Abstract
Paracoccidioides species are dimorphic fungi and are the etiologic agents of paracoccidioidomycosis, which is a serious disease that involves multiple organs. The many tissues colonized by this fungus suggest a variety of surface molecules involved in adhesion. A surprising finding is that most enzymes in the glycolytic pathway, tricarboxylic acid (TCA) cycle and glyoxylate cycle in Paracoccidioides spp. have adhesive properties that aid in interacting with the host extracellular matrix and thus act as ‘moonlighting’ proteins. Moonlighting proteins have multiple functions, which adds a dimension to cellular complexity and benefit cells in several ways. This phenomenon occurs in both eukaryotes and prokaryotes. For example, moonlighting proteins from the glycolytic pathway or TCA cycle can play a role in bacterial pathogenesis by either acting as proteins secreted in a conventional pathway and/or as cell surface components that facilitate adhesion or adherence. This review outlines the multifunctionality exhibited by many Paracoccidioides spp. enzymes, including aconitase, aldolase, glyceraldehyde-3-phosphate dehydrogenase, isocitrate lyase, malate synthase, triose phosphate isomerase, fumarase, and enolase. We discuss the roles that moonlighting activities play in the virulence characteristics of this fungus and several other human pathogens during their interactions with the host.
Keywords: Paracoccidioides spp., moonlighting proteins, virulence, glycolytic pathway and tricarboxylic acid cycle, glyoxylate cycle, adhesins
INTRODUCTION
A great challenge in studying proteins is understanding how encoded proteins function and interact with each other to coordinate essential cellular processes. Although many protein roles can be inferred by homology-based function predictions, this approach may be complicated for multifunctional proteins. The notion that one gene encodes one protein and results in only one function is outdated because proteins may have multiple functions (including on a single polypeptide chain), and the function may change based on external signals (Kirschner and Bisswanger, 1976; Jeffery, 1999; Wolff and Arnau, 2002; Jeffery, 2009; Pandini et al., 2012; Wienkers and Rock, 2014).
Multiple binding sites or changes in unusable regions of a protein structure may produce a new function because many proteins seem larger than necessary to perform only one function at a single binding site. These multifunctional proteins may benefit an organism because synthesizing fewer proteins may save cellular energy for additional functions, such as growth and reproduction (Jeffery, 1999).
Moonlighting proteins are exceptional multifunctional proteins; these multifunctional proteins can perform several additional functions that are often unrelated. These functions are typically independent, which means that if one function is inactivated, due a mutation, for example, the second function is unaffected (Huberts and van der Klei, 2010). The function of a moonlighting protein can vary based on changes in cellular location or expression, cell type, association between two or more polypeptide chains and the cellular levels of a ligand, substrate, cofactor, product, or different binding sites (Jeffery, 2003a); moonlighting cannot be attributed to hybrid genes, which are single genes that code for multiple proteins or polypeptides that express different functions after protease cleavage (Kainulainen and Korhonen, 2014). Moonlighting functions have been demonstrated by multiple independent studies with unexpected phenotypes, locations, and binding partners (Copley, 2012).
The steady increase in new proteins characterized as multifunctional supports the potential importance of in-depth studies on the mechanism underlying these moonlighting functions in the same cell. (Chung et al., 1999; Jeffery, 1999, 2003a). Moonlighting may be due to joint engineering of communication and cooperation for various functions and paths in a complex cell or different cell types in an organism (Jeffery, 2003b).
Multifunctional proteins are present in prokaryotes and eukaryotes, such as mammals, which compound the protein arsenal of these organisms (Clarke et al., 2001; James and Viola, 2002; Brilli and Fani, 2004; Orita et al., 2005). The moonlighting activities of one protein are typically in addition to their role in chemical metabolic reactions, which demonstrates that these proteins are highly variable; metabolic enzymes can perform double duty as transcription factors, participate in assembly or autophagy, or maintain the levels of oxidative phosphorylation in the cells through maintaining mitochondrial DNA, among other functions (Chen et al., 2005; Gancedo and Flores, 2008). Intriguingly, in many cases, these proteins are constitutively expressed at low levels and act as enzymes, but when they are expressed at high levels, they perform moonlighting functions (Baker, 1991; Gómez et al., 2011).
Although highly conserved proteins perform many moonlighting functions, moonlighting functions cannot be predicted based on sequence and structural comparisons. Researchers speculate that evolution produced proteins with almost identical structures but different functions because moonlighting may provide a means to expand the functional capabilities of an organism without a genome-wide expansion (Kelkar and Ochman, 2013). Researchers have proposed that a protein must have some inherent compatibility for a new function to develop a moonlighting function (Aharoni et al., 2005).
To identify the moonlighting site or sites, we must first study how the moonlighting protein evolved and how the moonlighting function is related to the original “active site” (Henderson and Martin, 2011). Certain moonlighting proteins are recruited to the cell surface and involved with pathogenic processes (Pancholi and Chhatwal, 2003; Wang et al., 2013). This process not only occurs in bacteria but also in fungi, including the Paracoccidioides genus. Moonlighting proteins may relocate to the bacterial surface and present adhesion activities specific for host cell targets. These adhesive activities in moonlighting proteins have been widely studied and seem to play important roles in bacterial adhesion and colonization (Wang et al., 2013). Most abundant moonlighting adhesins are proteins that interact with the adhesion complex through binding fibronectin, which is a protein present at high concentrations in the fluids between cells and in the extracellular matrix (ECM), and it links cells to the ECM through specific transmembrane receptors (Henderson and Martin, 2011).
Examples of secondary functions associated with catalysis have been reported in many organisms, including plants (Moore, 2004), animals (Sriram et al., 2005), yeast (Gancedo and Flores, 2008), and prokaryotes (Jeffery, 1999, 2004). In bacteria, the glycolytic enzymes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and enolase (ENO) as well as the chaperonin 60, Hsp70, and peptidyl-prolyl isomerase most commonly exhibit moonlighting functions during bacterial virulence, such as adhesion and modulation of host cell signaling processes (Henderson and Martin, 2011). Most examples have been demonstrated in yeast (Gancedo and Flores, 2008) likely because it is the best understood and an extensively studied model organism. The known moonlighting functions are extremely diverse and involved in several biological functions. Examples of well-characterized moonlighting proteins in Paracoccidioides spp. and other fungal species are shown in Table 1.
Table 1.
Moonlighting proteins characterized in different fungal species.
| Proteins | Metabolic pathway or function | Moonlighting function | Fungal species | Reference |
|---|---|---|---|---|
| Enolase | Glycolysis | Thermal tolerance and growth control | Saccharomyces cerevisiae | Sekita et al. (1985) |
| Invasion process; cell wall construction | Candida albicans | Walsh et al. (1991), van Deventer et al. (1994), Angiolella et al. (1996) | ||
| Invasion process (plasminogen binding) | C. albicans; Aspergillus fumigatus; Paracoccidioides spp. | Eroles et al. (1997), Fox and Smulian (2001), Jong et al. (2003), Nogueira et al. (2010), Marcos et al. (2012) | ||
| Adhesion process (fibronectin binding) | Paracoccidioides spp. | Donofrio et al. (2009), Marcos et al. (2012) | ||
| Malate synthase | Glyoxylate cycle | Adhesion process (type I and IV collagen and fibronectin binding) | Paracoccidioides spp. | da Silva Neto et al. (2009) |
| Aconitase | TCA cycle | Mitochondrial DNA maintenance | S. cerevisiae | Albring et al. (1977), Rouault et al. (1991), Lu et al. (2001), Chen et al. (2005), Myers (2009) |
| Iron regulatory protein | Paracoccidioides spp., S. cerevisiae | Narahari et al. (2000), da Fonseca et al. (2001), Barbosa et al. (2004) | ||
| Aldolase | Glycolysis | Invasion process (plasminogen binding) | C. albicans, Paracoccidioides spp. | Burucoa et al. (1995), McCarthy et al. (2002), Pitarch et al. (2002), Crowe et al. (2003), Chaves (2013) |
| GAPDH | Glycolysis | Adhesion and invasion processes (fibronectin, laminin and plasminogen-binding) | C. albicans | Pancholi and Fischetti (1992), Gil-Navarro et al. (1997), Delgado et al. (2003), Starnes et al. (2009) |
| Adhesion process (laminin, fibronectin and type I collagen binding) | Paracoccidioides spp. | Barbosa et al. (2006) | ||
| Isocitrate lyase | Glyoxylate cycle | Growth | Aspergillus fumigatus | Gainey et al. (1992), Valenciano et al. (1996), Ebel et al. (2006) |
| Adhesion process (fibronectin and type IV collagen binding) | Paracoccidioides spp. | Cruz et al. (2011) | ||
| Triose phosphate isomerase | Glycolysis | Adhesion process (laminin and fibronectin binding) | Paracoccidioides spp. | da Fonseca et al. (2001), Pereira et al. (2004, 2007) |
Many currently known moonlighting proteins are highly conserved enzymes present in many different organisms (Fothergill-Gilmore and Michels, 1993; Jeffery, 1999). Among these proteins, we highlight the enzymes involved in sugar metabolism (Hendriks et al., 1988; Wistow et al., 1988; Yuan et al., 1997; Chen et al., 2005; Decker and Wickner, 2006; Lu et al., 2007). Researchers have also suggested that most enzymes in the glycolytic pathway and tricarboxylic acid (TCA) cycle have moonlighting functions (Kim and Dang, 2005; Sriram et al., 2005). Moreover, Commichau et al. (2009) showed interactions between glycolytic enzymes and proteins involved in RNA degradation, which suggests the presence of additional moonlighting functions for these proteins.
