Abstract
Lentiviruses infect a wide range of mammal species. Much remains unknown about their deep history and host distribution. Here, we report the discovery of an endogenous lentivirus within the genome of the Sunda flying lemur (Galeopterus variegatus) (which we designate “Galeopterus variegatus endogenous lentivirus” [GvaELV]). We estimate the GvaELV genome invasion to have occurred more than 14 Ma, supporting an ancient origin of the lentivirus clade and an ancient lentiviral infection in colugo. Phylogenetic analyses show that GvaELV is a sister group of all previously known lentiviruses. The GvaELV genome appears to possess some primitive genomic features of a lentivirus, encoding not only a trans-activator of transcription (tat) gene but also two additional putative accessory genes that share no discernible similarity with other lentiviral accessory genes. The discovery of GvaELV provides novel insights into the prehistory and host distribution of lentivirus.
Keywords: Dermoptera, lentivirus, endogenous retrovirus, phylogenetics
Retroviruses employ a unique replication strategy: Their RNA genomes are reverse transcribed into DNA and the synthesized DNA is integrated into the host genome. Occasionally this process occurs in germ line cells. The integrated retroviruses, known as endogenous retroviruses (ERVs), become vertically inherited genomic loci and can become fixed in the host population. ERVs provide “molecular fossils” of past retroviral infections and are thus useful for the study of the prehistory of retroviruses (Katzourakis et al. 2007; Stoye 2012). Although ERVs are extremely abundant in vertebrate genomes, lentivirus endogenization seems to be extremely rare (Katzourakis et al. 2007; Gilbert et al. 2009). To date, only three endogenous lentiviruses have been reported: Rabbit endogenous lentivirus K (RELIK) in rabbits and hares (Katzourakis et al. 2007; Keckesova et al. 2009), prosimian immunodeficiency virus (PSIV) in Malagasy lemurs (Gifford et al. 2008; Gilbert et al. 2009), and Mustelidae endogenous lentivirus (MELV) in the weasel family (Cui and Holmes 2012; Han and Worobey 2012). However, knowledge of the host distribution and deep history of lentiviruses remain far from complete. The accumulating mammalian genomic sequences available provide great opportunities to discover novel endogenous lentiviruses.
We screened all currently available mammalian genome sequences for novel endogenous lentiviruses. The genome screening process identified sequences highly similar to lentiviral proteins within the genome sequence of the Sunda “flying lemur” (Galeopterus variegatus), a colugo (and not a true lemur) endemic throughout Southeast Asia. We designate this endogenous lentivirus “G. variegatus endogenous lentivirus” (GvaELV). A total of three full-length insertions, 25 partial insertions, and 155 solo-long-terminal repeats (LTRs) were identified (supplementary tables S1 and S2, Supplementary Material online). Phylogenetic analysis of retroviral Pol protein sequences shows that GvaELV and other lentiviruses form a monophyletic group with strong statistical support (Bayesian posterior probability = 1.0), indicating that GvaELV indeed appears to be a lentivirus, albeit one that forms a sister group to previously known variants (supplementary fig. S1, Supplementary Material online).
