Abstract
Unresolved questions about evolution of the large and diverse legume family include the timing of polyploidy (whole-genome duplication; WGDs) relative to the origin of the major lineages within the Fabaceae and to the origin of symbiotic nitrogen fixation. Previous work has established that a WGD affects most lineages in the Papilionoideae and occurred sometime after the divergence of the papilionoid and mimosoid clades, but the exact timing has been unknown. The history of WGD has also not been established for legume lineages outside the Papilionoideae. We investigated the presence and timing of WGDs in the legumes by querying thousands of phylogenetic trees constructed from transcriptome and genome data from 20 diverse legumes and 17 outgroup species. The timing of duplications in the gene trees indicates that the papilionoid WGD occurred in the common ancestor of all papilionoids. The earliest diverging lineages of the Papilionoideae include both nodulating taxa, such as the genistoids (e.g., lupin), dalbergioids (e.g., peanut), phaseoloids (e.g., beans), and galegoids (=Hologalegina, e.g., clovers), and clades with nonnodulating taxa including Xanthocercis and Cladrastis (evaluated in this study). We also found evidence for several independent WGDs near the base of other major legume lineages, including the Mimosoideae–Cassiinae–Caesalpinieae (MCC), Detarieae, and Cercideae clades. Nodulation is found in the MCC and papilionoid clades, both of which experienced ancestral WGDs. However, there are numerous nonnodulating lineages in both clades, making it unclear whether the phylogenetic distribution of nodulation is due to independent gains or a single origin followed by multiple losses.
Keywords: nodulation, polyploidy, legume, symbiotic nitrogen fixation, Papilionoideae, Mimosoideae
Introduction
The legumes (Leguminosae, Fabaceae) are the third largest family of flowering plants, with over 750 genera and 19,500 species (Lewis et al. 2005). The most familiar legumes are the many cultivated “beans”—for example, common bean (Phaseolus vulgaris), soybean (Glycine max), mungbean (Vigna radiata)—and “peas”—for example, pea (Pisum sativum), pigeonpea (Cajanus cajan)—all of which are members of the largest subfamily, the Papilionoideae (fig. 1). The family is diverse in every way, including habit and ecology, floral structure, and biochemistry; legumes range from tiny annual herbs to giant forest trees, and are characteristic, often dominant members of many ecosystems in nearly every climate zone (Doyle and Luckow 2003; Lewis et al. 2005).
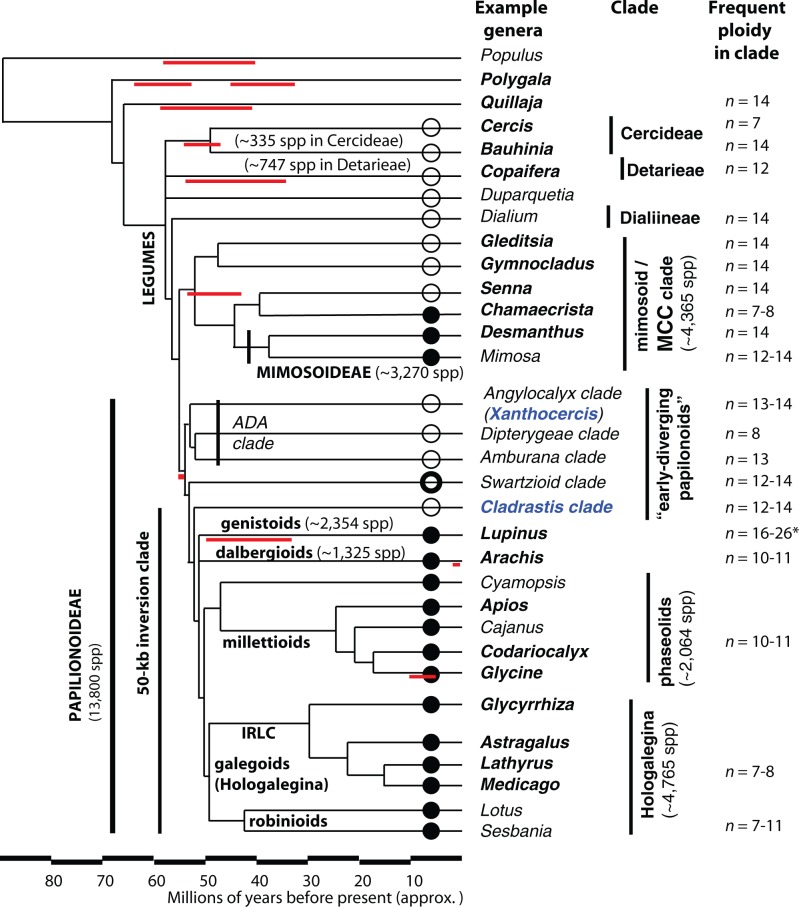
Fig. 1.
A summary phylogeny based on published trees for the Fabaceae (Wojciechowski et al. 2004; Cardoso et al. 2012; Manzanilla and Bruneau 2012), with approximate date estimates from Lavin et al. (2005). Estimates of species counts per clade are taken from Lewis et al. (2005). Nodulation status is shown with circles: Filled for “many species in this clade nodulate”; partly filled (Swartzieae) for “some species in this clade nodulate”; empty for “no species in this clade have been observed to nodulate.” Nodulation status is summarized from Sprent (2009). Chromosome counts on the right are predominant counts for each genus or clade. These are drawn from Doyle (2012) and supplementary file S5, Supplementary Material online. For the genistoids, the base chromosome count is likely x = 9, but chromosome counts within Lupinus (used in this project) are mostly in the range of n = 16–26 (Doyle 2012). Hypothesized placement of genome duplication events is indicated with horizontal red lines.
An attribute generally associated with legumes is the ability to form symbioses with diverse soil bacteria, collectively termed “rhizobia,” in which plant roots develop structures (nodules) where the bacteria are housed, nourished with plant carbon, and fix atmospheric nitrogen that is assimilated by the plant (Sprent 2009). However, nodulation is not unique to legumes, nor are all legumes capable of nodulating: Nodulation is phylogenetically scattered among families in the “nitrogen-fixing clade” of rosid angiosperms, and within legumes themselves it is found primarily in papilionoids and mimosoids, and is absent from the earliest diverging lineages of the family (reviewed in Doyle 2011; Werner et al. 2014). The number of origins of nodulation within the family remains unclear, for at least two reasons. For many genera, the ability of species to nodulate has not been assessed (Sprent 2009). Further, phylogenetic relationships in critical parts of the family, such as in lineages sister to the core clades within the mimosoids and papilionoids, remain unclear (Lavin et al. 2005). This situation is improving (Cardoso et al. 2012), but more resolution is needed (Legume Phylogeny Working Group 2013). Importantly, however, resolving relationships among major lineages within the Fabaceae and the phylogenetic distribution of currently nodulating species may not be sufficient for understanding the gain and loss of nodulation in the family.
The molecular and developmental pathways leading to the evolution of nodulation also remain unknown. Nodulation shares key portions of its program for interacting with symbionts with the much older mycorrhizal symbiosis (Gherbi et al. 2008; Madsen et al. 2010; Oldroyd et al. 2011; Op den Camp et al. 2011), and the concentration of nodulating plant families in the N-fixing clade (Soltis et al. 1995) suggests an evolutionary predisposition to nodulation, acquired by the ancestor of the clade (Gherbi et al. 2008; Doyle 2011; Werner et al. 2014). Whole-genome duplications (WGDs) have been common through angiosperm history (e.g., Cui et al. 2006; Van de Peer 2011) and hypothesized to promote diversification and innovation (e.g., Freeling and Thomas 2006; Soltis et al. 2009; Schranz et al. 2012). The ancestor of all seed plants experienced a WGD, and a WGD also occurred in the ancestor of all flowering plants (Jiao et al. 2011; Amborella Genome Project 2013). Additionally, the common ancestor of core eudicots (including legumes) was hexaploid (Jiao et al. 2012). Within the legumes, an ancient polyploidy event occurred in the progenitor of diverse papilionoid legumes, including Medicago truncatula, Lotus japonicus, G. max, and Arachis hypogaea (Blanc and Wolfe 2004; Schlueter et al. 2004; Pfeil et al. 2005; Cannon et al. 2006; Bertioli et al. 2009). Therefore, the ancestral WGD event that has been characterized in the M. truncatula, G. max, and other papilionoid genomes occurred at or near the origin of the papilionoid clade, around 55 Ma. Young et al. (2011) and others (Li et al. 2013) have hypothesized that this “papilionoid WGD (PWGD)” contributed to the evolution of key components in the early nodulation signaling cascade of papilionoids, enhancing their capacity for symbiotic nitrogen fixation (SNF) and contributing to their tremendous evolutionary success.
Uncertainty about the relationship of polyploidy and the origin of nodulation is due both to the presence of nonnodulating lineages within the Papilionoideae (Sprent 2009) and to the presence of nodulating species outside the Papilionoideae. In particular, Cannon et al. (2010) determined that Chamaecrista fasciculata, a member of the Mimosoideae–Caesalpinieae–Cassiinae (MCC; fig. 1) clade, does not share the ancestral PWGD event identified in synteny analyses of the Glycine (Schmutz et al. 2010) and Medicago (Young et al. 2011) genomes.
Here, we address lingering questions about the evolutionary history of the Fabaceae using a combination of available genomic data and whole transcriptome data for 20 species distributed across the family. We use phylogenomic approaches to analyze nuclear gene sequences in order to refine understanding of relationships among key legume lineages and to improve resolution of the timing of polyploidy events within the family. New transcriptome sequences enable elucidation of the timing of WGD events relative to the early papilionoid radiation, and the origins of the Mimosoideae, MCC, Detarieae, and Cercideae clades (fig. 1). With an improved understanding of the timing of WGDs across legume history, we consider how polyploidy may have influenced the evolution of nodulation within the legume family.
Results
Gene Families and Phylogenies
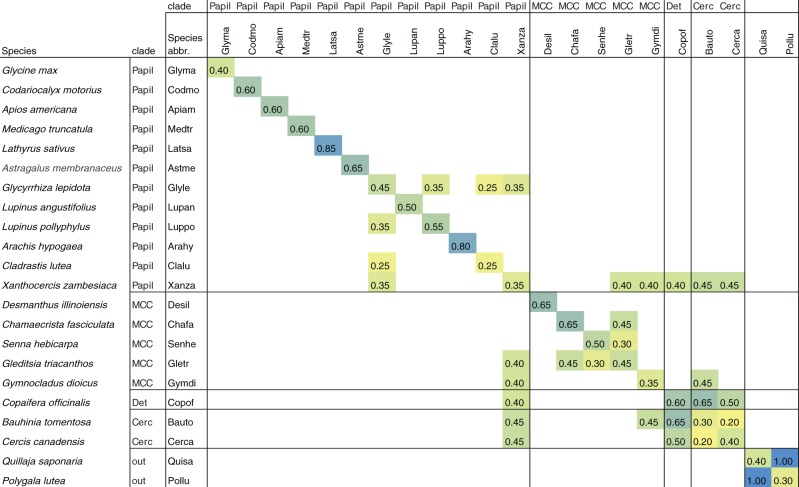
A phylogenomic analysis of WGDs was performed with 37 species (tables 1 and 2), including: 20 legumes; two near outgroups to the Fabaceae (Quillaja saponaria and Polygala lutea; both Fabales); representatives from four additional families within the large “nitrogen-fixing clade” of rosid angiosperms that contain nonlegume nodulators (Alnus serrulata—an actinorhizal nodulator, Cucumis sativus—within the Cucurbitales with the actinorhizal nodulator Datisca, and Fragaria vesca and Prunus persica—both within the Rosales with the rhizobial nodulator Parasponia); and 11 more distant outgroups, included to help span the eudicot and angiosperm phylogeny (table 1).
