Abstract
This paper examines the diffeomorphometry of MRI derived structural markers for the amygdala, in subjects with symptomatic Alzheimer’s disease (AD). Using linear mixed-effects models we show differences between those with symptomatic AD and controls. Based on template centered population analysis, the distribution of statistically significant change is seen in both the volume and shape of the amygdala in subjects with symptomatic AD compared to controls. We find that high-dimensional vertex based markers are statistically more significantly discriminating (p<.00001) than lower-dimensional markers and volumes, consistent with comparable findings in presymptomatic Alzheimer’s disease. Using a high-field 7T atlas, significant atrophy was found to be centered in the basomedial and basolateral subregions, with no evidence of centromedial involvement.
Keywords: amygdala, MCI, Alzheimer’s disease, shape, MRI
Introduction
Magnetic resonance imaging (MRI) studies have substantially advanced our knowledge of brain atrophy in Alzheimer’s Disease (AD). MRI measures are an indirect reflection of the neuronal injury that occurs in the brain as the AD pathophysiological process evolves. Several MRI measures are known to be altered among individuals with AD dementia or mild cognitive impairment (MCI). In the initial stages of AD, atrophy appears to have a predilection for brain regions in the medial temporal lobe with heavy deposits of neurofibrillary tangles (Arnold, et al., 1991, Braak and Braak, 1991, Price and Morris, 1999). Consistent with this pattern of neurofibrillary pathology, the volume of the entorhinal cortex and hippocampus have been shown to discriminate patients with AD dementia or MCI versus cognitively normal subjects and to be associated with likelihood of progression from MCI to AD dementia (Atiya, et al., 2003, Kantarci and Jack, 2004).
To date, most MRI studies of subcortical gray matter nuclei have defined a single measure of structural volume (Bossa, et al., 2011, McEvoy, et al., 2011, Roh, et al., 2011). While this has the advantage of being quantitative, it does not give specific information about subregions of atrophied nuclei. This information would be useful in order to determine whether morphometric results correlate with neuropathologic studies, to define better the subregional distribution of atrophy, and correlate pathologic changes with clinical features of AD.
Diffeomorphometry and geodesic positioning in computational anatomy (Miller, et al., 2014) for the study of the distribution of functional and structural change in neurodegeneration has already proved to be very powerful. Statistical shape analysis has been useful for studying normal age-related changes in subcortical nuclei, and for studying a number of other diseases (Ashburner, et al., 2003, Csernansky, et al., 1998, Csernansky, et al., 2000, Qiu, et al., 2010, Thompson, et al., 2004, Wang, et al., 2007). The study described here follows our previous work (Miller, et al., 2013) in which we used diffeomorphometry to measure subregional atrophy in three temporal lobe structures - entorhinal cortex (ERC), hippocampus and amygdala - in subjects with preclinical AD, i.e., individuals who were clinically and cognitively normal at the time of their MRI scans. This approach allows for a fine-scale, high-dimensional analysis of non-uniform change patterns in the structures, and complements coarser measures, such as measures of total volume. Despite its proximity to the hippocampus, relatively little is known about the role of amygdala in MCI and AD. Following earlier histopathological findings (Arriagada, et al., 1992, Herzog and Kemper, 1980, Scott, et al., 1991, Scott, et al., 1992, Tsuchiya and Kosaka, 1990), neuroimaging studies of AD patients suggest that amygdala volume may correlate with that of the hippocampus (Poulin, et al., 2011). Further, recent shape analysis (Cavedo, et al., 2011, Qiu, et al., 2009) suggests there is substantial atrophy within the amygdala in AD.
The study described here focuses on the examination of the amygdala via diffeomorphometry. By mapping features across coordinate systems it was possible to identify morphometric changes obtained in 1.5T scans within high-field 7T parcellations of the amygdala. We demonstrate that the location of statistically significant change is distributed across the core amygdala, including the basolateral and basomedial nuclei, in subjects with symptomatic AD compared to controls.
2. Subjects and Methods
2.1 Study Design
In the present study, known as the BIOCARD study, all subjects were cognitively normal when they were recruited. The mean age of the BIOCARD subjects at baseline was 57.1 years. MRI scans were acquired during the period 1995 – 2005. The participants have now been followed for up to 17 years. Table 1 provides summary of the demographic characteristics of the subjects.
Table 1.
Participant characteristics stratified by outcome status.
| Variable | Control Group (N=230) |
MCI During scan (N=9) |
AD Dementia- During Scan (N=7) |
|---|---|---|---|
| Age at time of baseline MRI scan, mean number of years (SD) | 55.4 (9.8) | 64.3 (9.96) | 63.8 (8.04) |
| Gender, females (%) | 61% | 33% | 57% |
2.2 Selection of Participants
A total of 354 individuals were initially enrolled in the study. Recruitment was conducted by the staff of the Geriatric Psychiatry branch of the Intramural Program of the National Institute of Mental Health (NIMH). Subjects were recruited via printed advertisements, articles in local or national media, informational lectures, or word-of-mouth. The study was designed to recruit and follow a cohort of cognitively normal individuals who were primarily in middle age. By design, approximately three quarters of the participants had a first degree relative with dementia of the Alzheimer type. The overarching goal was to identify variables among cognitively normal individuals that could predict the subsequent development of mild to moderate symptoms of AD. Toward that end, subjects were administered a comprehensive neuropsychological battery annually. Magnetic resonance imaging (MRI) scans, cerebrospinal fluid (CSF), and blood specimens were obtained approximately every two years. The study was initiated at the NIMH in 1995, and was stopped in 2005. In 2009, a research team at the Johns Hopkins School of Medicine was jointly funded by the National Institute on Aging (NIA) and NIMH to re-establish the cohort, continue the annual clinical and cognitive assessments, collect blood, and evaluate the previously acquired MRI scans, CSF and blood specimens. To the best of our knowledge, this is the only study in participants who were cognitively normal at entry, with this set of measures, and with such a long duration of follow-up.
At baseline, all participants completed a comprehensive evaluation at the Clinical Center of the National Institutes of Health (NIH). This evaluation consisted of a physical and neurological examination, an electrocardiogram, standard laboratory studies (e.g., complete blood count, vitamin B12, thyroid function, etc), and neuropsychological testing. Individuals were excluded from participation if they were cognitively impaired, as determined by cognitive testing, or had significant medical problems such as severe cerebrovascular disease, epilepsy or alcohol or drug abuse. Five subjects did not meet the entry criteria and were excluded at baseline, leaving a total 349 participants, who were followed over time.