In this review, we discuss multiple attributes of moonlighting proteins in Paracoccidioides spp. and indicate the studies that justify their inclusion in this category. Another goal of this review is to highlight the importance of this phenomenon and its wide implications for both basic and applied research.
MOONLIGHTING Paracoccidioides SPECIES COMPLEX PROTEINS
Studies have characterized the ECM components involved in interactions between Paracoccidioides spp. and the host. This genus is composed of two species, Paracoccidioides lutzii and P. brasiliensis; the latter is sub-classified into three different phylogenetic groups. The large number of tissues that this fungus can colonize and infect suggests that it includes many cell adhesins. Certain molecules from Paracoccidioides spp. were identified as ligands for ECM components (Mendes-Giannini et al., 2008). Gp43 was the first molecule identified as a laminin-binding protein (Vicentini et al., 1994; Hanna et al., 2000). Additional binding affinity assays have shown that gp43 can bind both fibronectin and laminin. In Paracoccidioides spp, certain additional adhesins have also been described and may play an important role in pathogenesis (Andreotti et al., 2005; González et al., 2005; Barbosa et al., 2006; Mendes-Giannini et al., 2006; Pereira et al., 2007; Donofrio et al., 2009).
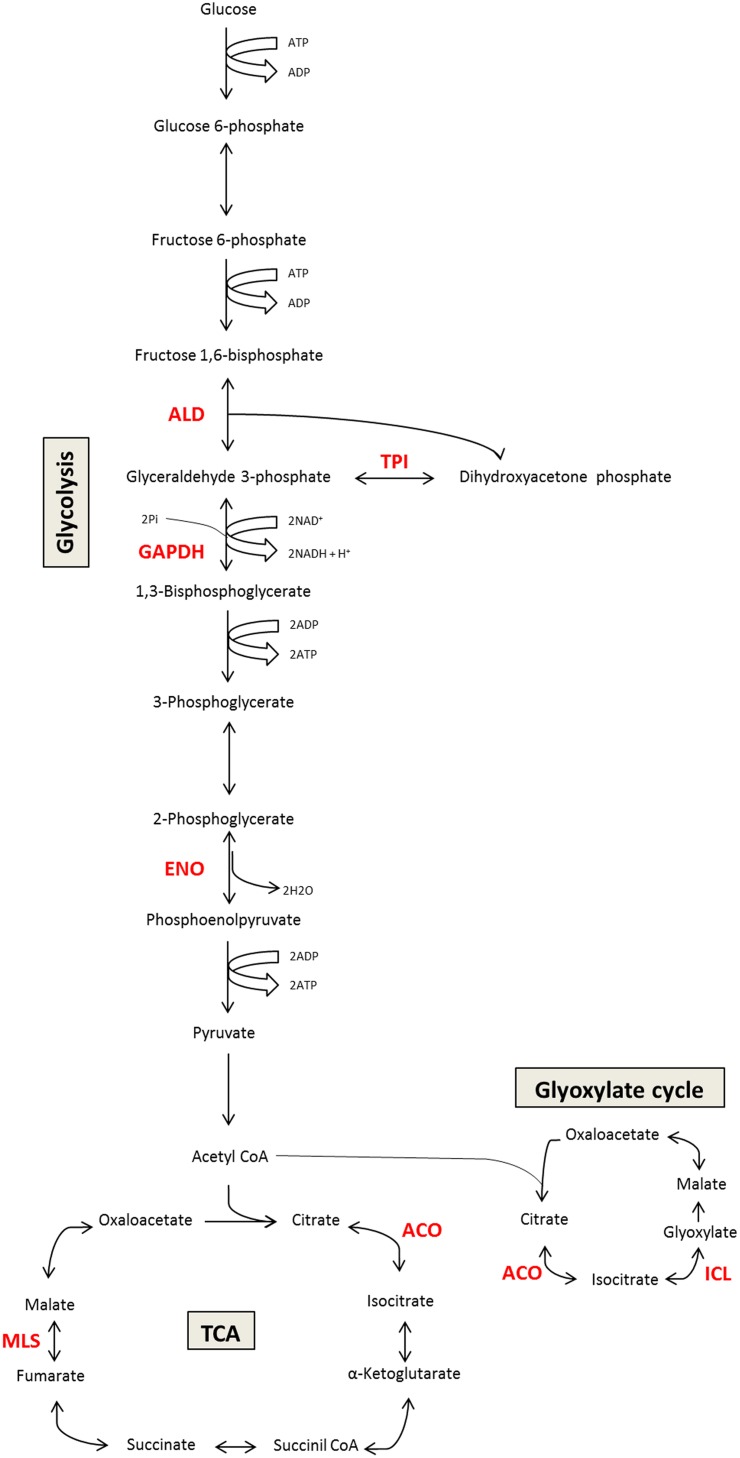
Several studies in Paracoccidioides spp. show that many metabolic enzymes may play a role in virulence. The most commonly identified moonlighting functions of Paracoccidioides spp. include functions related to adhesion and ECM-binding activity. Enzymes in the glycolytic pathway and TCA cycle act as moonlighting proteins in Paracoccidioides spp, including GAPDH, ENO, and fructose-1-6-bisphosphate aldolase (FBA); each displays different affinities for binding ECM components. Additionally, malate synthase (MLS) and isocitrate lyase (ICL) from the glyoxylate pathway as well as aconitase (ACO) from the TCA cycle may have multifunctional roles, including during the interaction between the fungus and host (da Silva Neto et al., 2009; Brito et al., 2011; Cruz et al., 2011). Figure 1 shows examples of moonlighting proteins described in Paracoccidioides spp.
FIGURE 1.
Schematic representation of moonlighting proteins involved in Paracoccidioides spp. carbon metabolism (glycolysis/TCA cycle/glyoxylate cycle). The figure summarizes the canonical function of the moonlighting proteins that are also involved in Paracoccidioides–host interactions. In red, we highlighted the enzymes with moonlighting functions that have been studied. ALD, aldolase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ENO, enolase; MLS, malate synthase; TPI, triose phosphate isomerase; ACO, aconitase; ICL, isocitrate lyase.
