Abstract
Naturally occurring polyketides and non-ribosomal peptides with broad and potent biological activities continue to inspire the discovery of new and improved analogs. The biosynthetic apparatus responsible for the construction of these natural products has been the target of intensive protein engineering efforts. Traditionally, engineering has focused on substituting individual enzymatic domains or entire modules with those of different building block specificity, or by deleting various enzymatic functions, in an attempt to generate analogs. This review highlights strategies based on site-directed mutagenesis of substrate binding pockets, semi-rational mutagenesis, and whole-gene random mutagenesis to engineer the substrate specificity, activity, and protein interactions of polyketide and non-ribosomal peptide biosynthetic machinery.
Introduction
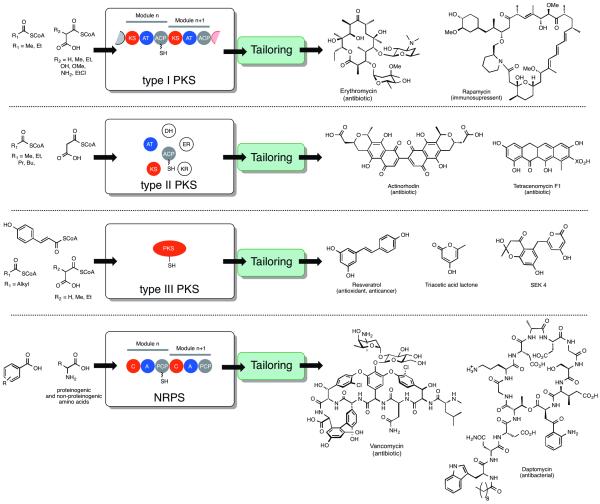
Polyketides and nonribosomal peptides are two large classes of natural products biosynthesized by polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs), respectively [1,2]. Many polyketides and nonribosomal peptides, and hybrids thereof, display potent and often clinically relevant biological activities, including anticancer (e.g. calicheamicin [3] and bleomycin [4]), immunosuppression (e.g. rapamycin [5]), and antibacterial (e.g. erythromycin [6] and vancomycin [7]) (Figure 1). Accordingly, natural products constitute a significant fraction of our current pharmacopeia. For example, of the currently approved anti-cancer drugs, including biologics and vaccines, 65% are natural products or small molecules derived or inspired from natural products [8]. Accordingly, there is significant interest in synthetic routes to such natural products and their analogs. Manipulation of PKS and NRPS machinery offers an attractive alternative to traditional synthetic and semi-synthetic strategies given the potential for combinatorial exploration of chemical space and high scale fermentation. Polyketide synthases are classified in a fashion reminiscent of fatty acid synthases (FASs) (Figure 1). The type I PKSs are multidomain assembly lines, where sets of requisite domains are organized into modules, each being responsible for a complete elongation step in the construction of the polyketide [9]. Substrates and intermediates are covalently tethered to an acyl carrier protein (ACP) domain within each module via a phosphopantetheine prosthetic arm. Fungal PKSs represent a class of important type I enzymes, whereby the requisite domains are housed in a single module that therefore acts in an iterative fashion [10,11]. Accordingly, the length and extent of modification of polyketides produced by fungal PKSs is “cryptically” encoded within the PKS. Conversely, type II PKSs employ discrete, monofunctional proteins that operate more or less independently [12]. Type III PKSs, which include the “chalcone synthase” family [13], use acyl-Coenzyme A (CoA) substrates directly without the use of an ACP, although exceptions are known [14]. Type III PKSs produce relatively simple aromatics with modest structural diversity, in comparison to the products of type I and II systems. The organization of NRPSs resembles that of type I PKSs and consists of one module for each amino acid incorporated into the peptide product (Figure 1). Increasingly, novel PKSs and NRPSs are being discovered that deviate from the canonical organization and can also include unusual domains [15,16]. The synthetic versatility of PKSs is determined in large by the range of starter and extender unit acyl-CoA thioesters available to and utilized by the enzymatic machinery (Figure 1), in addition to the number of extender unit condensations, variety of redox modifications, and cyclization mechanism for a given PKS. Similarly, the synthetic versatility of NRPSs is described by the range of starter and extender unit acids available to and utilized by NRPSs, in conjunction with the availability of other functions (epimerization, heterocyclizaton, oxidation, methylation) and the macrocyclization mechanism. Moreover, products of PKSs and NRPSs are often further decorated by various trans-acting enzymatic functions to furnish the mature natural product. In general, natural product diversification strategies that harness PKSs and NRPSs suffer from poor scope and utility due to restricted substrate specificity and modularity of biosynthetic components. Herein, recent advances related to engineering PKSs and NRPSs by point mutation, semi-random and random mutagenesis approaches will be highlighted. Several excellent reviews are available that summarize advances in combinatorial biosynthesis which aims to produce polyketide and non-ribosomal peptide analogs by substitutions of entire domains and modules [17,18].
Figure 1.
Structural organization and building block scope of PKSs and NRPSs. Typical selection of naturally occurring building blocks are shown.
Altering substrate specificity and protein interactions of PKSs/NRPSs Starter unit selection
Loading modules of type I PKSs catalyze selection and recruitment of starter units into polyketides. Accordingly, starter units are incorporated into polyketides at only one position, and have proved fruitful sites for interception by protein engineering. Notably, substitution of loading domains with those of broad specificity can be used to produce analogues and illustrate that downstream polyketide machinery is quite tolerant to non-natural or non-native starter unit side-chains. Recently, mutations have been discovered that improve production of erythromycin analogs from precursor-derived starter units. To achieve this, a plasmid based heterologous expression system in Escherichia coli (E. coli) was coupled with a colony bioassay [19]. Interestingly, functional mutations were found in the host vector rather than the polyketide synthase genes [20].