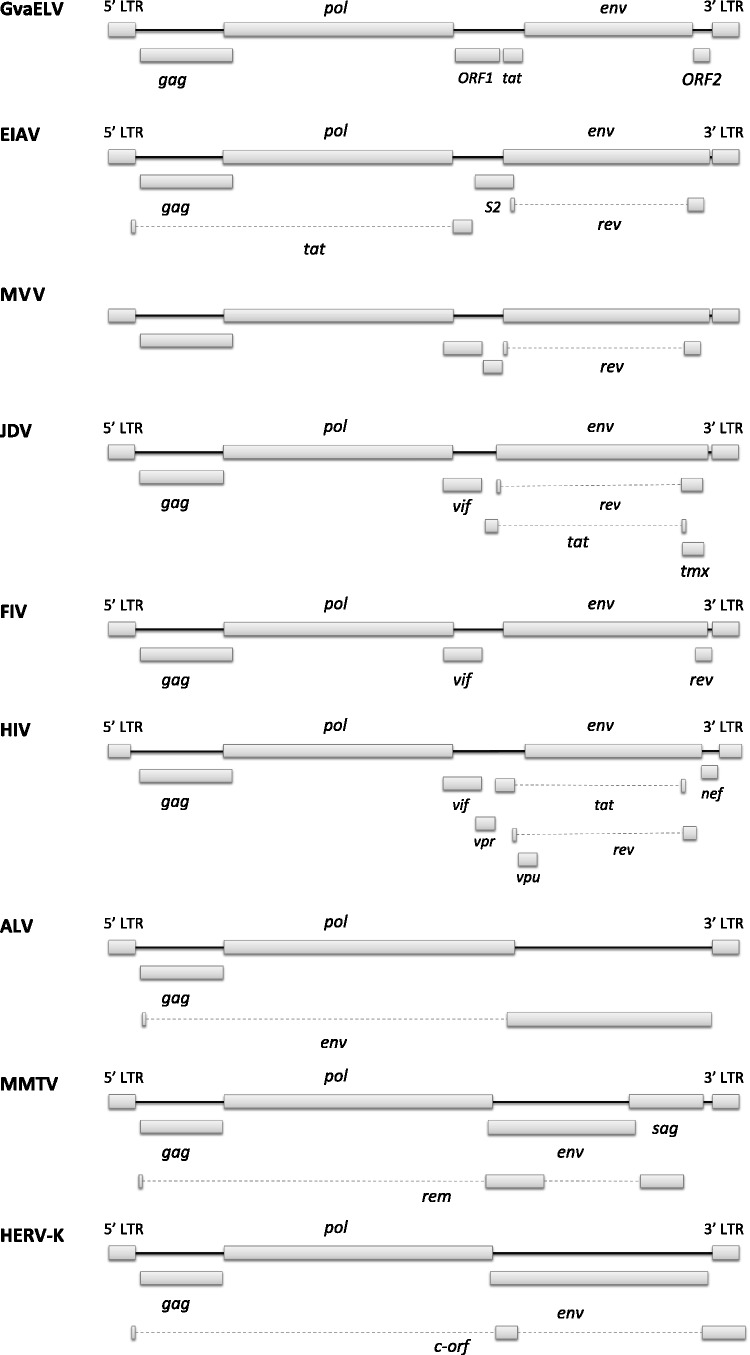
The reconstructed GvaELV genome contains the three major genes that are shared by all the retroviruses, that is, gag, pol, and env genes (fig. 1 and fig. S2, Supplementary Material online). The GvaELV genome also encodes at least three putative accessory genes, trans-activator of transcription (tat), ORF1, and ORF2. The proteins encoded by ORF1 and ORF2 do not share any significant similarity with other known retroviral accessory proteins.
Fig. 1.
Genome structures of representative lentiviruses and other retroviruses. ALV is an alpharetrovirus, MMTV and HERV-K are betaretroviruses, and the others are all lentiviruses. Virus names: ALV, avian leukemia virus; BIV, bovine immunodeficiency virus; HERV-K, human ERV K; HIV, human immunodeficiency virus; JDV, Jembrana disease virus; MMTV, mouse mammary tumor virus; MVV, maedi-visna virus; SRV-1, Simian retrovirus 1.