Table 1.
Species, Sequencing Methods, and Descriptive Statistics.
| Species Abbr. | Species | Sequence Type | Source | Assembly Method | No. of Unigenes | Contig N50 | Average Length | ||
|---|---|---|---|---|---|---|---|---|---|
| Glyma | Glycine max | Genomic | Phytozome | Genomic | 54,175 | 1,572 | 1,185.6 | ||
| Codmo | Codariocalyx motorius | NGS 1 | 1kp | Trinity | 40,430 | 1,217 | 819.0 | ||
| Apiam | Apios americana | NGS 2 | Cannon et al. (this study) | Trinity | 44,658 | 1,606 | 877.5 | ||
| Medtr | Medicago truncatula | Genomic | JCVI | Genomic | 62,383 | 1,326 | 871.8 | ||
| Latsa | Lathyrus sativus | NGS 1 | 1kp | Trinity | 61,935 | 1,146 | 576.6 | ||
| Astme | Astragalus membranaceus | NGS 1 | 1kp | Trinity | 76,669 | 1,059 | 691.8 | ||
| Glyle | Glycyrrhiza lepidota | NGS 1 | 1kp | Trinity | 70,070 | 1,105 | 711.1 | ||
| Lupan | Lupinus angustifolius | NGS 1 | 1kp | Trinity | 49,694 | 764 | 565.6 | ||
| Luppo | Lupinus polyphyllus | NGS 1 | 1kp | Trinity | 72,174 | 511 | 325.5 | ||
| Arahy | Arachis hypogaea | NGS 3 | GenBank | Trinity | 49,908 | 1,009 | 702.9 | ||
| Clalu | Cladrastis lutea | NGS 1 | 1kp | Trinity | 106,653 | 629 | 373.9 | ||
| Xanza | Xanthocercis zambesiaca | NGS 1 | 1kp | Trinity | 111,179 | 1,099 | 539.4 | ||
| Desil | Desmanthus illinoiensis | NGS 1 | 1kp | Trinity | 30,173 | 534 | 383.2 | ||
| Chafa | Chamaecrista fasciculata | NGS 1 | Cannon et al. (2010) | Trinity | 40,414 | 577 | 484.8 | ||
| Senhe | Senna hebicarpa | NGS 1 | 1kp | Trinity | 71,854 | 529 | 321.8 | ||
| Gletr | Gleditsia triacanthos | NGS 1 | 1kp | Trinity | 59,700 | 791 | 457.3 | ||
| Gymdi | Gymnocladus dioicus | NGS 1 | 1kp | Trinity | 93,331 | 636 | 368.1 | ||
| Copof | Copaifera officinalis | NGS 1 | 1kp | Trinity | 67,940 | 1,475 | 611.1 | ||
| Bauto | Bauhinia tomentosa | NGS 1 | 1kp | Trinity | 95,565 | 473 | 347.5 | ||
| Cerca | Cercis Canadensis | NGS 1 | 1kp | Trinity | 48,374 | 969 | 543.3 | ||
| Quisa | Quillaja saponaria | NGS 1 | 1kp | Trinity | 74,392 | 861 | 472.5 | ||
| Pollu | Polygala lutea | NGS 1 | 1kp | Trinity | 129,993 | 719 | 414.4 | ||
| Alnse | Alnus serrulata | NGS 1 | 1kp | Trinity | 86,216 | 929 | 468.8 | ||
| Cucsa | Cucumis sativus | Genomic | Phytozome | Genomic | |||||
| Frave | Fragaria vesca | Genomic | Phytozome | Genomic | |||||
| Prupe | Prunus persica | Genomic | Phytozome | Genomic | |||||
| Poptr | Populus trichocarpa | Genomic | Phytozome | Genomic | |||||
| Theca | Theobromo cacao | Genomic | Phytozome | Genomic | |||||
| Arath | Arabidopsis thaliana | Genomic | Phytozome | Genomic | |||||
| Carpa | Carica papaya | Genomic | Phytozome | Genomic | |||||
| Vitvi | Vitis vinifera | Genomic | Phytozome | Genomic | |||||
| Solly | Solanum lycopersicum | Genomic | Phytozome | Genomic | |||||
| Aqupu | Aquilegia formosa × pubescens | PlantGDB | Vmatch-PaCE-CAP3 | 19,615 | |||||
| Papso | Papaver somniferum | 1kp + SRA | MIRA-SOAP Trinity | 126,800 | 1,241 | 705.7 | |||
| Aspof | Asparagus officinalis | Leebens-Mack J, unpublished data | Trinity | 120,061 | 1,249 | 747.3 | |||
| Musac | Musa acuminata | Genomic | Global Musa Genomics Consortium | ||||||
| Ambtr | Amborella trichopoda | Genomic | AAGP | MIRA | 208,394 | ||||
Note.—Of the 37 species included in the study, 13 are represented by genomic-derived (CDS) gene models, and the remainder came from transcriptome assemblies: “NGS 1” consisting of assemblies generated using Illumina data from the Thousand Plant Transcriptomes (1KP) Project, “NGS 2” from another contributed Illumina-derived Trinity assembly (Cannon and Belamkar, this study), and “NGS 3” from an Illumina-derived SOAP Denovo assembly (Jiao et al. 2012). Data sources: 1KP-Consortium (2013); Amborella Genome Project (2013); Ancestral Angiosperm Genome Project (2013); NCBI Sequence Read Archive; PlantGDB (http://www.plantgdb.org).
Table 2.
Species, WGD Inferences (known and from this article), and Symbiotic Nitrogen Fixation Status.
| Species Abbr. | Clade | Tree Sequence Counts | Trees with This Species | Paralog Ks Peak | SNF | WGD/T | WGD | WGD |
|---|---|---|---|---|---|---|---|---|
| Glyma | Papilionoideae | 15,877 | 3,354 | 0.4 | Yes | Gamma3 | Papilionoid | Glycine |
| Codmo | Papilionoideae | 3,407 | 2,432 | 0.6 | Yes | Gamma3 | Papilionoid | |
| Apiam | Papilionoideae | 4,024 | 2,759 | 0.6 | Yes | Gamma3 | Papilionoid | |
| Medtr | Papilionoideae | 2,847 | 2,664 | 0.6 | Yes | Gamma3 | Papilionoid | |
| Latsa | Papilionoideae | 3,307 | 2,380 | 0.85 | Yes | Gamma3 | Papilionoid | |
| Astme | Papilionoideae | 3,430 | 2,444 | 0.65 | Yes | Gamma3 | Papilionoid | |
| Glyle | Papilionoideae | 3,797 | 2,582 | 0.55 | Yes | Gamma3 | Papilionoid | |
| Lupan | Papilionoideae | 2,345 | 1,814 | 0.5 | Yes | Gamma3 | Papilionoid | Lupinus |
| Luppo | Papilionoideae | 1,242 | 980 | 0.55 | Yes | Gamma3 | Papilionoid | Lupinus |
| Arahy | Papilionoideae | 3,118 | 2,048 | 0.05 | Yes | Gamma3 | Papilionoid | Arachis |
| Clalu | Papilionoideae | 1,921 | 1,617 | 0.25 | No | Gamma3 | Papilionoid | |
| Xanza | Papilionoideae | 3,174 | 2,398 | 0.35 | No | Gamma3 | Papilionoid | |
| Desil | MCC—Mimosoideae | 916 | 747 | 0.65 | Yes | Gamma3 | MCC | |
| Chafa | MCC—Cassia | 1,720 | 1,324 | 0.65 | Yes | Gamma3 | MCC | |
| Senhe | MCC—Cassia | 949 | 755 | 0.5 | No | Gamma3 | MCC | |
| Gletr | MCC—grade/Umtiza | 2,154 | 1,675 | 0.45 | No | Gamma3 | MCC | |
| Gymdi | MCC—grade/Umtiza | 1,662 | 1,313 | 0.35 | No | Gamma3 | MCC | |
| Copof | Detarieae | 2,950 | 2,154 | 0.6 | No | Gamma3 | Detarieae | |
| Bauto | Cercideae | 3,312 | 2,442 | 0.3 | No | Gamma3 | Cercideae? | |
| Cerca | Cercideae | 2,489 | 1,983 | 0.4 | No | Gamma3 | Cercideae? | |
| Quisa | Rosid—Fabales | 2,550 | 2,042 | 0.4 | No | Gamma3 | Quillaja | |
| Pollu | Rosid—Fabales | 2,955 | 2,079 | 0.3 | No | Gamma3 | Polygala | Polygala |
| Alnse | Rosid—Fagales | 2,925 | 2,189 | 0.4 | Yes | Gamma3 | ||
| Cucsa | Rosid—Cucurbitales | 2,916 | 2,872 | Yes* | Gamma3 | |||
| Frave | Rosid—Rosales | 4,909 | 2,945 | Yes* | Gamma3 | |||
| Prupe | Rosid—Rosales | 2,969 | 2,891 | Yes* | Gamma3 | |||
| Poptr | Rosid—Malpighiales | 9,620 | 3,140 | No | Gamma3 | Populus | ||
| Theca | Rosid—Malvales | 5,377 | 3,143 | No | Gamma3 | |||
| Arath | Rosid—Brassicales | 5,600 | 2,896 | No | Gamma3 | Brassicaceae | Brassica | |
| Carpa | Rosid—Brassicales | 4,284 | 2,905 | No | Gamma3 | |||
| Vitvi | Rosid—Vitales | 4,798 | 2,796 | No | Gamma3 | |||
| Solly | Asterid—Solanales | 5,793 | 2,995 | No | Gamma3 | Asterid | ||
| Aqupu | Ranunculales | 1,955 | 1,463 | No | Ranunc2? | |||
| Papso | Ranunculales | 3,804 | 2,393 | No | Ranunc2? | |||
| Aspof | Monocot | 3,103 | 2,065 | No | ||||
| Musac | Monocot | 7,519 | 2,663 | No | Monocot | Musa | Musa | |
| Ambtr | Angiosperm—out | 3,460 | 2,588 | No | ||||
Note.—Species counts (column 3) are the numbers of sequences for each species in the set of 3,360 evaluated trees. Presence of SNF is given in column 6, “SNF.” SNF is present for most of the papilionoid representatives, but not for the early-diverging Cladrastis or Xanthocercis. The “yes*” in this SNF column for Cucumis, Fragaria, and Prunus is to indicate that a type of SNF (utilizing an actinorhizal or rhizobial symbiont) is present in some species in these families (but not in these three species).