2.3 MRI Assessments
The MRI scans acquired at the NIH were obtained using a standard multi-modal protocol using GE 1.5T scanner. The scanning protocol included localizer scans, Axial FSE (Fast Spin Echo) sequence (TR = 4250, TE = 108, FOV = 512 × 512, thickness/gap = 5.0/0.0 mm, flip angle = 90, 28 slices), Axial Flair sequence (TR = 9002, TE = 157.5, FOV = 256 × 256, thickness/gap = 5.0/0.0 mm, flip angle = 90, 28 slices), Coronal SPGR (Spoiled Gradient Echo) sequence (TR = 24, TE = 2, FOV = 256 × 256, thickness/gap = 2.0/0.0 mm, flip angle = 20, 124 slices), Sagittal SPGR (Spoiled Gradient Echo) sequence (TR = 24, TE = 3, FOV = 256 × 256, thickness/gap 1.5/0.0 mm, flip angle = 45, 124 slices). The analyses described here used the coronal SPGR scans. A total of 805 scans were acquired from the participants, with a mean of 2.4 scans per person.
2.4 Clinical and Cognitive Assessment
The clinical and cognitive assessments of the participants have been described elsewhere (Moghekar, et al., 2013). The cognitive assessment consisted of a neuropsychological battery covering all major cognitive domains (i.e., memory, executive function, language, spatial ability, attention and processing speed). A clinical assessment was also conducted annually. This included the following: a physical and neurological examination, record of medication use, behavioral and mood assessments, family history of dementia, history of symptom onset, and a Clinical Dementia Rating (CDR), based on a semi-structured interview (Hughes, et al., 1982, Morris, 1993).
2.5 Consensus Diagnoses
Each case included in these analyses has received a consensus diagnosis by the staff of the BIOCARD Clinical Core at Johns Hopkins. This research team included: neurologists, neuropsychologists, research nurses and research assistants. During the study visit, each subject had received a comprehensive cognitive assessment and a Clinical Dementia Rating (CDR), as well as a comprehensive medical evaluation (including a medical, neurologic and psychiatric assessment). For the cases with evidence of clinical or cognitive dysfunction, a clinical summary was prepared that included information about demographics, family history of dementia, work history and past history of medical, psychiatric and neurologic disease, medication use and results from the neurologic and psychiatric evaluation. The reports of clinical symptoms from the subject and collateral sources, based on the CDR, were summarized, and the results of the neuropsychological testing were reviewed. Thus, the diagnostic process for each case was handled in a similar manner: (1) clinical data were examined pertaining to the medical, neurologic and psychiatric status of the subject, (2) reports of changes in cognition by the subject and by collateral sources were examined, and (3) decline in cognitive performance was established. These data were used to: (1) determine whether the subject had become cognitively impaired, (2) determine the likely etiology of any impairment, and (3) determine the age at which the clinical symptoms began, based primarily on the reports of clinical symptoms from the subject and from collateral sources. These diagnostic procedures are identical to those implemented in the Alzheimer’s Disease Research Centers program, supported by the NIA. It is acknowledged that, as this process is dependent on the clinical and cognitive data available at any one point in time, some subjects who are diagnosed as having mild cognitive impairment (MCI) may subsequently be diagnosed as normal or as Impaired not MCI. This represented 4 of the subjects in the total sample.
During the acquisition of the scans at the NIH, 7 subjects were diagnosed with dementia of Alzheimer’s type (DAT), and 9 subjects were diagnosed with mild cognitive impairment (MCI). We term this set of subjects the “symptomatic” cohort, i.e. subjects who had MCI or DAT at the time the scans were taken. Four or more years later, when the study was re-initiated at Johns Hopkins, an additional number of subjects had developed MCI or DAT. During the entire period 230 subjects continued to be cognitively normal, whom we defined as the control group. In the analyses described below (section 2.8.4), we included age and gender as covariates, in order to adjust for any differences in these parameters between the groups.
2.6 Region-of-Interest Analysis
Statistical shape analysis requires a preliminary alignment phase, which produces a high-dimensional representation in a fixed coordinate system. A common approach in this framework is to register all shapes to a single one, called the template, defining each anatomy by its position relative to the template. This is achieved via diffeomorphic mapping methods. It is important, in this context, to ensure that the template is as close as possible to the population, and it will be defined as the population average.
In each scan, the amygdala was segmented using the landmark region-of-interest (ROI) template-based LDDMM pipeline comparable to previous work (Csernansky, et al., 1998, Munn, et al., 2007). A representative elderly cognitively normal subject was selected as the template, and left and right structures were segmented manually. The principal axis was identified by placing the head landmark at the center of the inferior boundary of the entorhinal sulcus (on the most anterior coronal plane showing the limen insula), with the tail landmark placed at the center of the most caudal aspect of the basomedial nucleus (on the most caudal coronal plane showing the amygdala). Equidistant sections were selected perpendicular to this principal axis, with landmarks on each section placed at the dorsal lateral and dorsal medial extent of the amygdala, and at an intermediate point on the dorsal amygdala boundary, as well as at the ventral lateral and ventral medial extents of the amygdala, and intermediate point on ventral amygdala boundary. Please see http://caportal.cis.jhu.edu/wiki/tutorials/amygdala/amygdala.html for a description of the landmark placements. Inter-rater reliability for 9 scans yielded a mean kappa score of 80.95 for left and right segmentations.
Landmarks encompassing the amygdala were used to calculate a rigid transformation (Umeyama, 1991) between the template followed by LDDMM landmark matching (Joshi and Miller, 2000) and image matching (Beg, et al., 2005). These transformations were subsequently used to move the template segmentation onto each subject's MR scan yielding segmentations for each subject.