Moonlighting functions have been increasingly recognized in glycolytic pathway and TCA cycle enzymes. In particular, despite lacking identifiable secretion signals, glycolytic enzymes have been observed on the Paracoccidioides surface, where they exhibit various functions that are unrelated to glycolysis, including a direct interaction with the host’s soluble proteins and surface ligands. Certain Paracoccidioides spp. proteins translocate to the exterior through unconventional protein secretion mechanisms, such as an affinity between certain proteins that act as transport vesicle coat components, which eventually lead to adherence or internalization and delivery to a distinct endosomal compartment in secretory vesicles (Nombela et al., 2006). This process involves vesicles derived from inward membrane invagination (endosomes), which results in protein trafficking to the plasma membrane and/or extracellular space, controlling localization and/or activity (Keller et al., 2006; Nosanchuk et al., 2008; Oliveira et al., 2010; Rodrigues et al., 2014). Straus et al. (1996) first demonstrated protein transport in vesicles in P. brasiliensis. Longo et al. (2014) identified many P. brasiliensis surface proteins in extracellular vesicles, which suggests participation of these structures in the fungal secretome and cell wall metabolism. Extracellular vesicles produced by fungi mostly contain proteins related to diverse metabolic processes. In Paracoccidioides spp., GAPDH, ENO, TPI and FBA were identified in the vesicle proteome and were microscopically localized to the cell wall as well as implicated in adhesion to ECM components (Vallejo et al., 2012). These results explain the reports for numerous cytoplasmic proteins, wherein the proteins perform other functions outside the plasma membrane, both in the cell wall and extracellular environment.
Another aspect for consideration is the immunological role of fungal surface proteins; these proteins may interact with the host in numerous ways and modulate the immune response (Travassos and Taborda, 2012). For example, the recognition of cell wall-associated proteins by pre-activated T cells and/or antibodies may interfere with infection (Gow and Hube, 2012). In addition, secreted proteins have important functions, such as nutrient supply, cell-to-cell communication, environmental detoxification, killing potential competitors, and aiding survival in the host (Bonin-Debs et al., 2004; Nombela et al., 2006; Holbrook et al., 2011; Weber et al., 2012). One of the main characteristics of pathogenesis is inducing damage to the host, which can occur directly due to the fungus and its virulence factors when it invades deep into or through the host tissues. Damage may also result from over-activation of the immune system through, for example, massive neutrophil infiltration or an inappropriate and unbalanced systemic response, which produces life-threatening sepsis. Thus, immune recognition may not only be beneficial and crucial for fighting invading fungi but may also be an integral part of the disease process (Gow and Hube, 2012).
FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE
Fructose 1,6-bisphosphate aldolase catalyzes reversible cleavage of fructose 1-6, bisphosphate into two triose phosphates, dihydroxyacetone phosphate and glyceraldehyde 3-phosphate. The reaction is common to glycolysis and gluconeogenesis (Marsh and Lebherz, 1992). Paracoccidioides spp. contains two genes that encode different Class II FBAs. FBA gene duplication in Paracoccidioides spp. was supported by phylogenetic analysis and established a two-member family with potentially differing functions. In addition, expression analysis support differential expression of Pbfba1 and Pbfba2, which indicates distinct functions for the two proteins (Carneiro et al., 2005). The presence of a paralogous gene supports acquisition of a new function, even if the new function is related to the original function (Tatusov et al., 1997). In Paracoccidioides spp., the Pbfba2 transcript was only detected in mycelial form, whereas the Pbfba1 transcript was detected in yeast cells (Chaves, 2013), further suggesting distinct functions.
Interestingly, FBA was detected in the P. lutzii secretome and cell wall during macrophage infection (Chaves, 2013). In proteomic studies on the yeast and mycelial phases, FBA was detected in the cell wall and extracellular vesicles, exclusively in the P. brasiliensis yeast-phase (Longo et al., 2014). Data show that the cell surface FBA1 includes immunogenic properties because the native protein can be recognized by serum from patients infected with paracoccidioidomycosis (PCM; da Fonseca et al., 2001). In the Paracoccidioides genus, both FBA isoforms could bind human plasminogen and convert plasminogen into plasmin in the presence of tissue plasminogen activator (tPA), which may increase the fibrinolytic capacity of the fungus, demonstrating that FBA is involved in the adhesion and invasion processes (Chaves, 2013). FBA also seems important in host-fungus interactions (Burucoa et al., 1995; McCarthy et al., 2002; Starnes et al., 2009; Capodagli et al., 2014).
GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE
Glyceraldehyde 3-phosphate dehydrogenase is a glycolytic enzyme that catalyzes glyceraldehyde 3-phosphate conversion into 1,3-bisphosphoglycerate. The most common form in all organisms is the NAD+-dependent enzyme, which is typically located in the cytoplasm (Tunio et al., 2010).
In addition to the intracellular location of GAPDH, it is also present in the most external layer of the cell wall in Paracoccidioides spp. yeast cells at higher quantities than in the cytoplasm. The presence of GAPDH in the cell wall and extracellular vesicles of P. brasiliensis during the yeast and mycelium phases was demonstrated using proteomic analysis (Longo et al., 2014), which suggests that it is involved in pathogenesis (Barbosa et al., 2004). The involvement of surface GAPDH in the interaction between Paracoccidioides spp. and laminin, fibronectin, and type I collagen has been demonstrated (Barbosa et al., 2004, 2006). GAPDH binding to laminin, fibronectin, and type I collagen may indicate that it is a virulence factor.
Purified, recombinant GAPDH protein immunological reactivity with 70 human serum samples was tested using Western blot analysis. GAPDH reacted with antibodies in the PCM patient serum, but not in the control patients’ sera; thus, GAPDH is included in the arsenal of P. brasiliensis immunoreactive molecules (Santos et al., 2012).
The Paracoccidioides spp. GAPDH likely plays a role in the initial phases of fungal infection. A reduced interaction between Paracoccidioides spp. and epithelial cells was demonstrated using recombinant GAPDH protein and the polyclonal anti-GAPDH, which suggests that this protein may be important during the Paracoccidioides spp. infection process (Barbosa et al., 2006).
Bailão et al. (2006) used representational difference analysis (RDA) to identify genes induced during the infection process in experimental animal livers under conditions that mimic hematogenous dissemination of the fungus. The researchers showed that GAPDH was overexpressed during infection, and similar results were observed when P. brasiliensis yeast cells were incubated with human blood, which supports the notion that this molecule may be involved in pathogenesis. GAPDH mRNA was also identified in the P. brasiliensis transcriptome from mouse liver, which reinforces its potential role in the infection process (Costa et al., 2007).
In a study on the P. brasiliensis transcription response upon internalization by macrophages, GAPDH was down-regulated, which suggests a complex carbon-depleted environment in the macrophage that yields a similar adaptive response as in intracellular fungal pathogens (Tavares et al., 2007).
All of the data suggest that GAPDH includes adhesin characteristics and plays an important role in the fungus–host interaction, which triggers host cell processes involved in pathogenesis.
TRIOSE PHOSPHATE ISOMERASE (TPI)
Triose phosphate isomerase (TPI) is an enzyme that rapidly interconverts dihydroxyacetone phosphate and D-glyceraldehyde 3-phosphate in the glycolysis pathway (Wierenga et al., 2010). The Paracoccidioides spp. TPI was first identified by da Fonseca et al. (2001) through fractionating fungus extracts using two-dimensional electrophoresis and subsequent immunoblotting. Using a strategy to identify Paracoccidioides spp. proteins that react with PCM patient sera, TPI was characterized as an important immunogenic molecule (Pereira et al., 2004). TPI was identified in the cell wall and extracellular vesicles using liquid chromatography coupled with high-resolution mass-spectrometry (LC-MS/MS) in the yeast and mycelium forms of two different P. brasiliensis isolates, Pb18 and Pb03 (Longo et al., 2014).
Additionally, TPI expression is developmentally regulated in Paracoccidioides spp.; expression increases when the fungus adopts the pathogenic yeast-like morphology. TPI also plays a role in the fungus–host interaction because the recombinant protein interacts with pneumocytes through binding the ECM components laminin and fibronectin. Finally, P. brasiliensis pre-treatment with a TPI polyclonal antibody inhibits adhesion to pneumocytes (Pereira et al., 2007).
ENOLASE
Enolase is also referred to as phosphopyruvate hydratase and is one of the most abundantly expressed cytosolic proteins in many organisms. It is a key glycolytic enzyme that catalyzes 2-phosphoglycerate dehydration to phosphoenolpyruvate (Pancholi, 2001). ENO was identified in the cell wall and extracellular vesicles in both the P. brasiliensis mycelium and yeast phases; it is secreted from unconventional pathways, which was predicted using the Fungal Secretome Database (Longo et al., 2014). Unconventional extracellular export pathways include plasma membrane transporter actions and the use of vesicles that originate from the plasma membrane, lysosomal secretion, or exosome release (Nickel, 2010; Rodrigues et al., 2011).
