Abstract
The genus Burkholderia contains large number of diverse species which include many clinically important organisms, phytopathogens, as well as environmental species. However, currently, there is a paucity of biochemical or molecular characteristics which can reliably distinguish different groups of Burkholderia species. We report here the results of detailed phylogenetic and comparative genomic analyses of 45 sequenced species of the genus Burkholderia. In phylogenetic trees based upon concatenated sequences for 21 conserved proteins as well as 16S rRNA gene sequence based trees, members of the genus Burkholderia grouped into two major clades. Within these main clades a number of smaller clades including those corresponding to the clinically important Burkholderia cepacia complex (BCC) and the Burkholderia pseudomallei groups were also clearly distinguished. Our comparative analysis of protein sequences from Burkholderia spp. has identified 42 highly specific molecular markers in the form of conserved sequence indels (CSIs) that are uniquely found in a number of well-defined groups of Burkholderia spp. Six of these CSIs are specific for a group of Burkholderia spp. (referred to as Clade I in this work) which contains all clinically relevant members of the genus (viz. the BCC and the B. pseudomallei group) as well as the phytopathogenic Burkholderia spp. The second main clade (Clade II), which is composed of environmental Burkholderia species, is also distinguished by 2 identified CSIs that are specific for this group. Additionally, our work has also identified multiple CSIs that serve to clearly demarcate a number of smaller groups of Burkholderia spp. including 3 CSIs that are specific for the B. cepacia complex, 4 CSIs that are uniquely found in the B. pseudomallei group, 5 CSIs that are specific for the phytopathogenic Burkholderia spp. and 22 other CSI that distinguish two groups within Clade II. The described molecular markers provide highly specific means for the demarcation of different groups of Burkholderia spp. and they also offer novel and useful targets for the development of diagnostic assays for the clinically important members of the BCC or the pseudomallei groups. Based upon the results of phylogenetic analyses, the identified CSIs and the pathogenicity profile of Burkholderia species, we are proposing a division of the genus Burkholderia into two genera. In this new proposal, the emended genus Burkholderia will correspond to the Clade I and it will contain only the clinically relevant and phytopathogenic Burkholderia species. All other Burkholderia spp., which are primarily environmental, will be transferred to a new genus Paraburkholderia gen. nov.
Keywords: Burkholderia, Burkholderia cepacia complex, conserved signature indels, phylogenetic trees, molecular signatures
Introduction
The genus Burkholderia is a morphologically, metabolically, and ecologically diverse group of gram-negative bacteria (Yabuuchi et al., 1992; Coenye and Vandamme, 2003; Mahenthiralingam et al., 2005; Palleroni, 2005; Compant et al., 2008). Burkholderia species are ubiquitous in the environment (Coenye and Vandamme, 2003). They inhabit a wide range of ecological niches, ranging from soil to the human respiratory tract (Coenye and Vandamme, 2003). A group of 17 closely related Burkholderia species, the Burkholderia cepacia complex (BCC), are responsible for prevalent and potentially lethal pulmonary infections in immunocompromised individuals, such as individuals with cystic fibrosis (Mahenthiralingam et al., 2002, 2005; Biddick et al., 2003; Hauser et al., 2011). Burkholderia pseudomallei, a Burkholderia species related to the BCC, is the causative agent for the disease melioidosis, a potentially lethal septic infection which accounts for up to 20% of all community-acquired septicemias in some regions (White, 2003; Limmathurotsakul and Peacock, 2011). Other species related to the BCC are the causative agents of major infections in both animals (Burkholderia mallei) and plants (Burkholderia glumae and Burkholderia gladioli) (Whitlock et al., 2007; Nandakumar et al., 2009).