Synteny analysis of the G. max genome identified 12,196 paralog pairs traceable to the early-PWGD (Glycine PWGD paralogs; supplementary file S1, Supplementary Material online). These paralog pairs were analyzed within the context of their respective gene families, and used to help resolve the timing of the PWGD relative to papilionoid speciation events. Of 53,136 orthoMCL-based gene families (see Materials and Methods of the Amborella Genome Project 2013), 4,518 contained at least one Glycine PWGD paralog pair, and 1,601 orthogroups contained multiple PWGD paralog pairs. Related orthogroups were combined for 2,884 PWGD paralog pairs that were split in the original orthogroup gene family circumscription. In total, gene trees were estimated for 3,360 circumscribed gene families, and the timing of gene duplications associated with the PWGD was inferred relative to speciation events for each gene tree. These trees were also analyzed for relative timing of duplication and speciation events outside of papilionoid gene clades in order to infer ancient WGDs in nonpapilionoid legume lineages (described below).
Phylogenomic Inference of Species Relationships
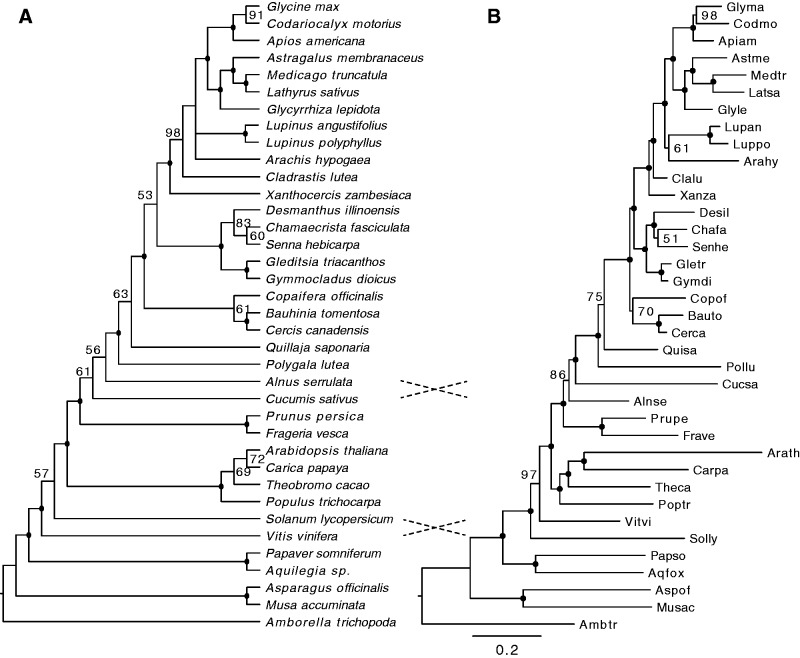
We used 99 putatively single-copy orthogroups (see Materials and Methods) to estimate relationships among major legume lineages using a coalescence-based analysis of gene trees (fig. 2A), implemented in the MP-EST program (Liu et al. 2010) and 101 orthogroups for a “supermatrix” analysis of a concatenated gene sequence alignment (fig. 2B). The topologies recovered using the two methods were generally concordant, suggesting that despite the short internodes on some parts of the tree, variation in gene histories due to incomplete lineage sorting at some loci did not greatly impact phylogenetic inference based on the concatenated data matrix. There were two weakly supported conflicts involving the placement of lineages outside of the Fabales (fig. 2), both of which may be an artifact of sparse sampling. With respect to ordinal relationships within the nitrogen-fixing clade, both trees are discordant with the large analysis of Wang et al. (2009), which places Fagales and Cucurbitales in a clade sister to Rosales, with Fabales as an outgroup. The MP-EST tree shows an unexpected placement for Vitis but with weak support, whereas the supermatrix tree places Vitis in its expected position with high support (fig. 2). Well-supported and congruent nodes in the resulting topologies were identical and similar to other published phylogenies (e.g., Lavin et al. 2005; Bell et al. 2010; Cardoso et al. 2012), but several features are worth noting. First, as described by Lavin et al. (2005) and Cardoso et al. (2012) for trees estimated on plastid gene alignments, our analyses of 99–101 nuclear genes were unable to robustly infer relationships among the three subclades sampled here from the 50-kb inversion clade: Dalbergioids (Arachis), genistoids (Lupinus), and the nonprotein amino acid clade that includes the majority of other core papilionoid legumes, notably millettioids (Glycine) and the inverted repeat loss clade (IRLC) of the Hologalegina (Medicago). This is consistent with rapid diversification early in the Papilionoideae, as reported by Lavin et al. (2005). Second, consistent with the Cardoso et al. (2012) analysis of plastome-encoded matK genes, Xanthocercis and Cladrastis were resolved as successive sister lineages to the core papilionoids, albeit with short branch lengths among these three lineages (fig. 2B). Third, the coalescence and supermatrix analyses both recovered a clade with Copaifera and the Cercideae, with 61% and 70% bootstrap support, respectively. This is similar to the Wojciechowski et al. (2004) chloroplast matK phylogeny, and the more recent Cardoso et al. (2012) phylogeny, but differs from the Bruneau et al. (2008) analysis, which placed Copaifera and its relatives (Detarieae) sister to all other legumes, albeit with low bootstrap support. Finally, a well-supported Mimosoid–Cassiinae–Caesalpinieae (MCC) clade includes a Gleditsia + Gymnocladus clade that is sister to a clade with the mimosoid representative (Desmanthus) and the two Cassiineae (Chamaecrista and Senna). This result is consistent with previous comprehensive legume phylogeny reconstructions (e.g., Wojciechowski et al. 2004; Bruneau et al. 2008; Manzanilla and Bruneau 2012).
Fig. 2.
Species relationships estimated from 101 single copy nuclear genes using coalescence-based MP-EST (Liu et al. 2010) (A) and RAxML analysis (Stamatakis et al. 2008) of the concatenated alignments (B). Bootstrap support values are shown adjacent to each node, or as black dots for nodes with 100% support. Branch lengths in the RAxML tree are substitutions per site. Dotted lines highlight discordances between the two trees. Species abbreviations in (B) use the first three letters of the genus and first two letters of the species. These abbreviations are used in phylogenies in figures 3–5.
Also in agreement with previous analyses, branch lengths in the supermatrix tree (fig. 2B) suggest an increased substitution rate in some of the “cool-season” species (the Hologalegina-IRLC clade: Glycyrrhiza, Astragalus, Medicago, and Lathyrus), for example, Lavin et al. (2005). This has also been reported for M. truncatula, where the per-site rate of synonymous substitutions following the PWGD (1.08 × 10−8 substitutions/year) is estimated to be approximately 1.8 times faster than in G. max (Schmutz et al. 2010; Young et al. 2011). At the same time, terminal branches leading to Cladrastis, Xanthocercis, Gymnocladus, Gleditsia, and Cercis are much shorter than branches leading to the core papilionoids, suggesting decreased substitution rates in these tree species relative to most of the sampled annual crop and model legume species. This rate variation across major lineages within the legumes is generally consistent with the plastid matK trees (Lavin et al. 2005).
Timing of the PWGD Relative to Divergence of Major Legume Lineages
To assess the relative timings of the PWGD and speciation history among the sampled legumes, we assessed gene trees for species representation within clades defined by the last common ancestor (LCA) of G. max homoeolog sets mapping to syntenic blocks derived from the PWGD. Two rounds of genome duplication are evident in synteny analysis of the G. max genome: The PWGD event and a more recent, Glycine-specific genome duplication that is estimated to have occurred 5–13 Ma (Schlueter et al. 2004; Pfeil et al. 2005; Schmutz et al. 2010). Many homoeolog sets identified in syntenic blocks derived from the PWGD and subsequent Glycine WGD included all four duplicate genes derived from these two WGDs and their expected relationships were recovered in the gene phylogenies. For example, consider a simplified phylogenetic representation of G. max (Gm) paralogs, ((Gm1,Gm2),(Gm3,Gm4)), where the Gm1/Gm2 and Gm3/Gm4 homoeolog pairs were derived from the Glycine WGD, and the ancestral genes for these two clades were derived from the earlier PWGD. Even with gene loss, many paralog sets with two and three genes still contain Glycine homoeologs derived from the PWGD and can therefore be used to mark the divergence time for the parental species of the PWGD allopolyploid. For example, the LCA of either the (Gm1, Gm4) or (Gm2, Gm3) paralog sets can be ascribed to the PWGD based on membership in synteny blocks that have been diverging since the PWGD.
The PWGD syntenic Glycine homoeolog pairs (supplementary file S1, Supplementary Material online) allowed us to identify a total of 2,432 nodes representing the LCA of PWGD homoeologs with bootstrap values (BSVs) of 50 or greater in their respective gene trees. Of these, 721 LCA nodes (with 2,058 homoeologous pairs) met the criteria outlined in Materials and Methods for congruence to the species tree topology (fig. 2) with unambiguous rooting to a noneudicot gene. These were used to test alternate hypotheses for the placement of the PWGD along the backbone of the species tree (fig. 3).
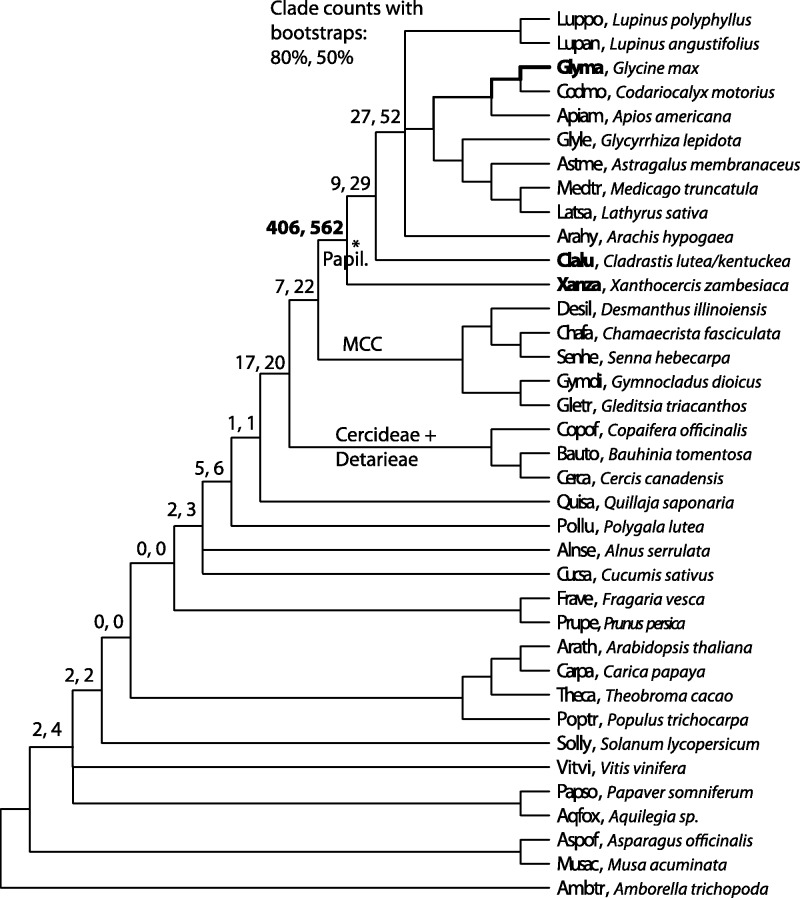
Fig. 3.
