Background: The Musashi family of RNA-binding proteins promotes progenitor cell proliferation.
Results: The majority of Musashi sequence specificity comes from a UAG motif. Sequences outside of UAG make minor contributions to affinity.
Conclusion: UAG forms the core Musashi recognition element.
Significance: Delineating the sequences that contribute to binding is critical to understanding RNA target selection.
Keywords: Cell Proliferation, Fluorescence Anisotropy, RNA, RNA-binding Protein, RNA-Protein Interaction, Binding Specificity, RNA Recognition, RRM
Abstract
Musashi (MSI) family proteins control cell proliferation and differentiation in many biological systems. They are overexpressed in tumors of several origins, and their expression level correlates with poor prognosis. MSI proteins control gene expression by binding RNA and regulating its translation. They contain two RNA recognition motif (RRM) domains, which recognize a defined sequence element. The relative contribution of each nucleotide to the binding affinity and specificity is unknown. We analyzed the binding specificity of three MSI family RRM domains using a quantitative fluorescence anisotropy assay. We found that the core element driving recognition is the sequence UAG. Nucleotides outside of this motif have a limited contribution to binding free energy. For mouse MSI1, recognition is determined by the first of the two RRM domains. The second RRM adds affinity but does not contribute to binding specificity. In contrast, the recognition element for Drosophila MSI is more extensive than the mouse homolog, suggesting functional divergence. The short nature of the binding determinant suggests that protein-RNA affinity alone is insufficient to drive target selection by MSI family proteins.
Introduction
The life of an RNA molecule is controlled by a suite of RNA-binding proteins (RBPs)2 that associate with specific regions of the RNA at specific times in its life. During transcription, the nascent pre-mRNA associates with components of the splicing machinery to ensure correct splicing patterns. Subsequently, the mRNA may be stabilized, destabilized, edited, or localized to a particular region of the cell, again due to specific protein associations. To carry out its task, an RBP must be able to identify and bind its precise target sequence in a heterogeneous mixture of nucleic acids. The fact that most RBP target sequences consist of short, often degenerate motifs makes the task even more complex.
The Musashi family of RNA-binding proteins is a widespread and highly conserved family that governs stem cell self-renewal and maintenance (1, 2). They are overexpressed in several tumor types, and their expression correlates with poor prognosis (3–7). There are two mammalian MSI family members: MSI1 and MSI2. MSI1 is expressed in neural and glial progenitor cells (8–10) as well as in progenitors of several epithelial tissues (11–13). MSI2 is 69% identical to MSI1 and is expressed in a partially overlapping set of tissues (14). MSI family members have also been identified in flies (15), worms (16), and frogs (17, 18) among other species. MSI proteins contain two RRMs that provide the surface for interaction with target RNA. The first RRM has a higher affinity for RNA than the second (19, 20). The C-terminal region of the protein interacts with poly(A)-binding protein, inhibiting translation of bound RNAs by competing with eIF4G (21).
Several different groups have investigated the RNA binding specificity of MSI family members. Okano and co-workers (22) used the in vitro evolution method SELEX to identify optimal binding motifs for mouse MSI1 and Drosophila MSI. Mouse MSI1 bound the sequence (G/A)U1–3AGU. The Xenopus protein has been shown to interact with a similar sequence (17). Additionally, the NMR structure of MSI1 in complex with a five-nucleotide RNA of the sequence GUAGU has been recently solved, demonstrating how this sequence interacts with MSI1 (23). Contrasting with these observations, the co-immunoprecipitation and microarray-based approach RNAcompete identified a strong preference for the sequence UAG followed by a weaker preference for UUAG for mouse MSI1; this is substantially different from the SELEX sequence (24). A similar discrepancy exists for the Drosophila protein. SELEX experiments identified the sequence GUUU(G/AG) (25), whereas RNAcompete identified GUAG with a slight preference for an upstream A and a downstream G for Drosophila MSI (24). We wished to understand which of these motifs forms the optimal Musashi binding determinant.
A number of MSI mRNA targets have been identified, and MSI can regulate their translation positively or negatively. Most of these targets contain the MSI1 consensus sequence within their 3′-UTRs. In neural lineage cells, MSI1 binds the Numb 3′-UTR and inhibits its translation (22). By inhibiting NUMB activity, MSI1 activates the NOTCH pathway, promoting stem cell self-renewal. In contrast, MSI1 has been shown to up-regulate expression of NUMB in gastric tissue (26). MSI1 has also been shown to bind Robo3 mRNA and up-regulate protein production, controlling midline crossing of precerebellar neurons (27). In contrast to many of its other targets, MSI1 is thought to interact with a portion of the Robo3 mRNA coding region that does not contain the MSI1 consensus sequence (27). Other mRNA targets of MSI include doublecortin (28), CDKN1A (29), and c-mos (17).
To better understand the Musashi binding determinant, we set out to determine which nucleotides make the strongest thermodynamic contribution to binding of MSI to its target RNA. We used a mutational series of RNAs and analyzed the interaction between the RNAs and three MSI family members using fluorescence polarization assays. Our results demonstrate that all three MSI proteins bind a core UAG motif. In contrast to mouse MSI1 and MSI2, additional G nucleotides both upstream and downstream of UAG contribute to the interaction with Drosophila MSI. Our results reveal the core motif that is essential for binding. Additionally, they explain why different high throughput approaches have revealed seemingly different Musashi binding motifs.
EXPERIMENTAL PROCEDURES
RNAs
Synthetic RNAs labeled with fluorescein amidite at the 3′-end were purchased from Integrated DNA Technologies.
Plasmids
For the mouse dual RRM construct (RRM1-2), amino acids 7–192 of mouse MSI1 were amplified from a mammalian genome ORF clone (100014969; Invitrogen) using primers 5′-cgcgcggatcccagcccggcctcgcctcccc-3′ and 5′-gcgcgaagcttcggggacatcacctcctttg-3′. The fragment was digested with BamHI and HindIII and cloned into a modified version of expression vector pET-22b in which the leader sequence was replaced with a His tag and a tobacco etch virus protease site. To produce the single RRM construct, the dual RRM construct was modified by QuikChange (Stratagene) mutagenesis to replace Met-104 with a stop codon. For human MSI2, amino acids 8–198 were amplified from Mammalian Gene Collection ORF clone 3505639 using primers 5′-cgcgcggatccggcacctcgggcagcgccaa-3′ and 5′-gcgcgaagctttcatgggaacatgacttctttcg-3′. The fragment was digested with BamHI and HindIII and cloned into the modified pET-22b described above. For the Drosophila construct, amino acids 162–364 were amplified from a Drosophila Genome Resource Center cDNA clone (LD31631) using primers 5′-gggggagctccccagcctgagcggaggc-3′ and 5′-gggggtcgacttctacggtgtgactgcttcctt-3′. The fragment was digested with SacI and SalI and cloned into the His-modified PET-22b vector. For the Drosophila RRM1 construct, Gln-258 was changed to a stop codon using QuikChange mutagenesis.
Protein Purification
Bacterial expression vectors were transformed into BL21(DE3), and protein expression was induced with 1 mm isopropyl β-d-thiogalactopyranoside. After 3 h at 37 °C, cells were pelleted and lysed in 50 mm NaH2PO4, 300 mm NaCl, 20 mm imidazole, and 5 mm β-mercaptoethanol with a microfluidizer (IDEX Health and Science). The soluble lysate was applied to a nickel-nitrilotriacetic acid column (Qiagen); washed with 50 mm NaH2PO4, 300 mm NaCl, 50 mm imidazole, and 5 mm β-mercaptoethanol; and eluted with 50 mm NaH2PO4, 300 mm NaCl, 300 mm imidazole, and 5 mm β-mercaptoethanol. Fractions containing recombinant protein were then purified over a HiTrap SP column (GE Healthcare) in a buffer containing 50 mm MOPS, pH 6.0, 20 mm NaCl, and 2 mm DTT. Proteins were eluted with a 0.1–1.0 m NaCl gradient in the same buffer. Drosophila RRM1 was eluted with a 0.02–2 m NaCl gradient. Fractions containing recombinant protein were then further purified over a HiTrap Q column (GE Healthcare) in a buffer containing 50 mm Tris, pH 8.8, 20 mm NaCl, and 2 mm DTT, eluting with a 0.1–1.0 m NaCl gradient. The Drosophila proteins were recovered from the flow-through. Recombinant proteins were stored in 50 mm Tris, pH 8.0, 20 mm NaCl, and 2 mm DTT at 4 °C.
Fluorescence Polarization
Fluorescence polarization (FP) assays were conducted as described elsewhere in detail except that the carrier bovine γ-globulin (Invitrogen) was added to the buffer (30). Briefly, a dilution series of recombinant protein was prepared in a buffer containing 50 mm Tris, pH 8.0, 100 mm NaCl, 0.01% IGEPAL CA-630, 0.01 mg/ml tRNA, and 0.1 mg/ml bovine γ-globulin. RNA was diluted in the same buffer to achieve a final concentration of 2 nm in the binding reaction. RNA was added to the protein dilutions and allowed to equilibrate for 3 h at room temperature. Polarization was detected with a Victor3 multilabel counter (PerkinElmer Life Sciences). For the dual RRM proteins, the maximal protein concentration in the dilution series was 2 μm. For the single RRM protein, the maximal concentration was increased to 4 μm.