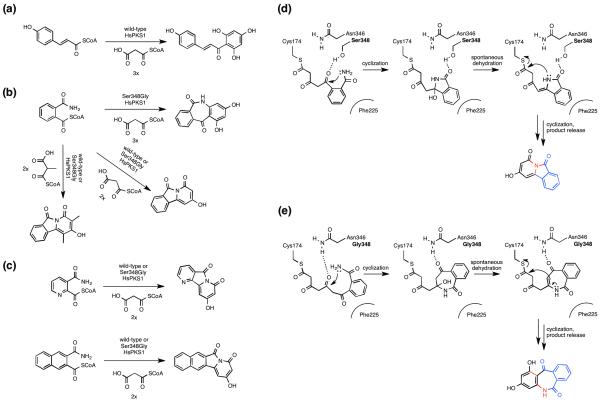
The volume and shape of CHS type III PKS active sites dictate substrate specificity, in addition to controlling polyketide chain length and cyclization pathway. Notably, many CHS type III PKSs display remarkable substrate and catalytic promiscuity [21••,22]. Subsequently, point mutations readily alter the substrate and product specificity of CHS PKSs, including starter unit preference. For example, building on the remarkable substrate tolerance of HsPKS1 from Huperzia serrata, a structure-based mutant (Ser348Gly) extended product chain length and also changed the cyclization mechanism [21••]. The authors speculated that the mutation expanded the space neighboring the catalytic residue, allowing condensation with up to three malonyl-CoA”s, versus two for the wild-type enzyme (Figure 2).
Figure 2.
Wild-type and engineered type II PKS HsPKS1 for the synthesis of unnatural alkaloids. (a) Native reaction catalyzed by the wild-type HsPKS1. (b) Substrate and catalytic versatility of the wild-type and mutant HsPKS1. (c) Starter unit promiscuity of HsPKS1. (d) Proposed catalytic mechanism for formation of the 6.5.6-fused tricyclic ring system by wild-type HsPKS1. (e) Proposed catalytic mechanism for formation of the 6.7.6-fused ring system by the engineered HsPKS1.
Extender unit selection and chain elongation
The substitution of acyltransferase (AT) domains with those of alternate specificity has been used to generate regioselectively-modified polyketide analogues. In many cases however, such analogs involve fairly conservative structural modifications, and product yields are often significantly reduced to the wild-type PKS [23]. Ultimately, less invasive strategies, that involve introduction of amino acid substitutions, either by rational redesign or directed evolution, may offer a more effective strategy for altering polyketide structure. Substrate promiscuity is often a useful prerequisite to successful directed evolution campaigns [24]. Accordingly, promiscuity of polyketide biosynthetic machinery towards non-native and non-natural extenders could prove very useful. While a modest selection of acyl-CoA extender units is cumulatively available to polyketide biosynthesis [25,26], most polyketide producing organisms provide biosynthetic routes to only a small number of unique extender units. Thus, many PKSs need to discriminate between the few extender units naturally provided by the host, yet emerging data suggests that non-native and non-natural extender units can be processed by PKSs [27,28]. For example, PikAIV from pikromycin biosynthesis was shown to display a hydrolytic editing mechanism to remove some non-native extender units that are provided by the pikromycin producing host [29•]. This knowledge allowed in vitro precursor-directed biosynthesis of a fully extended and cyclized C2-ethyl narbonolide analog. Recently, engineered malonyl-CoA synthetases with expanded substrate specificity were created for the chemo-enzymatic synthesis of a broad panel of natural and non-natural extender units, ultimately leading to the discovery of promiscuity in a unique trans-acyltransferase [30•,31]. In addition, the terminal module and thioesterase domain of the 6-deoxyerythronolide B synthase (DEBS) was revealed to be remarkably tolerant to a range of extender units [32]. Cumulatively, these studies suggest that AT and KS promiscuity could provide a platform for further protein engineering. Illustrative of this, computational redesign was recently employed to alter the extender unit specificity of the AT domain from DEBS module 6 [33]. Several mutants were designed and tested for their ability to utilize a non-natural propargyl extender unit. One mutation (Val295Ala) was subsequently shown to produce a mixture of erythromycin and the desired propargyl analogue. It remains to be seen whether further mutagenesis could improve the rather poor synthetic conversions of the Val295Ala mutant in order to provide a synthetically useful platform for polyketide modification. To achieve this, directed evolution strategies might be most effective, given our still incomplete understanding of the molecular determinants of substrate specificity and catalysis in PKSs. In an example of PKS directed evolution, a two-tier screening strategy was used to identify improved variants of the type III PKS phloroglucinol synthase PhlD [34]. A colony-based colorimetric screen using Gibb”s reagent for detection of the expected phenolic product, was followed by screening in microtiter plates using a more quantitative colorimetric assay for phloroglucinol. After shuffling 52 PhlD homologs, clones with multiple amino acid substitutions were obtained, and subsequent site-directed mutagenesis and saturation mutagenesis identified optimal mutations at key positions. More recently, this system was also used to improve the thermostability of this enzyme [35]. Engineering the ligand specificity of regulatory proteins [36] might afford more general tools for carrying out high-throughput screens or selections for PKS activities. For example, directed evolution has been used to generate AraC mutants that regulate reporter gene expression in response to binding non-natural ligands [37,38], including triacetic acid lactone (TAL), the product of the type III PKS, 2-pyrone synthase (2-PS) (PC Cirino et al., submitted). Subsequently, an in vivo based reporter system enabled directed evolution of the 2-PS from Gerbera hybrida, affording a mutant that supported 18-fold improved TAL production, compared to the wild-type PKS.