Illustration of tests for timing of the PWGD versus speciation times. Numbers at each node indicate the numbers of observed clades (at ≥80% and 50% BSVs, respectively) that are both consistent with the phylogeny in figure 2 and are rooted by the most recent common ancestor of Glycine paralogs (bold line) derived from the PWGD. The highest count (406 at ≥80% bootstrap) is at the papilionoid node itself, supporting a model in which the PWGD occurred just prior to the papilionoid diversification and thus gave rise to two clades with paralogous Glycine, Xanthocercis, and Cladrastis genes.
Figure 3 includes counts of gene tree clades (across all gene trees) that are both 1) consistent with the consensus species phylogeny (fig. 2) and 2) rooted by the LCA of Glycine paralogs mapping to synteny blocks for PWGD. For example, focusing on the nodes of Xanthocercis, Cladrastis, and core papilionoid diversification, we can frame two alternate hypotheses: Hypothesis 1, “early PWGD”—with the PWGD predating the divergence of lineages leading to Xanthocercis and the other papilionoids (Cladrastis, Medicago, Glycine, etc.); and Hypothesis 2, “later PWGD”—with the PWGD occurring after the divergence of lineages leading to Xanthocercis and the remaining papilionoids (i.e., species from the 50-kb inversion clade). The taxon representation within clades defined by the 721 rooted LCA nodes strongly supports placement of the PWGD just prior to the earliest diversification of all papilionoid lineages including Xanthocercis (fig. 3). Support for the “early PWGD” hypothesis requires that no nonpapilionoid genes appear within the PWGD clades in the gene trees and that no papilionoid genes (including Xanthocercis and Cladrastis) be found outside PWGD clades. With an 80% BSV threshold for a PWGD clade, 406 LCA nodes support Hypothesis 1 and 36 LCA nodes support Hypothesis 2 (fig. 3: 36 = 27 + 9 from the last two papilionoid clades with counts). Imposing a 50% BSV threshold, 562 LCA nodes support Hypothesis 1 and 81 (52 + 29) trees support Hypothesis 2. Cases were also found that supported alternative placements of the WGD, but with much lower frequencies (counts at other nodes). Gene trees with typical arrangements of Xanthocercis and Cladrastis genes are shown in figure 4 and supplementary figure S2, Supplementary Material online, with locations of hypothesized WGDs marked with red asterisks. Alternative placements for the PWGD events within the gene trees may be due to incomplete sorting of ancestral gene lineages between the rapid series of speciation events (Jones et al. 2013) that occurred early in papilionoid history, or errors in gene tree estimation.
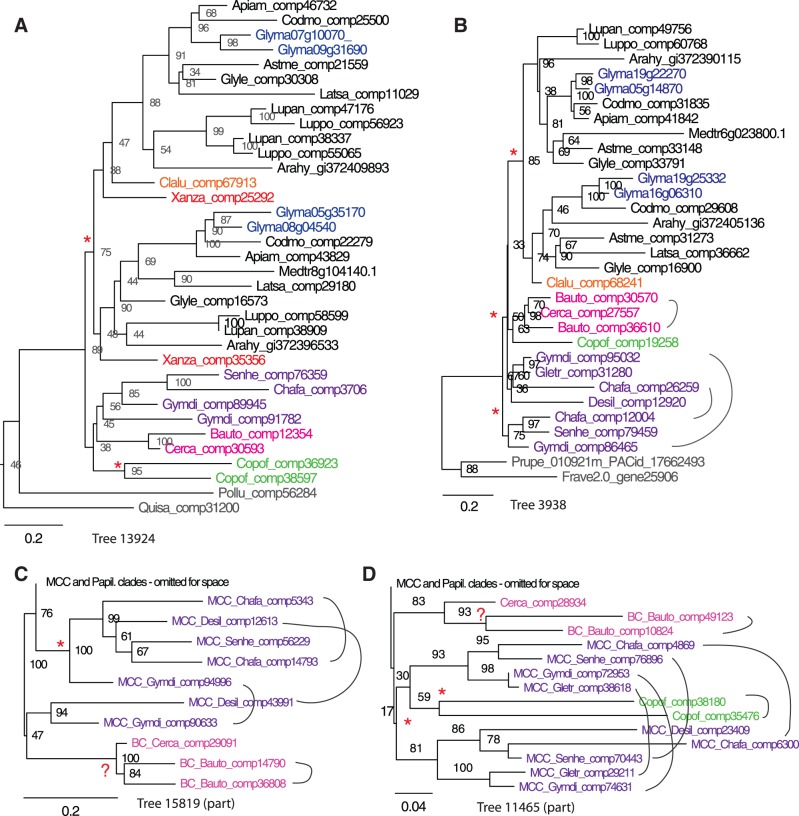
Fig. 4.
Sample gene trees, showing typical patterns seen among the 3,360 trees including Glycine homoeologs derived from the PWGD. Species names for each abbreviation are shown in figure 2 and table 1. Gene trees (A) and (B) are consistent with the PWGD predating divergence of Xanthocercis, Cladrastis, and other papilionoid lineages. Trees (B), (C), and (D) are consistent with an early WGD in the MCC clade and in the Cercideae. Outgroup taxa more distantly related than Polygala and Quillaja have been pruned for clarity. Trees are colored for ease of interpretation: Glycine max in blue; Xanthocercis and Cladrastis in red and orange, respectively; members of the MCC clade in purple, members of the Cercideae in pink, and the representative of the Detarieae (Copaifera) in green.
Analysis of Synonymous Substitutions
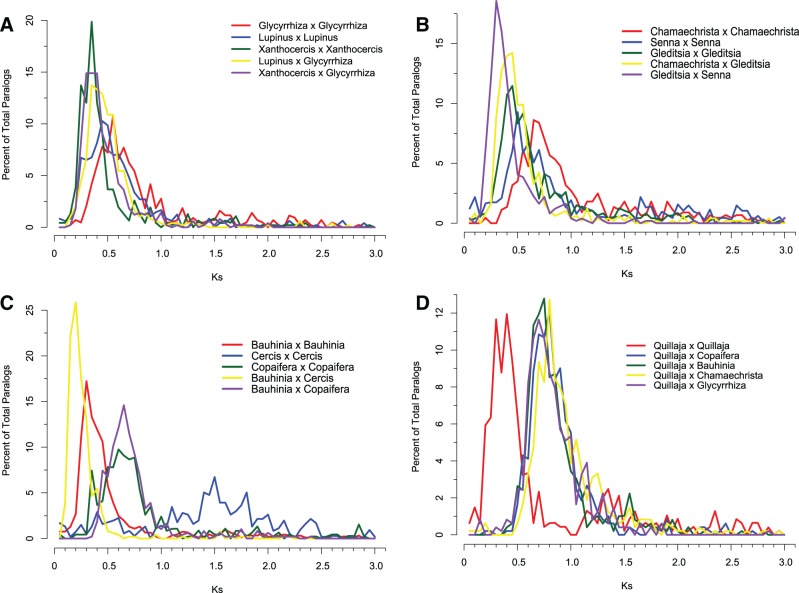
Plots of synonymous substitutions for legume paralogs and several outgroup species are shown in figure 5 and supplementary file S3, Supplementary Material online, and the modal Ks values are given in figure 6. Strikingly, all legumes (with the possible exception of Cercis; fig. 5) have Ks peaks in the range of 0.3–0.85. Many Ks plots also show a smaller, more diffuse peak in the range of approximately 1.5–2 (fig. 5) that may be attributable to the core eudicot WGD (Jiao et al. 2011, 2012). Given strong phylogenetic evidence for the placement of PWGD in a common ancestor of papilionoid species, strong peaks around 0.3–0.6 nonpapilionoid legumes may provide evidence for independent WGDs in these lineages. Gene tree analyses and comparisons of Ks distributions for intraspecific paralog pairs with interspecific best blast hits (homologs) were assessed to infer the timing of these other putative WGDs.
Fig. 5.
Plots of synonymous substitution frequencies by Ks class. Also see modal Ks values in figure 8. Selected comparisons: (A) Papilionoid examples: i) Glycyrrhiza paralog pairs (mode = 0.45), ii) Lupinus paralogs (mode = 0.50), iii) Xanthocercis paralogs (mode = 0.35), iv) Glycyrrhiza/Lupinus homolog pairs (mode = 0.35), and v) Glycyrrhiza/Xanthocercis homolog pairs (mode = 0.35). Comparisons suggest that PWGD (0.50–0.55 in Lupinus and 0.55 in Glycyrrhiza) predates Glycyrrhiza/Lupinus divergence (0.35). The Ks numbers for Cladrastis and Xanthocercis are ambiguous in terms of relative speciation versus PWGD timing. (B) MCC examples: i) Chamaecrista paralog pairs (mode = 0.65), ii) Senna paralog pairs (mode = 0.5), iii) Gleditsia paralog pairs (mode = 0.45), iv) Chamaecrista/Gleditsia ortholog pairs (mode = 0.45), and v) Senna/Gleditsia ortholog pairs (mode = 0.3). Comparisons among Senna and Gleditsia paralog and best BLAST hit homolog Ks plots suggest that the MCC WGD predates earliest diversification of MCC clade, but timing of WGD is ambiguous in comparison of the Chamaecrista and Gleditsia Ks plots. (C) Cercideae and Copaifera examples: Bauhinia paralog pairs (mode = 0.3), Cercis paralog pairs (mode = 0.4), Copaifera paralog pairs (mode = 0.6), Bauhinia/Cercis ortholog pairs (mode = 0.2), and Bauhinia/Copaifera ortholog pairs (mode = 0.65). These comparisons suggest separate Cercideae and Copof WGDs. (D) Comparisons with outgroup taxa, Quillaja: Quillaja paralog pairs (modes = 0.4, 1.4), Quillaja/Copaifera, Quillaja/Bauhinia, Quillaja/Chamaecrista, and Quillaja/Glycyrrhiza ortholog pairs (modes all 0.7–0.8).
Fig. 6.