The capacity to bind to plasminogen and fibronectin as well as superficial localization have been linked to the pathogenic role of ENO in Paracoccidioides spp. (Donofrio et al., 2009; Nogueira et al., 2010; Marcos et al., 2012). Certain non-glycolytic ENO properties described above, particularly the properties related to surface expression and plasminogen binding, indicate that ENO may play an important role in initiating the infection process through modulating the pericellular and intravascular fibrinolytic system. Additionally, the internal 254FYKADEKK262 motif may be responsible for plasminogen binding, especially through the C-terminal lysine. P. brasiliensis ENO also includes an RGD motif (Arg-Gly-Asp), which is a sequence motif that mediates cell attachment. Marcos et al. (2012) demonstrated that ENO attachment to pneumocytes was inhibited, which suggests that the RGD peptide competes with the ENO binding site in pneumocytes.
Nogueira et al. (2010) demonstrated that treating epithelial cells and phagocytes with recombinant P. brasiliensis ENO (rPbEno) increases the effectiveness of the Paracoccidioides spp. interaction with host components because rPbEno enhances the exposure of surface N-acetylglucosamine residues, which Paracoccidioides spp. uses as a surface site for adherence to host cells (Coltri et al., 2006; Ganiko et al., 2007; Donofrio et al., 2009; dos Reis Almeida et al., 2010).
These data indicate that the Paracoccidioides spp. ENO may also have different subcellular locations (i.e., the cytoplasm or cell wall). Further, ENO has other functions in addition to its metabolic role that contributes to the virulence of this fungus.
MALATE SYNTHASE
The glyoxylate cycle is a TCA cycle anaplerotic pathway that facilitates growth on C (2) compounds through bypassing the CO (2)-generating TCA cycle steps. MLS converts glyoxylate and acetyl-CoA to malate (Dunn et al., 2009).
In Paracoccidioides spp., MLS participates in the glyoxylate cycle and allantoin degradation, which allows the cell to use purine as a nitrogen source (Zambuzzi-Carvalho et al., 2009); MLS is likely important for infection because its transcript is up-regulated during the mycelium to yeast transition, during the infectious phase (Bastos et al., 2007), and, in yeast cells, during phagocytosis by murine macrophages (Derengowski et al., 2008). da Silva Neto et al. (2009) showed that the Paracoccidioides spp. MLS is located on the cell surface and binds certain ECM components, such as type I and IV collagens, fibronectin and pneumocytes. Anti-MLS Paracoccidioides spp. antibodies inhibit the interaction with epithelial cells in vitro, which suggests that this protein contributes to adhesion between the fungus and host tissues.
ISOCITRATE LYASE
Isocitrate lyase is a glyoxylate cycle enzyme that converts isocitrate to glyoxylate and succinate; it is important for maintaining the TCA cycle afforded by the glyoxylate cycle when pyruvate generation from glycolysis is lower and fatty acid β-oxidation provides the major carbon source (Gould et al., 2006).
Isocitrate lyase protein was observed in the Paracoccidioides spp. culture filtrate; it is actively secreted to the Paracoccidioides spp. cell surface (Cruz et al., 2011). Troian (2009) demonstrated that recombinant ICL from P. brasiliensis (PbICL) and its polyclonal antibody inhibited interactions between P. brasiliensis and epithelial cells, which suggest a role in adhesion to host tissue. Cruz et al. (2011) reported that recombinant ICL binds fibronectin and type IV collagen, which reinforces the importance of this protein during the Paracoccidioides–host interaction.
Isocitrate lyase transcripts from Paracoccidioides spp. are induced during the yeast phase, during infection in a murine model (Felipe et al., 2005; Costa et al., 2007), and during the mycelium to yeast transition (Goldman et al., 2003; Bastos et al., 2007). Additionally, the gene that encodes ICL was induced during the fungus–macrophage interaction upon carbon starvation (Lima et al., 2014).
Argentilactone, which is a natural constituent of essential oil from Hyptis avalifolia, and its semi-synthetic derivate inhibited ICL activity in the presence of acetate, which affects P. lutzii yeast growth and mycelium to yeast differentiation (Prado et al., 2014). This finding suggests a significant role for Paracoccidioides spp. ICL in the host–pathogen interaction because the transition to the yeast phase of the fungus is essential for establishing infection and disease (San-Blas et al., 2002; Santana et al., 2012). Considering that pathogenic microorganisms utilize different carbon sources during pathogenesis (Barelle et al., 2006) and considering that PbICL is regulated by carbon sources (Prado et al., 2014), it is notable that ICL inhibition can affect cell growth and differentiation due a change in the carbon source used by the pathogen.
These studies indicate an adhesin behavior for Paracoccidioides spp. ICL and that it plays an important role in adhesion and colonization of this fungus in host tissue.
ACONITASE
Aconitase catalyzes the second step of the TCA cycle, which includes stereo-specific isomerization of citrate to isocitrate via cis-aconitate. In addition to its role in energy generation, the TCA cycle generates essential precursors for amino acid, fatty acid, and carbohydrate biosynthesis (da Fonseca et al., 2001).
Brito et al. (2011) used Western blot and immunocytochemistry analysis to demonstrate P. brasiliensis ACO (PbACO) in the extracellular fluid and that is associated with the cell wall, mitochondria, cytosol, and peroxisomes in yeast cells. Additionally, the researchers observed that PbACO was overexpressed when the cells were grown with ethanol and acetate as carbon sources and at higher iron levels, which suggests a potential role for PbACO in iron metabolism.
FINAL STATEMENTS
Studies on bacterial and fungal moonlighting proteins are in an early stage. One startling discovery demonstrated that the majority of proteins in the bacterial glycolytic pathway have certain adhesive functions (Henderson and Martin, 2011), which was also observed in Paracoccidioides spp. In this review, we collect the results for moonlighting enzyme activities in the glycolytic pathway as well as in the TCA and glyoxylate cycles, which have been described as ECM ligands in Paracoccidioides spp. In addition to adding to the number and types of known moonlighting proteins, these new examples also add to current information on the general importance of moonlighting proteins (Chung et al., 1999; Jeffery, 1999, 2003a, 2009).