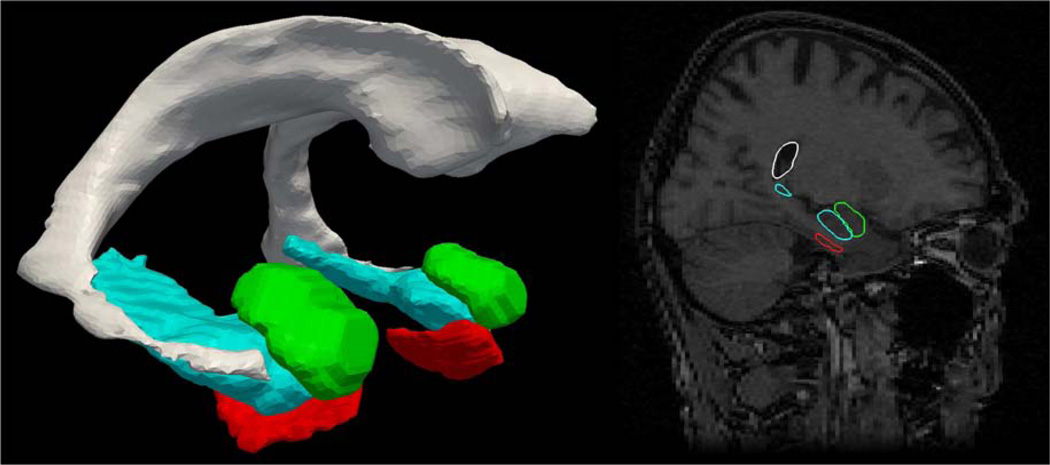
Segmentations were individually inspected, manually corrected where necessary and then converted into triangulated surfaces using the open source iso2mesh software. Fig. 1 shows the amygdala relative to the hippocampus and entorhinal cortex and their embedding in a section in one subject.
Figure 1.
Left: surface reconstruction of amygdala (green), entorhinal cortex (red), hippcampus (blue), ventricle (gray) from one BIOCARD subject. Right: Corresponding MRI section with reconstructed structures embedded in the MRI volume.
2.7 Amygdalar Nuclei Parcellation
An isotropic 7T MRI scan of resolution 0.8mm was used to reconstruct high-field parcellation of the population amygdala. The 7T subject is a 42 year old male who is healthy by self-report. The subject was scanned using a standard MPRAGE protocol in a Philips Achieva 7.0T scanner (TR =4.3 ms, TE=1.95ms, flip=7, FOV=220×220×180). The amygdala was subdivided into four nuclei: lateral, basolateral, basomedial and centromedial nuclei using definitions based on the Paxino Atlas of the Human Brain (Mai, et al., 2004) and illustrated in detail at http://caportal.cis.jhu.edu/wiki/tutorials/amygdala/amygdala.html.
2.8 Diffeomorphometry Shape Analysis via Surface-Based Morphometry (SBM)
2.8.1 Template averaging
Using rigid registration (rotation and translation) each amygdalar surface (from section 2.6) was aligned to a common spatial position. Rigid registration computes an optimal transformation between vertices of two surfaces S0 and S1, by minimizing a score combining registration and soft assignment, given by
Here R and T are a rotation matrix and a translation vector, respectively; σ1 and σ2 are area forms on S1 and S2 respectively and w is a soft assignment function defined on S1× S2, which is positive and satisfies ∫S1 w(x′, y)dσ1(x = ∫S2 w(x, y′)dσ2(y′) = 1 for all (x, y) ∈ S1 × S2 Right subvolumes were flipped before alignment to facilitate comparison of left and right structures in fixed coordinates.
The rigidly aligned volumes were used to generate a population template (Ma, et al., 2010) based on a generative shape model, in which an observed surface is modeled as a random deformation of a template with additive noise. Given this model, the population template is estimated from the surface population data using the mode approximation to the EM algorithm, subject to the topology constraints that the population template is a diffeomorphic transformation of a fixed reference shape, called the hypertemplate. In the Bayesian viewpoint, the population template is considered as a random deformation of the hypertemplate. The population template becomes the coordinate system and is computed by applying the algorithm to the population of 173 baseline scans and is therefore blind to labels.
2.8.2 LDDMM Surface Registration
Non-rigid registration between the population template and all surfaces is done via LDDMM surface registration (Vaillant and Glaunes, 2005) which computes a smooth, invertible, transformation that deforms the template to a surface that is very close to the target. More precisely, it minimizes a two-term energy function taking the form
where Stemp is the population template surface, Sots is the observed surface, and Sdef is the deformed population template. The first term, distshape, is a geodesic distance in shape space, which computes and optimizes a least-deformation path between two surfaces, the distance being given by the optimal deformation cost (Vaillant and Glaunes, 2005). The error term, computes a norm between surfaces. The construction is based on the representation of surfaces as geometric currents.
2.8.3 Shape Statistics
During the preprocessing phase, each subject’s left structure was registered to the template, resulting in the computation, at each vertex k of the template surface, and for each subject s, of a deformation marker Jk(S) that measures the expansion/atrophy at vertex k in subject s relative to the template. This is defined as the logarithm of the local expansion/reduction in surface area around vertex k entailed by template registration, and can be interpreted mathematically as a log-jacobian on the population template surface. See Figs. 3–5 for examples of such markers. These markers were then transformed into a sequence of shape indicators at different resolution as follows:
High resolution - The vertex Jacobian marker Jk(s) which has 750 dimensions associated to the number of vertices.
Projection on Laplace-Beltrami eigenbasis - The first 25 eigenvectors of the surface Laplacian are computed (on the population template) and the 25-dimensional projection of the surface log-Jacobian of the deformation.
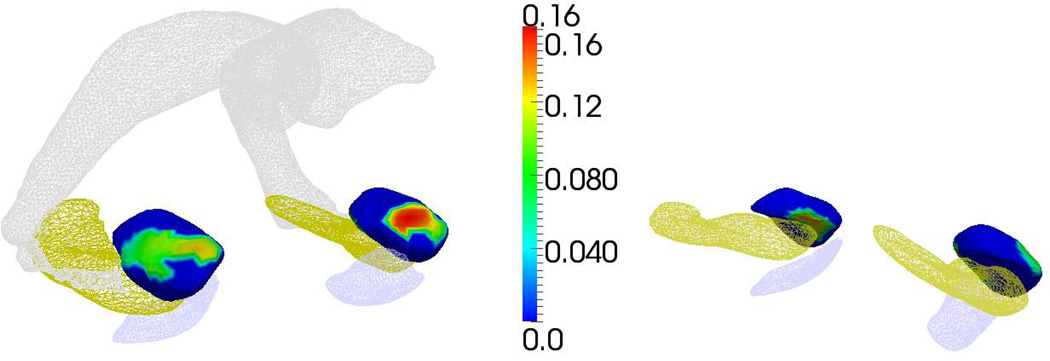
High-field defined subnuclei markers - Four amygdala subfields are defined from the 7T, high-resolution images, which are mapped onto the population template using LDDMM surface matching. This transports the labelled high-field template to the population template allowing the labeling of the vertices. The diffeomorphic surface mapping generates the bijection between the 1.5T population template and the high-field 7T atlas on the continuum of the 3D coordinate system allowing us to transfer the labeled information between the two coordinate systems. The templates were highly sampled so that vertices are small, thereby allowing precise transfer of information between the vertices of the two coordinate systems as defined by the diffeomorphic bijection as shown in Figure 2. The labeled vertices of the subfields are used to develop an integrated Jacobian marker for each of the four nuclei lateral, basolateral, basomedial, and centromedial. This is a four-dimensional marker shown in Figure 3. Note that the values are all positive, indicating that group difference is only associated with atrophy.