In spite of the large diversity and varied pathogenicity among the >70 members of the group, all Burkholderia species are currently placed within one genus (Coenye and Vandamme, 2003; Palleroni, 2005). The phylogeny and taxonomy of the genus Burkholderia is primarily defined on the basis of 16S rRNA sequence analysis (Yabuuchi et al., 1992; Palleroni, 2005; Yarza et al., 2008). The inferences obtained from 16S rRNA analysis have been further substantiated by other phylogenetic methods, including recA gene based analysis (Payne et al., 2005), acdS gene based analysis (Onofre-Lemus et al., 2009), DNA–DNA hybridization (Gillis et al., 1995), whole cell fatty acid analysis (Stead, 1992), multilocus sequence analysis (Tayeb et al., 2008; Spilker et al., 2009; Estrada-de los Santos et al., 2013), gene gain/loss analysis (Zhu et al., 2011), and whole genome phylogenetic analysis (Ussery et al., 2009; Segata et al., 2013). In many of these phylogenetic studies, the members of the genus Burkholderia can be divided into two or more distinct phylogenetic groups, with one group consisting of members of the BCC and related species (Payne et al., 2005; Tayeb et al., 2008; Yarza et al., 2008; Spilker et al., 2009; Ussery et al., 2009; Gyaneshwar et al., 2011; Vandamme and Dawyndt, 2011; Zhu et al., 2011; Estrada-de los Santos et al., 2013; Segata et al., 2013). Although there are some commonly shared features among closely related groups of Burkholderia species, there is no known morphological, biochemical, or molecular characteristic specific to the larger phylogenetic groups within the genus (ex. the BCC and related species).
The advent of next generation sequencing methods has led to a rapid increase in the number of genome sequences available for bacterial species (Mardis, 2008). The availability of these sequences for members of the genus Burkholderia provides us better means to evaluate the phylogenetic relationships among different species (Ciccarelli et al., 2006; Wu et al., 2009). Importantly, the large data sets of sequences allows for the use of comparative genomic techniques to discover novel molecular markers that can provide independent evidence for different phylogenetic groups within the genus Burkholderia (Gupta, 1998, 2014; Gao and Gupta, 2012). In this work, we describe one type of molecular marker, conserved sequence insertions or deletions (CSIs), which are uniquely present in protein sequences from a defined group of organisms, that can be used to delineate different phylogenetic groups of Burkholderia species independently of traditional phylogenetic methods (Gupta, 1998, 2001; Gao and Gupta, 2012). Our comparative analysis of Burkholderia genomes has led to the identification of 42 unique CSIs that delineate different phylogenetic groups within the genus in clear molecular terms. A clade of Burkholderia containing the BCC and related organisms (Clade I) was supported by both phylogenetic evidence and 6 identified CSIs. We have also identified 3 CSIs specific for the BCC, 4 CSIs specific for the B. pseudomallei group, and 5 CSIs specific for the plant pathogenic Burkholderia spp. The remaining members of the genus Burkholderia formed another monophyletic clade (Clade II) in our phylogenetic trees which was supported by 2 CSIs. Within Clade II, we identified two smaller clades of Burkholderia that were supported by 16 and 6 CSIs. The grouping of members of the genus Burkholderia into at least two large, monophyletic groups has also been observed in a large body of prior phylogenetic research (Payne et al., 2005; Tayeb et al., 2008; Yarza et al., 2008; Spilker et al., 2009; Ussery et al., 2009; Gyaneshwar et al., 2011; Zhu et al., 2011; Estrada-de los Santos et al., 2013; Segata et al., 2013). Based on the phylogenetic evidence and our identified CSIs, we propose division of the genus Burkholderia into two genera: an emended genus Burkholderia containing clinically important and phytopathogenic members of the genus and a new genus Paraburkholderia gen. nov. harboring the environmental species.