Recently, a mass spectrometry-based investigation of the substrate specificity of KS domains from the bacillaene and psymberin PKSs revealed distinct substrate specificity profiles that provided a platform for engineering [39•]. The BaeL KS5 was found to be quite tolerant to unbranched short acyl-thioesters of N-acetylcysteamine (SNAc) yet could not process a β-branched analog, a feature congruent with the location of KS5 immediately upstream of a β-branching step. Psy A ketosynthase (KS)-1 and KS2 each displayed marked promiscuity and could tolerate both unbranched and branched substrates. While promiscuity of KS2 was correctly predicted, that of KS1 was rationalized by the presence of a specificity-conferring GNAT domain preceding the ketosynthase. Subsequently, homology models revealed that the residue preceding the active site cysteine could play a role in determining specificity towards branched substrates. Gratifyingly, introduction of the Met237Ala mutation into BaeL KS5 proved sufficient to afford activity towards the branched substrate. Although the condensation activities of these wild-type and mutant KS”s were not examined, this study nonetheless represents a rare example of successfully altering the substrate specificity of a polyketide synthase, and sets the stage for further bioengineering.
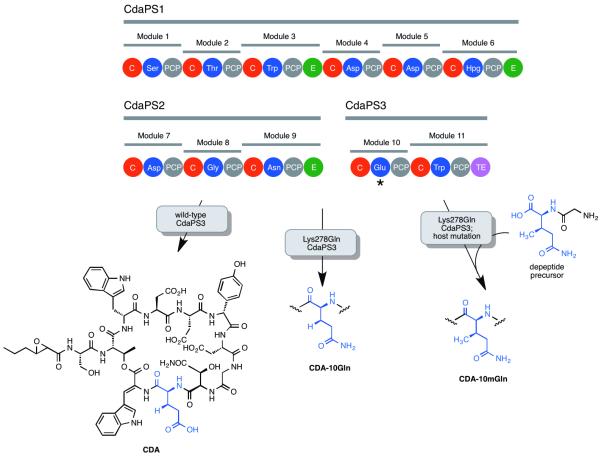
Traditionally, nonribosomal peptide analogs have been generated by substituting individual adenylation (A) domains with those of non-native specificity [18]. Such chimera”s often display reduced product yield compared to wild-type NRPSs, and directed evolution has been used to rescue the activity of chimeric NRPSs [40]. Perhaps a more efficient approach could involve directed mutagenesis of the target A-domain, a strategy that is also not necessarily limited to the introduction of proteinogenic amino acids. The “NRPS code” has proven an effective device for predicting the substrate specificity and corresponding non-ribosomal peptides [41], and more recently is now also emerging as a useful platform for engineering the specificity of NRPS A-domains. For example, targeting the module 10 A-domain of CdaPS3 from the biosynthesis of calcium dependent antibiotic (CDA), Micklefield and co-workers used site-directed mutagenesis to shift activity towards the incorporation of (2S,3R)-3-methyl glutamine (mGln) and Gln over the natural substrates (2S,3R)-3-methyl glutamic acid (mGlu) and Glu [42••]. Sequence alignments of Glu- and Gln-activating A-domains revealed that Glu-activating A-domains often have Lys or His at positions 239 or 278, while Gln-activating A-domains often have Gln at the these two positions. Thus, the two single mutants Lys278Gln and Gln236Glu were each generated, and subsequently Lys278Gln was shown to effect the CDA product distribution in the Streptomyces coelicolor host. In fact, the Lys278Gln mutant produced the desired glutamine-containing CDA analog as the major product, with only minor yields of the glutamic acid and methyl glutamic acid containing analogs (Figure 3). To incorporate the non-native substrate mGln, a host strain was employed that was not able to biosynthesize mGlu, which would otherwise compete with mGln. A hydrolytically stable dipeptide precursor was used to generate the desired mGln in vivo following feeding to liquid cultures and proteolysis. The Lys278Gln mutation was sufficient to generate the desired mGln containing CDA analog, although as the minor product. Notably, this work stands as the first example of the designed incorporation of a synthetic non-natural amino acid into a nonribosomal peptide product. Other published examples of A-domain engineering have involved isolated A-domains in vitro, and/or somewhat conservative exchanges of proteinogenic amino acids. For example, three residues of the L-valine-activating A-domain from andrimid biosynthesis AdmK were targeted for combinatorial mutagenesis on the basis of sequence alignments and examination of the ten sites known to be involved in the NRPS code [43]. Rather than completely saturating each site, codons were selected that were predicted to favor the incorporation of nonpolar substrates into andrimid, which yielded 1404 unique mutants. To ensure 95% coverage, ~14,000 members of the library were screened for analog production by LC-MS analysis of crude cell extracts, resulting in the identification of four clones that produced andrimid analogs. Two clones produced a mixture of isoleucine or isoleucine substituted andrimid analogs, while the other two clones both produced a mixture of alanine and phenylalanine substituted analogs. Improvement in selectivity and product yield was achieved when each of these four amino acids were supplied in excess, such that close to wild-type production levels were obtained in some cases. This study is notable in that several new andrimid analogs were obtained in a single experiment via the combinatorial exploration of multiple solutions to the challenge of altering substrate specificity in the context of the complex intracellular environment of the native producer. Using successive saturation mutagenesis at 8 positions defined by the NRPS code, and a high-throughput ATP/PPi-exchange assay, Hollfelder et al. identified mutants of the L-phenylalanine specific tyrocidine synthetase 1 A-domain (TycA) that displayed specificity changes of 105 towards L-alanine [44]. Remarkably, this significant change in specificity was achieved via the introduction of only three amino acid substitutions. Yeast surface display was recently used to engineer the specificity of aryl-acid activating A-domains from bacillibactin biosynthesis [45]. Chemically stable bisubstrate analogs of the acyl-AMP adenylate were designed that included non-native structural modifications to the aryl acid portion, in addition to a biotin-linker to enable enrichment of variant A-domains that were able to bind the substrate mimics. As might be expected for a selection strategy that relied only on binding affinity, and one that omitted the carrier protein, improvements to Km were largely responsible for large specificity shifts of selected variants. New A-domain substrate specificities have also been created using the more invasive mutagenic approach of chimeragenesis, using A-domains from the biosynthesis of hormaomycin [46]. Inspired by bioinformatic analysis that suggested A-domains acquired new substrate specificities by recombination of A-domain fragments during evolution, chimeric A-domains were created by replacing the core active site regions of a [β-Me]Phe activating “scaffold” A-domain (HrmO3A) with core regions derived from three A-domains which each activated (3-Ncp)Ala, threonine, and valine, respectively. Three out of five chimeras displayed substrate specificity profiles that were almost identical to that of the domains from which the core regions were chosen. Notably, these chimera”s also displayed high levels of activity, as compared to the wild-type scaffold proteins.