Data Analysis
FP data were analyzed as described previously (30). Briefly, polarization (millipolarization) was plotted against the protein concentration (m). The data were then fit to the Hill equation to obtain the Kd,app. Each RNA was assayed in triplicate. In one case, where an accurate fit could not be achieved due to low affinity, the Hill coefficient was constrained to 1.0 during curve fitting. Krel was determined from Equation 1.
 |
where Kd,app2 is the mutant value and Kd,app1 is the Kd,app of the control RNA. The change in free energy change (ΔΔG) due to the mutation was calculated according to Equation 2.
where T is the temperature and R is the gas constant.
NMR
Labeling with 15N was performed by growing cells in isotopically enriched M9 medium with 1 g of 15NH4Cl/liter. Two-dimensional 1H-15N heteronuclear single quantum coherence spectra were collected using samples of U-15N-labeled MSI1 in a 90% H2O and 10% D2O buffer solution of 50 mm Tris, pH 7.0. A series of 1H-15N heteronuclear single quantum coherence spectra of 15N-labeled MSI1 were collected at different concentrations of unlabeled RNA (either 5′-UUUAUUUUAGUUUUU-3′ or 5′-UUUAUAGUUUUU-3′). The titrations were terminated when changes of amide group chemical shift positions were no longer observed. All experiments were performed at 600 MHz on a Varian Inova spectrometer equipped with a triple resonance cold probe at 298 K. Data processing was performed using NMRPipe (31) and SPARKY (32) software.
UAG Frequency Determination
The UTRs of RNAs determined to be associated with MSI by Vo et al. (3) were downloaded from the hg38 assembly of the human genome via the University of California at Santa Cruz genome browser. For the random background sets, all UTRs were retrieved from the University of California at Santa Cruz genome browser, and 50 UTRs were randomly selected using a random number generator. All sequence processing was conducted using custom Perl scripts.
For the trimer analysis, the number of occurrences of each three-nucleotide motif was determined, and the frequency of each motif in the set of UTRs was calculated. The MSI-associated set and the three random sets were processed separately. The frequency change was determined by subtracting the mean of the frequency in the random sets from the frequency in the MSI-associated set. Data were ordered for ease of visualization before plotting. Z-scores were calculated as described in Yeo et al. (33) using the random sets for expected values.
For the cluster analysis, data were analyzed in 50-nucleotide overlapping windows. Windows overlapped by one nucleotide. For each window, the number of UAGs was counted. The fraction of windows at each UAG content level was calculated. Each data set was analyzed independently. Statistical significance was determined using a t test, comparing the fraction of windows in the MSI-associated set with each of the three random sets. Statistical tests and population distribution calculations were conducted using IGOR Pro (Wavemetrics).
Homology Modeling
The RNA-bound structure of MSI1 RRM1 (Protein Data Bank code 2RS2) was downloaded from the Protein Data Bank. The composite file was split into separate entries, and the structures were visualized using PyMOL (Schrödinger, LLC). The homology model of Drosophila RRM1 was created using SWISS-MODEL (34).
RESULTS
Binding of Mouse MSI1 RRM1-2 to Mutant RNAs
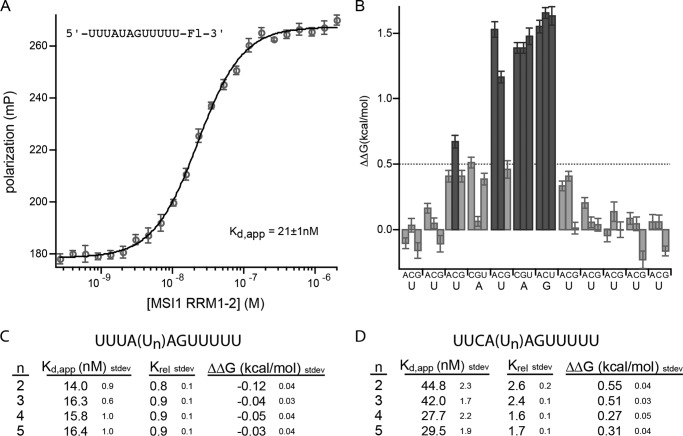
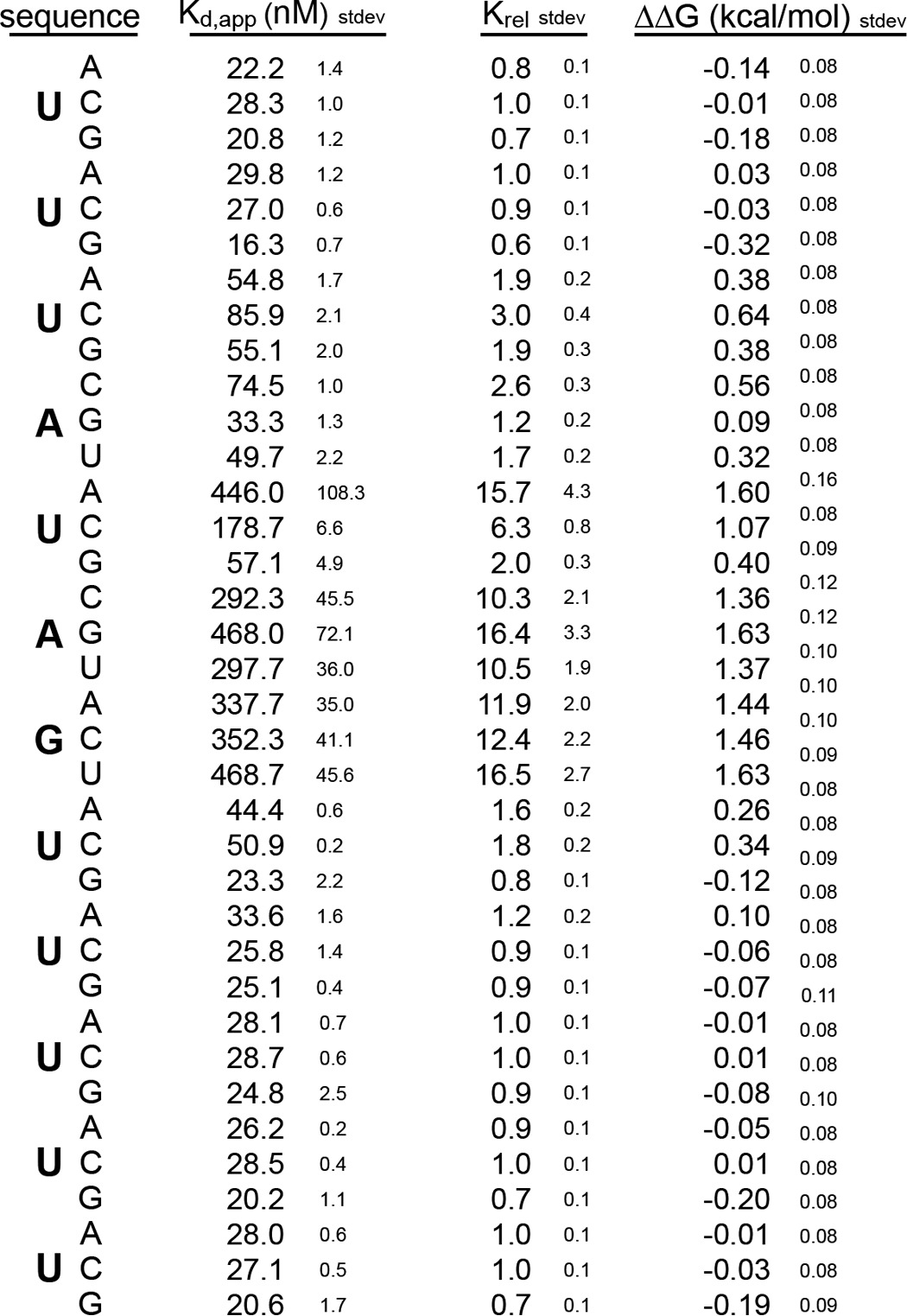
Based upon the binding aptamer identified by Okano and co-workers (22) using SELEX, we inserted the sequence AUAGU into a background of U to produce a 12-nucleotide RNA fragment of the sequence 5′-UUUAUAGUUUUU-3′. This was the control RNA sequence that served as the starting point for the mutational series. We analyzed the interaction between the control RNA and mouse MSI RRM1-2 with fluorescence polarization. The Kd,app of the interaction is 21 ± 1 nm (Fig. 1A).
FIGURE 1.