Materials and methods
Phylogenetic analysis
A concatenated sequence alignment of 21 highly conserved proteins (viz. ArgRS, EF-G, GyrA, GyrB, Hsp60, Hsp70, IleRS, RecA, RpoB, RpoC, SecY, ThrRS, TrpS, UvrD, ValRS, 50S ribosomal proteins L1, L5 and L6, and 30S ribosomal proteins S2, S8 and S11) was used to perform phylogenetic analysis. Due to their presence in most bacteria, these proteins have been extensively utilized for phylogenetic studies (Gupta, 1998, 2009; Kyrpides et al., 1999; Harris et al., 2003; Charlebois and Doolittle, 2004; Ciccarelli et al., 2006). The amino acid sequences for these conserved proteins were obtained from NCBI database for all of the species/strains listed in Table 1, which includes 45 sequenced species of the genus Burkholderia. Furthermore, three genomes from other members of class Betaproteobacteria (viz. Cupriavidus necator N-1, Bordetella pertussis Tohama I, and Neisseria meningitides MC58), serving as outgroups in our analysis, were also retrieved from NCBI database. Depending on genome availability, type strains were selected for most of the species. Multiple sequence alignments for these proteins were created using Clustal_X 1.83 and concatenated into a single alignment file (Jeanmougin et al., 1998). Poorly aligned regions from the alignment file were removed using Gblocks 0.91b and the resulting alignment, which contained 7688 aligned characters, was ultimately utilized for phylogenetic analysis (Castresana, 2000). A maximum likelihood (ML) tree based on 100 bootstrap replicates of this alignment was constructed using MEGA 6.0 while employing Jones-Taylor–Thornton substitution model (Jones et al., 1992; Tamura et al., 2013).
Table 1.
Genome characteristics of the sequenced members of the genus Burkholderia.
| Organism | BioProject | Size (Mb) | GC% | Chromosomes | Proteins | References |
|---|---|---|---|---|---|---|
| Burkholderia cenocepacia J2315 | PRJNA57953 | 8.06 | 66.9 | 3 | 7116 | Holden et al., 2009 |
| Burkholderia pseudomallei K96243 | PRJNA57733 | 7.25 | 68.1 | 2 | 5727 | Holden et al., 2004 |
| Burkholderia mallei ATCC 23344 | PRJNA57725 | 5.84 | 68.5 | 2 | 5022 | Nierman et al., 2004 |
| Burkholderia thailandensis E264 | PRJNA58081 | 6.72 | 67.6 | 2 | 5632 | Kim et al., 2005 |
| Burkholderia oklahomensis C6786 | PRJNA54789 | 6.99 | 67.0 | – | 6954 | NMRCb |
| Burkholderia multivorans ATCC 17616 | PRJNA58909 | 7.01 | 66.7 | 3 | 6111 | DOEd |
| Burkholderia ambifaria AMMD | PRJNA58303 | 7.53 | 66.8 | 3 | 6610 | Coenye et al., 2001b |
| Burkholderia glumae BGR1 | PRJNA59397 | 7.28 | 67.9 | 2 | 5773 | Lim et al., 2009 |
| Burkholderia xenovorans LB400 | PRJNA57823 | 9.73 | 62.6 | 3 | 8702 | Chain et al., 2006 |
| Burkholderia sp. CCGE1002 | PRJNA42523 | 7.88 | 63.3 | 3 | 6889 | Ormeno-Orrillo et al., 2012 |
| Burkholderia sp. CCGE1001 | PRJNA42975 | 6.83 | 63.6 | 2 | 5965 | DOEd |