Based on all of the data, the Paracoccidioides spp. moonlighting proteins include different functions in addition to their conventional metabolism roles due to their surface location. Clearly, moonlighting, or the ability to perform biological functions unrelated to the canonical function assigned to the protein, is common in fungal proteins in addition to bacteria (Kainulainen and Korhonen, 2014). A general conclusion is that moonlighting proteins seem to communicate with the environment and in response to environmental changes or stress. The moonlighting proteins present on the cell surface of Paracoccidioides species and released through vesicles are thought to function in host interactions. The specific roles of most cell surface proteins remain unclear, but a few such proteins are involved in cell wall biosynthesis/remodeling, adaptation to different environmental conditions, and PCM pathogenesis. Thus, moonlighting proteins may be potential targets for designing drugs against systemic mycosis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
- Aharoni A., Gaidukov L., Khersonsky O., Mcq Gould S., Roodveldt C., Tawfik D. S. (2005). The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 37 73–76. [DOI] [PubMed] [Google Scholar]
- Albring M., Griffith J., Attardi G. (1977). Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc. Natl. Acad. Sci. U.S.A. 74 1348–1352 10.1073/pnas.74.4.1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreotti P. F., Monteiro Da Silva J. L., Bailão A. M., Soares C. M., Benard G., Soares C. P., et al. (2005). Isolation and partial characterization of a 30 kDa adhesin from Paracoccidioides brasiliensis. Microbes Infect. 7 875–881 10.1016/j.micinf.2005.02.005 [DOI] [PubMed] [Google Scholar]
- Angiolella L., Facchin M., Stringaro A., Maras B., Simonetti N., Cassone A. (1996). Identification of a glucan-associated enolase as a main cell wall protein of Candida albicans and an indirect target of lipopeptide antimycotics. J. Infect. Dis. 173 684–690 10.1093/infdis/173.3.684 [DOI] [PubMed] [Google Scholar]
- Bailão A. M., Schrank A., Borges C. L., Dutra V., Walquíria Inês Molinari-Madlum E. E., Soares Felipe M. S., et al. (2006). Differential gene expression by Paracoccidioides brasiliensis in host interaction conditions: representational difference analysis identifies candidate genes associated with fungal pathogenesis. Microbes Infect. 8 2686–2697 10.1016/j.micinf.2006.07.019 [DOI] [PubMed] [Google Scholar]
- Baker H. V. (1991). GCR1 of Saccharomyces cerevisiae encodes a DNA binding protein whose binding is abolished by mutations in the CTTCC sequence motif. Proc. Natl. Acad. Sci. U.S.A. 88 9443–9447 10.1073/pnas.88.21.9443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa M. S., Báo S. N., Andreotti P. F., De Faria F. P., Felipe M. S., Dos Santos Feitosa L., et al. (2006). Glyceraldehyde-3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect. Immun. 74 382–389 10.1128/IAI.74.1.382-389.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa M. S., Cunha Passos D. A., Felipe M. S., Jesuíno R. S., Pereira M., De Almeida Soares C. M. (2004). The glyceraldehyde-3-phosphate dehydrogenase homologue is differentially regulated in phases of Paracoccidioides brasiliensis: molecular and phylogenetic analysis. Fungal Genet. Biol. 41 667–675 10.1016/j.fgb.2004.02.002 [DOI] [PubMed] [Google Scholar]
- Barelle C. J., Priest C. L., Maccallum D. M., Gow N. A. R., Odds F. C., Brown A. J. P. (2006). Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol. 8 961–971 10.1111/j.1462-5822.2005.00676.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos K. P., Bailão A. M., Borges C. L., Faria F. P., Felipe M. S., Silva M. G., et al. (2007). The transcriptome analysis of early morphogenesis in Paracoccidioides brasiliensis mycelium reveals novel and induced genes potentially associated to the dimorphic process. BMC Microbiol. 7:29 10.1186/1471-2180-7-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin-Debs A. L., Boche I., Gille H., Brinkmann U. (2004). Development of secreted proteins as biotherapeutic agents. Expert Opin. Biol. Ther. 4 551–558 10.1517/14712598.4.4.551 [DOI] [PubMed] [Google Scholar]
- Brilli M., Fani R. (2004). The origin and evolution of eucaryal HIS7 genes: from metabolon to bifunctional proteins? Gene 339 149–160 10.1016/j.gene.2004.06.033 [DOI] [PubMed] [Google Scholar]
- Brito W. E. A., Rezende T. C., Parente A. F., Ricart C. A., Sousa M. V., Báo S. N., et al. (2011). Identification, characterization and regulation studies of the aconitase of Paracoccidioides brasiliensis. Fungal Biol. 115 697–707 10.1016/j.funbio.2011.02.011 [DOI] [PubMed] [Google Scholar]
- Burucoa C., Frémaux C., Pei Z., Tummuru M., Blaser M. J., Cenatiempo Y., et al. (1995). Nucleotide sequence and characterization of peb4A encoding an antigenic protein in Campylobacter jejuni. Res. Microbiol. 146 467–476 10.1016/0923-2508(96)80292-0 [DOI] [PubMed] [Google Scholar]
- Capodagli G. C., Sedhom W. G., Jackson M., Ahrendt K. A., Pegan S. D. (2014). A noncompetitive inhibitor for Mycobacterium tuberculosis’s class IIa fructose 1,6-bisphosphate aldolase. Biochemistry 53 202–213 10.1021/bi401022b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro L. C., De Faria F. P., Felipe M. S., Pereira M., De Almeida Soares C. M. (2005). Paracoccidioides brasiliensis presents two different cDNAs encoding homologues of the fructose 1,6-biphosphate aldolase: protein isolation, cloning of the cDNAs and genes, structural, phylogenetic, and expression analysis. Fungal Genet. Biol. 42 51–60 10.1016/j.fgb.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Chaves E. G. A. (2013). Identificaç ao e Análise de Protenías Ligantes de Plasminogênio de Paracoccidioides. Master, Universidade Federal de Goiás, Goiás. [Google Scholar]
- Chen X., Wang X., Kaufman B., Butow R. (2005). Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 307 714–717 10.1126/science.1106391 [DOI] [PubMed] [Google Scholar]
- Chung S., Mclean M. R., Rymond B. C. (1999). Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA 5 1042–1054 10.1017/S1355838299990635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. L., Scopes D. A., Sodeinde O., Mason P. J. (2001). Glucose-6-phosphate dehydrogenase-6-phosphogluconolactonase. A novel bifunctional enzyme in malaria parasites. Eur. J. Biochem. 268 2013–2019 10.1046/j.1432-1327.2001.02078.x [DOI] [PubMed] [Google Scholar]
- Coltri K. C., Casabona-Fortunato A. S., Gennari-Cardoso M. L., Pinzan C. F., Ruas L. P., Mariano V. S., et al. (2006). Paracoccin, a GlcNAc-binding lectin from Paracoccidioides brasiliensis, binds to laminin and induces TNF-alpha production by macrophages. Microbes Infect. 8 704–713 10.1016/j.micinf.2005.09.008 [DOI] [PubMed] [Google Scholar]
- Commichau F. M., Rothe F. M., Herzberg C., Wagner E., Hellwig D., Lehnik-Habrink M., et al. (2009). Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol. Cell. Proteom. 8 1350–1360 10.1074/mcp.M800546-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley S. D. (2012). Moonlighting is mainstream: paradigm adjustment required. Bioessays 34 578–588 10.1002/bies.201100191 [DOI] [PubMed] [Google Scholar]
- Costa M., Borges C. L., Bailão A. M., Meirelles G. V., Mendonça Y. A., Dantas S. F., et al. (2007). Transcriptome profiling of Paracoccidioides brasiliensis yeast-phase cells recovered from infected mice brings new insights into fungal response upon host interaction. Microbiology 153 4194–4207 10.1099/mic.0.2007/009332-0 [DOI] [PubMed] [Google Scholar]
- Crowe J. D., Sievwright I. K., Auld G. C., Moore N. R., Gow N. A., Booth N. A. (2003). Candida albicans binds human plasminogen: identification of eight plasminogen-binding proteins. Mol. Microbiol. 47 1637–1651 10.1046/j.1365-2958.2003.03390.x [DOI] [PubMed] [Google Scholar]