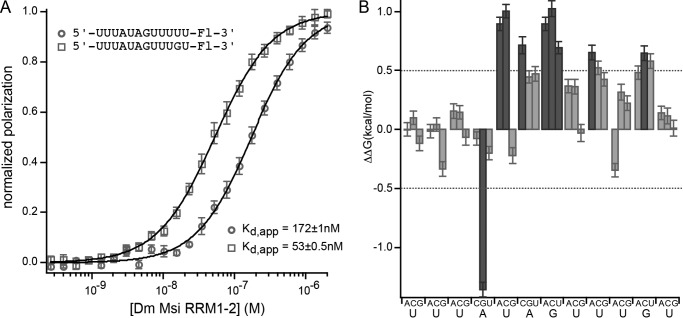
A, FP of the RNA 5′-UUUAUAGUUUUU-Fl-3′ (where Fl indicates fluorescein) bound to a series of concentrations of mouse MSI1 RRM1-2. Data were fit to the Hill equation for calculation of the Kd,app. B, the change in ΔΔG due to the indicated mutation at each position of the RNA depicted in A. Mutations causing a ΔΔG of greater than 0.5 kcal/mol are shaded dark gray. C, binding of mutant RNAs to MSI1 RRM1-2. The indicated number of uridines (n) was incorporated into the RNA upstream of the central AG. No change in the affinity of the protein for the RNA was observed. D, binding of mutant RNAs that contain a C at position 3 to MSI1 RRM1-2. The indicated number of uridines (n) were incorporated into the RNA as in C. The presence of the C at position 3 reduced the affinity of the interaction in the presence of additional uridines. Error bars represent the S.D. mP, millipolarization.
To analyze the contribution of each nucleotide position to the interaction, we designed a mutational series of RNAs in which each position is individually mutated to each of the three remaining nucleotides, generating 36 mutant variants in total. The most significant changes upon binding were due to mutation of U5, A6, or G7. Mutation of U5 to A or C and mutation of A6 and G7 to any of the other nucleotides caused a change in the standard ΔΔG of more than 1 kcal/mol with a Krel of greater than 10 (Fig. 1B and Table 1). Mutation of position U3 to a C had a less severe effect on the interaction (ΔΔG = 0.67 ± 0.05 kcal/mol). All other mutations had little to no effect on binding free energy. The mutational analysis shows that binding is energetically driven by the sequence UAG at positions 5 through 7 with a slight additional contribution from the absence of a C at position 3.
TABLE 1.
Binding of mouse MSI1 RRM1-2 to mutant RNAs
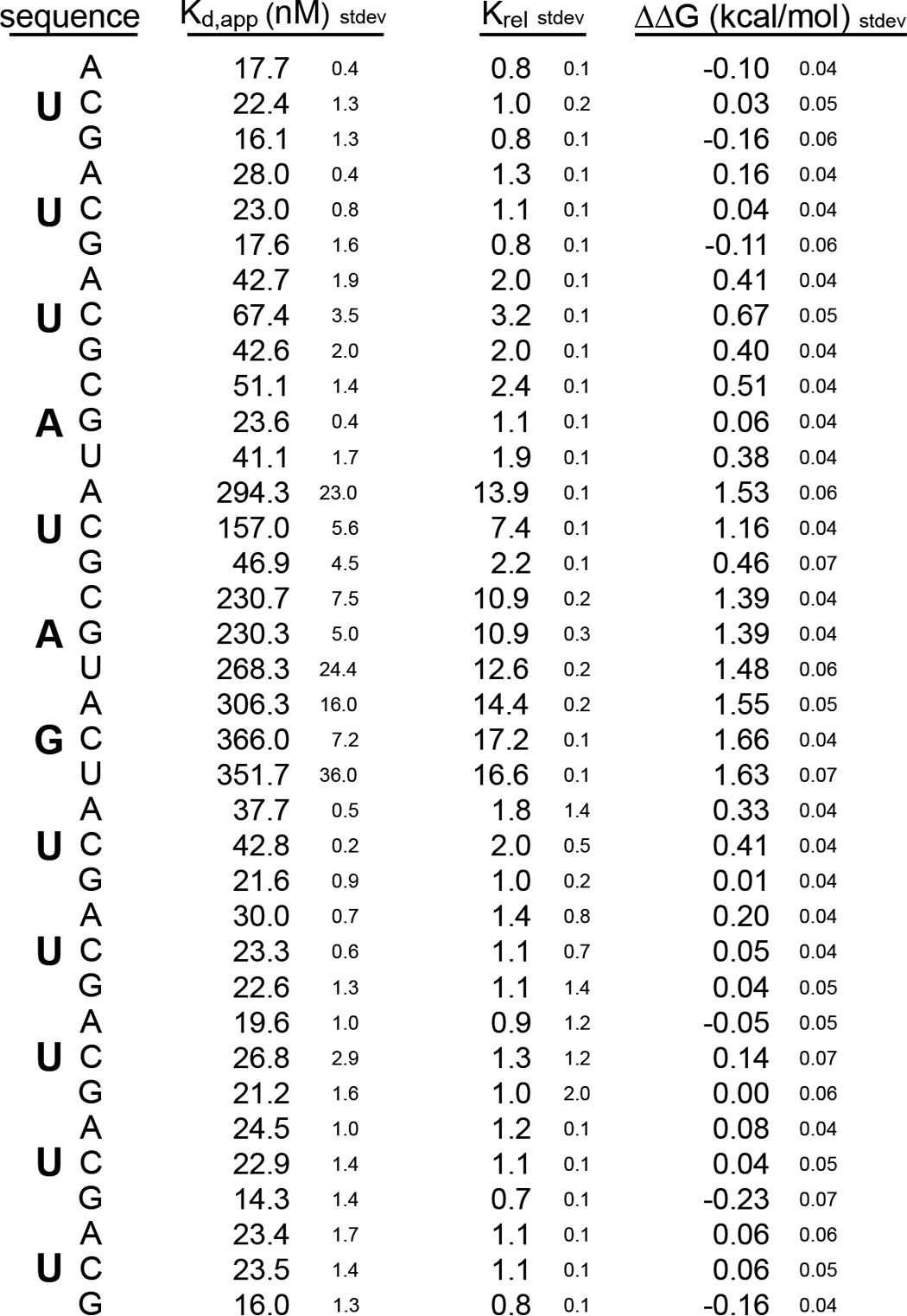
We noticed that mutation of U5 to G introduced a new UAG motif within the RNA. To determine whether the lack of an effect of this mutation is due to the presence of the new UAG, we analyzed an RNA of the sequence 5′-UUGAGAGUUUUU-3′. This RNA contains a G at position 3 in addition to position 5. The mutations at position 3 or 5 alone had little to no effect on binding (Fig. 1B). However, the double mutation had a more severe effect with a ΔΔG of 1.35 ± 0.04 kcal/mol. Together, the data indicate that UAG provides the critical associations between MSI RRM1-2.
Binding of Mouse MSI1 RRM1-2 Tolerates Spacing Flexibility
SELEX experiments performed by Okano and co-workers (22) suggest that one to three uridines are tolerated in the MSI1 binding motif between positions 3 and 5. To determine whether an increase in the number of uridines alters the free energy of binding, we generated a series of spacing mutants in which the position of the U is expanded to accommodate up to five uridines. We found that the interaction of MSI RRM1-2 was insensitive to insertions in this position (Fig. 1C). Moreover, mutation of U to C in position 3 caused a decrease in the affinity of the interaction regardless of the presence of the insertion (Fig. 1D). The data confirm that insertion of uridines upstream of the UAG is tolerated and show that as many as five (if not more) uridines can be accommodated with no impact on binding. The data are consistent with a model in which a flexible stretch of bases may loop out of the recognition site.