| Burkholderia sp. CCGE1003 | PRJNA46253 | 7.04 | 63.2 | 2 | 5988 | DOEd |
| Burkholderia sp. Ch1-1 | PRJNA48975 | 8.74 | 62.4 | – | 7742 | DOEd |
| Burkholderia sp. H160 | PRJNA55101 | 7.89 | 62.9 | – | 7460 | Ormeno-Orrillo et al., 2012 |
| Burkholderia sp. 383 | PRJNA58073 | 8.68 | 66.3 | 3 | 7716 | DOEd |
| Burkholderia sprentiae WSM5005 | PRJNA66661 | 7.76 | 63.2 | – | – | DOEd |
| Burkholderia sp. YI23 | PRJNA81081 | 8.90 | 63.3 | 3 | 7804 | Lim et al., 2012 |
| Burkholderia sp. SJ98 | PRJNA160003 | 7.88 | 61.4 | – | 7268 | Kumar et al., 2012 |
| Burkholderia sp. WSM2230 | PRJNA165309 | 6.31 | 63.1 | – | – | DOEd |
| Burkholderia sp. KJ006 | PRJNA165871 | 6.63 | 67.2 | 3 | 6024 | Kwak et al., 2012 |
| Burkholderia sp. TJI49 | PRJNA179699 | 7.38 | 66.9 | – | 8940 | Khan et al., 2013 |
| Burkholderia sp. BT03 | PRJNA180532 | 10.64 | 61.9 | – | 10126 | Oak Ridgec |
| Burkholderia sp. WSM2232 | PRJNA182741 | 7.21 | 63.1 | – | – | DOEd |
| Burkholderia sp. WSM3556 | PRJNA182743 | 7.68 | 61.8 | – | – | DOEd |
| Burkholderia sp. URHA0054 | PRJNA190816 | 7.24 | 62.8 | – | – | DOEd |
| Burkholderia sp. WSM4176 | PRJNA199219 | 9.07 | 62.9 | – | 8336 | DOEd |
| Burkholderia sp. JPY251 | PRJNA199221 | 8.61 | 63.1 | – | 7873 | DOEd |
| Burkholderia sp. JPY347 | PRJNA199222 | 6.39 | 63.1 | – | 5963 | DOEd |
| Burkholderia sp. RPE64 | PRJNA205541 | 6.96 | 63.1 | 3 | 6498 | Shibata et al., 2013 |
| Burkholderia vietnamiensis G4 | PRJNA58075 | 8.39 | 65.7 | 3 | 7617 | DOEd |
| Burkholderia dolosa AUO158 | PRJNA54351 | 6.42 | 66.8 | – | 4795 | Broad Institutea |
| Burkholderia phymatum STM815 | PRJNA58699 | 8.68 | 62.3 | 2 | 7496 | Vandamme et al., 2002b |
| Burkholderia phytofirmans PsJN | PRJNA58729 | 8.21 | 62.3 | 2 | 7241 | Weilharter et al., 2011 |
| Burkholderia ubonensis Bu | PRJNA54793 | 6.93 | 67.3 | – | 7181 | NMRCb |
| Burkholderia graminis C4D1M | PRJNA54887 | 7.48 | 62.9 | – | 6747 | DOEd |
| Burkholderia rhizoxinica HKI 454 | PRJNA60487 | 3.75 | 60.7 | 1 | 3870 | Lackner et al., 2011 |
| Burkholderia gladioli BSR3 | PRJNA66301 | 9.05 | 67.4 | 2 | 7411 | Seo et al., 2011 |
| Burkholderia cepacia GG4 | PRJNA173858 | 6.47 | 66.7 | 2 | 5825 | Hong et al., 2012 |
| Candidatus Burkholderia kirkii UZHbot1 | PRJNA74017 | 4.01 | 62.9 | – | 2069 | Van Oevelen et al., 2002b |
| Burkholderia mimosarum LMG 23256 | PRJNA163559 | 8.41 | 63.9 | – | – | DOEd |
| Burkholderia terrae BS001 | PRJNA168186 | 11.29 | 61.8 | – | 10234 | Nazir et al., 2012 |
| Burkholderia pyrrocinia CH-67 | PRJNA199595 | 8.05 | 67.4 | – | 7324 | Song et al., 2012 |
| Burkholderia kururiensis M130 | PRJNA199910 | 7.13 | 65.0 | – | 6311 | Coutinho et al., 2013 |
| Burkholderia phenoliruptrix BR3459a | PRJNA176370 | 7.65 | 63.1 | 2 | 6496 | Oliveira Cunha et al., 2012 |
| Burkholderia bryophila 376MFSha3.1 | PRJNA201182 | 7.38 | 61.9 | – | 6722 | DOEd |
The Broad Institute Genome Sequencing Platform (Broad Institute).