Ks modes for selected species comparisons. Values correspond to the primary peaks shown in figure 7. Higher Ks values are more blue; lower are more yellow. Example interpretation: WGDs occurred recently in each of Quillaja and Polygala (0.4 and 0.3), long after divergence of their respective lineages (1.0). The primary peak in the Xanthocercis Ks plot exhibits more recent divergence (0.35) than the peaks in plots for most species from the MCC, Detarieae, and Cercideae (0.4–0.45), suggesting old WGDs in those clades.
Evidence for Additional WGDs within the Papilionoideae
The G. max genome exhibits a very high number of retained duplicate genes, including terminal duplicates (i.e., sister genes in the gene trees) derived from the Glycine WGD. Other papilionoid taxa with relatively large numbers of terminal duplicates include Arachis (100) and Lupinus (81, including both Lupinus polyphyllus, Luppo and L. angustifolius, Lupan). Arachis hypogaea is a recent allopolyploid (Moretzsohn et al. 2013), so it may be surprising that we did not see more terminal duplicates. The low count may be due to contig collapses in this transcriptome assembly for this species (Zhang et al. 2012). For Lupinus, gene trees that include genes sampled from both species duplication events within a clade for the genus imply that the duplication event occurred before the divergence of New World (L. polyphyllus) and Old World (L. angustifolius) clades, for example, ((L.ang.1, L.pol.1), (L.ang.2, L.pol.2)), or (L.ang.2, (L.ang.1, L.pol.1)). This is consistent with reports of a WGD in Lupinus or in the genisteae (Kroc et al. 2014).
Evidence for WGDs in the Cercideae and in Copaifera
Analyses of Ks plots and gene trees such as those in figure 4 and supplementary figure S2, Supplementary Material online, provide evidence for an ancient WGD within the Cercideae. There is a strong Ks peaks in the paralog plot for Bauhinia at 0.3 and possibly a weak peak for Cercis at Ks = 0.4 (fig. 5C). Both of these values are greater than the Ks peak for interspecific ortholog pairs for these species (Ks = 0.2), suggesting either that a WGD could have predated the divergence of Bauhinia and Cercis or that a WGD was present only in Bauhinia, and the higher rate of substitutions in Bauhinia (fig. 2B) produces a Bauhinia–Cercis ortholog Ks peak that appears more “recent” (0.2) than the Bauhinia–Bauhinia paralogous peak (0.3). Analyses of gene trees with Bauhinia paralog pairs showing modal Ks values (0.25–0.45) were largely consistent with duplication in the Cercideae before divergence of Bauhinia and Cercis, for example, (Bauhinia, (Bauhinia, Cercis)); supplementary file S4, Supplementary Material online. However, the sample of informative trees was small; just 45 LCA nodes for these Bauhinia paralog pairs were supported with bootstrap support ≥80%, of which 20 (44.4%) support placement of a WGD within the Cercideae. The best-supported alternative hypothesis, with 11 trees (24.4%) exhibiting BSV ≥80%, places a WGD in a common ancestor of Copaifera and the Cercideae. This hypothesis will be tested most rigorously once a genome has been sequenced within the Cercideae and phylogenetic analyses can be performed on homeologs identified in synteny analyses as we describe for the PWGD described above.
Our phylogenetic analyses place Copaifera as sister to the Cercideae clade, albeit with weak bootstrap support (61% and 70% in fig. 2). The Copaifera Ks plot also exhibited a peak at Ks = 0.6, but in contrast to the Ks analysis of Bauhinia and Cercis paralog pairs, Ks peaks for Copaifera–Bauhinia and Copaifera–Cercis homolog pairs were both centered at Ks = 0.65, suggesting independent genome-scale duplication events in Copaifera and the Cercideae. Placement of an independent WGD within the Copaifera lineage is supported by a count of 142 terminal Copaifera duplicates (i.e., Copaifera, Copaifera) in the gene trees. In sum, these analyses are in agreement with Ks plots (fig. 5C) and terminal duplicate counts, supporting the inference of independent WGDs within the Cercideae and Copaifera lineages, though with uncertain timing of the inferred WGD within the Cercideae.
Evidence for a WGD Early in the MCC Clade
Members of the MCC clade (Chamaecrista, Senna, Desmanthus, Gymnocladus, and Gleditsia) show Ks peaks ranging from 0.35 (Gymnocladus and Gleditsia) to 0.65 Chamaecrista and Desmanthus) (figs. 5 and 6). These Ks peak values generally correlate with substitution rates as reflected in the branch lengths of the 101 gene supermatrix tree (fig. 2B): The low Ks modal values for Gymnocladus and Gleditsia correspond with short branches in the species tree, whereas Chamaecrista, Senna, and Desmanthus have larger Ks paralogous peaks and longer branch lengths in the species tree. Analysis of interspecific homolog pair Ks distributions for all species pairs within the MCC clade supports a shared WGD on the lineage leading to the LCA of the clade. A WGD early in the MCC clade is consistent with the trees shown in figure 4 and supplementary figure S2, Supplementary Material online, each of which has more than two MCC species with retained paralog pairs derived from duplications predating the early diversification of MCC lineages. In Ks comparisons of all but two of the MCC species pairs, Ks peaks for the intraspecific paralog pairs are greater than Ks peaks for the corresponding interspecific homolog pairs (fig. 6). The two exceptions (Desmanthus–Gymnocladus and Chamaecrista–Gleditsia) involve comparisons of slowly and rapidly evolving lineages (fig. 6). As discussed by Cui et al. (2006), variation in substitution rates complicates interpretation of among species comparisons of Ks plots.
We visually examined 264 trees containing duplicate genes sampled from the MCC clades in order to evaluate evidence for a hypothesized early-MCC WGD (supplementary file S4, Supplementary Material online). The mimosoid duplications primarily occur early, and they often are seen in two or more mimosoid species: For example, Chamaecrista and Desmanthus both show duplicates mapping to the MCC ancestral lineage in 19 gene trees, and Gymnocladus and Gleditsia paralog pairs occurring within 23 gene trees exhibit relationships consistent with the hypothesized early-MCC WGD. However, as discussed above for the Cercideae, this hypothesis will be tested most rigorously once a genome has been sequenced within the MCC clade and phylogenetic analyses can be performed on homeologs identified in synteny analyses.
Evidence for WGDs in Quillaja and Polygala
For the two closest outgroups to the legumes sampled in this study, Quillaja and Polygala, there are Ks peaks at 0.4 for Quillaja, and 0.3 and 0.8 for Polygala duplicate gene pairs. Plots of Ks values for interspecific homolog pairs for Quillaja and Polygala exhibit a clear peak at Ks = 1.0, suggesting that the duplication peaks seen in paralog plots for these two species represent lineage-specific events that postdated their LCA (figs. 4 and 5 and supplementary fig. S2, Supplementary Material online). Additional evidence for WGDs in these lineages comes from counts of terminal duplicates in the gene trees. Such paralogous or “terminal” duplicates appear, for example, as (Polygala, Polygala) or (Quillaja, Quillaja) in a simplified tree string (supplementary fig. S2A and C, Supplementary Material online, shows example of such a terminal duplicates for Polygala and Quillaja). There are 280 terminal duplicates for Polygala and 61 terminal duplicates for Quillaja, in 3,360 trees with genes from these species. This compares, for example, with terminal duplication counts of 0, 13, and 8 for Cladrastis, Xanthocercis, and Medicago, respectively; all of these taxa lack a lineage-specific WGD.
Chromosome Counts and Polyploidy
We compiled chromosome counts from 428 legume taxa (supplementary file S5, Supplementary Material online; Bennett and Leitch 2012). Typical chromosome counts for each clade are summarized in figure 1. Legume chromosome counts are also summarized and reviewed in Doyle (2012). A wide range of base chromosome numbers is seen in the Papilionoideae. The most common counts in the MCC clades are x = 12–14, with an exception in Chamaecrista, with counts of n = 7, 8, and 14 (supplementary file S5, Supplementary Material online; Bennett and Leitch 2012). (Here and below, we use “x” to indicate the inferred “base” chromosome number of a group or genus and “n” to indicate the ploidy number of an individual species or accession.) Through the rest of the Caesalpinoideae lineages (Cercideae, Detarieae, Dialiinae), n = 14 is most common (also 12 in the Detarieae), with an exception of n = 7 for Cercis (Doyle 2012). The identification of independent WGDs in each of the major legume clades suggests that the ancestral chromosome number for the Fabaceae may have been x = 7 consistent with Goldblatt’s (1981) speculations. Additional sampling of key lineages (e.g., Duparquetiinae; Bruneau et al. 2008) of the caesalpinioid grade is required to test this hypothesis.
Discussion
The initial focus of this study was to determine whether the PWGD, known to have occurred before the diversification of the “50-kb inversion” clade (Doyle 2012; Legume Phylogeny Working Group 2013), also predated the sequential divergence of species-poor papilionoid lineages that form a grade subtending the speciose “50-kb inversion” clade. We find clear support for this hypothesis, based on preponderance of genes with topologies consistent with a shared duplication between two such early-diverging papilionoids (Xanthocercis and Cladrastis) and the remaining papilionoids sampled here (fig. 2). These taxa share with other papilionoids a peak of gene pairs in the Ks distribution consistent with such a polyploidy event (table 2 and fig. 6). Our results also support the conclusion of Cannon et al. (2010) that the ancestry of Chamaecrista and thus the MCC clade does not include the PWGD. Our analyses also provide new evidence that there were additional independent ancient WGD events within the Fabaceae but outside the papilionoid clade. Ks plots and gene tree analyses suggest separate genome-wide duplications on lineages leading to Copaifera (Detarieae), the Cercideae, the MCC clade, and the papilionoids (figs. 5 and 6).
Legume Phylogeny
Conclusions about the timings of WGDs with respect to speciation events depend critically on the species phylogeny. Although some nodes remain unresolved, the species phylogeny estimated from 101 nuclear genes (fig. 2) matches other current matK and rbcL phylogenies (Cardoso et al. 2012) in most respects, including relationships among early diverging papilionoid lineages: Xanthocercis resolves as sister (100%) to Cladrastis plus the remaining papilionoid legumes; and Cladrastis is in turn sister (100%) to an unresolved polytomy (the 50-kb inversion clade) involving Arachis (dalbergioid clade), Lupinus (genistoid clade), and the clade with other derived papilionoid legumes. In addition, the MCC clade is well supported (100%). This has been observed before (Bruneau et al. 2008; Manzanilla and Bruneau 2012), and the term MCC was coined in Doyle (2011) to describe this group. Cercis and Bauhinia form a well-supported Cercideae clade (100%). The weaker placement of Copaifera (Detarieae) with the Cerciceae (61–70%) is worth noting, as the relative placements of the Detarieae and Cercideae are generally poorly supported or conflicting in recent molecular phylogenies (Lavin et al. 2005; Bruneau et al. 2008); but this relationship is not crucial for the placements of WGDs described above.