Binding Specificity Is Determined by RRM1
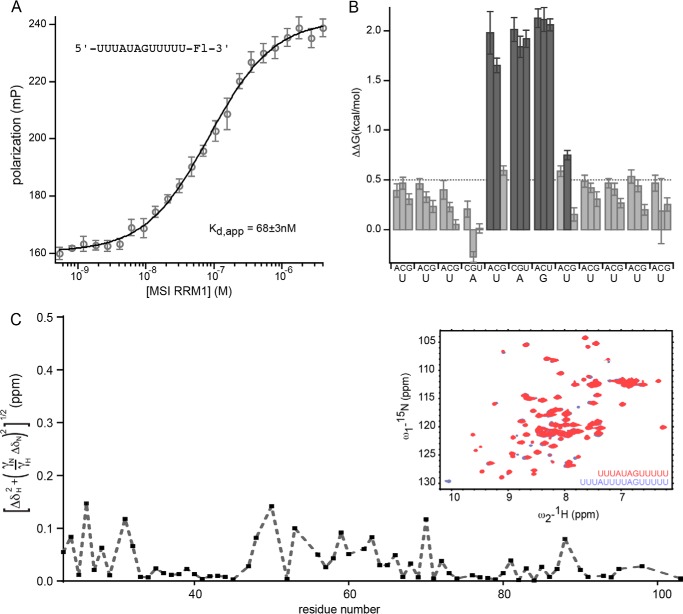
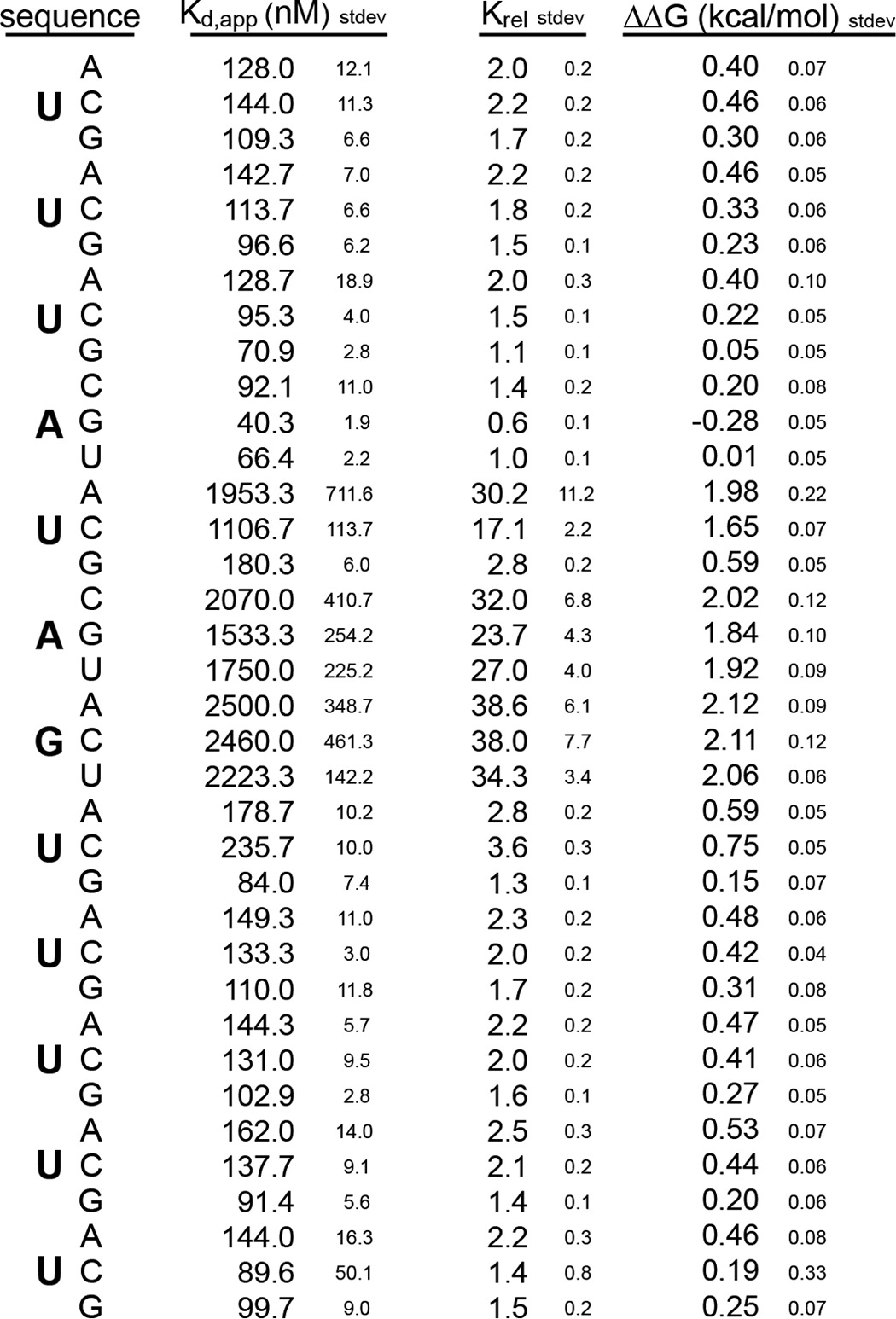
NMR experiments conducted by Katahira and co-workers (23) have suggested that many of the amino acids that contact RNA in RRM2 are similar to those making contacts in RRM1. We were interested in determining whether the presence of RRM2 alters the specificity of the MSI RRM1 protein-RNA interaction. We therefore analyzed the mutation series to determine whether the specificity of RRM1 is the same. The single RRM protein interacts with the control RNA of sequence 5′-UUUAUAGUUUUU-3′ with a Kd,app of 68 ± 3 nm (Fig. 2A). The affinity is 3-fold weaker than that of the dual RRM construct (Fig. 1A). Analysis of the mutant series revealed that, just as for the two RRM construct, the nucleotides that have the strongest contribution to the affinity of the interaction are the central UAG (Fig. 2B and Table 2). Therefore, the second RRM domain contributes to the affinity of the interaction but does not alter its specificity.
FIGURE 2.
A, FP of the RNA 5′-UUUAUAGUUUUU-FAM-3′ bound to a series of concentrations of mouse MSI1 RRM1. Data were fit to the Hill equation for calculation of the Kd,app. B, the change in ΔΔG due to the indicated mutation at each position of the RNA depicted in A. Mutations causing a ΔΔG of greater than 0.5 kcal/mol are shaded dark gray. C, chemical shift changes in MSI1 upon binding of two different RNA sequences: 5′-UUUAUAGUUUUU-3′ and 5′-UUUAUUUUAGUUUUU-3′. ΔδH and ΔδN are the chemical shift changes observed for the 1H and 15N resonances in the two RNA complexes, respectively. The inset shows the overlay of the 1H-15N heteronuclear single quantum coherence spectra of MSI1 bound to 5′-UUUAUAGUUUUU-3′ (red) and 5′-UUUAUUUUAGUUUUU-3′ (purple). Error bars represent the S.D. mP, millipolarization.
TABLE 2.
Binding of mouse MSI1 RRM1 to mutant RNAs
MSI1 RRM1-2 has a similar affinity for RNAs with uridine insertions upstream of the UAG motif (Fig. 1C). We wished to determine whether RNAs with and without uridine insertions are recognized in the same way. Analysis of the 1H-15N heteronuclear single quantum coherence spectra of MSI1 RRM1 bound to two different RNA sequences (5′-UUUAUAGUUUUU-3′ and 5′-UUUAUUUUAGUUUUU-3′) shows that the signals of the amide groups of MSI1 in the two complexes mostly overlap (Fig. 2C). The average chemical shift difference between corresponding residues in the two complexes is 0.033 ± 0.036 ppm. In addition, the residues that show the largest chemical shift changes agree with those reported for binding with the RNA sequence 5′-AGCGUUAGUUAUUUAGUUCG-3′ (35). Because the chemical shift of a nucleus is very sensitive to its chemical environment, this result indicates that the mode of RNA recognition is the same in the three RNA-bound complexes and that the RNA-bound structure of MSI1 is very similar when bound to either 5′-UUUAUAGUUUUU-3′ or 5′-UUUAUUUUAGUUUUU-3′.
UAG Motifs Are Enriched in Samples Associated with MSI1
Several reports have been published recently by others that identify mRNAs that co-immunoprecipitate with MSI protein. We decided to make use of these data to assess whether UAG motifs are enriched in MSI targets. For the analysis, we used the 3′-UTRs of a set of RNAs found by Vo et al. (3) to be associated with MSI1 in Daoy cells. Vo et al. (3) noted the presence of GUAG motifs with a nearby UAG in targets.
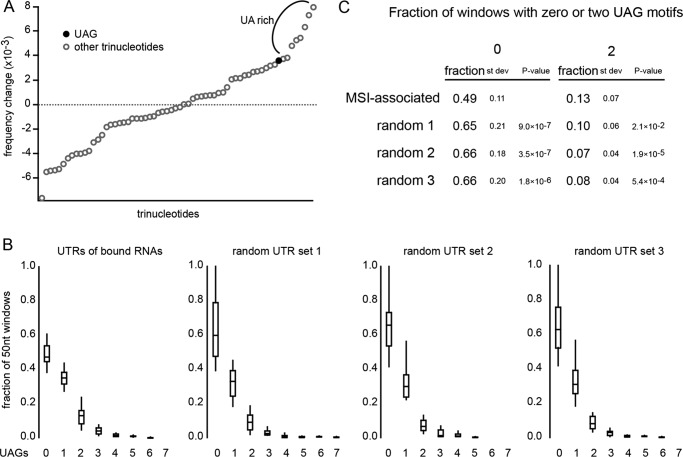
We first asked whether the motif UAG is more frequent in MSI target mRNAs than expected. For a background set, we produced three different sets of 50 randomly selected UTRs from the human genome. We counted the number of occurrences of each trinucleotide motif and determined its frequency in each set. We then examined the difference in the frequency of each motif in the MSI-associated set compared with the background sets. We found that UAG with a frequency change of 3.6 × 10−3 is one of the top enriched motifs in the MSI-associated set (Fig. 3A). The top seven enriched motifs each include only combinations of U and A, possibly pointing to an unstructured quality of the MSI-associated UTRs (Fig. 3A). The only remaining trinucleotide with an enrichment greater than that of UAG in the set is CUA, which has a frequency change of 3.7 × 10−3. The binding data presented here do not explain why CUA trinucleotides are enriched in mRNAs that co-immunoprecipitate with MSI1.