Figure 3.
Statistically significant family wise error rate (FWER) at 5% significance for the mixed effects modeling of the symptomatic group compared to the controls, demonstrating atrophy for left and right amygdala. Left panel shows statistics depicted from medial-rostral aspect; right panel shows depiction from medial-caudal aspect. Statistically significant vertex coloring given by the natural log of the surface Jacobian β + β′ā indexed over the template between the control vs symptomatic groups corrected at the average age. For reference only, shown are entorhinal cortex, hippocampus and lateral ventricle.
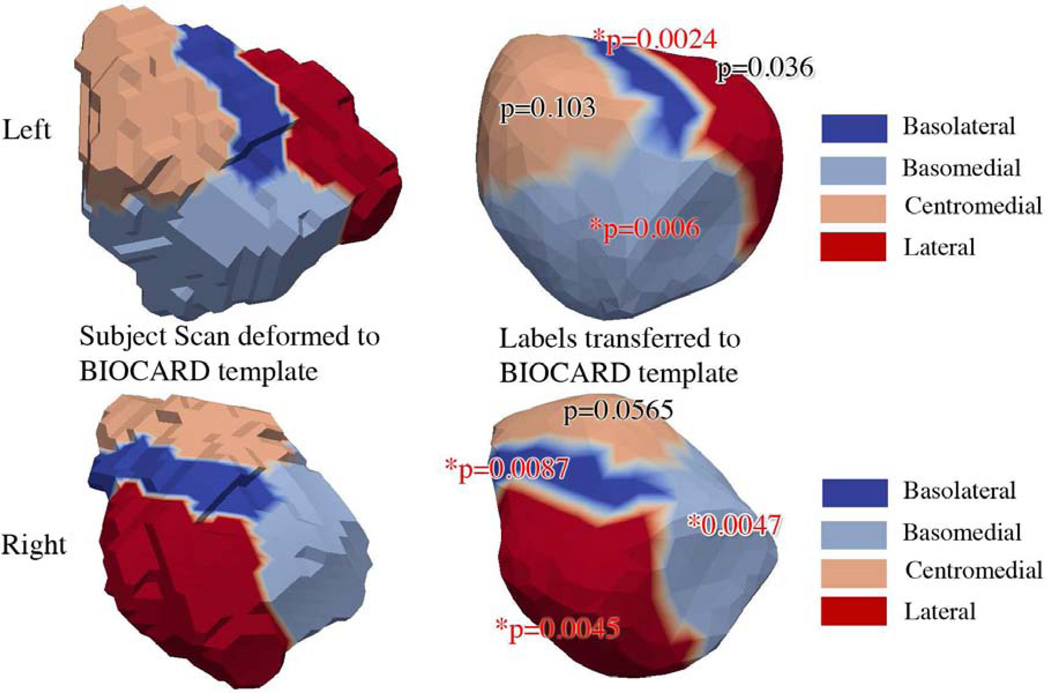
Figure 5.
Results of mixed effects modeling of the symptomatic group compared to the controls for the nuclei of the amygdala. The figures on the left show the 7T high-field left amygdala template (top row) with four subfields defined from the 0.8mm isotropic 7T MRI; the bottom row shows right high-field amygdala. The figures on the right show the subfields transferred to the 1.5T population template showing statistically significant subregions. Significance is determined at .05 FWER, based on Bonferroni bound p<.0125. The top row shows the basomedial (p=.0024) and basolateral (p=.006), with no significance lateral (p=.036) and centromedial (p=.103) for the left amygdala. The bottom row, right column, shows statistically significant subregions, basomedial (p=.0087), basolateral (p=.0047), lateral (p=.0045), with no significance centromedial (p=.0565).
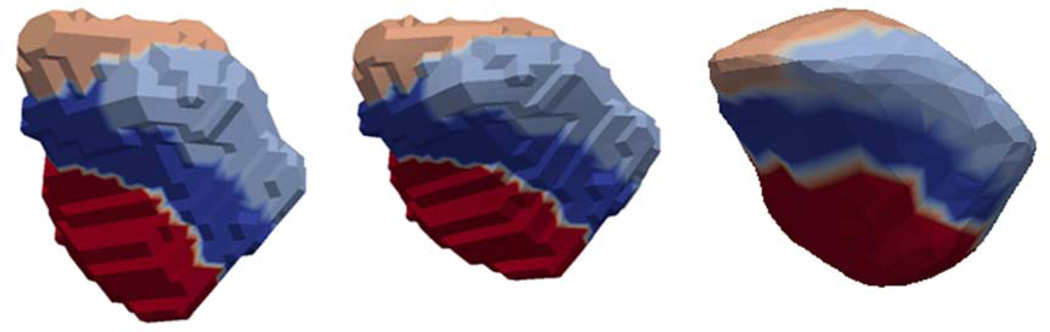
Figure 2.
High-field 7T amygdala template (left) mapped into the 1.5T population coordinates (middle); regions correspond to parcellation into four subnuclei: lateral (red), basolateral (blue) baso-medial (sky-blue), centromedial (caramel). The figure on the right shows a labeling into the 4 subfield parcellation of the 1.5 T population atlas generated by transferring the labels in the closest vertex (middle) to the 1.5T coordinate system.
Global: logarithm of the structure volume.
2.8.4 Linear mixed-effects model
The statistical analyses compared subjects with symptomatic AD (i.e., subjects with MCI and AD dementia) versus cognitively normal individuals. The analyses included age, gender and log intracranial volumes as covariates, and computed statistics at each vertex of the template surface, corrected for multiple comparisons, using permutation tests.
Our statistical model is a linear, mixed-effects model in which the noise associated with the scans from the same subject is modeled as different than the noise associated cross-sectionally in averaging to the common template. Hence the term “mixed-effects”. So we introduce group variables Y(s) equal to 1 if subject s belonged a disease group, and 0 otherwise. These disease groups will be MCI, AD, MCI+AD, for the symptomatic classification. We then use the linear statistical model defined as
where s denotes subjects/structure; j represents scan order in the subject’s time series; v indexes the multivariate marker (v is between 1 and 1, 4, 7, 25 or 750); Jvj(s) is the vth coordinate of deformation marker for subject s at scan j;aj(s) is the subject’s age at scan j; g(s) is the subject’s group (control or disease); c(s) and d(s) are covariates and respectively denote the subject’s intracranial volume and gender; (εvj(s) represents the noise, modeled as εvj(s) = ηv(s) + ζvj(s) where ηv(s) measures between-subject variation and ζvj(s) measures within-subject variation. Both noise are assumed to be centered Gaussian, with variance and , respectively.
