor domains of kalirin in regulation of neurite growth and neuronal morphology. J. Neurosci. 21, 8426–8434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsieh-Wilson L. C., Benfenati F., Snyder G. L., Allen P. B., Nairn A. C., Greengard P. (2003) Phosphorylation of spinophilin modulates its interaction with actin filaments. J. Biol. Chem. 278, 1186–1194 [DOI] [PubMed] [Google Scholar]
- 26. Horton A. C., Ehlers M. D. (2004) Secretory trafficking in neuronal dendrites. Nat. Cell Biol. 6, 585–591 [DOI] [PubMed] [Google Scholar]
- 27. Ehlers M. D. (2007) Secrets of the secretory pathway in dendrite growth. Neuron 55, 686–689 [DOI] [PubMed] [Google Scholar]
- 28. Pierce J. P., Mayer T., McCarthy J. B. (2001) Evidence for a satellite secretory pathway in neuronal dendritic spines. Curr. Biol. 11, 351–355 [DOI] [PubMed] [Google Scholar]
- 29. Horton A. C., Rácz B., Monson E. E., Lin A. L., Weinberg R. J., Ehlers M. D. (2005) Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron 48, 757–771 [DOI] [PubMed] [Google Scholar]
- 30. Zhou W., Chang J., Wang X., Savelieff M. G., Zhao Y., Ke S., Ye B. (2014) GM130 is required for compartmental organization of dendritic golgi outposts. Curr. Biol. 24, 1227–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Horton A. C., Ehlers M. D. (2003) Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J. Neurosci. 23, 6188–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Remmers C., Sweet R. A., Penzes P. (2014) Abnormal kalirin signaling in neuropsychiatric disorders. Brain Res. Bull. 103, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cahill M. E., Jones K. A., Rafalovich I., Xie Z., Barros C. S., Muller U., Penzes P. (2012) Control of interneuron dendritic growth through NRG1/erbB4-mediated kalirin-7 disinhibition. Mol. Psychiatry 10.1038/mp.2011.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xie Z., Photowala H., Cahill M. E., Srivastava D. P., Woolfrey K. M., Shum C. Y., Huganir R. L., Penzes P. (2008) Coordination of synaptic adhesion with dendritic spine remodeling by AF-6 and kalirin-7. J. Neurosci. 28, 6079–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ho A., Morishita W., Atasoy D., Liu X., Tabuchi K., Hammer R. E., Malenka R. C., Südhof T. C. (2006) Genetic analysis of Mint/X11 proteins: essential presynaptic functions of a neuronal adaptor protein family. J. Neurosci. 26, 13089–13101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ho A., Morishita W., Hammer R. E., Malenka R. C., Sudhof T. C. (2003) A role for Mints in transmitter release: Mint 1 knockout mice exhibit impaired GABAergic synaptic transmission. Proc. Natl. Acad. Sci. U.S.A. 100, 1409–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng C. Y., Petralia R. S., Wang Y. X., Kachar B., Wenthold R. J. (2010) SAP102 is a highly mobile MAGUK in spines. J. Neurosci. 30, 4757–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jaskolski F., Mayo-Martin B., Jane D., Henley J. M. (2009) Dynamin-dependent membrane drift recruits AMPA receptors to dendritic spines. J. Biol. Chem. 284, 12491–12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alam M. R., Johnson R. C., Darlington D. N., Hand T. A., Mains R. E., Eipper B. A. (1997) Kalirin, a cytosolic protein with spectrin-like and GDP/GTP exchange factor-like domains that interacts with peptidylglycine α-amidating monooxygenase, an integral membrane peptide-processing enzyme. J. Biol. Chem. 272, 12667–12675 [DOI] [PubMed] [Google Scholar]
- 40. Hayashi-Takagi A., Takaki M., Graziane N., Seshadri S., Murdoch H., Dunlop A. J., Makino Y., Seshadri A. J., Ishizuka K., Srivastava D. P., Xie Z., Baraban J. M., Houslay M. D., Tomoda T., Brandon N. J., Kamiya A., Yan Z., Penzes P., Sawa A. (2010) Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat. Neurosci. 13, 327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ho A., Liu X., Südhof T. C. (2008) Deletion of Mint proteins decreases amyloid production in transgenic mouse models of Alzheimer's disease. J. Neurosci. 28, 14392–14400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Minami S. S., Sung Y. M., Dumanis S. B., Chi S. H., Burns M. P., Ann E. J., Suzuki T., Turner R. S., Park H. S., Pak D. T., Rebeck G. W., Hoe H. S. (2010) The cytoplasmic adaptor protein X11α and extracellular matrix protein Reelin regulate ApoE receptor 2 trafficking and cell movement. FASEB J. 24, 58–69 [DOI] [PMC free article] [PubMed] [Google Scholar]