FIGURE 3.
A, change in the frequency of each trinucleotide in the 3′-UTRs of MSI1-associated RNAs as determined by Vo et al. (3). Data were obtained by subtracting the mean frequency of each trinucleotide in three random sets of 50 UTRs from the frequency of each trimer in the MSI-associated UTR set. Data were ordered by value for ease of visualization. UAG is indicated with a filled circle. Other trinucleotides are indicated with a hollow circle. B, box plots indicating the population distribution of 50-nucleotide (nt) windows containing the indicated number of UAG motifs. The random sets are the same as in A. C, statistical values for 50-nucleotide windows containing zero and two UAGs for the data presented in B. Data are the mean, S.D., and p value of the MSI-associated set compared with each of the random sets using a t test. Error bars represent the S.D.
Because RNAs with multiple MSI binding sites may be more likely to associate with MSI in vivo, we next examined whether there are clusters of UAG in the MSI-associated set. Again we compared the data for the MSI-associated RNAs with data obtained from the same three randomly selected background UTR sets used in Fig. 3A. Each UTR was divided into 50-nucleotide overlapping tiles, and the number of UAG motifs within each tile was counted. For each number of motifs, the fraction of tiles with that number was determined. We found a significant decrease in the number of tiles with no UAG motifs in the MSI-associated set (Fig. 3, B and C), whereas the number of tiles with a single UAG was unchanged. We found a significant increase in the number of tiles with two UAG motifs in the MSI-associated set (Fig. 3, B and C). No other numbers of motifs were significantly changed in comparison with all three random sets.
To compare the enrichment of UAG in the MSI-associated UTR set with the enrichment of another RBP that has a larger binding site with a well defined specificity, FOX2, we calculated the Z-score. The Z-score for UAG was 2.79, indicating that UAG enrichment is unlikely to occur by chance. In comparison, the reported Z-score for the target sequence of FOX2 (UGCAUG) is 25.16 (33), indicating a greater significance of FOX2 binding site enrichment. It is important to note, however, that differences between the two data sets (3′-UTRs of MSI-associated RNAs versus cross-link immunoprecipitation sequence clusters) make direct comparison difficult.
In summary, we found a decrease in the number of 50-nucleotide windows with no UAG motifs and an increase in the number of 50-nucleotide windows with two UAG motifs in the MSI1-associated set. Additionally, we found that UAG was one of the most enriched trimers in the MSI-associated data set. These observations support our finding that UAG is the core motif driving association.
Human MSI2 Recognizes the Same Sequence as Mouse MSI1
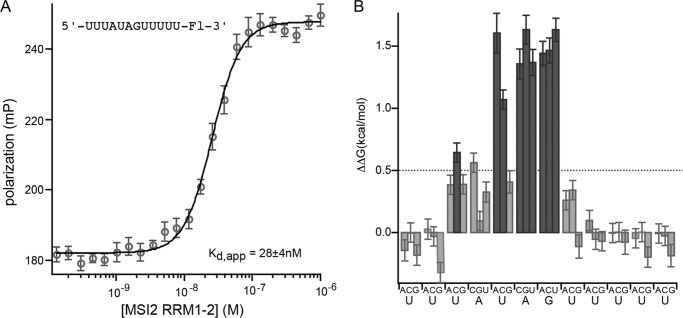
The high degree of conservation within the MSI protein family suggests that their recognition sites may be the same. To test this with another family member, we assessed the binding of human MSI2 RRM1-2 to the same library of mutant RNAs used for mouse MSI1. The two RNA binding domains are 87% identical (Fig. 4). Human MSI2 bound the control sequence 5′-UUUAUAGUUUUU-3′ with a Kd,app of 28 ± 4 nm. Mutational analysis revealed that, as for MSI1, the largest contributions to binding energy come from the UAG element (Fig. 5 and Table 3).
FIGURE 4.

Alignment of the RNA binding domains from Drosophila MSI, human MSI2, and mouse MSI1 RNA. Conserved amino acids are shaded gray. Uppercase letters correspond to amino acids that are conserved in at least two of the three proteins. Lowercase letters represent amino acids that are not conserved.
FIGURE 5.
A, FP of the RNA 5′-UUUAUAGUUUUU-FAM-3′ bound to a series of concentrations of human MSI2 RRM1-2. Data were fit to the Hill equation for calculation of the Kd,app. B, the change in ΔΔG due to the indicated mutation at each position of the RNA depicted in A. Mutations causing a ΔΔG of greater than 0.5 kcal/mol are shaded dark gray. Error bars represent the S.D. mP, millipolarization.
TABLE 3.
Binding of human MSI2 RRM1-2 to mutant RNAs
Drosophila MSI Recognizes a Broader Region of RNA than Mouse MSI1
MSI family members are highly conserved among many different animal phyla. One of the most divergent members is the namesake of the family, Drosophila MSI. Drosophila has another MSI family member, known as RBP6, that is more similar to the mammalian proteins (36). We wished to determine whether Drosophila MSI has the same preference for UAG as mouse MSI1. Previous SELEX experiments by Okano and co-workers (25) have identified a motif consisting of GU3–6(G/AG), suggesting that the motif identified by this protein is different from the mouse MSI1 motif.
A sequence alignment of RRM1 from Drosophila and mouse reveals that many of the amino acids that are involved in RNA recognition are conserved between the two proteins, suggesting that the mode of RNA recognition may be the same (Fig. 6A). To assess the possibility that binding requirements may differ for Drosophila MSI, we prepared a homology model of Drosophila MSI RRM1 based upon the recently published structure of mouse RRM1 bound to RNA of the sequence GUAGU (23). In the homology model, many features of the association, including both hydrophobic and electrostatic interactions, are preserved (Fig. 6B). The model therefore predicts that the sequence specificity of the two proteins should be similar.
FIGURE 6.
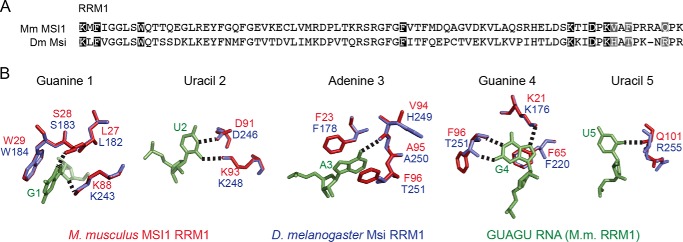
A, alignment of RRM1 of mouse (Mm) MSI1 and Drosophila (Dm) MSI. Conserved amino acids that interact with RNA are highlighted black. Unconserved amino acids are in gray. Interacting amino acids were as determined by Ohyama et al. (23). B, homology model of Drosophila MSI RRM1 and mouse MSI1 RRM1. Bases and amino acids are as indicated in the figure. The model was based upon the structure 2RS2 in the Protein Data Bank.
We therefore analyzed the affinity of Drosophila RRM1-2 for the control RNA used in the mouse mutational analysis. Drosophila RRM1-2 recognized this RNA with a Kd,app of 172 ± 1 nm (Fig. 7A) in contrast to mouse RRM1-2, which interacted with a Kd,app of 21 ± 1 nm. Because the published consensus sequence of Drosophila MSI is the sequence GU3–6(G/AG) (25), we also analyzed the mutant that contained a G at position 11, 5′-UUUAUAGUUUGU-3′. This RNA interacted with Drosophila MSI RRM1-2 with a Kd,app of 53 ± 1 nm (Fig. 7A). The same mutation had no effect on the affinity of mouse RRM1-2 (Fig. 1A). Sequence requirements of the two proteins are therefore not identical.
FIGURE 7.
A, FP of the RNAs 5′-UUUAUAGUUUGU-FAM-3′ and 5′-UUUAUAGUUUUU-FAM-3′ bound to a series of concentrations of Drosophila (Dm) MSI RRM1-2. Data were fit to the Hill equation for calculation of the Kd,app. Data are normalized for display purposes. B, the change in ΔΔG due to the indicated mutation at each position of the RNA depicted in A. Mutations causing a ΔΔG of greater than 0.5 kcal/mol are shaded dark gray. Error bars represent the S.D.
To determine which nucleotides within the RNA make the strongest contributions to binding affinity with Drosophila RRM1-2, we designed a mutation series of RNAs based upon the control sequence 5′-UUUAUAGUUUGU-3′. As we would predict from the homology model, the UAG motif contributes binding energy to the interaction. Mutation of G7 to any other nucleotide caused a change in the free energy of association of greater than 0.5 kcal/mol (Fig. 7B and Table 4). Likewise, mutation of U5 to A or C caused a ΔΔG of more than 0.5 kcal/mol (Fig. 7B and Table 4). It is likely that mutation of this position to a G has no apparent effect as it creates a second UAG, similar to the mouse protein. Mutation of A6 to C caused a ΔΔG of more than 0.5 kcal/mol, whereas mutation to G or U caused a change of ∼0.5 kcal/mol (Fig. 7B and Table 4).