Naval Medical Research Center/ Biological Defense Research Directorate (NMRC).
Oak Ridge National Lab (Oak Ridge).
DOE Joint Genome Institute (DOE).
A maximum likelihood 16S rRNA gene sequence consensus tree was also created for 101 sequences, which included 97 representative strains from the genus Burkholderia and four outgroup sequences from the genera Cupriadivus and Ralstonia. The sequences utilized in the study were obtained from the Ribosomal Database Project (RDP III) (Cole et al., 2009) and NCBI. All the sequences were aligned using MAAFT 7 (Katoh and Standley, 2013) and a ML tree based upon 1000 bootstrap replicates of this alignment was constructed using the General Time Reversible Model (Tavaré, 1986) in MEGA 6.0 (Tamura et al., 2013).
Identification of molecular markers (CSIs)
BLASTp searches were conducted for all proteins from chromosomes 2 and 3 (accession numbers NC_008061 and NC_008061) of Burkholderia cenocepacia J2315 (Holden et al., 2009) to identify CSIs that are shared by different members of the genus Burkholderia. Species that appeared as top hits with high scoring homologs (E values < 1e−20) from the genus Burkholderia and other outgroups were selected. Multiple sequence alignments were created using the Clustal_X 1.83 (Jeanmougin et al., 1998). These alignments were visually inspected for the presence of insertions or deletions (indels) restricted to either some or all members of the genus Burkholderia and flanked by at least 5–6 conserved amino acid residues on both sides in the neighboring 30–40 amino acids. Indel queries that were not flanked by conserved regions were not further evaluated. The species specificity of the indel queries meeting the above criterion was further evaluated by performing BLASTp searches on short sequence segments containing the insertions or deletions, and their flanking conserved regions (60–100 amino acids long). The searches were conducted against the NCBI non-redundant (nr) database and a minimum of 250 BLAST hits were examined for the presence or absence of CSIs. The results of these analyses were evaluated as described in detail in our recent work (Gupta, 2014). Signature files for the CSIs that were specific for members of the genus Burkholderia were created and formatted using the programs SIG_CREATE and SIG_STYLE (accessible from Gleans.net) as described by Gupta (2014). The sequence alignment files presented here contain information for all detected insertions or deletions from the Burkholderia group of interest, but only a limited number from species that are serving as outgroups. Sequence information for different strains of various species is not shown, but they all exhibited similar pattern. Lastly, unless otherwise indicated, the CSIs shown here are specifically found in the indicated groups and similar CSIs were not detected in the 250 Blast hits with the query sequences.