Figure 3.
Organization of the NRPS responsible for CDA biosynthesis. Shown are the products of the wild-type and engineered NRPS. The module targeted for mutagenesis is highlighted with an asterisk.
Other PKS and NRPS functions
Reductive domains of PKSs have been targeted for engineering in order to alter specificity. Aside from inactivation studies [47], stereospecificity of reductive domains has also been altered to produce analogs with different stereochemistry [48,49]. However, a complete understanding of the molecular basis for stereocontrol is still missing [50,51]. Directed evolution approaches that use small molecule surrogates of the usually ACP-tethered intermediates [52] may be better suited to identify stereochemistry-determining residues both proximal and distal to the ketoreductase (KR) active site. Most PKS thioesterase (TE) domains are highly substrate dependent and the molecular basis for macrocyclization specificity is poorly understood [53]. An improved understanding of TE specificity could lead to the construction of custom TE domains with tailored substrate-, regio-, and stereospecificity that might prove powerful catalysts for synthesis of cyclized polyketides and their analogs. Although a wealth of structural and mechanistic details of NRPS TE domains is now cumulatively available [54], there are few reports of altering the specificity or activity of NRPS TE domains, and a potential high-throughput screen for NRPS TE has yet to be utilized [55].
Protein:protein interactions
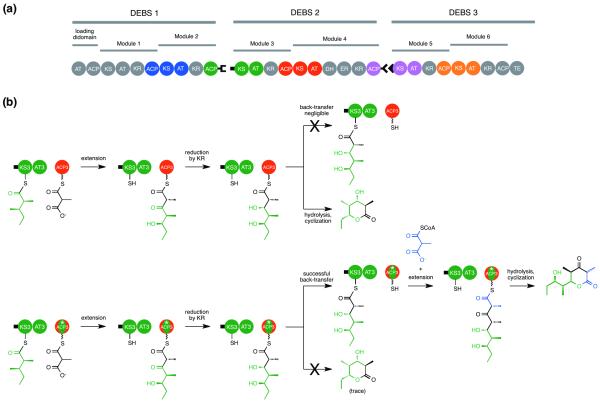
Protein-protein interactions play crucial roles in intramodular and intermodular polyketide chain transfer between the KS and ACP in type I modular PKSs. Intramodular KS:ACP interactions are responsible for polyketide chain elongation whereby the KS catalyzes Claisen condensation and transfers the extended polyketide intermediate to the ACP located within the same module. On the other hand, intermodular KS:ACP interactions are responsible for chain translocation between modules, whereby the KS accepts the polyketide intermediate from the ACP of the upstream module prior to elongation. Crucially, polyketide chain transfer in type I modular PKSs is unidirectional and specific protein interactions between ACP and KS likely somehow prevents back-transfer of the elongated intermediate [56-59]. To account for this behavior, Khosla et al. recently used a set of kinetic assays combined with a panel of designed DEBS ACP chimera”s to identify two orthogonal protein interaction surfaces on the ACP, one region being responsible for chain elongation and the other for chain translocation [60]. A molecular docking model also provided a description of how the ACP interacts with the KS and AT during chain elongation. Gratifyingly, this model was in complete agreement with the data provided by analysis of the ACP chimera”s and other earlier work [56]. As confirmation of this model, mutation of two ACP residues that appeared particularly important for the interactions significantly reduced chain elongation activity. A subsequent molecular modeling and docking study focused on chain transfer between ACP4 and the KS5-AT5 didomain of the downstream module and revealed that ACP4 interacts with the same deep cleft of the didomain as ACP5, but in a different position and orientation as required for intramodular chain elongation [61••]. These studies by the Khosla group are a tour de force of enzymology and protein engineering, and culminated in the successful reprogramming of a normally non-iterative DEBS module to catalyze an additional round of chain elongation (Figure 4) [61••], attesting to the quality and accuracy of the protein interaction models. Mechanism-based crosslinkers [62-64] and more recently, the ability to photocrosslink ACP and KS domains via unnatural amino acid mutagenesis [65] are likely to continue to contribute to our understanding of protein interactions among various PKSs and trans-acting domains, particularly those that are poorly structurally characterized compared to the DEBS system, and those that lack convenient kinetic assays.
Figure 4.
Engineering protein:protein interactions in DEBS. (a) Unidirectional translocation of the elongating polyketide chain is ensured by the presence of matching protein interaction interfaces between the ACP of a given module and the KS:AT didomain of the immediately downstream module. (b) Normally, back-transfer of the triketide intermediate produced by one round of elongation catalyzed by the KS:AT didomain fragment (KS3:AT3, green) and ACP (ACP3, red) is negligible, and the expected triketide ketolactone is produced. In contrast, an engineered chimeric ACP (ACP3*, red/green) supported robust back-transfer to affect an additional round of elongation and production of the tetraketide ketolactone.