The parameters and ρ are estimated by maximum-likelihood. The estimation procedure is iterative and alternates the following two steps until convergence (which usually happens after a small number of iterations).
Step 1: Least square estimation, updating all parameters with fixed ρ. Let n denote the number of subjects, Ns the number of scans for subject s and N the total number of scans (the sum of all Ns). Let d be the number of variables in the linear model (d = 6 in our case) and K the dimension of the shape marker. Rewrite the linear model in the form: J(s) = X(s)θ + ε (s) where J(s) is a Ns × K matrix, X(s) is Ns × d and θ is d × K. Define the matrices and . Define also where Xj(s) is the jth row of X(s), and similarly to set
and
Then, the least square estimator of θ is given by
To estimate the variance, define the residual R(s) = Y(s) − X(s)θ, which is an Ns × K matrix.
For a given v, let . Then
Step 2: Update ρ with all other parameters fixed. Compute, with the notation above
Then, ρ̂ is defined as the minimizer of
This minimization has no closed-form solution and must be performed numerically.
Note that this model assumes that ρ is independent of the shape coordinate, v. Although extending this algorithm to a coordinate-dependent parameter would be straightforward, we have preferred not to do so to avoid the computational burden of performing K non-linear optimization procedures at each step above, especially in the context of permutation tests that are used to estimate p-values.
After convergence, the log-likelihood is given by (up to an additive constant)
2.8.5 Analysis with Family-Wise Error Rates (FWER)
For each type of shape marker, we perform an omnibus test for the significance of the group variables, by testing the null hypothesis . The test statistic is the likelihood ratio between the compared models. The set of coordinates v for which the null hypothesis is rejected is computed via permutation tests, and corrected for multiple comparisons within the shape marker (no correction is made across shape markers, since these are highly correlated).
Letting F denote the log-likelihood ratio, the maximum value over all vertices, , is compared to those obtained by performing the same computation several times, with group labels randomly assigned to subjects. The p-value is given by the fraction of times the values of F* computed after permutation is larger than the value obtained with the true groups. Similarly, p-values are obtained for the volume except that no multiple testing correction is needed.
Permutation testing provides a (conservative) estimate of the set of vertices k on which the null hypothesis is not valid. This set is defined by D = {k : Fk ≥ q*} where q* is the 95 percentile of the observed value of F* over the permutations (Nichols and Hayasaka, 2003). These results are visualized by coloring the vertices that were significant with an atrophy measure defined as , where βk,0, βk are the coefficients associated to the group variable in the regression model for vertex k, and is the average age in the considered disease group population.
Results
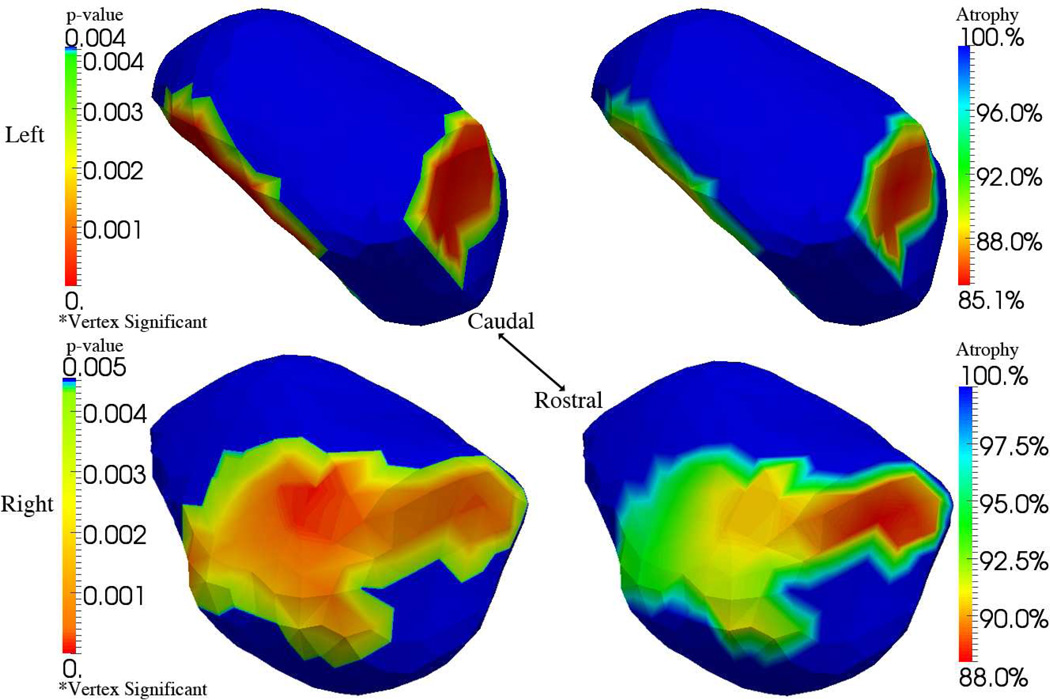
The results of the mixed effects models comparing the controls and the symptomatic cases, taken together as a group, are shown in Table 2, which provides the p-values for shape change between the groups, as measured by the high-dimensional shape markers, based on the Jacobian statistics at the vertices and Laplace Beltrami expansions, and the low-dimensional volume markers. As can be seen, the high-dimensional vertex based markers are statistically more significantly discriminating (p<.00001) than lower-dimensional markers and volumes. In order to more easily visualize the location of these significant differences, the p-values are superimposed on a template of the amygdala, showing the location of maximum difference between the groups (see Figure 4). Note that the images show the amount of surface atrophy relative to the template, which is expressed in percentages, 100% meaning no difference with template. The red value of 85% means, in terms of the atrophy measure, there has been a change (a net decrease) of 15% from the original template to the disease group at 85%.
Table 2.