The 5′-LTR and 3′-LTR of a nascent ERV are identical at endogenization but then accumulate mutations independently through time. Thus, the sequence divergence between the 5′-LTR and 3′-LTR sequences of an ERV is roughly proportional to the ERV’s genome integration time and can be used to estimate the integration time (Johnson and Coffin 1999; Stoye 2012). In this study, we identified three full-length GvaELV insertions within contigs G_variegatus-3.0.2-377.4, G_variegatus-3.0.2-2016.3, and G_variegatus-3.0.2-5697.1 (table 1 and supplementary table S1, Supplementary Material online). Because an estimate of the neutral evolutionary rate is not available for G. variegatus, we used human and mouse lineage neutral substitution rates as conservative upper and lower rate bounds (Waterston et al. 2002; Katzourakis et al. 2007; Gifford et al. 2008). The oldest invasion event was estimated to occur at least 14.3 Ma (table 1). However, these estimates should be taken with caution for three reasons: 1) G. variegatus might have a higher or lower rate of neutral evolution than the rate bounds we used here, 2) we do not know for certain whether LTRs evolve neutrally after endogenization, and 3) LTRs might undergo gene conversion, which could lead to underestimation of the integration time (Johnson and Coffin 1999). The variation in integration date estimates across the three pairs of 5′-LTR and 3′-LTR (table 1) might be caused by gene conversion in LTRs. Otherwise it might reflect separate lentiviral endogenization events taking place of several million years, which indicates continual circulation of exogenous lentiviruses in these animals over several millions of years.
Table 1.
Estimates of the GvaELV Genome Invasion Time.
| Contig | Genetic Distance of 5′- and 3′-LTRs (subs/site) | Invasion Time (Ma) |
|
|---|---|---|---|
| Mouse Neutral Rate (4.5 × 10−9 subs/site/year) | Human Neutral Rate (2.2 × 10−9 subs/site/year) | ||
| G_variegatus-3.0.2-377.4 | 0.1289 | 14.3 | 29.3 |
| G_variegatus-3.0.2-5697.1 | 0.0955 | 10.6 | 21.7 |
| G_variegatus-3.0.2-2016.3 | 0.0404 | 4.5 | 9.2 |
Lentiviruses were once thought to have a relatively recent origin (Gifford 2012). Molecular clock analyses of contemporary viral sequences suggest a timeframe of several hundred or thousand years for some lentivirus groups, such as simian immunodeficiency virus (SIV) and feline immunodeficiency virus (FIV) (Biek et al. 2006; Wertheim and Worobey 2009; Worobey et al. 2011). The previously described endogenous lentiviruses (RELIK, PSIV, and MELV) establish that the corresponding lentiviral groups are at least several million years old. Our analyses here show that lentiviruses had been infecting colugo at least 14 Ma (possibly as far as 30 Ma), confirming that lentivirus is ancient in evolutionary origin (Katzourakis et al. 2007; Gilbert et al. 2009; Han and Worobey 2012).