The peptidyl carrier protein (PCP) of NRPSs must deliver its cargo to multiple domains within each module of a NRPS. Several crystal structures have contributed to understanding the interdomain interactions in NRPSs, and have highlighted the role of alternating catalytic states of the A-domain and that of large conformational changes, although these studies have yet to identify the PCP:A domain interface [54,66]. Furthermore, alanine-scanning mutagenesis of the enterobactin PCP successfully identified residues involved in interaction between the phosphopantetheinyl transferase EntD and the condensation domain EntF [67,68], but not that between the cognate A-domain EntE. In order to gain insight into the interactions that take place between PCP and A-domains, a chimera between the free-standing A-domain EntE and dual ArCP/isochorismate domain EntB from enterobactin biosynthesis was constructed [69]. An inhibitor was also designed and included in crystallization trials with the hope of stabilizing and trapping the thioesterification step. The resulting structure proved to be an accurate model for adenylation-PCP interactions and was used to guide mutation of a non-cognate EntE homolog, BasE from acinetobactin biosynthesis in Acinetobacter baumannii. Notably, one mutant BasE was improved 53-fold in terms of the kcat/KM with EntB and DHB [69].
Conclusions
Although the incredible success of rational redesign and directed evolution strategies for tailoring the activities of enzymes has been slow to transfer specifically to the specialized and exceedingly complex cases of PKSs and NRPSs, increasingly, such approaches are now being employed to alter the substrate specificity of PKSs and NRPSs. Although the successful manipulation of PKS substrate specificity is somewhat lagging behind that of NRPSs, promiscuous substrate specificities of PKSs and related biosynthetic machinery are rapidly being discovered which in turn provides a crucial blueprint for engineering. Recently, even manipulation of the delicate protein:protein interactions that determine the direction and order of building block condensation has not proven immune to successful engineering. Ultimately, protein engineering examples that focus on altering the specificity of PKSs and NRPSs will likely improve our understanding of the molecular basis for substrate specificity and catalysis, and will inspire further refinements of our engineering algorithms. Future developments will likely hinge upon the combination of engineered PKS and NRPS components with the development of new synthetic biology tools for building block generation, identification and recombination of modular parts, control of gene expression, and biosensing of key intermediates and product analogs. Cumulatively, such advances are now poised to result in potentially efficient strategies to produce polyketide and non-ribosomal peptide analogs with significant alterations to structure compared to their natural product counterparts.
Acknowledgements
Research on natural product chemical and synthetic biology in our lab is supported by a National Science Foundation CAREER Award (CHE-1151299), NIH grant 1R01GM104258-01, NC State University Faculty Research and Development Awards (2010 and 2012), and NC State University start-up funds. The author would like to thank Ms. Irina Koryakina and Ms. Zhixia Ye for critical reading of this manuscript.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- [1].Walsh CT, Fischbach MA. Natural products version 2.0: connecting genes to molecules. J Am Chem Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Meier JL, Burkart MD. The chemical biology of modular biosynthetic enzymes. Chem Soc Rev. 2009;38:2012–2045. doi: 10.1039/b805115c. [DOI] [PubMed] [Google Scholar]
- [3].Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, et al. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- [4].Du LC, Sanchez C, Chen M, Edwards DJ, Shen B. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem Biol. 2000;7:623–642. doi: 10.1016/s1074-5521(00)00011-9. [DOI] [PubMed] [Google Scholar]
- [5].Aparicio JF, Molnar I, Schwecke T, Konig A, Haydock SF, Khaw LE, Staunton J, Leadlay PF. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene. 1996;169:9–16. doi: 10.1016/0378-1119(95)00800-4. [DOI] [PubMed] [Google Scholar]
- [6].Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- [7].Solenberg PJ, Matsushima P, Stack DR, Wilkie SC, Thompson RC, Baltz RH. Production of hybrid glycopeptide antibiotics in vitro and in Streptomyces toyocaensis. Chem Biol. 1997;4:195–202. doi: 10.1016/s1074-5521(97)90288-x. [DOI] [PubMed] [Google Scholar]
- [8].Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- [10].Cox RJ, Simpson TJ. Fungal type I polyketide synthases. Methods Enzymol. 2009;459:49–78. doi: 10.1016/S0076-6879(09)04603-5. [DOI] [PubMed] [Google Scholar]
- [11].Cox RJ. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- [12].Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat Prod Rep. 2007;24:162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- [13].Abe I, Morita H. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat Prod Rep. 2010;27:809–838. doi: 10.1039/b909988n. [DOI] [PubMed] [Google Scholar]
- [14].Gruschow S, Buchholz TJ, Seufert W, Dordick JS, Sherman DH. Substrate profile analysis and ACP-mediated acyl transfer in Streptomyces coelicolor Type III polyketide synthases. ChemBioChem. 2007;8:863–868. doi: 10.1002/cbic.200700026. [DOI] [PubMed] [Google Scholar]
- [15].Gulder TA, Freeman MF, Piel J. The Catalytic Diversity of Multimodular Polyketide Synthases: Natural Product Biosynthesis Beyond Textbook Assembly Rules. Top Curr Chem. 2011:1–53. doi: 10.1007/128_2010_113. [DOI] [PubMed] [Google Scholar]
- [16].Condurso HL, Bruner SD. Structure and noncanonical chemistry of nonribosomal peptide biosynthetic machinery. Nat Prod Rep. 2012;29:1099–1110. doi: 10.1039/c2np20023f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wong FT, Khosla C. Combinatorial biosynthesis of polyketides--a perspective. Curr Opin Chem Biol. 2012;16:117–123. doi: 10.1016/j.cbpa.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Giessen TW, Marahiel MA. Ribosome-independent biosynthesis of biologically active peptides: Application of synthetic biology to generate structural diversity. FEBS Lett. 2012;586:2065–2075. doi: 10.1016/j.febslet.2012.01.017. [DOI] [PubMed] [Google Scholar]
- [19].Lee HY, Khosla C. Bioassay-guided evolution of glycosylated macrolide antibiotics in Escherichia coli. PLoS Biology. 2007;5:243–250. doi: 10.1371/journal.pbio.0050045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee HY, Harvey CJ, Cane DE, Khosla C. Improved precursor-directed biosynthesis in E. coli via directed evolution. J Antibiot (Tokyo) 2011;64:59–64. doi: 10.1038/ja.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Morita H, Yamashita M, Shi SP, Wakimoto T, Kondo S, Kato R, Sugio S, Kohno T, Abe I. Synthesis of unnatural alkaloid scaffolds by exploiting plant polyketide synthase. Proc Natl Acad Sci USA. 2011;108:13504–13509. doi: 10.1073/pnas.1107782108. •• Here the authors capitalized on the substrate and catalytic promiscuity of a type II PKS to synthesize a range of unnatural hybrid polyketide-alkaloid products. Remarkably, a structure-guided mutation led to the alteration of extender unit acyl-CoA’s incorporated and altered the cyclization mechanism.