TABLE 4.
Binding of Drosophila MSI RRM1-2 to mutant RNAs
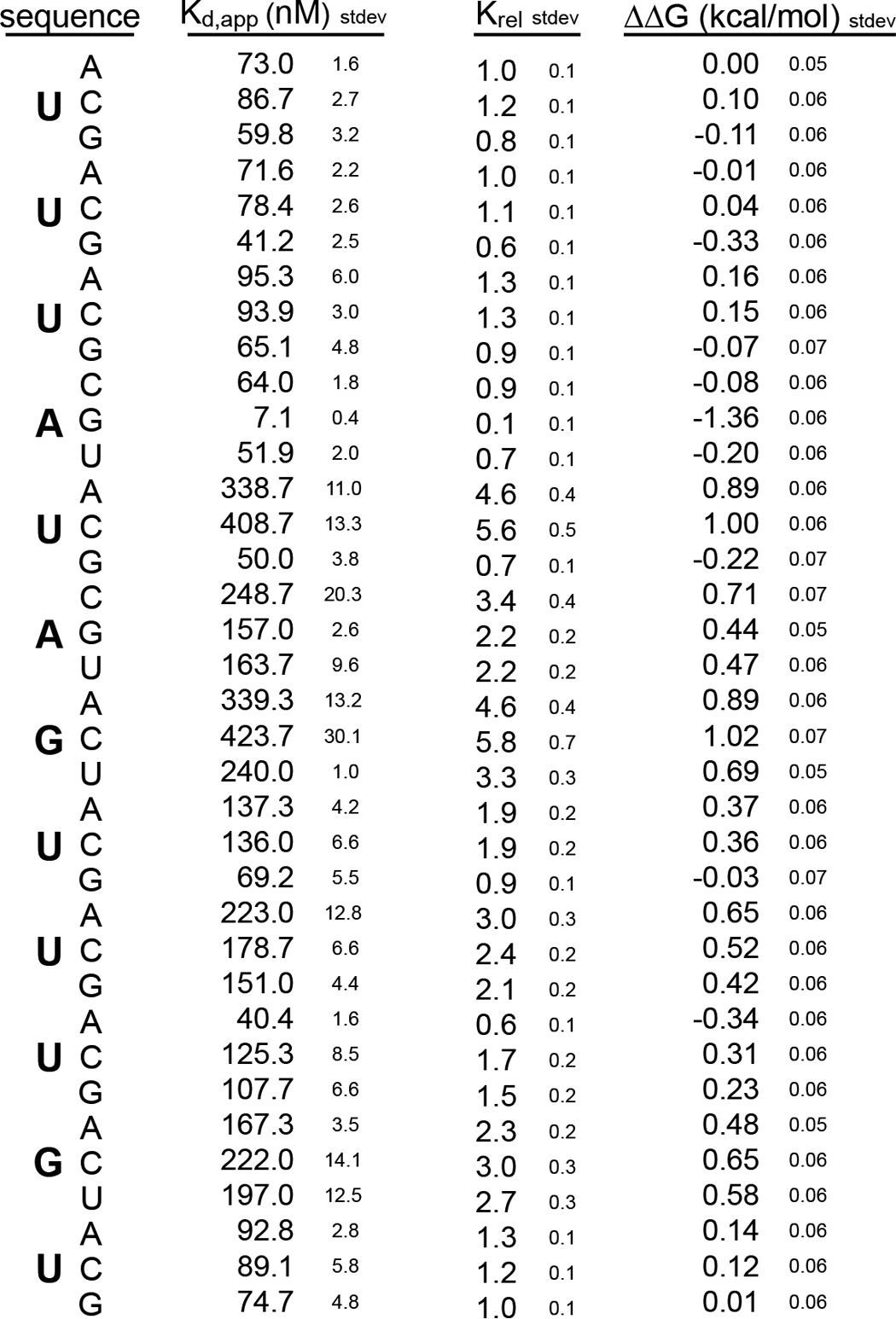
We found several differences in the sequence specificity of the Drosophila protein. The first is a strong preference for a G in position 4 (Fig. 7B). Although the NMR structure of the mouse protein was solved with an RNA that contained a G in this position (23), our mutational analysis showed an equal preference for an A at this site (Fig. 1B). Given that the amino acids predicted to contact this nucleotide are conserved between the two proteins, it is unclear why the mouse protein would have equal preference for A and G whereas the Drosophila protein strongly prefers a G. The structure of mouse MSI1 bound to RNA shows that G1 is recognized via hydrogen bonds to the N7 and 2′-OH and by stacking with tryptophan 29. These contacts should equally accommodate an A or G as we observe for the mouse protein but not the fly protein. It is likely that additional contacts enforce the G preference in the Drosophila variant. The second is that we observed that mutations at positions 9 and 11 also produced changes of ∼0.5 kcal/mol (Fig. 7B). Because the homology model does not predict an extended interface, there must be aspects of the interaction between the Drosophila protein and the RNA that are not predicted by the mouse interaction.
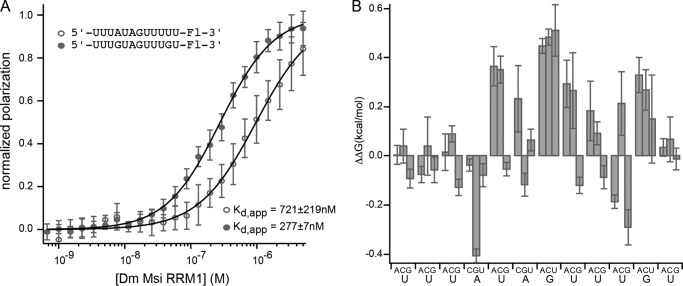
The observation that Drosophila MSI interacts with a longer nucleotide sequence than the mouse protein suggests the possibility that RRM2 contributes to the specificity of the Drosophila protein unlike its mouse homologs. To test this idea, we prepared Drosophila MSI RRM1 and analyzed its interaction with the same series of RNAs analyzed for Drosophila RRM1-2. The affinity of RRM1 for the starting sequence, 5′-UUUAUAGUUUGU-3′, was 558 ± 23 nm, which is 10-fold weaker than that of RRM1-2 (53 ± 1 nm) (Table 5), suggesting that RRM2 contributes to the affinity of MSI for RNA as it does for mouse. We observed similar sequence requirements for RRM1 at positions 4 and 11 as for RRM1-2, suggesting that RRM1 defines the specificity of the interaction and that Drosophila RRM1 makes different contacts with RNA than mouse RRM1 (Fig. 8A and Table 5). Despite the similarities in nucleotide requirements, the magnitude of the change in affinity upon mutation was not as large as that of RRM1-2 (Figs. 7B and 8B). The reduction is likely due to the inherent difficulties of measuring sequence-specific binding of weaker interactions, such as nonspecific binding at high protein concentration and underestimation of the Kd of the weakest binders during curve fitting.
TABLE 5.
Binding of Drosophila MSI RRM1 to mutant RNAs
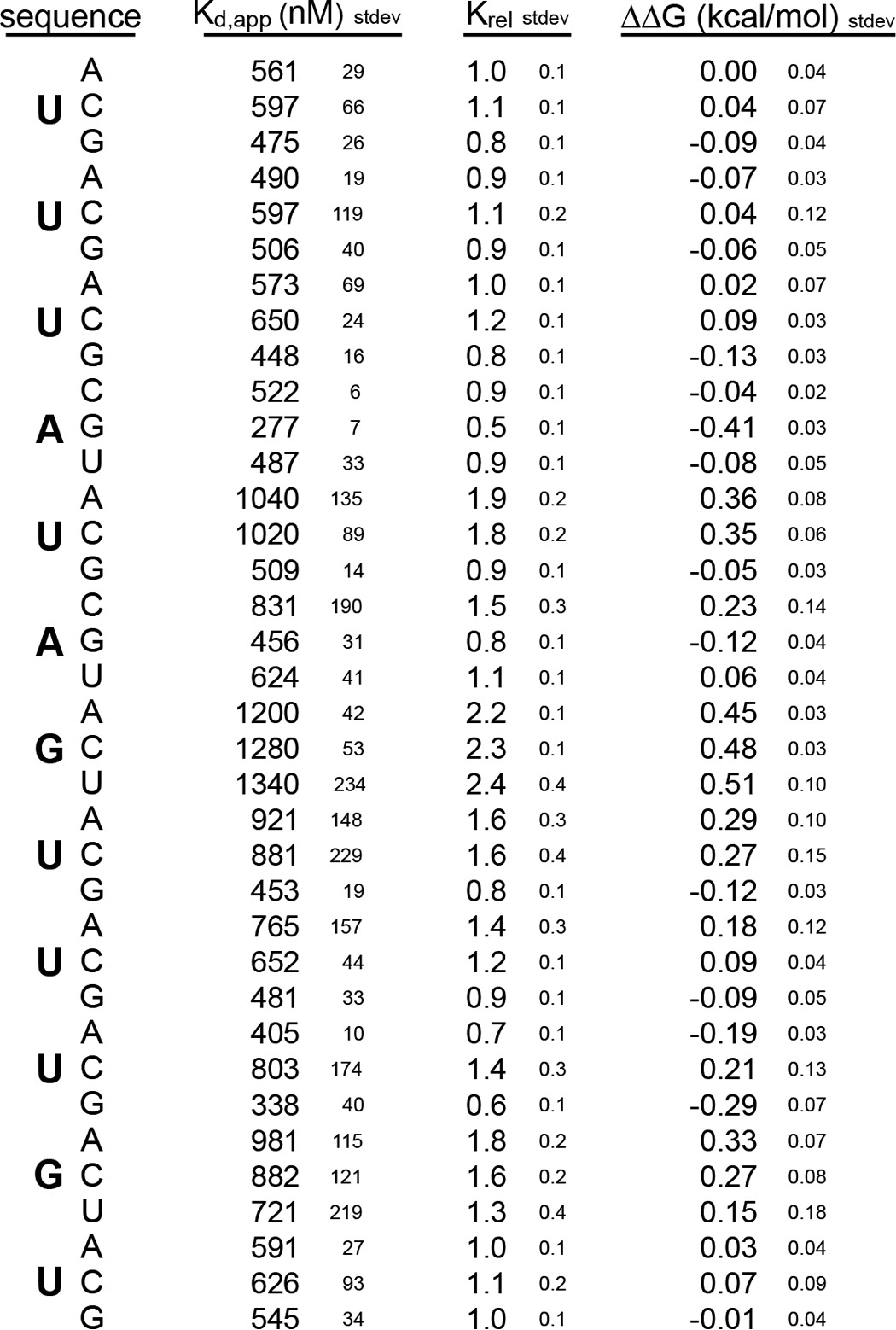
FIGURE 8.