- Cruz A. H., Brock M., Zambuzzi-Carvalho P. F., Santos-Silva L. K., Troian R. F., Góes A. M., et al. (2011). Phosphorylation is the major mechanism regulating isocitrate lyase activity in Paracoccidioides brasiliensis yeast cells. FEBS J. 278 2318–2332 10.1111/j.1742-4658.2011.08150.x [DOI] [PubMed] [Google Scholar]
- da Fonseca C. A., Jesuino R. S., Felipe M. S., Cunha D. A., Brito W. A., Soares C. M. (2001). Two-dimensional electrophoresis and characterization of antigens from Paracoccidioides brasiliensis. Microbes Infect. 3 535–542 10.1016/S1286-4579(01)01409-5 [DOI] [PubMed] [Google Scholar]
- da Silva Neto B. R., De Fátima Da Silva J., Mendes-Giannini M. J., Lenzi H. L., De Almeida Soares C. M., Pereira M. (2009). The malate synthase of Paracoccidioides brasiliensis is a linked surface protein that behaves as an anchorless adhesin. BMC Microbiol. 9:272 10.1186/1471-2180-9-272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker B. L., Wickner W. T. (2006). Enolase activates homotypic vacuole fusion and protein transport to the vacuole in yeast. J. Biol. Chem. 281 14523–14528 10.1074/jbc.M600911200 [DOI] [PubMed] [Google Scholar]
- Delgado M. L., Gil M. L., Gozalbo D. (2003). Candida albicans TDH3 gene promotes secretion of internal invertase when expressed in Saccharomyces cerevisiae as a glyceraldehyde-3-phosphate dehydrogenase-invertase fusion protein. Yeast 20 713–722 10.1002/yea.993 [DOI] [PubMed] [Google Scholar]
- Derengowski L. S., Tavares A. H., Silva S., Procópio L. S., Felipe M. S., Silva-Pereira I. (2008). Upregulation of glyoxylate cycle genes upon Paracoccidioides brasiliensis internalization by murine macrophages and in vitro nutritional stress condition. Med. Mycol. 46 125–134 10.1080/13693780701670509 [DOI] [PubMed] [Google Scholar]
- Donofrio F. C., Calil A. C., Miranda E. T., Almeida A. M., Benard G., Soares C. P., et al. (2009). Enolase from Paracoccidioides brasiliensis: isolation and identification as a fibronectin-binding protein. J. Med. Microbiol. 58 706–713 10.1099/jmm.0.003830-0 [DOI] [PubMed] [Google Scholar]
- dos Reis Almeida F. B., De Oliveira L. L., Valle De Sousa M., Barreira M. C., Hanna E. S. (2010). Paracoccin from Paracoccidioides brasiliensis; purification through affinity with chitin and identification of N-acetyl-beta-D-glucosaminidase activity. Yeast 27 67–76 10.1002/yea.1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn M. F., Ramírez-Trujillo J. A., Hernández-Lucas I. (2009). Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology 155 3166–3175 10.1099/mic.0.030858-0 [DOI] [PubMed] [Google Scholar]
- Ebel F., Schwienbacher M., Beyer J., Heesemann J., Brakhage A. A., Brock M. (2006). Analysis of the regulation, expression, and localisation of the isocitrate lyase from Aspergillus fumigatus, a potential target for antifungal drug development. Fungal Genet. Biol. 43 476–489 10.1016/j.fgb.2006.01.015 [DOI] [PubMed] [Google Scholar]
- Eroles P., Sentandreu M., Elorza M. V., Sentandreu R. (1997). The highly immunogenic enolase and Hsp70p are adventitious Candida albicans cell wall proteins. Microbiology 143 313–320 10.1099/00221287-143-2-313 [DOI] [PubMed] [Google Scholar]
- Felipe M. S., Andrade R. V., Arraes F. B., Nicola A. M., Maranhão A. Q., Torres F. A., et al. (2005). Transcriptional profiles of the human pathogenic fungus Paracoccidioides brasiliensis in mycelium and yeast cells. J. Biol. Chem. 280 24706–24714 10.1074/jbc.M500625200 [DOI] [PubMed] [Google Scholar]
- Fothergill-Gilmore L. A., Michels P. A. (1993). Evolution of glycolysis. Prog. Biophys. Mol. Biol. 59 105–235 10.1016/0079-6107(93)90001-Z [DOI] [PubMed] [Google Scholar]
- Fox D., Smulian A. G. (2001). Plasminogen-binding activity of enolase in the opportunistic pathogen Pneumocystis carinii. Med. Mycol. 39 495–507 10.1080/mmy.39.6.495.507 [DOI] [PubMed] [Google Scholar]
- Gainey L. D., Connerton I. F., Lewis E. H., Turner G., Ballance D. J. (1992). Characterization of the glyoxysomal isocitrate lyase genes of Aspergillus nidulans (acuD) and Neurospora crassa (acu-3). Curr. Genet. 21 43–47 10.1007/BF00318653 [DOI] [PubMed] [Google Scholar]
- Gancedo C., Flores C. L. (2008). Moonlighting proteins in yeasts. Microbiol. Mol. Biol. Rev. 72 197–210 10.1128/MMBR.00036-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganiko L., Puccia R., Mariano V. S., Sant’anna O. A., Freymüller E., Roque-Barreira M. C., et al. (2007). Paracoccin, an N-acetyl-glucosamine-binding lectin of Paracoccidioides brasiliensis, is involved in fungal growth. Microbes Infect. 9 695–703 10.1016/j.micinf.2007.02.012 [DOI] [PubMed] [Google Scholar]
- Gil-Navarro I., Gil M. L., Casanova M., O’Connor J. E., Martínez J. P., Gozalbo D. (1997). The glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is a surface antigen. J. Bacteriol. 179 4992–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman G. H., Dos Reis Marques E., Duarte Ribeiro D. C., De Souza Bernardes L. A., Quiapin A. C., Vitorelli P. M., et al. (2003). Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot. Cell 2 34–48 10.1128/EC.2.1.34-48.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez A., Hernández S., Amela I., Piñol J., Cedano J., Querol E. (2011). Do protein-protein interaction databases identify moonlighting proteins? Mol. Biosyst. 7 2379–2382 10.1039/c1mb05180f [DOI] [PubMed] [Google Scholar]
- González A., Gómez B. L., Diez S., Hernández O., Restrepo A., Hamilton A. J., et al. (2005). Purification and partial characterization of a Paracoccidioides brasiliensis protein with capacity to bind to extracellular matrix proteins. Infect. Immun. 73 2486–2495 10.1128/IAI.73.4.2486-2495.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould T. A., Van De Langemheen H., Muñoz-Elías E. J., Mckinney J. D., Sacchettini J. C. (2006). Dual role of isocitrate lyase 1 in the glyoxylate and methylcitrate cycles in Mycobacterium tuberculosis. Mol. Microbiol. 61 940–947 10.1111/j.1365-2958.2006.05297.x [DOI] [PubMed] [Google Scholar]
- Gow N. A. R., Hube B. (2012). Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 15 406–412 10.1016/j.mib.2012.04.005 [DOI] [PubMed] [Google Scholar]
- Hanna S. A., Monteiro Da Silva J. L., Giannini M. J. (2000). Adherence and intracellular parasitism of Paracoccidioides brasiliensis in Vero cells. Microbes Infect. 2 877–884 10.1016/S1286-4579(00)00390-7 [DOI] [PubMed] [Google Scholar]
- Henderson B., Martin A. (2011). Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79 3476–3491 10.1128/IAI.00179-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks W., Mulders J. W., Bibby M. A., Slingsby C., Bloemendal H., De Jong W. W. (1988). Duck lens epsilon-crystallin and lactate dehydrogenase B4 are identical: a single-copy gene product with two distinct functions. Proc. Natl. Acad. Sci. U.S.A. 85 7114–7118 10.1073/pnas.85.19.7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook E. D., Edwards J. A., Youseff B. H., Rappleye C. A. (2011). Definition of the extracellular proteome of pathogenic-phase Histoplasma capsulatum. J. Proteome Res. 10 1929–1943 10.1021/pr1011697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberts D. H., van der Klei I. J. (2010). Moonlighting proteins: an intriguing mode of multitasking. Biochim. Biophys. Acta 1803 520–525 10.1016/j.bbamcr.2010.01.022 [DOI] [PubMed] [Google Scholar]
- James C. L., Viola R. E. (2002). Production and characterization of bifunctional enzymes. Substrate channeling in the aspartate pathway. Biochemistry. 41 3726–3731 10.1021/bi0159074 [DOI] [PubMed] [Google Scholar]
- Jeffery C. J. (1999). Moonlighting proteins. Trends Biochem. Sci. 24 8–11 10.1016/S0968-0004(98)01335-8 [DOI] [PubMed] [Google Scholar]
- Jeffery C. J. (2003a). Moonlighting proteins: old proteins learning new tricks. Trends Genet. 19 415–417 10.1016/S0168-9525(03)00167-7 [DOI] [PubMed] [Google Scholar]