Differences between controls and symptomatic subjects.. Rows 2 and 3 show p-value resulting from linear mixed effects model analysis of controls versus the combined group of symptomaticcases listed from high to low-dimension. Columns list p-value for the vertex (750 dimensions per structure), and Laplace-Beltrami eigenvectors (25 dimensions per structure), the four 7T regions, and volume biomarkers (1 dimension per structure). Regions in the 7T template are significant at .05 FWER after correcting for multiple comparisons. P-value testing based on geometry segment clustering gives p-values of .028 and .002 for left and right, respectively.
| Groups | Side | Vertex | Laplace | 7T regions | Volume |
|---|---|---|---|---|---|
| Controls vs. | |||||
| Symptomatic | Left | <0.00001 | 0.002 | 0.0047 (1,2) | 0.004 |
| Controls vs. | |||||
| Symptomatic | Right | 0.0001 | 0.005 | 0.01 (1,2,4) | 0.002 |
Figure 4.
Results of mixed effects modeling of the symptomatic group compared to the controls. The top and bottom rows portray the left and right amygdala respectively, showing the p-values (left) and degree of atrophy shown as a percentage decrease relative to the template (right) for the 7T regions projected onto the significant high-field regions, i.e., the basolateral and basomedial nuclei. The blue in the figure on the left implies vertex statistics that are not-significant for FWER 5%;. The blue in the figure on the right implies no atrophy relative to template (determinant Jacobian=1).
In addition, Figure 5 shows the location of the significant differences between the controls and the symptomatic cases in relation to the nuclei of the amygdala, using a high-field 7T atlas. Significance is determined at .05 FWER, based on Bonferroni bound p<.0125. The top row shows the left amygdala subregions with the basomedial (p=.0024) and basolateral (p=.006) significant, with no significance for the lateral (p=.036) and centromedial (p=.103) subregions. The bottom row shows the right amygdala, showing statistically significant subregions, basomedial (p=.0087), basolateral (p=.0047), lateral (p=.0045), with no significance centromedial (p=.0565). Thus, significant atrophy was centered in the basomedial and basolateral subregions, with no evidence of centromedial involvement.
Table 3 provides the p-values for the MCI and the AD dementia cases, taken separately, compared to the controls. The differences between the AD cases and the controls are statistically significant for both left and right hemisphere (p=0.24 and .002, respectively). The differences between the MCI cases and the controls, while in the same direction, as not statistically significant.
Table 3.
Differences between Controls (NC) and MCI subjects (rows 2, 3) and AD dementia subjects (rows 4, 5)/p-value testing based on geometric segments was significant for the AD dementia cases, left and right p-value .024 and .002 respectively.
| Groups | Side | Vertex | Laplace | 7T regions | Volume |
|---|---|---|---|---|---|
| MCI vs | |||||
| NC | Left | 0.01242 | 0.004 | 0.0695 | 0.118 |
| MCI vs | |||||
| NC | Right | 0.067325 | 0.030 | 0.0746 | 0.097 |
| AD vs | |||||
| NC | Left | <0.00001 | 0.005 | 0.0003 (1,2) | 0.002 |
| AD vs | |||||
| NC | Right | 0.0002 | 0.005 | 0.0089 (1,4) | 0.002 |
Discussion
The examination of amygdalar shape analysis in this cohort has revealed volumetric changes as well as non-uniform shape changes in the amygdala in subjects with symptomatic AD. By lever-aging a high-field amygdala atlas, significantly atrophied subregions were located specifically in the basomedial and lateral nuclei. This is consistent with the histopathological findings with greatest cell packing density loss in the medial region and some loss in the lateral region (Herzog and Kemper, 1980). It is also interesting that the region of significant atrophy illustrated in Figs. 3, 4 and 5 is identical to that observed in autopsied amygdala with AD (Scott, et al., 1991) i.e. superiorly adjacent to the paralaminar region. The same study showed great loss of "large nerve cells" in the magnocellular regions of the amygdala which is deep inside the basomedial nucleus. This is consistent with the hypothesis that the observed changes in the surface of the basomedial nucleus reflect the underlying changes.
Our results can be interpreted in terms of the core and non-core amygdalar parcellations by Price (1987, 2003) based on subregions sharing developmental, structural, and functional characteristics. Core amgydala consists of the lateral, basal, and accessory basal nuclei, and the non-core amygdala consists of remaining nuclei including the central, medial, and cortical nuclei (Munn, et al., 2007, Sheline, et al., 1998). From a functional standpoint, subnuclei in the core amygdala are associated with a role in emotional processing and storage of emotional memories (LeDoux and Schiller, 2009, Price, 1981) which provide clues as to how they may be affected in MCI and AD.
The volumetric changes are consistent with those observed in a recent study (Poulin, et al., 2011) of two large samples of MCI and AD cases. The non-uniform changes in shape may explain the variation in volume in the relatively few neuroimaging studies of amygdala in MCI and AD. The lateral part of the amgydala has also been noted in recent shape analysis by Cavedo et al. (2011) and Qiu et al. (2009). The former study is pertinent since a diffeomorphic approach was applied to a small sample and a parcellated amygdala atlas albeit at 3T was used.
The diffeomorphometry methods described here enable transfer of the high-field amygdala parcellation ensuring anatomical consistency. This also allows for the explicit testing of statistical significance by a sub-region by sub-region basis allowing us to demonstrate the significant changes in the affected basolateral regions. The fact that there are significant projections from these regions in amygdala to hippocampus and entorhinal cortex also make it significant in light of our other findings in those structures in previous work (Miller, et al., 2013). Given the temporal lobe circuitry model of the degenerative processes in MCI and AD, future studies will be necessary to correlate shape changes in the amygdala with that in the hippocampus, entorhinal cortex and other structures. Such expanded studies provide additional biomarkers to that of the hippocampus in the early stages of MCI and AD.
We have developed several high field atlases based on 7T and 11T datasets. At these resolutions we are able to define the subfields of the hippocampus including the three partitions of cornu ammons, dentate gyrus and subiculum. These atlases serve as partitions that can be utilized in interpreting the statistically significant vertex based shape change. By mapping our 1.5T population template diffeomorphically to the high field atlas the partitions can be used to segregate the vertices of the template. This serves as another method for developing a lower dimensional representation, intermediate between the 25 dimensional Laplace Beltrami basis and the single volume number of the entire structure. The importance of this parcellation is it is biologically meaningful.
Conclusion
In summary, application of advanced computational anatomy diffeomorphometry methods detected atrophy in basomedial and basolateral regions of the amygdala in patients with symptomatic AD. Despite the small sample size, atrophied subregions of the amygdala could be detected with p-values based on family-wise error rates which unlike false discovery rates have the advantage of not requiring additional assumptions on the data such as independence or positive dependence. These findings warrant further investigation in a larger dataset.
Highlights.