Results
Branching pattern of Burkholderia species in concatenated protein and 16S rRNA trees
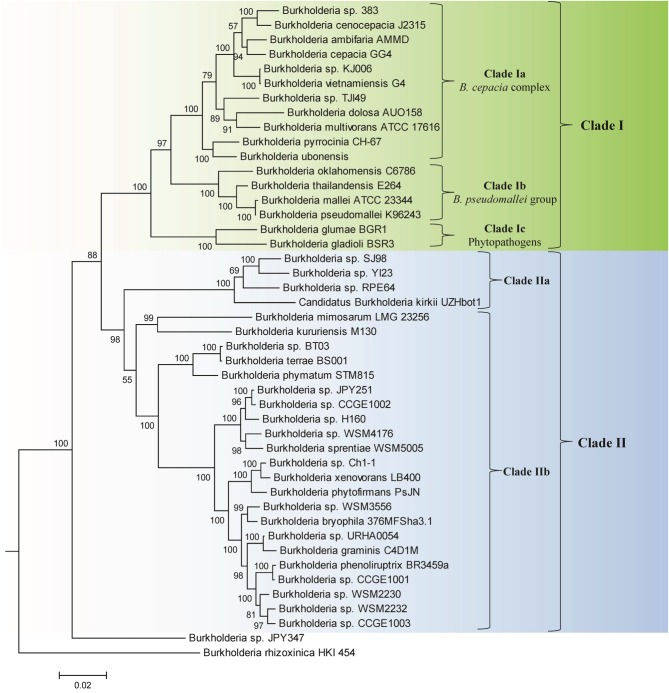
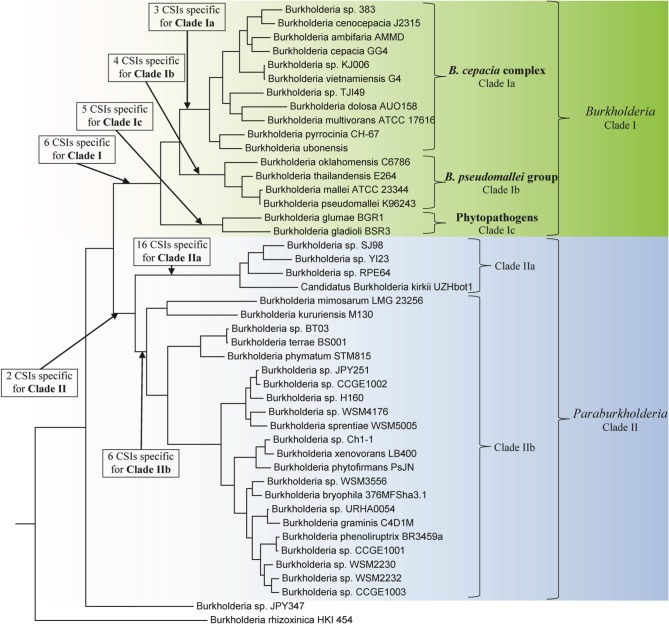
Genome sequences of 45 species of Burkholderia were available from the NCBI genome database at the time of this work (NCBI, 2014). Some characteristics of these genomes are listed in Table 1. The genome sizes of the sequenced Burkholderia species show large variation (from 3.75–11.29 Mb) and the numbers of proteins in them also varied in a similar proportion. In this work we have produced a ML phylogenetic tree based on the concatenated amino acid sequences of 21 conserved housekeeping and ribosomal proteins obtained from 45 sequenced Burkholderia species (Figure 1). The Burkholderia species formed two large clades in the protein based ML tree: One consisting of the BCC and related organisms (Clade I) and another comprised mainly of environmental or poorly characterized Burkholderia species (Clade II). Within Clade I, three smaller, distinct clades are also observed. The first of these clades (Clade Ia) is wholly comprised of the sequenced BCC species, the second clade (Clade Ib) groups B. pseudomallei and closely related species, and the third clade (Clade Ic) consists of the plant pathogenic species, B. glumae and B. gladioli. Clade II could also be divided into two smaller clades, Clade IIa and Clade IIb. Clade IIa is separated from Clade IIb by a long branch, suggesting that a large amount of genetic divergence has occurred between the two groups. In addition to the two main clades of Burkholderia, two species, Burkholderia sp. JPY347 and Burkholderia rhizoxinica, branched early in the tree and did not associate with either Clade I or II.
Figure 1.
A maximum likelihood phylogenetic tree of the genome sequenced members of the genus Burkholderia based upon concatenated sequences of 21 conserved proteins. The tree was rooted using Cupriavidus necator N-1, Bordetella pertussis Tohama I, and Neisseria meningitides MC58. Bootstrap analysis scores are indicated for each node. The major Burkholderia clades (Clades I and II) and their main sub-clades are indicated by brackets.