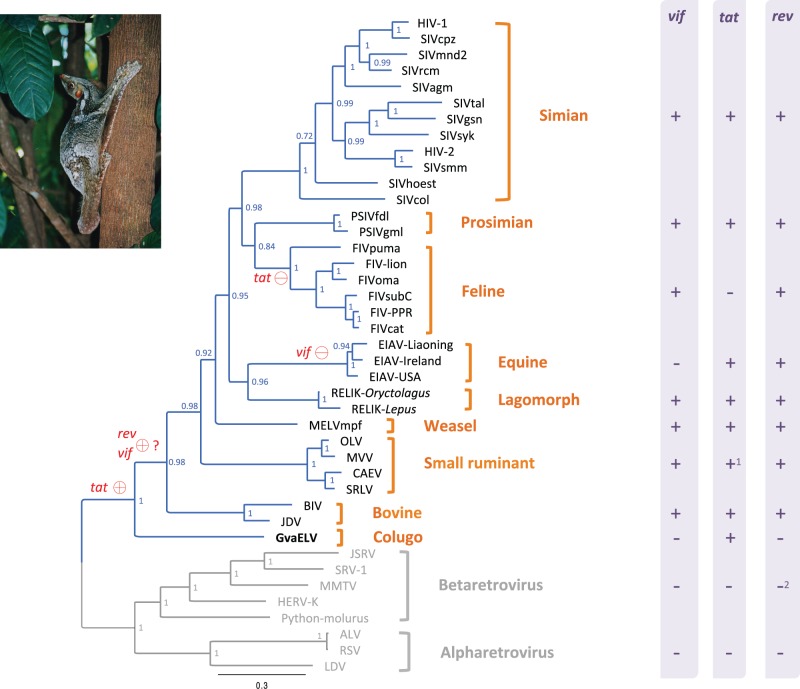
Lentivirus was thought to infect eight groups of mammals, that is, simian, prosimian, feline, bovine, equine, small ruminant, and lagomorph species (Gifford 2012; Han and Worobey 2012). The discovery of GvaELV now provides evidence for ancient lentiviral infection in a colugo. GvaELV might constitute a new lentiviral group. The discovery of GvaELV also suggests that the host distribution of lentivirus might be much wider than previously thought (Han and Worobey 2012). Colugos are the closest living relative of primates (Janecka et al. 2007; Perelman et al 2011). However, our phylogenetic analysis shows that GvaELV does not group with lentiviruses of either simian or prosimian origin (fig. 2). This phylogenetic pattern might be readily explained by frequent cross species transmission throughout lentivirus evolutionary history, as lentiviruses can frequently switch hosts (Franklin et al. 2007; Wertheim and Worobey 2007; Minardi da Cruz et al. 2013).
Fig. 2.
Phylogeny and genomic structure diversity among exogenous and endogenous lentiviruses. The phylogeny was reconstructed based on the concatenated sequences of Gag and Pol proteins. The phylogeny is a 50% majority-rule consensus tree. Node labels are posterior probabilities. The lentivirus lineage is highlighted in blue. The presence and absence of accessory genes (vif, tat, and rev) in lentiviruses, alpharetroviruses, and betaretroviruses are indicated by + and − in the right column, respectively. Red circles on branches indicate the most parsimonious scenario of birth (+) and death (−) of related accessory genes. Note 1: Small ruminant lentivirus (SLRV) encodes a tat ORF that shares discernible similarity with tat gene of other lentiviruses, although the SLRV Tat protein is not functionally similar to that of other lentiviruses (Villet et al. 2003). Note 2: Although some betaretroviruses, such as mouse mammary tumor virus and human ERV K, encode functional homologs of rev genes (Yang et al. 1999; Mertz et al. 2005; Gifford 2012), these betaretrovirus proteins do not share discernible sequence similarity with lentiviral Rev protein (Yang et al. 1999). Virus names: ALV, avian leukemia virus; BIV, bovine immunodeficiency virus; CAEV, caprine arthritis–encephalitis virus; HERV-K, human ERV K; HIV, human immunodeficiency virus; JDV, Jembrana disease virus; JSRV, jaagsiekte sheep retrovirus; LDV, lymphoproliferative disease virus; MMTV, mouse mammary tumor virus; MVV, maedi-visna virus; OLV, olive lentivirus; Python-molurus, Python molurus ERV; RSV, Rous sarcoma virus; SRLV, small ruminant lentivirus; SRV-1, Simian retrovirus 1. Colugo photo courtesy of Nina Holopainen.