- [22].Wakimoto T, Mori T, Morita H, Abe I. Cytotoxic tetramic acid derivative produced by a plant type-III polyketide synthase. J Am Chem Soc. 2011;133:4746–4749. doi: 10.1021/ja2006737. [DOI] [PubMed] [Google Scholar]
- [23].Hans M, Hornung A, Dziarnowski A, Cane DE, Khosla C. Mechanistic analysis of acyl transferase domain exchange in polyketide synthase modules. J Am Chem Soc. 2003;125:5366–5374. doi: 10.1021/ja029539i. [DOI] [PubMed] [Google Scholar]
- [24].Gatti-Lafranconi P, Hollfelder F. Flexibility and Reactivity in Promiscuous Enzymes. ChemBioChem. 2013;14:285–292. doi: 10.1002/cbic.201200628. [DOI] [PubMed] [Google Scholar]
- [25].Wilson MC, Moore BS. Beyond ethylmalonyl-CoA: The functional role of crotonyl-CoA carboxylase/reductase homologs in expanding polyketide diversity. Nat Prod Rep. 2011;29:72–86. doi: 10.1039/c1np00082a. [DOI] [PubMed] [Google Scholar]
- [26].Chan YA, Podevels AM, Kevany BM, Thomas MG. Biosynthesis of polyketide synthase extender units. Nat Prod Rep. 2009;26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lechner A, Wilson MC, Ban YH, Hwang J-y, Yoon YJ, Moore BS. Designed Biosynthesis of 36-Methyl-FK506 by Polyketide Precursor Pathway Engineering. ACS Synth Biol. 2013 doi: 10.1021/sb3001062. DOI: 10.1021/sb3001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mo S, Kim DH, Lee JH, Park JW, Basnet DB, Ban YH, Yoo YJ, Chen SW, Park SR, Choi EA, et al. Biosynthesis of the allylmalonyl-CoA extender unit for the FK506 polyketide synthase proceeds through a dedicated polyketide synthase and facilitates the mutasynthesis of analogues. J Am Chem Soc. 2011;133:976–985. doi: 10.1021/ja108399b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bonnett SA, Rath CM, Shareef AR, Joels JR, Chemler JA, Hakansson K, Reynolds K, Sherman DH. Acyl-CoA Subunit Selectivity in the Pikromycin Polyketide Synthase PikAIV: Steady-State Kinetics and Active-Site Occupancy Analysis by FTICR-MS. Chem Biol. 2011;18:1075–1081. doi: 10.1016/j.chembiol.2011.07.016. • Here the authors provide evidence that the PikAIV system supports substrate loading of a non-native extender unit acyl-CoA followed by slow off-loading by hydrolysis. This model suggests a strategy for future engineering efforts by targeting non-natural extender units that are only slowly removed from the PKS.
- [30].Koryakina I, McArthur J, Randall S, Draelos MM, Musiol EM, Muddiman DC, Weber T, Williams GJ. Poly specific trans-acyltransferase machinery revealed via engineered acyl-CoA synthetases. ACS Chem Biol. 2013;8:200–208. doi: 10.1021/cb3003489. • This study employed mutant acyl-CoA synthetases for the synthesis of a panel of diverse natural and non-natural extender unit acyl-CoA’s. Subsequently, this panel was used to probe the promiscuity of several standalone trans-ATs, revealing promiscuity of the enzyme KirCII. As an example of the first promiscuous trans-AT, this work sets the stage for further improving the substrate promiscuity of KirCII and other trans-ATs, and will lead to the development of strategies that harness this promiscuity for diversification of polyketides.