A, FP of the RNAs 5′-UUUGUAGUUUGU-FAM-3′ and 5′-UUUAUAGUUUUU-FAM-3′ bound to a series of concentrations of Drosophila (Dm) MSI RRM1. Data were fit to the Hill equation for calculation of the Kd,app and were normalized for display purposes. B, the change in ΔΔG due to the indicated mutation at each position of the RNA depicted in A. Error bars represent the S.D.
DISCUSSION
We have delineated the contribution of each nucleotide to binding affinity for three MSI family members, mouse MSI1, human MSI2, and Drosophila MSI. For mouse MSI1 and human MSI2, the three-nucleotide core motif UAG is responsible for the majority of the binding affinity. There are two major differences between binding of the Drosophila and the mammalian proteins with RNA. First, Drosophila MSI requires a more extended motif, including the presence of a downstream guanosine. This guanosine appears to be specified by RRM1. Second, mouse MSI1 is able to recognize the sequences GUAGU and AUAGU with similar affinities, whereas Drosophila MSI has a higher affinity for the sequence GUAGU. The reason for these differences is unclear given that the majority of the amino acids contacting RNA in the mouse structure are conserved with the Drosophila protein. However, visual inspection of the sequence of RRM1 in the two species reveals a lysine at position 244 that is a threonine in the mouse (Fig. 5). This amino acid is adjacent to Lys-243, which is predicted to make contacts with G1. Lys-244 is therefore a candidate factor that may contribute to guanosine specification by Drosophila MSI RRM1. Additionally, the RNAcompete Drosophila binding site indicates a weaker preference for an A upstream of the G, further suggesting that there may be aspects of the interaction between Drosophila MSI and RNA that differ from the mouse interaction.
We have also determined that insertion of a stretch of up to four additional uridines immediately upstream of the UAG but downstream of the G or A has no effect on binding affinity. Although the identity of the nucleotide at position 4 has a relatively small impact on affinity, the presence of a cytidine at position 3 reduced affinity by ∼0.5 kcal/mol even when up to three uridines were present. The effect was reduced in mutants that contained more than three uridines. Identification of cross-link immunoprecipitation tags for MSI would reveal whether MSI-bound RNAs in cells contain insertions or other sequence variations within this region.
The results of our analyses provide an explanation for the differences between MSI consensus sequences determined by different methods. For the mouse protein, our data provide support for the motif identified by SELEX, (G/A)U1–3AGU (22). Additionally, our data are consistent with the recently published NMR structure of MSI bound to GUAGU (23). Finally, our observation that UAG is the critical binding element provides an explanation as to why the sequence UAGUUAG was identified by RNAcompete with a strong preference for the first UAG and a weak preference for the following four nucleotides (24). The second UAG is likely to be favored due to avidity effects caused by the presence of a second interaction site. For the Drosophila protein, our data expand upon the motif identified by SELEX, GUUU(G/AG) (25), as it reveals the need for a UAG and the similarity between the mouse and Drosophila interactions. Finally, our data expand upon the consensus sequences identified by SELEX by revealing the relative contribution of each nucleotide to the interaction affinity. Such information is critical in the identification of suboptimal binding sites, which may be more accessible than optimal sites for interaction with competing regulatory factors within a cell.
The preference of the Musashi family proteins appears to be unique among RNA-binding proteins. According to the resource provided by RNAcompete, the RNA-binding proteins with the most closely related binding determinants specifying UAG are pufferfish heterogeneous nuclear ribonucleoprotein AB and human DAZAP1 (24). Each of these contains a UAG within the context of a larger motif. Several other proteins also contain UAGs within larger motifs, including heterogeneous nuclear ribonucleoproteins A1 and A2B1 (24, 37). More work is needed to assess the relative contribution of individual nucleotides within each motif.
Recently published RNA immunoprecipitation experiments using human MSI1 have identified recognition sites for the two RRMs within targets, that is the presence of a GUAG for RRM1 and a UAG for RRM2 (3). However, NMR experiments have shown that RRM2 has limited affinity for RNA due to its surface electrostatic potential and its relative rigidity (20). Our experiments demonstrate that for 12-nucleotide RNAs the presence of RRM2 increases the affinity of interaction without changing the nucleotide requirements. In other words, the central UAG is the major binding determinant in the presence and absence of RRM2. Despite this fact, when we analyzed the number of UAG motifs in RNAs that have been shown previously to associate with MSI (3), we were able to detect an increase in the frequency of 50-nucleotide windows that contain two UAG motifs. There are two possible explanations for this observation. First, recovery after MSI1 immunoprecipitation is more probable for RNAs that contain more UAG motifs and may be bound to multiple MSI1 proteins due to avidity. Second, multiple UAG motifs could be required for in vivo association in a way not described by our study.
RRMs are highly versatile RNA-binding platforms. They have been shown to interact with nucleotide sequences that vary in length from two nucleotides for the second RRM of nucleolin and TDP-43 (38, 39) and three nucleotides for deleted in azoospermia-like (DAZL) (40) to eight nucleotides for the first RRM of U2B″ (41). The lengths of the binding determinants for both mouse and Drosophila Musashi proteins are within this range. Many RRM-containing proteins have two or more tandem RRMs. Recognition of RNA by both RRMs would be expected to increase affinity and specificity. For TDP-43, this has been shown to be the case (39). However, for both mouse and Drosophila Musashi proteins, the presence of the second domain does not affect the specificity of the interaction.
Understanding the forces that drive RBPs to select specific RNA targets from a heterogeneous pool of nucleic acids is central to understanding how they control gene expression. The relative importance of the three-nucleotide UAG MSI motif is seemingly at odds with its relative frequency in the pool of cellular RNAs. For MSI and other proteins with short recognition elements, it is likely that other factors in addition to binding site recognition contribute to the process of target selection. Understanding the nature of these factors and their contribution to target selection will be an exciting area of research in the future.
Acknowledgment
We thank Phil Zamore for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM098763 (to F. M.) and GM081422 and GM098643 (to S. P. R.).
- RBP
- RNA-binding protein
- FP
- fluorescence polarization
- MSI
- Musashi
- RRM
- RNA recognition motif
- SELEX
- systematic evolution of ligands by exponential enrichment.