Our phylogenetic analysis shows that GvaELV is a sister group of all previously known lentiviruses (fig. 2). Given its unique phylogenetic position, the GvaELV genome might possess some primitive genomic features of lentivirus. Lentiviruses are complex retroviruses, typically encoding many accessory genes with a variety of functions. Most of these accessory genes are found in only one specific lentiviral group (e.g., negative factor (nef) and viral protein R (vpr) genes are simian lentiviral group specific) (Gifford 2012). However, three accessory genes are widely distributed: regulator of virion expression (rev) gene is encoded by all lentiviruses, viral infectivity factor (vif) gene is encoded by all except equine infectious anemia virus (EIAV), and tat gene is encoded by all except FIV (Gifford 2012; Han and Worobey 2012; fig. 2). We did not find any significant Rev and Vif protein homologs in the GvaELV genome (fig. 1). Based on the phylogenetic analysis and retroviral genome structure information, a parsimonious scenario for the gain and loss of these three accessory genes could be conceived: The most recent common ancestor of the nine lentiviral groups acquired the tat gene; lentiviruses might acquire rev and vif genes after diverging from the colugo lentivirus; the tat and vif genes were later lost in FIV and EIAV, respectively (fig. 2). However, the hypothesized scenario of the acquisition of the rev and vif genes should be taken with caution for two reasons: 1) The rev and vif genes might be specifically lost in GvaELV and 2) the similarity between related proteins of GvaELV and other lentiviruses (figs. S3 and S4, Supplementary Material online) might be eroded due to rapid evolution and/or a long timeframe.
GvaELV encodes two additional putative accessory genes (ORF1 and ORF2) with unknown function. Functional characterization of these accessory genes might provide further insights into the early evolution of lentivirus genome structure and would present a fascinating window on the functional biology of a very old retrovirus. Lentiviruses and host restriction factors (such as APOBEC3G, SAMHD1, and TRIM5α) are locked in evolutionary “arms races,” some of which have been ongoing for million years (Duggal and Emerman 2012; Compton et al. 2013). As GvaELV might have been infecting colugo more than 14 Ma, evolutionary and functional analyses of colugo restriction factor and GvaELV interactions would have important implications in understanding the deep history of lentivirus-host interface.
Materials and Methods
Screening and Genome Reconstruction
The tBLASTn algorithm with representative lentiviral protein sequences (supplementary table S3, Supplementary Material online) as queries was employed to screen all available Whole Genome Shotgun sequences from GenBank for endogenous lentivirus. Sequences highly similar to lentiviral proteins were aligned using MUSCLE (Edgar 2004). We reconstructed the GvaELV genome based on the alignment. Ancestral states of the GvaELV genome sequence were reconstructed using maximum-likelihood method available in MEGA5 (Tamura et al. 2011). The solo-LTRs were identified using the BLASTn algorithm with reconstructed GvaELV LTR as a query and E value of 10−10 as cut-off value.
Phylogenetic Analysis
To assess the relationship between GvaELV and other retroviruses, representative retroviral Pol protein sequences (supplementary table S3, Supplementary Material online) were aligned using MUSCLE (Edgar 2004). To assess the relationship among exogenous and endogenous lentiviruses, Gag and Pol protein sequences were concatenated and aligned using MUSCLE (Edgar 2004). The ambiguous regions in the alignments were removed using Gblocks 0.91b and then manually edited (Talavera and Castresana 2007). Phylogenetic analyses were performed using MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003) with 500,000–1,000,000 generations in four chains sampling posterior trees every 100 generations. The first 25% of the posterior trees were discarded.
Dating Analysis
Three full-length GvaELV insertions were identified in this study. For each of them, the 5′-LTR and 3′-LTR sequences were aligned using MUSCLE (Edgar 2004). The genetic distance between 5′-LTR and 3′-LTR was calculated with Kimura two-parameter substitution model (Kimura 1980).
Supplementary Material
Supplementary figures S1–S4 and tables S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Richard K. Wilson and the Genome Institute, Washington University School of Medicine for releasing the Galeopterus variegatus 3.0.2 sequence assembly and one anonymous reviewer for extremely constructive suggestions. This work was supported by National Institute of Allergy and Infectious Diseases (NIAID) grant R01 AI084691 and the David and Lucile Packard Foundation.