The PWGD Predated Radiation of the Papilionoideae
The finding that the PWGD occurred before the divergence of Xanthocercis and Cladrastis from the other core papilionoids suggests that the PWGD is ancestral to the LCA of all papilionoid species. In the most comprehensive published molecular phylogeny of the papilionoid legumes (Cardoso et al. 2012), Xanthocercis and Cladrastis fall into two “early-diverging” papilionoid clades (fig. 1): Xanthocercis in the Angylocalyx clade within the “ADA” clade, consisting of the Angylocalyx, Dipterygae, and Amburana clades; and Cladrastis in the Cladrastis clade, sister to the 50-kb inversion clade (which contains all of the remaining papilionoid legumes in our study). The only remaining early-diverging papilionoid clade that was not sampled in this study is the Swartzioid clade.
Multiple Viable Models of Evolution of Symbiotic Nitrogen Fixation in the Papilionoid Legumes
The Swartzioid clade in both the Cardoso et al. (2012) and Lavin et al. (2005) topologies (fig. 1) resolves as sister to Cladrastis + the 50-kb inversion clade, with Xanthocercis and the ADA species being sister to both of these, although this conclusion should be tempered by moderate bootstrap support (75%) for the Swartzioid placement in Cardoso et al. (2012). It would be valuable to test both the placement and WGD status of the Swartzioid sequences in gene families. The Swartzioid clade is of particular interest because it is the only clade among the early-branching papilionoid groups placed outside the 50-kb inversion clade that is known to contain species which nodulate and possess SNF, an ability it shares with the majority of the subclades in the 50-kb inversion clade. Interestingly, Cladrastis and other members of the Cladrastis clade—the sister group of the 50-kb inversion clade—do not possess SNF. Unfortunately, relationships of the early radiation of the 50-kb inversion clade remain poorly resolved in chloroplast phylogenies (Cardoso et al. 2012), and because these clades vary in their ability to nodulate, it is difficult to make robust inferences about the origin(s) of nodulation in papilionoid legumes. The current data remain consistent with various models, including multiple gains of SNF in the Papilionoideae subsequent to the divergence of the nonnodulating ADA clade: In the Swartzeae and one or more times independently within the 50-kb inversion clade. Alternatively, there could have been a single origin of nodulation in papilionoids after the divergence of the ADA clade, with loss of the trait in the Cladrastis clade and one or more times within the 50-kb inversion clade (see discussion of SNF evolution below).
Evidence for Additional WGDs outside the Papilionoideae
This study’s finding of three additional WGDs early in the legume radiation—in the Cercideae, the Detarieae, and in the MCC clade—is supported to varying degrees by several lines of evidence: Strong Ks peaks in each species (excepting Cercis, with a weak peak; fig. 5), repeated phylogenetic patterns consistent with WGDs near the base of each of the three clades (fig. 4), and analyses of terminal duplicates within the gene phylogenies.
Our species sampling, though obviously minimal for a family the size of the legumes, contains key early-diverging lineages in the Papilionoideae and MCC—and therefore provides good confidence for our conclusions that polyploidies occurred early in these clades, as well as in the Cercideae (Bauhinia) and the Detarieae (Copaifera). We also note, however, the sparseness of our taxon sampling and acknowledge that the timing of duplications in the MCC and Cercideae, and the numbers of origins and losses of nodulation in the MCC and Papilionoideae will require further investigation.
In addition to the four early WGDs that we report from the legumes, we also see evidence for early WGDs in Quillaja and Polygala. The Ks values and phylogenetic placements of these six WGDs place these events in the time frame of about 60–40 Ma, per the dates in Lavin et al. (2005): Approximately 58 Ma at the base of the papilionoids, approximately 55 Ma at the base of the MCC clade, and less well-circumscribed dates for the WGDs in Bauhinia, Copaifera, Quillaja, and Polygala. Intriguingly, a wave of genome duplications has been observed across many lineages in this time frame. Vanneste et al. (2014) and Fawcett et al. (2009) propose that polyploidization paved the way for adaptive evolution of duplicated genes following the Cretaceous–Paleogene extinction event at approximately 66 Ma. Vanneste et al. (2014) report independent WGDs in 20 lineages in the last approximately 100 Ma, with significant concentration of these events near the time of the Cretaceous–Paleogene extinction. Our results suggest four more WGD instances within this time frame (fig. 1)—though with considerable uncertainty about the exact timing.
The finding of WGD early in the MCC clade lineages was unexpected, given our earlier conclusion (Cannon et al. 2010) that there was no evidence of any WGD more recent than the eudicot triplication (Jiao et al. 2011) affecting Chamaecrista. Inclusion of five MCC species in this study, as well as better assemblies and many more gene trees, has allowed for much stronger inference than was possible in the earlier study. The other primary result from Cannon et al. (2010), that the Chamaecrista lineage did not share the PWGD with papilionoids, is strongly supported in this study.
Chromosome Counts and WGDs in the Legumes
Although the gene tree and Ks signals are arguably the strongest indicators of WGDs in the nonpapilionoid legumes, the chromosome counts are also interesting and appear to corroborate a model of WGDs having occurred within each represented legume lineage. The most common chromosome number across the legumes is n = 12–14 (reviewed in Doyle 2012; also fig. 1 and supplementary file S5, Supplementary Material online). Numbers in this range are seen in the Cercideae (excepting Cercis itself, with n = 7), in the Detarieae, Dialiineae, the MCC clade (excepting some Chamaecrista, with species showing n = 7, 8, and 14), and in many of the early-diverging papilionoids: Xanthocercis is n = 13 and Cladrastis is n = 14—which we now have shown to have diverged from each other and the core papilionoids after the PWGD. Doyle (2012) concluded from these observations that the base number of legumes was likely to be x = 12. Taking into account our finding of additional WGD events, we infer that the ancestral legume chromosomal state was instead most likely x = 6–7. This is consistent with the hypothesis posed by Raven (1975) and Goldblatt (1981), based on extensive (but prephylogenetic) analyses of chromosomal data, that the legumes may have arisen from a progenitor with x = 7, and then undergone early polyploidy in several lineages—followed by scattered aneuploidy in some lineages (e.g., Chamaecrista). Subsequent to the PWGD, it appears that chromosome reductions occurred in several papilionoid lineages. Indeed, given the rapid radiation of papilionoids (Lavin et al. 2005), within a few million years base chromosome numbers in all of the major lineages of the 50-kb inversion clade decreased, for example to x = 9 in genistoids, x = 10 in dalbergioids, x = 10–11 in millettioids, and x = 7–8 in Hologalegina (Doyle 2012, fig. 1). Although synteny analyses of available papilionoid genomes (Cannon et al. 2006; Schmutz et al. 2010; Young et al. 2011) indicate that the PWGD was indeed a polyploidy event involving all chromosomes, we do not have genome sequences for any legume taxon outside of the papilionoids.
Gene tree and Ks evidence of a WGD early in Lupinus (prior to divergence of Old World L. angustifolius and New World L. polyphyllus) is consistent with chromosome counts. As noted above, the genistoid clade is ancestrally x = 9. However, within that clade the tribe Genisteae, to which Lupinus belongs, is characterized by considerable chromosomal variation, often within individual genera, and often involving high chromosome numbers (Doyle 2012); the longstanding hypothesis that polyploidy occurred early in the evolution of the Genisteae (Goldblatt 1981) has been supported as phylogenies for the group became available (Doyle 2012), and by a recent synteny analysis between L. angustifolius and M. truncatula (Kroc et al. 2014). The chromosome number for L. angustifolius is n = 20, and L. polyphyllus is typical of the New World radiation of this large genus in being x = 24, with cytotypes of n = 24 and n = 48 reported in the Index of Plant Chromosome Numbers (IPCN: http://www.tropicos.org/Project/IPCN, last accessed December 2013). In the phylogeny of Lavin et al. (2005), the radiation of core Genisteae is dated at 19–35 Ma, with Lupinus diverging shortly after this. The Ks in the Lupinus species is 0.50–0.55 (figs. 5 and 6), suggesting WGD early within the Genisteae; however, further sampling in the genistoids is required to pinpoint the time of polyploidy in the group.
Models of the Evolution of SNF in the Legumes
Given lack of strong resolution in key portions of legume phylogeny, the unknown nodulation status of critical taxa, and the inability to distinguish ancestral absence of nodulation from secondary loss of the trait, it has been difficult to identify when, where, and how often nodulation evolved in legumes (Doyle 2011). Werner et al. (2014), using a new, comprehensive angiosperm phylogeny (Zanne et al. 2014), an updated list of nodulating taxa, and newly developed modeling approaches, hypothesized three key stages in the origin and evolution of nodulation. In an initial “potentiating” stage, one or more mutations in a still unknown developmental or biochemical pathway led to a “precursor” of nodulation in the ancestor of the N-fixing clade, an idea that (called “predisposition”) has been hypothesized for many years (Soltis et al. 1995; Kistner and Parniske 2002; Doyle 2011). The origin of the predisposition state appears to have been a single event, in contrast to the second, “actualizing” phase, in which the ability to nodulate was realized independently around eight different times in the preferred model of Werner et al. (2014), including several times within legumes. In their models, both the precursor and nodulation states appear to be evolutionarily labile, the former estimated to have been lost around 17 times in the N-fixing clade as a whole, the latter around ten times. In the model of Werner et al. (2014), a third, “refinement,” phase constitutes a shift from a nodulation state to a “stable fixer” state. They hypothesize that shifts to the “stable fixer” state have occurred frequently—24 times in their best model, and numerous times within legumes alone—but are almost never lost. SNF evolved independently from the precursor state once in the papilionoid lineage, and at least twice in the MCC clade in the analysis of Werner et al. (2014), and the vast majority of the papilionoid clade lineages are optimized as “stable fixers.” The notion of “potentiating,” “actualizing,” and “refinement” phases of SNF evolution is interesting, but care should be taken in evaluating the timing and frequency of shifts among these phases as inferred by Werner et al. (2014) because the phylogeny on which they mapped these shifts includes some clear anomalies with respect to legume relationships (e.g., Cardoso et al. 2012, 2013, fig. 2) that may affect their optimizations.
Could polyploidy be involved in shifts to any of these three phases modeled by Werner et al. (2014)? Polyploidy does not appear to be coincident with the origin of the nitrogen fixing clade, and thus is not implicated in the origin of the precursor. Within the N-fixing clade and within legumes, polyploidy is not sufficient for nodulation to evolve from the precursor condition. Our results suggest that several nonnodulating taxa have experienced WGD events: Independent polyploidy in the nonlegumes Polygala and Quillaja, as well as in the legume tribe Detarieae. Moreover, none of these taxa is descended from nodulating ancestors in the trees of Werner et al. (2014). In the preferred model of Werner et al. (2014), the actualization of nodulation in the MCC clade occurs multiple times, long after the polyploidy event. It is possible that WGD provided a second potentiating innovation that was not actualized until much later in the evolution of this clade.