- Jeffery C. J. (2003b). Multifunctional proteins: examples of gene sharing. Ann. Med. 35 28–35 10.1080/07853890310004101 [DOI] [PubMed] [Google Scholar]
- Jeffery C. J. (2004). Molecular mechanisms for multitasking: recent crystal structures of moonlighting proteins. Curr. Opin. Struct. Biol. 14 663–668 10.1016/j.sbi.2004.10.001 [DOI] [PubMed] [Google Scholar]
- Jeffery C. J. (2009). Moonlighting proteins–an update. Mol. Biosyst. 5 345–350 10.1039/b900658n [DOI] [PubMed] [Google Scholar]
- Jong A. Y., Chen S. H., Stins M. F., Kim K. S., Tuan T. L., Huang S. H. (2003). Binding of Candida albicans enolase to plasmin(ogen) results in enhanced invasion of human brain microvascular endothelial cells. J. Med. Microbiol. 52 615–622 10.1099/jmm.0.05060-0 [DOI] [PubMed] [Google Scholar]
- Kainulainen V., Korhonen T. K. (2014). Dancing to another tune-adhesive moonlighting proteins in bacteria. Biology 3 178–204 10.3390/biology3010178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar Y. D., Ochman H. (2013). Genome reduction promotes increase in protein functional complexity in bacteria. Genetics 193 303–307 10.1534/genetics.112.145656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S., Sanderson M. P., Stoeck A., Altevogt P. (2006). Exosomes: from biogenesis and secretion to biological function. Immunol. Lett. 107 102–108 10.1016/j.imlet.2006.09.005 [DOI] [PubMed] [Google Scholar]
- Kim J. W., Dang C. V. (2005). Multifaceted roles of glycolytic enzymes. Trends Biochem. Sci. 30 142–150 10.1016/j.tibs.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Kirschner K., Bisswanger H. (1976). Multifunctional proteins. Annu. Rev. Biochem. 45 143–166 10.1146/annurev.bi.45.070176.001043 [DOI] [PubMed] [Google Scholar]
- Lima P. E. S., Casaletti L., Bailão A. M., De Vasconcelos A. T., Fernandes G. A. R., Soares C. M. (2014). Transcriptional and proteomic responses to carbon starvation in Paracoccidioides. PLoS Negl. Trop. Dis. 8:e2855 10.1371/journal.pntd.0002855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo L. V. G., Da Cunha J. P. C., Sobreira T. J. P., Puccia R. (2014). Proteome of cell wall-extracts from pathogenic Paracoccidioides brasiliensis: comparison among morphological phases, isolates, and reported fungal extracellular vesicle proteins. EuPA Open Proteom. 3 216–228 10.1016/j.euprot.2014.03.003 [DOI] [Google Scholar]
- Lu M., Ammar D., Ives H., Albrecht F., Gluck S. L. (2007). Physical interaction between aldolase and vacuolar H+-ATPase is essential for the assembly and activity of the proton pump. J. Biol. Chem. 282 24495–24503 10.1074/jbc.M702598200 [DOI] [PubMed] [Google Scholar]
- Lu M., Holliday L. S., Zhang L., Dunn W. A., Jr., Gluck S. L. (2001). Interaction between aldolase and vacuolar H+-ATPase: evidence for direct coupling of glycolysis to the ATP-hydrolyzing proton pump. J. Biol. Chem. 276 30407–30413 10.1074/jbc.M008768200 [DOI] [PubMed] [Google Scholar]
- Marcos C. M., De Fátima Da Silva J., De Oliveira H. C., Moraes Da Silva R. A., Mendes-Giannini M. J., Fusco-Almeida A. M. (2012). Surface-expressed enolase contributes to the adhesion of Paracoccidioides brasiliensis to host cells. FEMS Yeast Res. 12 557–570 10.1111/j.1567-1364.2012.00806.x [DOI] [PubMed] [Google Scholar]
- Marsh J. J., Lebherz H. G. (1992). Fructose-bisphosphate aldolases: an evolutionary history. Trends Biochem. Sci. 17 110–113 10.1016/0968-0004(92)90247-7 [DOI] [PubMed] [Google Scholar]
- McCarthy J. S., Wieseman M., Tropea J., Kaslow D., Abraham D., Lustigman S., et al. (2002). Onchocerca volvulus glycolytic enzyme fructose-1,6-bisphosphate aldolase as a target for a protective immune response in humans. Infect. Immun. 70 851–858 10.1128/IAI.70.2.851-858.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes-Giannini M. J., Andreotti P. F., Vincenzi L. R., Da Silva J. L., Lenzi H. L., Benard G., et al. (2006). Binding of extracellular matrix proteins to Paracoccidioides brasiliensis. Microbes Infect. 8 1550–1559 10.1016/j.micinf.2006.01.012 [DOI] [PubMed] [Google Scholar]
- Mendes-Giannini M. J., Monteiro Da Silva J. L., De Fátima Da Silva J., Donofrio F. C., Miranda E. T., Andreotti P. F., et al. (2008). Interactions of Paracoccidioides brasiliensis with host cells: recent advances. Mycopathologia 165 237–248 10.1007/s11046-007-9074-z [DOI] [PubMed] [Google Scholar]
- Moore B. (2004). Bifunctional and moonlighting enzymes: lighting the way to regulatory control. Trends Plant Sci. 9 221–228 10.1016/j.tplants.2004.03.005 [DOI] [PubMed] [Google Scholar]
- Myers M. G., Jr. (2009). Cell biology. Moonlighting in mitochondria. Science 323 723–724 10.1126/science.1169660 [DOI] [PubMed] [Google Scholar]
- Narahari J., Ma R., Wang M., Walden W. E. (2000). The aconitase function of iron regulatory protein 1. Genetic studies in yeast implicate its role in iron-mediated redox regulation. J. Biol. Chem. 275 16227–16234 10.1074/jbc.M910450199 [DOI] [PubMed] [Google Scholar]
- Nickel W. (2010). Pathways of unconventional protein secretion. Curr. Opin. Biotechnol. 21 621–626 10.1016/j.copbio.2010.06.004 [DOI] [PubMed] [Google Scholar]
- Nogueira S. V., Fonseca F. L., Rodrigues M. L., Mundodi V., Abi-Chacra E. A., Winters M. S., et al. (2010). Paracoccidioides brasiliensis enolase is a surface protein that binds plasminogen and mediates interaction of yeast forms with host cells. Infect. Immun. 78 4040–4050 10.1128/IAI.00221-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nombela C., Gil C., Chaffin W. L. (2006). Non-conventional protein secretion in yeast. Trends Microbiol. 14 15–21 10.1016/j.tim.2005.11.009 [DOI] [PubMed] [Google Scholar]
- Nosanchuk J. D., Nimrichter L., Casadevall A., Rodrigues M. L. (2008). A role for vesicular transport of macromolecules across cell walls in fungal pathogenesis. Commun. Integr. Biol. 1 37–39 10.4161/cib.1.1.6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira D. L., Nakayasu E. S., Joffe L. S., Guimarães A. J., Sobreira T. J., Nosanchuk J. D., et al. (2010). Biogenesis of extracellular vesicles in yeast: many questions with few answers. Commun. Integr. Biol. 3 533–535 10.4161/cib.3.6.12756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita I., Yurimoto H., Hirai R., Kawarabayasi Y., Sakai Y., Kato N. (2005). The archaeon Pyrococcus horikoshii possesses a bifunctional enzyme for formaldehyde fixation via the ribulose monophosphate pathway. J. Bacteriol. 187 3636–3642 10.1128/JB.187.11.3636-3642.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancholi V. (2001). Multifunctional alpha-enolase: its role in diseases. Cell. Mol. Life Sci. 58 902–920 10.1007/PL00000910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancholi V., Chhatwal G. S. (2003). Housekeeping enzymes as virulence factors for pathogens. Int. J. Med. Microbiol. 293 391–401 10.1078/1438-4221-00283 [DOI] [PubMed] [Google Scholar]
- Pancholi V., Fischetti V. A. (1992). A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176 415–426 10.1084/jem.176.2.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandini A., Fornili A., Fraternali F., Kleinjung J. (2012). Detection of allosteric signal transmission by information-theoretic analysis of protein dynamics. FASEB J. 26 868–881 10.1096/fj.11-190868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L. A., Báo S. N., Barbosa M. S., Da Silva J. L., Felipe M. S., De Santana J. M., et al. (2007). Analysis of the Paracoccidioides brasiliensis triosephosphate isomerase suggests the potential for adhesin function. FEMS Yeast Res. 7 1381–1388 10.1111/j.1567-1364.2007.00292.x [DOI] [PubMed] [Google Scholar]
- Pereira L. A., Pereira M., Felipe M. S., Zancopé-Oliveira R. M., Soares C. M. (2004). Proteomic identification, nucleotide sequence, heterologous expression and immunological reactivity of the triosephosphate isomerase of Paracoccidioides brasiliensis. Microbes Infect. 6 892–900 10.1016/j.micinf.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Pitarch A., Sánchez M., Nombela C., Gil C. (2002). Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol. Cell. Proteomics 1 967–982 10.1074/mcp.M200062-MCP200 [DOI] [PubMed] [Google Scholar]