References
- Biek R, Drummond AJ, Poss M. A virus reveals population structure and recent demographic history of its carnivore host. Science. 2006;311:538–541. doi: 10.1126/science.1121360. [DOI] [PubMed] [Google Scholar]
- Cui J, Holmes EC. Endogenous lentiviruses in the ferret genome. J Virol. 2012;86:3383–3385. doi: 10.1128/JVI.06652-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton AA, Malik HS, Emerman M. Host gene evolution traces the evolutionary history of ancient primate lentiviruses. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120496. doi: 10.1098/rstb.2012.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal NK, Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol. 2012;12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin SP, Troyer JL, Terwee JA, Lyren LM, Boyce WM, Riley SP, Roelke ME, Crooks KR, Vandewoude S. Frequent transmission of immunodeficiency viruses among bobcats and pumas. J Virol. 2007;81:10961–10969. doi: 10.1128/JVI.00997-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford RJ. Viral evolution in deep time: lentiviruses and mammals. Trends Genet. 2012;28:89–100. doi: 10.1016/j.tig.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Gifford RJ, Katzourakis A, Tristem M, Pybus OG, Winters M, Shafer RW. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc Natl Acad Sci U S A. 2008;105:20362–20367. doi: 10.1073/pnas.0807873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Maxfield DG, Goodman SM, Feschotte C. Parallel germline infiltration of a lentivirus in two Malagasy lemurs. PLoS Genet. 2009;5:e1000425. doi: 10.1371/journal.pgen.1000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GZ, Worobey M. Endogenous lentiviral elements in the weasel family (Mustelidae) Mol Biol Evol. 2012;29:2905–2908. doi: 10.1093/molbev/mss126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janecka JE, Miller W, Pringle TH, Wiens F, Zitzmann A, Helgen KM, Springer MS, Murphy WJ. Molecular and genomic data identify the closest living relative of primates. Science. 2007;318:792–794. doi: 10.1126/science.1147555. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Coffin JM. Constructing primate phylogenies from ancient retrovirus sequences. Proc Natl Acad Sci U S A. 1999;96:10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. Proc Natl Acad Sci U S A. 2007;104:6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z, Ylinen LM, Towers GJ, Gifford RJ, Katzourakis A. Identification of a RELIK orthologue in the European hare (Lepus europaeus) reveals a minimum age of 12 million years for the lagomorph lentiviruses. Virology. 2009;384:7–11. doi: 10.1016/j.virol.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Minardi da Cruz JC, Singh DK, Lamara A, Chebloune Y. Small ruminant lentiviruses (SRLVs) break the species barrier to acquire new host range. Viruses. 2013;5:1867–1884. doi: 10.3390/v5071867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JA, Simper MS, Lozano MM, Payne SM, Dudley JP. Mouse mammary tumor virus encodes a self-regulatory RNA export protein and is a complex retrovirus. J Virol. 2005;79:14737–14747. doi: 10.1128/JVI.79.23.14737-14747.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol. 2012;10:395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villet S, Bouzar BA, Morin T, Verdier G, Legras C, Chebloune Y. Maedi-visna virus and caprine arthritis encephalitis virus genomes encode a Vpr-like but no Tat protein. J Virol. 2003;77:9632–9638. doi: 10.1128/JVI.77.17.9632-9638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Worobey M. A challenge to the ancient origin of SIVagm based on African green monkey mitochondrial genomes. PLoS Pathog. 2007;3:e95. doi: 10.1371/journal.ppat.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Worobey M. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput Biol. 2009;5:e1000377. doi: 10.1371/journal.pcbi.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M, Telfer P, Souquière S, Hunter M, Coleman CA, Metzger MJ, Reed P, Makuwa M, Hearn G, Honarvar S, et al. Island biogeography reveals the deep history of SIV. Science. 2011;329:1487. doi: 10.1126/science.1193550. [DOI] [PubMed] [Google Scholar]
- Yang J, Bogerd HP, Peng S, Wiegand H, Truant R, Cullen BR. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc Natl Acad Sci U S A. 1999;96:13404–13408. doi: 10.1073/pnas.96.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.