- [31].Koryakina I, Williams GJ. Mutant malonyl-CoA synthetases with altered specificity for polyketide synthase extender unit generation. ChemBioChem. 2011;12:2289–2293. doi: 10.1002/cbic.201100383. [DOI] [PubMed] [Google Scholar]
- [32].Koryakina I, McArthur JB, Draelos MM, Williams GJ. Promiscuity of a modular polyketide synthase towards natural and non-natural extender units. Org Biomol Chem. 2013 doi: 10.1039/c3ob40633d. DOI: 10.1039/c1033ob40633d. [DOI] [PubMed] [Google Scholar]
- [33].Sundermann U, Bravo-Rodriguez K, Klopries S, Kushnir S, Gomez H, Sanchez-Garcia E, Schulz F. Enzyme-Directed Mutasynthesis: A Combined Experimental and Theoretical Approach to Substrate Recognition of a Polyketide Synthase. ACS Chem Biol. 2013;8:443–450. doi: 10.1021/cb300505w. [DOI] [PubMed] [Google Scholar]
- [34].Zha W, Rubin-Pitel SB, Zhao H. Exploiting genetic diversity by directed evolution: molecular breeding of type III polyketide synthases improves productivity. Mol Biosyst. 2008;4:246–248. doi: 10.1039/b717705d. [DOI] [PubMed] [Google Scholar]
- [35].Rao G, Lee JK, Zhao H. Directed evolution of phloroglucinol synthase PhID with increased stability for phloroglucinol production. Appl Microbiol Biotechnol. 2013 doi: 10.1007/s00253-013-4713-4. DOI: 10.1007/s00253-00013-04713-00254. [DOI] [PubMed] [Google Scholar]
- [36].Gredell JA, Frei CS, Cirino PC. Protein and RNA engineering to customize microbial molecular reporting. Biotechnol J. 2012;7:477–499. doi: 10.1002/biot.201100266. [DOI] [PubMed] [Google Scholar]
- [37].Tang SY, Cirino PC. Design and application of a mevalonate-responsive regulatory protein. Angew Chem Int Ed Engl. 2011;50:1084–1086. doi: 10.1002/anie.201006083. [DOI] [PubMed] [Google Scholar]
- [38].Tang SY, Fazelinia H, Cirino PC. AraC regulatory protein mutants with altered effector specificity. J Am Chem Soc. 2008;130:5267–5271. doi: 10.1021/ja7109053. [DOI] [PubMed] [Google Scholar]
- [39].Jenner M, Frank S, Kampa A, Kohlhaas C, Poplau P, Briggs GS, Piel J, Oldham NJ. Substrate specificity in ketosynthase domains from trans-AT polyketide synthases. Angew Chem Int Ed Engl. 2013;52:1143–1147. doi: 10.1002/anie.201207690. • The authors developed a mass spectrometry based method to probe the specificity of KS domains from trans-AT PKS clusters. This data highlighted key differences in substrate specificity that provided a blueprint for subsequent successful KS engineering, providing a rare example of altering the substrate specificity of a PKS component.
- [40].Fischbach MA, Lai JR, Roche ED, Walsh CT, Liu DR. Directed evolution can rapidly improve the activity of chimeric assembly-line enzymes. Proc Natl Acad Sci USA. 2007;104:11951–11956. doi: 10.1073/pnas.0705348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- [42].Thirlway J, Lewis R, Nunns L, Al Nakeeb M, Styles M, Struck A-W, Smith C, Micklefield J. Introduction of a non-natural amino acid into a nonribosomal peptide antibiotic by modification of adenylation domain specificity. Angew Chem Int Ed Engl. 2012;51:7181–7184. doi: 10.1002/anie.201202043. •• Here the authors provided the first example of engineering an NRPS for the introduction of a non-proteinogenic amino acid into a non-ribosomal peptide. This work stands as a rare example of engineering non-ribosomal peptide products in vivo, and affords a valuable opportunity to produce peptide analogs with improved pharmacological properties.
- [43].Evans BS, Chen Y, Metcalf WW, Zhao H, Kelleher NL. Directed evolution of the nonribosomal peptide synthetase AdmK generates new andrimid derivatives in vivo. Chem Biol. 2011;18:601–607. doi: 10.1016/j.chembiol.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Villiers B, Hollfelder F. Directed evolution of a gatekeeper domain in nonribosomal peptide synthesis. Chem Biol. 2011;18:1290–1299. doi: 10.1016/j.chembiol.2011.06.014. [DOI] [PubMed] [Google Scholar]
- [45].Zhang K, Nelson KM, Bhuripanyo K, Grimes KD, Zhao B, Aldrich CC, Yin J. Engineering the Substrate Specificity of the DhbE Adenylation Domain by Yeast Cell Surface Display. Chem Biol. 2013;20:92–101. doi: 10.1016/j.chembiol.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Crusemann M, Kohlhaas C, Piel J. Evolution-guided engineering of nonribosomal peptide synthetase adenylation domains. Chem Sci. 2013;4:1041–1045. [Google Scholar]
- [47].Ding W, Lei C, He Q, Zhang Q, Bi Y, Liu W. Insights into bacterial 6-methylsalicylic acid synthase and its engineering to orsellinic acid synthase for spirotetronate generation. Chem Biol. 2010;17:495–503. doi: 10.1016/j.chembiol.2010.04.009. [DOI] [PubMed] [Google Scholar]
- [48].Kwan DH, Leadlay PF. Mutagenesis of a modular polyketide synthase enoylreductase domain reveals insights into catalysis and stereospecificity. ACS Chem Biol. 2010;5:829–838. doi: 10.1021/cb100175a. [DOI] [PubMed] [Google Scholar]
- [49].Baerga-Ortiz A, Popovic B, Siskos AP, O’Hare HM, Spiteller D, Williams MG, Campillo N, Spencer JB, Leadlay PF. Directed mutagenesis alters the stereochemistry of catalysis by isolated ketoreductase domains from the erythromycin polyketide synthase. Chem Biol. 2006;13:277–285. doi: 10.1016/j.chembiol.2006.01.004. [DOI] [PubMed] [Google Scholar]
- [50].Kwan DH, Tosin M, Schlager N, Schulz F, Leadlay PF. Insights into the stereospecificity of ketoreduction in a modular polyketide synthase. Org Biomol Chem. 2011;9:2053–2056. doi: 10.1039/c1ob00022e. [DOI] [PubMed] [Google Scholar]