In contrast, the relationship between polyploidy and “refinement” of nodulation in papilionoid evolution may be more direct. Recent authors (Young et al. 2011; Li et al. 2013) have speculated that the PWGD led to the refinement of a pre-existing nodulation symbiosis in papilionoids, and, following these ideas, Werner et al. (2014) suggest that the PWGD could be associated with the origin of stable nodulation in the subfamily. However, our placement of the PWGD in the papilionoid common ancestor coincides precisely with the actualization of nodulation in the preferred model of Werner et al. (2014), suggesting that polyploidy may have been associated not with refinement, but with the transition from the precursor phase to a unique origin of nodulation.
If polyploidy provided the raw material for the genetic innovations that allowed the development of SNF in the papilionoids as has been hypothesized by Li et al. (2013) and Young et al. (2011), then evolution of duplicate gene function (neofunctionalization or subfunctionalization) for SNF-related processes may have occurred after Cladrastis and Xanthocercis lineages diverged from the core papilionoid clade. Such a delay in neofunctionalization or subfunctionalization following a WGD event may account for lags between polypoidy events and the origin of evolutionary innovations contributing to increased diversification (Soltis et al. 2009; Schranz et al. 2012).
Conclusions
Our main findings are that WGDs occurred near the origins of the major legume clades: In the Cercideae, in the Detarieae, in the MCC clade, and in the Papilionoideae. The strongest evidence for these is in the Papilionoideae, where a combined analysis of phylogeny and known synteny-derived paralogous genes places the WGD prior to the origin of the Papilionoideae, and after divergence of the Papilionoideae from the MCC clade. We cannot say that every legume has a WGD in its history after the origin of this family, as we have not sampled species within the Dialiineae or Duparquetia, nor at the base of every clade—and indeed, there is no clear evidence for a WGD in Cercis. However, the WGDs that we have observed affect the large majority of species in the legume family. Within the papilionoid legume stem lineage, the PWGD predated the diversification of early-diverging nodulators (much of the large 50-kb inverted repeat clade and the Swartzioid clade) and nonnodulators (the Cladrastis clade, the ADA clade—represented here by Xanthocercis—and some subclades of the 50-kb inversion clade). Given independent WGDs affecting at least four legume lineages, together with the predominant chromosomal state of n = 14 in each, we hypothesize an ancestral-legume genome with x = 7 chromosomes, a reduction from the n = 21 state following the eudicot triplication (Salse 2012). As for the relationship between polyploidy and nodulation, it is possible that polyploidy contributed to the evolution of the complex modern symbiotic rhizobial nodule, but the evolutionary relationship between polyploidy and nodulation is neither sufficient nor simple. There have most likely been several independent origins of nodulation in the legumes, and it appears that most legumes are fundamentally polyploid, and many of these lack the capacity for SNF.
Materials and Methods
Taxon Selection and Data Acquisition
Species were selected for transcriptome sequencing primarily on the bases of taxonomic coverage and nodulation status (see table 1): Two Lupinus species as New- and Old World representatives of the genistoid clade; Cladrastis and Xanthocercis as early-diverging papilionoid legumes; Desmanthus and Chamaecrista, as diverse (nodulating) representatives from the MCC, and Gleditsia, Gymnocladus and Senna as (nonnodulating) representatives from the MCC; Copaifera as a representative from the Detarieae; Bauhinia and Cercis as representatives from the Cercideae; Quilaja and Polygala as close outgroups to the legumes; and other nonlegume species to provide representatives from the Cucurbitaceae and Rosaceae (families that include nodulating genera)—and more distant outgroups to help provide context for phylogenetic tree interpretation. For the legume species, tissue for RNA sequencing (RNA-Seq) was collected in 2010–2011 as described in the Thousand Plant Transcriptomes project, http://www.onekp.com/samples/list.php?set = angiosperm, then shipped on dry ice for sequencing at Beijing Genomics Institute.
De Novo Transcriptome Assembly
Raw 100 nt paired-end Illumina mRNA sequence reads were obtained and 3′-ends of reads were quality trimmed using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), removing any bases with Phred scores less than 20. We also discarded any low quality read less than 40 nt long or with less than 80% of bases having a Phred score greater than 20. Illumina adapter sequences and primers were trimmed using Flexible Adapter Remover version 2.0 software (Dodt et al. 2012), searching the entire read and retaining the longer segment if trimmed into two pieces.
The cleaned sequence reads were assembled using default parameters in Trinity (Haas et al. 2013; version r2012-06-08). To filter lowly expressed or potentially artificial transcripts from a given Trinity graph component (roughly considered a gene), we aligned all cleaned reads to the assembly with Bowtie (Langmead and Salzberg 2012; v 0.12.8), wrapped by the “alignReads.pl” script in Trinity. We quantified transcript abundances of properly sorted read pairs with RSEM (Li and Dewey 2011), removing any transcript isoforms with less than 1% of the per-component FPKM (fragments per kilobase of exon per million fragments mapped) expression. This filtered assembly of high confidence transcripts was used for all downstream analysis.
Gene Family Estimations
Filtered assemblies were subject to BLASTX searches (1e-10 cutoff) against 22 plant genome gene annotations used by the Amborella Genome Project (2013) to circumscribe gene families. To infer conceptual protein translations in the proper reading frame, assembled transcripts and their best BLASTX hits to the 22-genome protein sequence sets were aligned in Genewise using a custom Perl pipeline. Custom cleaning scripts then identify the longest Genewise translation, remove stop codons, and produce a filtered coding sequence (CDS) and corresponding protein sequence.
Previously, the protein sequences from the 22 genome sequences were subject to an all-by-all BLASTP and clustered into estimated gene families using OrthoMCL (Amborella Genome Project 2013) Using the best BLASTX hit to the protein sequences, transcript assemblies were sorted to the 22 genome gene family circumscriptions. Each CDS inferred from the transcript sequences was sorted into the gene family containing the best BLASTX hit; gene models from the M. truncatula, P. persica, and Cucumis sativa were sorted into orthogroups using best HMMER hits by E value (1e-30 cutoff) rather than best BLAST hit. To correct for oversplitting in the OrthoMCL gene family circumscription, two gene families were collapsed if they contained paralogs identified in synteny analyses of the G. max genome (i.e., syntelogs; supplementary file S1, Supplementary Material online).
Gene Family Alignments and Gene Tree Construction
Peptide sequences for each gene family were aligned using MUSCLE v3.8.31 (Edgar 2004), and CDSs were forced onto the protein sequence alignments using PAL2NAL v13 (Suyama et al. 2006). To quality filter the CDS alignments, taxa were removed if their sequence was shorter than 30% of the full length gene family alignment. Further, an alignment column was removed if it contained gaps in greater than 90% of sequences. Maximum-likelihood trees for each gene family were constructed in RAxML (Stamatakis et al. 2008) using the GTR + Γ model over 500 bootstrap replicates, and rooted to outgroups Amborella trichopoda, Musa acuminata, or Asparagus officinalis.
Species Tree Construction
Species trees were estimated using concatenated and coalescent-based approaches. The set of 970 single/low copy gene families of Duarte et al. (2010) was used as a starting point for identification of single copy orthogroups in the data analyzed. Orthogroup trees were analyzed for copy count for each species. If multiple transcript assemblies for a species were sorted to a putative single-copy gene family and the transcripts comprised a clade of only that species with a BSV of at least 50 (i.e., splice variants, alleles or terminal duplicates), then a consensus sequence was generated. Otherwise, the orthogroup was removed from the single-copy analysis. With this strict filtering we were left with 101 orthogroups that were judged as single copy in all taxa included in our analysis. These were used for species tree estimation.
In order to perform a supermatrix analysis, each aligned gene family was concatenated for a species. If the species did not have a representative in the gene family, N’s of alignment length were added. All 101 orthogroups were represented in the concatenated analysis. The topology was reconstructed using RAxML with the GTP + Γ model over 500 bootstrap replicates and rooted to A. trichopoda.
The coalescence-based analysis was conducted using MP-EST (Liu et al. 2010) on the STRAW web-served (Shaw et al. 2013). Gene trees that were used in the MP-EST analysis were generated in RAxML as above. MP-EST requires the outgroup to be present in all gene trees, which was set as A. trichopoda, so only 99 gene families were used for the analyses. A total of 500 bootstrap replicates were used with default web-server parameters.
Identification of Glycine PWGD Homoeologs
Peptides from the G. max cv. Williams 82, assembly version 1, annotation version 1.1 were first reduced to the longest splice variant (Phytozome file Gmax_v1.1_189_peptide_primaryOnly.fa). These were compared with one another using BLAST + v2.2.27 (Camacho et al. 2009). Matches between protein sequences were filtered to e value ≤ 1e-10 and top reciprocal best hits per chromosome pair. Synteny blocks were then calculated from this input using DAGchainer (Haas et al. 2004), with four minimum aligned pairs (-A) per diagonal. Values of synonymous substitution per synonymous site (Ks) were then calculated for each gene pair in a synteny block, using a perl script (dag_ks.pl; Cannon SB, unpublished data) that aligns CDSs in protein sequence space, then calculates Ks using the Codeml method from the PAML package, version 4.4 (Yang 2007). Finally, using reports of median Ks per synteny block based on Ks frequency analyses, blocks with median Ks ≥ 0.35 and <1.5 were taken as having derived from the PWGD. The Glycine PWGD paralogs are in supplementary file S1, Supplementary Material online.
Tree-Based Hypothesis Testing
Gene trees were estimated and queried to test hypothesized timing of gene duplications relative to speciation events. Given the incomplete nature of RNA-Seq data, a number of sequence and gene tree filtering steps were implemented to avoid phylogenetic estimation artifacts attributable to long branches and missing data. The 3,360 orthogroups identified to contain paralog pairs were reduced to 2,764 by removing those that did not have designated outgroups (Amborella, Musa, or Asparagus). Remaining gene family trees were queried using the syntenically identified paralog pairs from Glycine. For each tree with a syntenic, duplicate gene pair, the LCA node was identified and the timing of the duplication event was inferred by assessing the species represented in the clade defined by the homoeologs’ LCA. Support for the placement of the homoeolog LCA (=PWGD) was assessed as the BSV for the clade defined by the LCA. We report the number of LCA nodes that had BSVs ≥80% and ≥50%.
The timing of gene duplication events attributed to the PWGD was assessed for each node in the species tree (fig. 2). For a node to be counted as supporting a specific placement of the PWGD event in the species tree, two criteria had to be met: 1) All taxa represented in the clade defined by the homoeolog LCA node in a gene tree could only be those taxa found above the corresponding node in the species tree and 2) the sister clade to the homoeolog LCA node in the gene tree had to contain taxa from the sister lineage in the species tree and could not have taxa found above the node. These criteria provided a bracketed estimate for the timing of divergence between parents contributing to the papilionoid allopolyploidy event that we refer to as the PWGD.