- Prado R. S., Alves R. J., Oliveira C. M., Kato L., Silva R. A., Quintino G. O., et al. (2014). Inhibition of Paracoccidioides lutzii Pb01 isocitrate lyase by the natural compound argentilactone and its semi-synthetic derivatives. PLoS ONE 9:e94832 10.1371/journal.pone.0094832.g001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M. L., Nakayasu E. S., Almeida I. C., Nimrichter L. (2014). The impact of proteomics on the understanding of functions and biogenesis of fungal extracellular vesicles. J. Proteom. 97 177–186 10.1016/j.jprot.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M. L., Nosanchuk J. D., Schrank A., Vainstein M. H., Casadevall A., Nimrichter L. (2011). Vesicular transport systems in fungi. Future Microbiol. 6 1371–1381 10.2217/fmb.11.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault T. A., Stout C. D., Kaptain S., Harford J. B., Klausner R. D. (1991). Structural relationship between an iron-regulated RNA-binding protein (IRE-BP) and aconitase: functional implications. Cell 64 881–883 10.1016/0092-8674(91)90312-M [DOI] [PubMed] [Google Scholar]
- San-Blas G., Niño-Vega G., Iturriaga T. (2002). Paracoccidioides brasiliensis and paracoccidioidomycosis: molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med. Mycol. 40 225–242 10.1080/mmy.40.3.225.242 [DOI] [PubMed] [Google Scholar]
- Santana L. A. D. P., Vainstein M. H., Tomazett P. K., Santos-Silva L. K., Góes A. M., Schrank A., et al. (2012). Distinct chitinases are expressed during various growth phases of the human pathogen Paracoccidioides brasiliensis. Mem. Inst. Oswaldo Cruz 107 310–316 10.1590/S0074-02762012000300004 [DOI] [PubMed] [Google Scholar]
- Santos R. D. S., Martelli De Paula N., Barbosa M., De Almeida Soares C. M. (2012). Caracterização imunológica da proteína recombinante gliceraldeído-3-fosfato desidrogenase do patógeno humano Paracoccidioides brasiliensis. SaBios: Rev. Saúde Biol. 7 35–45. [Google Scholar]
- Sekita S., Yoshihira K., Natori S., Harada F., Iida K., Yahara I. (1985). Structure-activity relationship of thirty-nine cytochalasans observed in the effects on cellular structures and cellular events and on actin polymerization in vitro. J. Pharmacobiodyn. 8 906–916. [DOI] [PubMed] [Google Scholar]
- Sriram G., Martinez J. A., Mccabe E. R., Liao J. C., Dipple K. M. (2005). Single-gene disorders: what role could moonlighting enzymes play? Am. J. Hum. Genet. 76 911–924 10.1086/430799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starnes G. L., Coincon M., Sygusch J., Sibley L. D. (2009). Aldolase is essential for energy production and bridging adhesin-actin cytoskeletal interactions during parasite invasion of host cells. Cell Host Microbe 5 353–364 10.1016/j.chom.2009.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus A. H., Freymüller E., Travassos L. R., Takahashi H. K. (1996). Immunochemical and subcellular localization of the 43 kDa glycoprotein antigen of Paracoccidioides brasiliensis with monoclonal antibodies. J. Med. Vet. Mycol. 34 181–186 10.1080/02681219680000301 [DOI] [PubMed] [Google Scholar]
- Tatusov R. L., Koonin E. V., Lipman D. J. (1997). A genomic perspective on protein families. Science 278 631–637 10.1126/science.278.5338.631 [DOI] [PubMed] [Google Scholar]
- Tavares A. H., Silva S. S., Dantas A., Campos E. G., Andrade R. V., Maranhão A. Q., et al. (2007). Early transcriptional response of Paracoccidioides brasiliensis upon internalization by murine macrophages. Microbes Infect. 9 583–590 10.1016/j.micinf.2007.01.024 [DOI] [PubMed] [Google Scholar]
- Travassos L. R., Taborda C. P. (2012). Paracoccidioidomycosis vaccine. Hum. Vaccin. Immunother. 8 1450–1453 10.4161/hv.21283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troian R. F. (2009). Caracterização dA Isocitrato Liase e Metilisocitrato Liase do Fungo Patogênico Humano Paracoccidioides brasiliensis. Mestrado, Instituto de Patologia Tropical e Saúde Pública, Universidade Federal de, Goiás. [Google Scholar]
- Tunio S. A., Oldfield N. J., Ala’aldeen D. A., Wooldridge K. G., Turner D. P. (2010). The role of glyceraldehyde 3-phosphate dehydrogenase (GapA-1) in Neisseria meningitidis adherence to human cells. BMC Microbiol. 10:280 10.1186/1471-2180-10-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenciano S., Lucas J. R., Pedregosa A., Monistrol I. F., Laborda F. (1996). Induction of beta-oxidation enzymes and microbody proliferation in Aspergillus nidulans. Arch. Microbiol. 166 336–341 10.1007/s002030050392 [DOI] [PubMed] [Google Scholar]
- Vallejo M. C., Nakayasu E. S., Matsuo A. L., Sobreira T. J., Longo L. V., Ganiko L., et al. (2012). Vesicle and vesicle-free extracellular proteome of Paracoccidioides brasiliensis: comparative analysis with other pathogenic fungi. J. Proteome Res. 11 1676–1685 10.1021/pr200872s [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deventer A. J., van Vliet H. J., Hop W. C., Goessens W. H. (1994). Diagnostic value of anti-Candida enolase antibodies. J. Clin. Microbiol. 32 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini A. P., Gesztesi J. L., Franco M. F., De Souza W., De Moraes J. Z., Travassos L. R., et al. (1994). Binding of Paracoccidioides brasiliensis to laminin through surface glycoprotein gp43 leads to enhancement of fungal pathogenesis. Infect. Immun. 62 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T. J., Hathorn J. W., Sobel J. D., Merz W. G., Sanchez V., Maret S. M., et al. (1991). Detection of circulating candida enolase by immunoassay in patients with cancer and invasive candidiasis. N. Engl. J. Med. 324 1026–1031 10.1056/NEJM199104113241504 [DOI] [PubMed] [Google Scholar]
- Wang G., Xia Y., Cui J., Gu Z., Song Y., Chen Y. Q., et al. (2013). The roles of moonlighting proteins in bacteria. Curr. Issues Mol. Biol. 16 15–22. [PubMed] [Google Scholar]
- Weber S. S., Parente A. F. A., Borges C. L., Parente J. A., Bailão A. M., De Almeida Soares C. M. (2012). Analysis of the secretomes of Paracoccidioides mycelia and yeast cells. PLoS ONE 7:e52470 10.1371/journal.pone.0052470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienkers L. C., Rock B. (2014). Multienzyme kinetics and sequential metabolism. Methods Mol. Biol. 1113 93–118 10.1007/978-1-62703-758-7_6 [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Kapetaniou E. G., Venkatesan R. (2010). Triosephosphate isomerase: a highly evolved biocatalyst. Cell. Mol. Life Sci. 67 3961–3982 10.1007/s00018-010-0473-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistow G. J., Lietman T., Williams L. A., Stapel S. O., De Jong W. W., Horwitz J., et al. (1988). Tau-crystallin/alpha-enolase: one gene encodes both an enzyme and a lens structural protein. J. Cell Biol. 107 2729–2736 10.1083/jcb.107.6.2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff A. M., Arnau J. (2002). Cloning of glyceraldehyde-3-phosphate dehydrogenase-encoding genes in Mucor circinelloides (Syn. racemosus) and use of the gpd1 promoter for recombinant protein production. Fungal Genet. Biol. 35 21–29 10.1006/fgbi.2001.1313 [DOI] [PubMed] [Google Scholar]
- Yuan W., Tuttle D. L., Shi Y. J., Ralph G. S., Dunn W. A. (1997). Glucose-induced microautophagy in Pichia pastoris requires the alpha-subunit of phosphofructokinase. J. Cell Sci. 110(Pt 16) 1935–1945. [DOI] [PubMed] [Google Scholar]
- Zambuzzi-Carvalho P. F., Cruz A. H., Santos-Silva L. K., Goes A. M., Soares C. M., Pereira M. (2009). The malate synthase of Paracoccidioides brasiliensis Pb01 is required in the glyoxylate cycle and in the allantoin degradation pathway. Med. Mycol. 47 734–744 10.3109/13693780802609620 [DOI] [PubMed] [Google Scholar]