Sir,
Infectious diseases are now the world's biggest killer of children and young adults, it is also the basis of debilitating illness all through the year keeping children away from school and preventing adults from working.[1] Conventional methods of culture are now being replaced by automated culture systems, owing to reduced time for culture and ease of laboratory work using machines and higher isolation rate against conventional methods.[2] Lot of information is available for culturing blood samples using BACTEC™ culture, but there is a paucity of data regarding sterile body fluids culture. Hence, this study was done to compare isolation rate and the time needed for the detection of microorganisms between conventional methods of culture and BACTEC™ culture.
This prospective study was conducted for a period of 2 years from July 2011 to July 2013 in the Department of Microbiology of a tertiary care hospital after obtaining the Institutional Ethical Committee approval. A total of 127 body fluid samples were inoculated into BACTEC™ peds Plus™/F vials, incubated in BACTEC™ machine. Direct plating of sample onto sheep blood agar, chocolate agar, MacConkey agar and incubated aerobically at 37°C for 48 h. Organisms isolated were identified using standard microbiological procedures and tested for antimicrobial susceptibility according to Clinical and Laboratory Standards Institute guidelines.[1,2] All data were analyzed using SPSS (IBM) Descriptive statistics were derived using frequency, percentage, and mean.
In this study, 21 samples out of 127, showed a positive result as compared with 4 out of 127 by conventional method. The average time taken for growth in a conventional method was 26.5 h, while that in BACTEC™ is 14.25 h. This significant increase in isolation rate with reduced time to positivity gives BACTEC™ culture edge over conventional culture methods. This is quite a significant time difference considering that presumptive identification of the pathogen was informed to the clinician 12.25 h earlier than a conventional method. This would significantly affect the change of empirical antibiotic therapy given to the patient, eventually aiding in therapy and the final clinical outcome.
Furthermore, the initial Gram's stain report of positive cultures and the association of it with the bacteria isolated were compared. Though few samples showed no bacteria, but polymorphonuclear leukocytes (PMNLs) and mononuclear cells on initial Gram's stain were seen. Alberts et al.,[3] states that presence of PMNLs and lymphocytes suggests infection, and it is responsible for phagocytosis and killing of invading bacteria. Consequently even low bacterial load in the sample, which is undetectable by conventional method, can be detected by BACTEC™.[4,5]
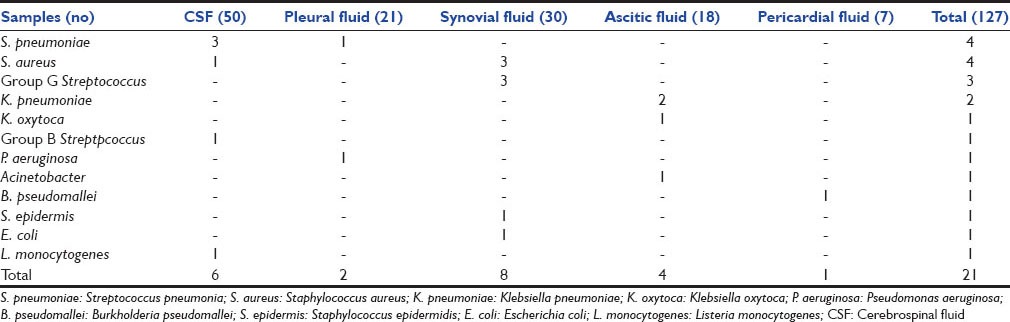
Staphylococcus aureus, Streptococcus pneumoniae, Klebsiella pneumoniae and Pseudomonas aeruginosa were the frequently isolated bacteria in both methods. The fastidious bacteria like Group B and G Streptococcus, Burkholderia pseudomallei, Listeria monocytogenes, S. pneumoniae, Escherichia coli, Acinetobacter were isolated only from BACTEC™. Table 1 shows the distribution of bacteria isolated in various samples. L. monocytogenes, S. pneumoniae and Group B Streptococcus were also isolated from blood culture.
Table 1.
Distribution of organisms isolated from sterile body fluid samples
In conclusion, BACTEC™ automated culture has higher isolation rate of pathogenic bacteria and lesser time for identification even with less bacterial load or count in the sterile body fluid sample. It also helps in guiding antimicrobial therapy especially in critically ill patients and eventually affecting patient's outcome.
References
- 1.Collee JG, Marr W. Culture of bacteria. In: Collee JG, Fraser AG, Marmion BP, Simmons A, editors. Mackie and McCartney: Practical Medical Microbiology. 14th ed. New Delhi: Reed Elsevier; 2008. p. 124. [Google Scholar]
- 2.CLSI. CLSI Document M100-S21. Wayne, PA: Clinical and Laboratory Standards Institute; 2009. Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- 3.Alberts B, Johnson A, Lewis J, Raf M, Roberts K, Walter P. Molecular Biology of the Cell. 4th ed. New York: Garland Science; 2002. [Last accessed on 2013 Jul 25]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK21054/ [Google Scholar]
- 4.Velay A, Schramm F, Gaudias J, Jaulhac B, Riegel P. Culture with BACTEC Peds Plus bottle compared with conventional media for the detection of bacteria in tissue samples from orthopedic surgery. Diagn Microbiol Infect Dis. 2010;68:83–5. doi: 10.1016/j.diagmicrobio.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Daur AV, Klimak F, Jr, Cogo LL, Botão GD, Monteiro CL, Dalla Costa LM. Enrichment methodology to increase the positivity of cultures from body fluids. Braz J Infect Dis. 2006;10:372–3. doi: 10.1590/s1413-86702006000600002. [DOI] [PubMed] [Google Scholar]