Additional Data Available at Public Repositories
Transcriptome assemblies, peptide translations, alignments, phylogenetic trees, and related intermediate analyses are available at both http://mirrors.iplantcollaborative.org/legume_wgd and http://datadryad.org or through http://dx.doi.org/10.5061/dryad.ff1tq
Supplementary Material
Supplementary files S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was funded in part by The 1000 Plants (1KP) initiative, led by G.K.S.W. The 1KP initiative is funded by the Alberta Ministry of Innovation and Advanced Education, Alberta Innovates Technology Futures (AITF) Innovates Centres of Research Excellence (iCORE), Musea Ventures, and BGI-Shenzhen. This work was also supported in part by the National Science Foundation grants DEB 0830009 and IOS 0922742 to J.L.-M., IOS 0822258 to J.J.D. and S.B.C., DEB 1257522 to J.J.D., and NIH 1R01DA025197-02 to T.K. The authors thank Pam and Doug Soltis and Nicholas Miles for help in coordinating species selection and for manuscript comments, Mark Chase for contribution of the Quillaja sequence, Haibo Liu, David Hufnagel, and Wei Huang for assistance in evaluating phylogenetic tree patterns, and Nathan Weeks for IT support. They also thank Naim Matasci and the iPlant Collaborative for hosting their data. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Author contributions are as follows: Analysis, writing, and conceptualization: S.B.C., M.R.M., A.H., J.L.-M., J.J.D.; computing support and analysis: S.D.; contribution of data sets: S.B.C., M.K.D., M.N.N., M.R., T.K., Y.P., B.J., C.N.S.; project support: G.K.S.W., E.C.; and sequencing and transcriptome assembly: Y.Z., X.T., C.C.
References
- Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- Ancestral Angiosperm Genome Project. 2013. [Internet]. Available: http://ancangio.uga.edu.
- Bell CD, Soltis DE, Soltis PS. The age and diversification of the angiosperms re-revisited. Am J Bot. 2010;97:1296–1303. doi: 10.3732/ajb.0900346. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. 2012. Plant DNA C-values database (release 6.0, December 2012). [cited 2013 Dec]. Available from: http://www.kew.org/cvalues.
- Bertioli DJ, Moretzsohn MC, Madsen LH, Sandal N, Leal-Bertioli SC, Guimaraes PM, Hougaard BK, Fredslund J, Schauser L, Nielsen AM, et al. An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genomics. 2009;10:45. doi: 10.1186/1471-2164-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau A, Mercure M, Lewis GP, Herendeen PS. Phylogenetic patterns and diversification in the caesalpinioid legumes. Botany. 2008;86:697–718. [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Ilut D, Farmer AD, Maki SL, May GD, Singer SR, Doyle JJ. Polyploidy did not predate the evolution of nodulation in all legumes. PLoS One. 2010;5:e11630. doi: 10.1371/journal.pone.0011630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Sterck L, Rombauts S, Sato S, Cheung F, Gouzy J, Wang X, Mudge J, Vasdewani J, Schiex T, et al. Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc Natl Acad Sci U S A. 2006;103:14959–14964. doi: 10.1073/pnas.0603228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso D, de Queiroz LP, Pennington RT, de Lima HC, Fonty E, Wojciechowski MF, Lavin M. Revisiting the phylogeny of papilionoid legumes: new insights from comprehensively sampled early-branching lineages. Am J Bot. 2012;99:1991–2013. doi: 10.3732/ajb.1200380. [DOI] [PubMed] [Google Scholar]
- Cui L, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, Soltis PS, Carlson JE, Arumuganathan K, Barakat A, et al. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006;16:738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt M, Roehr JT, Ahmed R, Dieterich C. FLEXBAR-Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms. Biology (Basel) 2012;1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. Phylogenetic perspectives on the origins of nodulation. Mol Plant Microbe Interact. 2011;24:1289–1295. doi: 10.1094/MPMI-05-11-0114. [DOI] [PubMed] [Google Scholar]
- Doyle JJ. Polyploidy and genome evolution. Heidelberg/New York/Dordrecht/London: Springer; 2012. Polyploidy in legumes; pp. 147–180. [Google Scholar]
- Doyle JJ, Luckow MA. The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 2003;131:900–910. doi: 10.1104/pp.102.018150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte JM, Wall PK, Edger PP, Landherr L, Ma H, Pires JC, Leebens-Mack J, dePamphilis CW. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol Biol. 2010;10:61. doi: 10.1186/1471-2148-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci U S A. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- Gherbi H, Markmann K, Svistoonoff S, Estevan J, Autran D, Giczey G, Auguy F, Peret B, Laplaze L, Franche C, et al. SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankiabacteria. Proc Natl Acad Sci U S A. 2008;105:4928–4932. doi: 10.1073/pnas.0710618105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldblatt P. Cytology and the phylogeny of leguminosae. In: Polhill RM, Raven PH, editors. 1981. Advances in legume systematics, Part 2. Royal Botanic Gardens, Kew. p. 427–464. [Google Scholar]
- 1000 Green Plant Transcriptome Project [Internet]. 2013. Available from: http://www.onekp.com.
- Haas BJ, Delcher AL, Wortman JR, Salzberg SL. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20:3643–3646. doi: 10.1093/bioinformatics/bth397. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Leebens-Mack J, Ayyampalayam S, Bowers JE, McKain MR, McNeal J, Rolf M, Ruzicka DR, Wafula E, Wickett NJ, et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012;13:R3. doi: 10.1186/gb-2012-13-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Jones G, Sagitov S, Oxelman B. Statistical inference of allopolyploid species networks in the presence of incomplete lineage sorting. Syst Biol. 2013;62:467–478. doi: 10.1093/sysbio/syt012. [DOI] [PubMed] [Google Scholar]
- Kistner C, Parniske M. Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci. 2002;7:511–518. doi: 10.1016/s1360-1385(02)02356-7. [DOI] [PubMed] [Google Scholar]
- Kroc M, Koczyk G, Swiecicki W, Kilian A, Nelson MN. New evidence of ancestral polyploidy in the Genistoid legume Lupinus angustifolius L. (narrow-leafed lupin) Theor Appl Genet. 2014;127:1237–1249. doi: 10.1007/s00122-014-2294-y. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Legume Phylogeny Working Group. Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades. Taxon. 2013;62:217–248. [Google Scholar]
- Lewis G, Schrire B, Mackind B, Lock M. 2005 Legumes of the World. Royal Botanic Gardens, Kew. [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QG, Zhang L, Li C, Dunwell JM, Zhang YM. Comparative genomics suggests that an ancestral polyploidy event leads to enhanced root nodule symbiosis in the Papilionoideae. Mol Biol Evol. 2013;30:2602–2611. doi: 10.1093/molbev/mst152. [DOI] [PubMed] [Google Scholar]
- Liu L, Yu L, Edwards SV. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evol Biol. 2010;10:302. doi: 10.1186/1471-2148-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun. 2010;1:10. doi: 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanilla V, Bruneau A. Phylogeny reconstruction in the Caesalpinieae grade (Leguminosae) based on duplicated copies of the sucrose synthase gene and plastid markers. Mol Phylogenet Evol. 2012;65:149–162. doi: 10.1016/j.ympev.2012.05.035. [DOI] [PubMed] [Google Scholar]
- Moretzsohn MC, Gouvea EG, Inglis PW, Leal-Bertioli SC, Valls JF, Bertioli DJ. A study of the relationships of cultivated peanut (Arachis hypogaea) and its most closely related wild species using intron sequences and microsatellite markers. Ann Bot. 2013;111:113–126. doi: 10.1093/aob/mcs237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet. 2011;45:119–144. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- Op den Camp R, Streng A, De Mita S, Cao Q, Polone E, Liu W, Ammiraju JSS, Kudrna D, Untergasser A, Bisseling T, et al. LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume parasponia. Science. 2011;331:909–912. doi: 10.1126/science.1198181. [DOI] [PubMed] [Google Scholar]
- Pfeil BE, Schlueter JA, Shoemaker RC, Doyle JJ. Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst Biol. 2005;54:441–454. doi: 10.1080/10635150590945359. [DOI] [PubMed] [Google Scholar]
- Raven PH. The bases of angiosperm phylogeny: cytology. Ann Mo Bot Gard. 1975;62:724–764. [Google Scholar]
- Salse J. In silico archeogenomics unveils modern plant genome organisation, regulation and evolution. Curr Opin Plant Biol. 2012;15:122–130. doi: 10.1016/j.pbi.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC. Mining EST databases to resolve evolutionary events in major crop species. Genome. 2004;47:868–876. doi: 10.1139/g04-047. [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the paleopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Mohammadin S, Edger PP. Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Curr Opin Plant Biol. 2012;15:147–153. doi: 10.1016/j.pbi.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Shaw T, Ruan Z, Glenn T, Liu L. STRAW: a web server for species tree analysis. Nucleic Acids Res. 2013;41:W238–W241. doi: 10.1093/nar/gkt377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Sankoff D, Depamphilis CW, Wall PK, Soltis PS. Polyploidy and angiosperm diversification. Am J Bot. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Morgan DR, Swensen SM, Mullin BC, Dowd JM, Martin PG. Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc Natl Acad Sci U S A. 1995;92:2647–2651. doi: 10.1073/pnas.92.7.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent J. Legume nodulation: a global perspective. Oxford: Wiley-Blackwell; 2009. [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y. A mystery unveiled. Genome Biol. 2011;12:113. doi: 10.1186/gb-2011-12-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K, Baele G, Maere S, Van de Peer Y. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 2014;24(8):1334–1347. doi: 10.1101/gr.168997.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Moore MJ, Soltis PS, Bell CD, Brockington SF, Alexandre R, Davis CC, Latvis M, Manchester SR, Soltis DE. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc Natl Acad Sci U S A. 2009;106:3853–3858. doi: 10.1073/pnas.0813376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner GD, Cornwell WK, Sprent JI, Kattge J, Kiers ET. A single evolutionary innovation drives the deep evolution of symbiotic N2-fixation in angiosperms. Nat Commun. 2014;5:4087. doi: 10.1038/ncomms5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski MF, Lavin M, Sanderson MJ. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot. 2004;91:1846–1862. doi: 10.3732/ajb.91.11.1846. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Young ND, Debelle F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanne AE, Tank DC, Cornwell WK, Eastman JM, Smith SA, FitzJohn RG, McGlinn DJ, O’Meara BC, Moles AT, Reich PB, et al. Three keys to the radiation of angiosperms into freezing environments. Nature. 2014;506:89–92. doi: 10.1038/nature12872. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liang S, Duan J, Wang J, Chen S, Cheng Z, Zhang Q, Liang X, Li Y. De novo assembly and characterisation of the transcriptome during seed development, and generation of genic-SSR markers in peanut (Arachis hypogaea L.) BMC Genomics. 2012;13:90. doi: 10.1186/1471-2164-13-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.