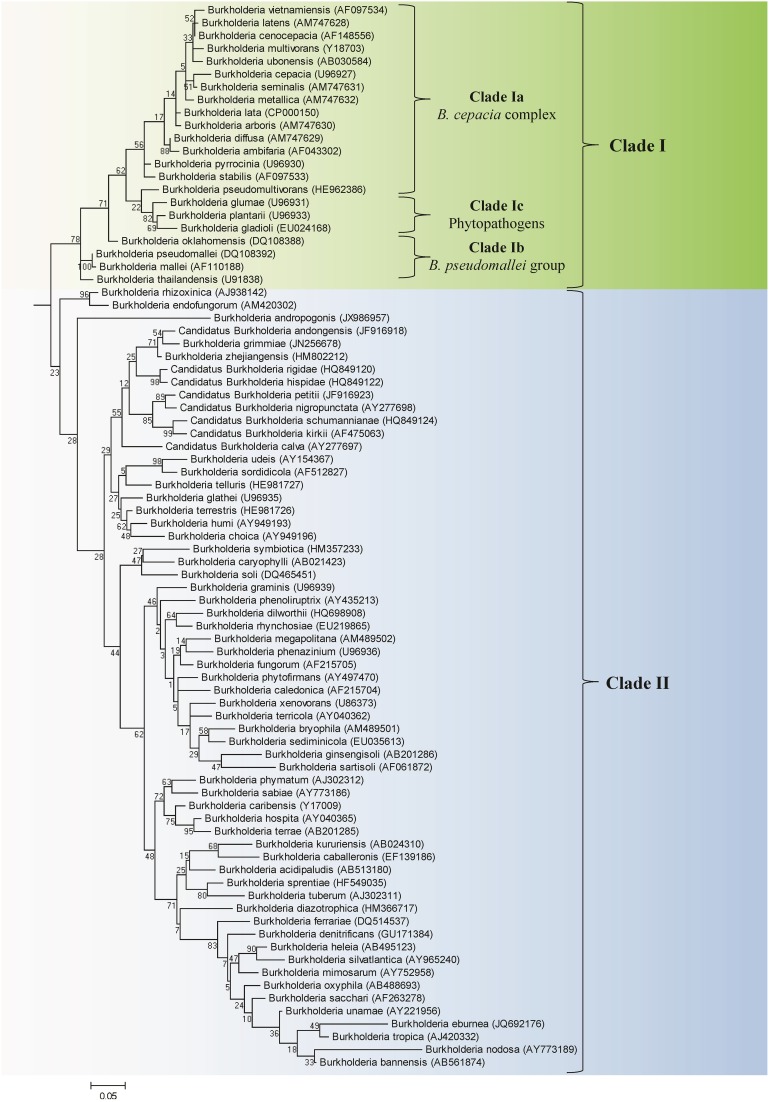
We have also constructed a 16S rRNA based ML phylogenetic tree for 97 Burkholderia strains and candidate species (Figure 2). In this 16S rRNA based phylogenetic tree we observed broadly similar patterns to our protein based phylogeny. A clade consisting of the BCC and related organisms (Clade I) was clearly resolved. The three subclades within Clade I, the BCC (Clade Ia), the B. pseudomallei group (Clade Ib), and the plant pathogenic species (Clade Ic) were well resolved, though some species exhibited aberrant branching (ex. B. oklahomensis and B. pseudomultivorans). A large assemblage of the remaining Burkholderia species, roughly corresponding to Clade II in our concatenated protein based phylogenetic tree, was also observed in the 16S rRNA tree. However, due to significant number of unsequenced Burkholderia species which are present in the 16S rRNA database it is difficult to accurately identify the groups within Clade II of the 16S rRNA tree which correspond to Clades IIa and IIb in our concatenated protein based phylogenetic tree. Bootstrap support for branches in the 16S rRNA based tree were also significantly lower than they were in the concatenated protein tree indicating that some of the observed branching patterns may not be reliable. However, the clade consisting of the BCC and related organisms (Clade I) has strong bootstrap support and has been identified in a large number of previous 16S rRNA based phylogenetic studies (Yabuuchi et al., 1992; Palleroni, 2005; Yarza et al., 2008; Suarez-Moreno et al., 2012).