We examine the amygdala using MRI diffeomorphometry
Differences in amygdala shape are shown between controls and symptomatic AD
Differences in amygdala volume are shown between controls and symptomatic AD
High dimensional markers are more discriminating than low dimensional markers
Atrophy is centered in the basomedial and basolateral amygdala subregions
Acknowledgements
This study is supported in part by grants from the Johns Hopkins Brain Sciences Institute and National Institutes of Health: U01-AG03365, P50-AG005146, R01 EB008171 and P41-RR015241. The BIOCARD Study consists of 7 Cores with the following members: (1) the Administrative Core (Marilyn Albert, Barbara Rodzon), (2) the Clinical Core (Ola Selnes, Marilyn Albert, Rebecca Gottesman, Ned Sacktor, Anja Soldan, Guy McKhann, Scott Turner, Leonie Farrington, Maura Grega, Daniel D’Agostino, Sydney Feagen, David Dolan, Hillary Dolan), (3) the Imaging Core (Michael Miller, Susumu Mori, Tilak Ratnanather, Timothy Brown, Hayan Chi, Anthony Kolasny, Kenichi Oishi, Thomas Reigel, Elizabeth Postell, William Schneider, Laurent Younes), (4) the Biospecimen Core (Richard O’Brien, Abhay Moghekar, Richard Meehan), (5) the Informatics Core (Roberta Scherer, Curt Meinert, David Shade, Ann Ervin, Jennifer Jones, Matt Toepfner, Lauren Parlett,April Patterson, Lisa Lassiter), the (6) Biostatistics Core (Mei-Cheng Wang, Shanshan Li, Yi Lu), and (7) the Neuropathology Core (Juan Troncoso, Barbara Crain, Olga Pletnikova, Gay Rudow, Karen Fisher).
We are grateful to the members of the BIOCARD Scientific Advisory Board who provide continued oversight and guidance regarding the conduct of the study including: Drs. John Csernansky, David Holtzman, David Knopman, Walter Kukull and John McArdle, as well as Drs. Neil Buckholtz, John Hsiao, Laurie Ryan and Jovier Evans, who provide oversight on behalf of the National Institute on Aging (NIA) and the National Institute of Mental Health (NIMH), respectively. We would also like to thank the members of the BIOCARD Resource Allocation Com-mittee who provide ongoing guidance regarding the use of the biospecimens collected as part of the study, including: Drs. Constantine Lyketsos, Carlos Pardo, Gerard Schellenberg, Leslie Shaw, Madhav Thambisetty, and John Trojanowski.
We would like to acknowledge the contributions of the Geriatric Psychiatry Branch (GPB) of the intramural program of the NIMH who initiated the study (PI: Dr. Trey Sunderland). We are particularly indebted to Dr. Karen Putnam, who has provided ongoing documentation of the GPB study procedures and the datafiles received from NIMH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement: All authors do not have conflicts of interest.
Data contained in the manuscript has not been submitted or been previously published, nor submitted elsewhere and will not be submitted elsewhere while under consideration at Neurobiology of Aging.
All authors have reviewed the contents of the manuscript being submitted, approve of its contents and validate the accuracy of the data.
References
- Arnold SE, Hyman BT, Flory J, Damasio AR, Van Hoesen GW. The topographical and neuroanatomical distribution of neurofibrillary tangles and neuritic plaques in the cerebral cortex of patients with Alzheimer's disease. Cereb Cortex. 1991;1(1):103–116. doi: 10.1093/cercor/1.1.103. [DOI] [PubMed] [Google Scholar]
- Arriagada PV, Growdon JH, Hedley-Whyte ET, Hyman BT. Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer's disease. Neurology. 1992;42(3 Pt 1):631–639. doi: 10.1212/wnl.42.3.631. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Csernansky JG, Davatzikos C, Fox NC, Frisoni GB, Thompson PM. Computer-assisted imaging to assess brain structure in healthy and diseased brains. Lancet Neurol. 2003;2(2):79–88. doi: 10.1016/s1474-4422(03)00304-1. [DOI] [PubMed] [Google Scholar]
- Atiya M, Hyman BT, Albert MS, Killiany R. Structural magnetic resonance imaging in established and prodromal Alzheimer disease: a review. Alzheimer disease and associated disorders. 2003;17(3):177–195. doi: 10.1097/00002093-200307000-00010. [DOI] [PubMed] [Google Scholar]
- Beg MF, Miller MI, Trouve A, Younes L. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. International Journal of Computer Vision. 2005;61(2):139–157. [Google Scholar]
- Bossa M, Zacur E, Olmos S. Statistical analysis of relative pose information of subcortical nuclei: application on ADNI data. NeuroImage. 2011;55(3):999–1008. doi: 10.1016/j.neuroimage.2010.12.078. doi:S1053-8119(11)00002-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Cavedo E, Boccardi M, Ganzola R, Canu E, Beltramello A, Caltagirone C, Thompson PM, Frisoni GB. Local amygdala structural differences with 3T MRI in patients with Alzheimer disease. Neurology. 2011;76(8):727–733. doi: 10.1212/WNL.0b013e31820d62d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csernansky J, Joshi S, Wang L, Haller J, Gado M, Miller J, Grenander U, Miller M. Hippocampal morphometry in schizophrenia by high dimensional brain mapping. Proc Natl Acad Sci U S A. 1998;95:11406–11411. doi: 10.1073/pnas.95.19.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csernansky J, Wang L, Joshi S, Miller J, Gado M, Kido D, McKeel D, Morris J, Miller M. Early {DAT} is distinguished from aging by high-dimensional mapping of the hippocampus. Neurology. 2000;55:1636–1643. doi: 10.1212/wnl.55.11.1636. [DOI] [PubMed] [Google Scholar]
- Herzog AG, Kemper TL. Amygdaloid changes in aging and dementia. Arch Neurol. 1980;37(10):625–629. doi: 10.1001/archneur.1980.00500590049006. [DOI] [PubMed] [Google Scholar]
- Hughes CP, Berg L, Danziger WL, Coben LA, Martin RL. A new clinical scale for the staging of dementia. Br J Psychiatry. 1982;140:566–572. doi: 10.1192/bjp.140.6.566. [DOI] [PubMed] [Google Scholar]
- Joshi S, Miller MI. Landmark Matching Via Large Deformation Diffeomorphisms. IEEE Transactions on Image Processing. 2000;9(8):1357–1370. doi: 10.1109/83.855431. [DOI] [PubMed] [Google Scholar]