- [51].Javidpour P, Das A, Khosla C, Tsai SC. Structural and biochemical studies of the hedamycin type II polyketide ketoreductase (HedKR): molecular basis of stereo- and regiospecificities. Biochemistry. 2011;50:7426–7439. doi: 10.1021/bi2006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].O’Hare HM, Baerga-Ortiz A, Popovic B, Spencer JB, Leadlay PF. High-throughput mutagenesis to evaluate models of stereochemical control in ketoreductase domains from the erythromycin polyketide synthase. Chem Biol. 2006;13:287–296. doi: 10.1016/j.chembiol.2006.01.003. [DOI] [PubMed] [Google Scholar]
- [53].Pinto A, Wang M, Horsman M, Boddy CN. 6-Deoxyerythronolide B synthase thioesterase-catalyzed macrocyclization is highly stereoselective. Org Lett. 2012;14:2278–2281. doi: 10.1021/ol300707j. [DOI] [PubMed] [Google Scholar]
- [54].Hur GH, Vickery CR, Burkart MD. Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology. Nat Prod Rep. 2012;29:1074–1098. doi: 10.1039/c2np20025b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Grunewald J, Kopp F, Mahlert C, Linne U, Sieber SA, Marahiel MA. Fluorescence resonance energy transfer as a probe of peptide cyclization catalyzed by nonribosomal thioesterase domains. Chem Biol. 2005;12:873–881. doi: 10.1016/j.chembiol.2005.05.019. [DOI] [PubMed] [Google Scholar]
- [56].Chen AY, Schnarr NA, Kim CY, Cane DE, Khosla C. Extender unit and acyl carrier protein specificity of ketosynthase domains of the 6-deoxyerythronolide B synthase. J Am Chem Soc. 2006;128:3067–3074. doi: 10.1021/ja058093d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kim CY, Alekseyev VY, Chen AY, Tang YY, Cane DE, Khosla C. Reconstituting modular activity from separated domains of 6-deoxyerythronolide B synthase. Biochemistry. 2004;43:13892–13898. doi: 10.1021/bi048418n. [DOI] [PubMed] [Google Scholar]
- [58].Wu N, Cane DE, Khosla C. Quantitative analysis of the relative contributions of donor acyl carrier proteins, acceptor ketosynthases, and linker regions to intermodular transfer of intermediates in hybrid polyketide synthases. Biochemistry. 2002;41:5056–5066. doi: 10.1021/bi012086u. [DOI] [PubMed] [Google Scholar]
- [59].Wu N, Tsuji SY, Cane DE, Khosla C. Assessing the balance between protein-protein interactions and enzyme-substrate interactions in the channeling of intermediates between polyketide synthase modules. J Am Chem Soc. 2001;123:6465–6474. doi: 10.1021/ja010219t. [DOI] [PubMed] [Google Scholar]
- [60].Kapur S, Chen AY, Cane DE, Khosla C. Molecular recognition between ketosynthase and acyl carrier protein domains of the 6-deoxyerythronolide B synthase. Proc Natl Acad Sci USA. 2011;107:22066–22071. doi: 10.1073/pnas.1014081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kapur S, Lowry B, Yuzawa S, Kenthirapalan S, Chen AY, Cane DE, Khosla C. Reprogramming a module of the 6-deoxyerythronolide B synthase for iterative chain elongation. Proc Natl Acad Sci USA. 2012;109:4110–4115. doi: 10.1073/pnas.1118734109. •• This study is the culmination of several years of work by the Khosla group aiming at understanding the mechanism by which the 6-deoxyerythronolide B synthase catalyzes unidirectional polyketide biosynthesis. Ultimately, a ratchet mechanism was proposed as a model to explain unidirectional translocation of the growing polyketide chain. The accuracy of this model was verified by successfully engineering module 3 of the synthase to catalyze two successive rounds of chain elongation, rather than one.
- [62].Blatti JL, Beld J, Behnke CA, Mendez M, Mayfield SP, Burkart MD. Manipulating fatty acid biosynthesis in microalgae for biofuel through protein-protein interactions. PLoS One. 2012;7:e42949. doi: 10.1371/journal.pone.0042949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Worthington AS, Hur GH, Burkart MD. Activity-guided engineering of natural product carrier proteins. Mol Biosyst. 2011;7:365–370. doi: 10.1039/c0mb00251h. [DOI] [PubMed] [Google Scholar]
- [64].Worthington AS, Porter DF, Burkart MD. Mechanism-based crosslinking as a gauge for functional interaction of modular synthases. Org Biomol Chem. 2010;8:1769–1772. doi: 10.1039/b925966j. [DOI] [PubMed] [Google Scholar]
- [65].Ye Z, Bair M, Desai H, Williams GJ. A photocrosslinking assay for reporting protein interactions in polyketide and fatty acid synthases. Mol BioSyst. 2011;7:3152–3156. doi: 10.1039/c1mb05270e. [DOI] [PubMed] [Google Scholar]
- [66].Tanovic A, Samel SA, Essen LO, Marahiel MA. Crystal structure of the termination module of a nonribosomal peptide synthetase. Science. 2008;321:659–663. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- [67].Zhou Z, Lai JR, Walsh CT. Interdomain communication between the thiolation and thioesterase domains of EntF explored by combinatorial mutagenesis and selection. Chem Biol. 2006;13:869–879. doi: 10.1016/j.chembiol.2006.06.011. [DOI] [PubMed] [Google Scholar]
- [68].Lai JR, Fischbach MA, Liu DR, Walsh CT. Localized protein interaction surfaces on the EntB carrier protein revealed by combinatorial mutagenesis and selection. J Am Chem Soc. 2006;128:11002–11003. doi: 10.1021/ja063238h. [DOI] [PubMed] [Google Scholar]
- [69].Sundlov JA, Shi C, Wilson DJ, Aldrich CC, Gulick AM. Structural and functional investigation of the intermolecular interaction between NRPS adenylation and carrier protein domains. Chem Biol. 2012;19:188–198. doi: 10.1016/j.chembiol.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]