REFERENCES
- 1. Okano H., Kawahara H., Toriya M., Nakao K., Shibata S., Imai T. (2005) Function of RNA-binding protein Musashi-1 in stem cells. Exp. Cell Res. 306, 349–356 [DOI] [PubMed] [Google Scholar]
- 2. Siddall N. A., McLaughlin E. A., Marriner N. L., Hime G. R. (2006) The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc. Natl. Acad. Sci. U.S.A. 103, 8402–8407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vo D. T., Subramaniam D., Remke M., Burton T. L., Uren P. J., Gelfond J. A., de Sousa Abreu R., Burns S. C., Qiao M., Suresh U., Korshunov A., Dubuc A. M., Northcott P. A., Smith A. D., Pfister S. M., Taylor M. D., Janga S. C., Anant S., Vogel C., Penalva L. O. (2012) The RNA-binding protein Musashi1 affects medulloblastoma growth via a network of cancer-related genes and is an indicator of poor prognosis. Am. J. Pathol. 181, 1762–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Byers R. J., Currie T., Tholouli E., Rodig S. J., Kutok J. L. (2011) MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood 118, 2857–2867 [DOI] [PubMed] [Google Scholar]
- 5. Kharas M. G., Lengner C. J., Al-Shahrour F., Bullinger L., Ball B., Zaidi S., Morgan K., Tam W., Paktinat M., Okabe R., Gozo M., Einhorn W., Lane S. W., Scholl C., Fröhling S., Fleming M., Ebert B. L., Gilliland D. G., Jaenisch R., Daley G. Q. (2010) Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 16, 903–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ito T., Kwon H. Y., Zimdahl B., Congdon K. L., Blum J., Lento W. E., Zhao C., Lagoo A., Gerrard G., Foroni L., Goldman J., Goh H., Kim S.-H., Kim D.-W., Chuah C., Oehler V. G., Radich J. P., Jordan C. T., Reya T. (2010) Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 466, 765–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Sousa Abreu R., Sanchez-Diaz P. C., Vogel C., Burns S. C., Ko D., Burton T. L., Vo D. T., Chennasamudaram S., Le S. Y., Shapiro B. A., Penalva L. O. (2009) Genomic analyses of Musashi1 downstream targets show a strong association with cancer-related processes. J. Biol. Chem. 284, 12125–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dobson N. R., Zhou Y.-X., Flint N. C., Armstrong R. C. (2008) Musashi1 RNA-binding protein regulates oligodendrocyte lineage cell differentiation and survival. Glia 56, 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaneko Y., Sakakibara S., Imai T., Suzuki A., Nakamura Y., Sawamoto K., Ogawa Y., Toyama Y., Miyata T., Okano H. (2000) Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev. Neurosci. 22, 139–153 [DOI] [PubMed] [Google Scholar]
- 10. Sakakibara S., Imai T., Hamaguchi K., Okabe M., Aruga J., Nakajima K., Yasutomi D., Nagata T., Kurihara Y., Uesugi S., Miyata T., Ogawa M., Mikoshiba K., Okano H. (1996) Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev. Biol. 176, 230–242 [DOI] [PubMed] [Google Scholar]
- 11. Asai R., Okano H., Yasugi S. (2005) Correlation between Musashi-1 and c-hairy-1 expression and cell proliferation activity in the developing intestine and stomach of both chicken and mouse. Dev. Growth Differ. 47, 501–510 [DOI] [PubMed] [Google Scholar]
- 12. Wang X.-Y., Yin Y., Yuan H., Sakamaki T., Okano H., Glazer R. I. (2008) Musashi1 modulates mammary progenitor cell expansion through proliferin-mediated activation of the Wnt and Notch pathways. Mol. Cell. Biol. 28, 3589–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kayahara T., Sawada M., Takaishi S., Fukui H., Seno H., Fukuzawa H., Suzuki K., Hiai H., Kageyama R., Okano H., Chiba T. (2003) Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 535, 131–135 [DOI] [PubMed] [Google Scholar]
- 14. Sakakibara S., Nakamura Y., Satoh H., Okano H. (2001) RNA-binding protein Musashi2: developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J. Neurosci. 21, 8091–8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakamura M., Okano H., Blendy J. A., Montell C. (1994) Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron 13, 67–81 [DOI] [PubMed] [Google Scholar]
- 16. Yoda A., Sawa H., Okano H. (2000) MSI-1, a neural RNA-binding protein, is involved in male mating behaviour in Caenorhabditis elegans. Genes Cells 5, 885–895 [DOI] [PubMed] [Google Scholar]
- 17. Charlesworth A., Wilczynska A., Thampi P., Cox L. L., MacNicol A. M. (2006) Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 25, 2792–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Richter K., Good P. J., Dawid I. B. (1990) A developmentally regulated, nervous system-specific gene in Xenopus encodes a putative RNA-binding protein. New Biol. 2, 556–565 [PubMed] [Google Scholar]
- 19. Kurihara Y., Nagata T., Imai T., Hiwatashi A., Horiuchi M., Sakakibara S., Katahira M., Okano H., Uesugi S. (1997) Structural properties and RNA-binding activities of two RNA recognition motifs of a mouse neural RNA-binding protein, mouse-Musashi-1. Gene 186, 21–27 [DOI] [PubMed] [Google Scholar]
- 20. Miyanoiri Y. (2003) Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, Musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics. J. Biol. Chem. 278, 41309–41315 [DOI] [PubMed] [Google Scholar]
- 21. Kawahara H., Imai T., Imataka H., Tsujimoto M., Matsumoto K., Okano H. (2008) Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Biol. 181, 639–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Imai T., Tokunaga A., Yoshida T., Hashimoto M., Mikoshiba K., Weinmaster G., Nakafuku M., Okano H. (2001) The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 21, 3888–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ohyama T., Nagata T., Tsuda K., Kobayashi N., Imai T., Okano H., Yamazaki T., Katahira M. (2012) Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions. Nucleic Acids Res. 40, 3218–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ray D., Kazan H., Cook K. B., Weirauch M. T., Najafabadi H. S., Li X., Gueroussov S., Albu M., Zheng H., Yang A., Na H., Irimia M., Matzat L. H., Dale R. K., Smith S. A., Yarosh C. A., Kelly S. M., Nabet B., Mecenas D., Li W., Laishram R. S., Qiao M., Lipshitz H. D., Piano F., Corbett A. H., Carstens R. P., Frey B. J., Anderson R. A., Lynch K. W., Penalva L. O., Lei E. P., Fraser A. G., Blencowe B. J., Morris Q. D., Hughes T. R. (2013) A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Okabe M., Imai T., Kurusu M., Hiromi Y., Okano H. (2001) Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature 411, 94–98 [DOI] [PubMed] [Google Scholar]
- 26. Takahashi T., Suzuki H., Imai T., Shibata S., Tabuchi Y., Tsuchimoto K., Okano H., Hibi T. (2013) Musashi-1 post-transcriptionally enhances phosphotyrosine-binding domain-containing m-Numb protein expression in regenerating gastric mucosa. PLoS One 8, e53540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuwako K., Kakumoto K., Imai T., Igarashi M., Hamakubo T., Sakakibara S., Tessier-Lavigne M., Okano H. J., Okano H. (2010) Neural RNA-binding protein Musashi1 controls midline crossing of precerebellar neurons through posttranscriptional regulation of Robo3/Rig-1 expression. Neuron 67, 407–421 [DOI] [PubMed] [Google Scholar]
- 28. Horisawa K., Imai T., Okano H., Yanagawa H. (2009) 3′-Untranslated region of doublecortin mRNA is a binding target of the Musashi1 RNA-binding protein. FEBS Lett. 583, 2429–2434 [DOI] [PubMed] [Google Scholar]
- 29. Battelli C., Nikopoulos G. N., Mitchell J. G., Verdi J. M. (2006) The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol. Cell. Neurosci. 31, 85–96 [DOI] [PubMed] [Google Scholar]
- 30. Pagano J. M., Clingman C. C., Ryder S. P. (2011) Quantitative approaches to monitor protein-nucleic acid interactions using fluorescent probes. RNA 17, 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 32. Goddard T. D., Kneller D. G. (2008) SPARKY 3, University of California, San Francisco [Google Scholar]
- 33. Yeo G. W., Coufal N. G., Liang T. Y., Peng G. E., Fu X.-D., Gage F. H. (2009) An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 16, 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 35. Clingman C. C., Deveau L. M., Hay S. A., Genga R. M., Shandilya S. M., Massi F., Ryder S. P. (2014) Allosteric inhibition of a stem cell RNA-binding protein by an intermediary metabolite. Elife 3, e02848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Siddall N. A., Kalcina M., Johanson T. M., Monk A. C., Casagranda F., Been R. P., McLaughlin E. A., Hime G. R. (2012) Drosophila Rbp6 is an orthologue of vertebrate Msi-1 and Msi-2, but does not function redundantly with dMsi to regulate germline stem cell behaviour. PLoS One 7, e49810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burd C. G., Dreyfuss G. (1994) RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 13, 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Allain F. H., Bouvet P., Dieckmann T., Feigon J. (2000) Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 19, 6870–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lukavsky P. J., Daujotyte D., Tollervey J. R., Ule J., Stuani C., Buratti E., Baralle F. E., Damberger F. F., Allain F. H. (2013) Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat. Struct. Mol. Biol. 20, 1443–1449 [DOI] [PubMed] [Google Scholar]
- 40. Jenkins H. T., Malkova B., Edwards T. A. (2011) Kinked β-strands mediate high-affinity recognition of mRNA targets by the germ-cell regulator DAZL. Proc. Natl. Acad. Sci. U.S.A. 108, 18266–18271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Price S. R., Evans P. R., Nagai K. (1998) Crystal structure of the spliceosomal U2B″-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature 394, 645–650 [DOI] [PubMed] [Google Scholar]