- Kantarci K, Jack CR., Jr Quantitative magnetic resonance techniques as surrogate markers of Alzheimer's disease. NeuroRx : the journal of the American Society for Experimental NeuroTherapeutics. 2004;1(2):196–205. doi: 10.1602/neurorx.1.2.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux JE, Schiller D. The human amygdala: Insights from other Animals. In: Whalen PJ, Phelps EA, editors. The Human Amygdala. New York, NY: Guilford Press; 2009. pp. 43–60. [Google Scholar]
- Ma J, Miller M, Younes L. A Bayesian Generative Model for Surface Template Estimation. International journal of biomedical imaging. 2010;2010 doi: 10.1155/2010/974957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai JK, Assheuer J, Paxinos G. Atlas of the human brain. 2nd ed. Amsterdam; Boston: Elsevier Academic Press; 2004. [Google Scholar]
- McEvoy LK, Holland D, Hagler DJ, Jr, Fennema-Notestine C, Brewer JB, Dale AM. Mild cognitive impairment: baseline and longitudinal structural MR imaging measures improve predictive prognosis. Radiology. 2011;259(3):834–843. doi: 10.1148/radiol.11101975. doi:radiol.11101975 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI, Younes L, Ratnanather JT, Brown T, Trinh H, Postell E, Lee DS, Wang M-C, Mori S, O'Brien R, Albert M. The diffeomorphometry of temporal lobe structures in preclinical Alzheimer's disease. NeuroImage: Clinical. 2013;3:352–360. doi: 10.1016/j.nicl.2013.09.001. doi: http://dx.doi.org/10.1016/j.nicl.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI, Younes L, Trouvé A. Diffeomorphometry and geodesic positioning systems for human anatomy. TECHNOLOGY. 2014;02(01):36–43. doi: 10.1142/S2339547814500010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghekar A, Li S, Lu Y, Li M, Wang MC, Albert M, O'Brien R, Team BR. CSF biomarker changes precede symptom onset of mild cognitive impairment. Neurology. 2013;81(20):1753–1758. doi: 10.1212/01.wnl.0000435558.98447.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43(11):2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Munn MA, Alexopoulos J, Nishino T, Babb CM, Flake LA, Singer T, Ratnanather JT, Huang H, Todd RD, Miller MI, Botteron KN. Amygdala volume analysis in female twins with major depression. Biological Psychiatry. 2007;62(5):415–422. doi: 10.1016/j.biopsych.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols T, Hayasaka S. Controlling the family wise error rate in functional neuroimaging: a comparative review. Statistical Methods in Medical Research. 2003;12(5):419–446. doi: 10.1191/0962280203sm341ra. [DOI] [PubMed] [Google Scholar]
- Poulin SP, Dautoff R, Morris JC, Barrett LF, Dickerson BC Alzheimer's Disease Neuroimaging, I. Amygdala atrophy is prominent in early Alzheimer's disease and relates to symptom severity. Psychiatry Res. 2011;194(1):7–13. doi: 10.1016/j.pscychresns.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL. The efferent projections of the amygdaloid complex in the rat, cat and monkey. In: Ben-Ari Y, editor. The Amygdaloid Complex. Amsterdam: Elsevier/North-Holland; 1981. pp. 121–132. [Google Scholar]
- Price JL. The limbic region. II: The amygdaloid complex. In: Bjorklund A, Hokfelt T, Swanson LW, editors. Handbook of Chemical Neuroanatomy, Vol 5: Integrated Systems of the CNS, Part I. New York, NY: Elsevier; 1987. [Google Scholar]
- Price JL. Comparative aspects of amygdala connectivity. Amygdala in Brain Function: Bacic and Clinical Approaches. 2003;985:50–58. doi: 10.1111/j.1749-6632.2003.tb07070.x. [DOI] [PubMed] [Google Scholar]
- Price JL, Morris JC. Tangles and plaques in nondemented aging and "preclinical" Alzheimer's disease. Ann Neurol. 1999;45(3):358–368. doi: 10.1002/1531-8249(199903)45:3<358::aid-ana12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Qiu A, Adler M, Crocetti D, Miller M, Mostofsky S. Basal Ganglia Shapes Predict Social, Communication, and Motor Dysfunctions in Boys With Autism Spectrum Disorder. Journal of the American Academy of Child & Adolescent Psychiatry. 2010;49:539–551. doi: 10.1016/j.jaac.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Qiu A, Fennema-Notestine C, Dale AM, Miller MI. Regional shape abnormalities in mild cognitive impairment and Alzheimer's disease. NeuroImage. 2009;45:656–661. doi: 10.1016/j.neuroimage.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh JH, Qiu A, Seo SW, Soon HW, Kim JH, Kim GH, Kim MJ, Lee JM, Na DL. Volume reduction in subcortical regions according to severity of Alzheimer's disease. J Neurol. 2011;258(6):1013–1020. doi: 10.1007/s00415-010-5872-1. [DOI] [PubMed] [Google Scholar]
- Scott SA, DeKosky ST, Scheff SW. Volumetric atrophy of the amygdala in Alzheimer's disease: quantitative serial reconstruction. Neurology. 1991;41(3):351–356. doi: 10.1212/wnl.41.3.351. [DOI] [PubMed] [Google Scholar]
- Scott SA, DeKosky ST, Sparks DL, Knox CA, Scheff SW. Amygdala cell loss and atrophy in Alzheimer's disease. Ann Neurol. 1992;32(4):555–563. doi: 10.1002/ana.410320412. [DOI] [PubMed] [Google Scholar]
- Sheline YI, Gado MH, Price JL. Amygdala core nuclei volumes are decreased in recurrent major depression. Neuroreport. 1998;9(9):2023–2028. doi: 10.1097/00001756-199806220-00021. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, De Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer disease. NeuroImage. 2004;22(4):1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- Tsuchiya K, Kosaka K. Neuropathological study of the amygdala in presenile Alzheimer's disease. J Neurol Sci. 1990;100(1–2):165–173. doi: 10.1016/0022-510x(90)90029-m. [DOI] [PubMed] [Google Scholar]
- Umeyama S. Least-Squares Estimation of Transformation Parameters between 2 Point Patterns. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1991;13(4):376–380. [Google Scholar]
- Vaillant M, Glaunes J. Surface matching via currents. LNCS. 2005;3565:381–392. doi: 10.1007/11505730_32. [DOI] [PubMed] [Google Scholar]
- Wang L, Beg M, Ratnanather J, Ceritoglu C, Younes L, Morris J, Csernansky J, Miller M. Large deformation diffeomorphism and momentum based hippocampal shape discrimination in dementia of the Alzheimer type. IEEE transactions on medical imaging. 2007;26:462–470. doi: 10.1109/TMI.2005.853923. [DOI] [PMC free article] [PubMed] [Google Scholar]