Figure 2.
A maximum likelihood tree based on the 16S rRNA gene sequences of 97 members of the genus Burkholderia. Accession numbers for the 16S rRNA sequenced used for each organism are provided in the brackets following the name of the organism. The tree was rooted using four species from the genera Cupriadivus and Ralstonia. Bootstrap analysis scores are indicated for each node. The major Burkholderia clades (Clades I and II) and the subclades within Clade I are indicated by brackets.
Molecular signatures distinguishing the clade I and clade II Burkholderia
Rare genetic changes, such as insertions and deletions in essential genes/proteins, which occur in a common ancestor can be inherited by the various decedent species related to this common ancestor (Gupta, 1998; Rokas and Holland, 2000; Gogarten et al., 2002; Gupta and Griffiths, 2002). Due to the rarity and the specific presence of these rare genetic changes to a related group of organisms, they can serve as important molecular markers and provide a novel means to understand the evolutionary interrelationships between different closely related species (Gupta, 1998; Gupta and Griffiths, 2002; Gao and Gupta, 2012).
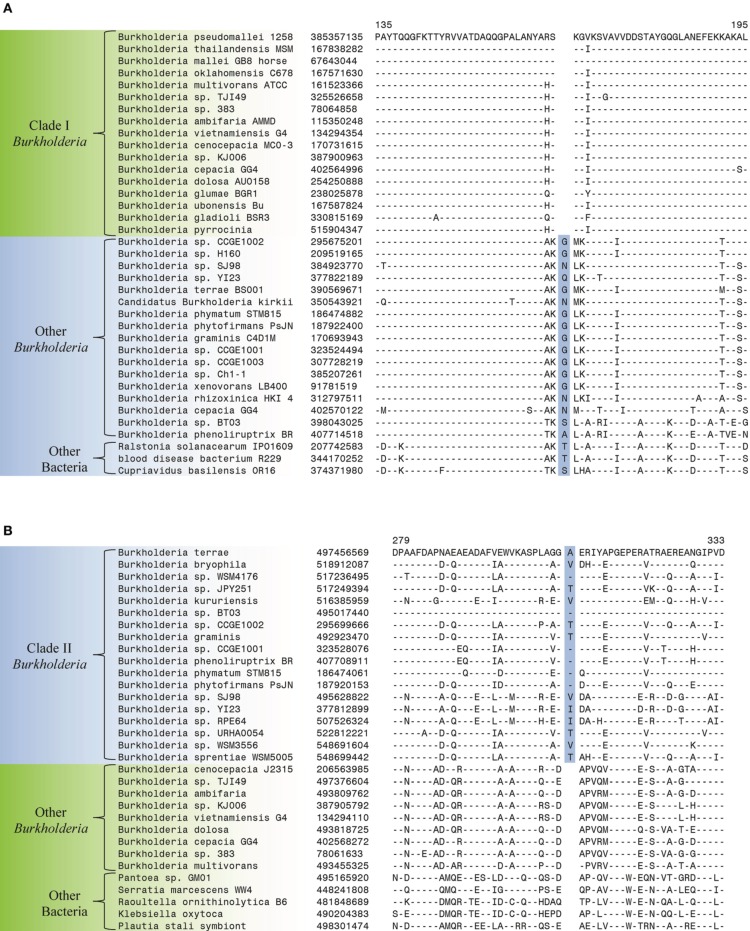
The comparative analysis of protein sequences from Burkholderia species that was carried out in the present work has identified a number of CSIs that serve to clearly distinguish a number of different clades within the genus Burkholderia. These studies have led to identification of 6 CSIs that are specific for the Clade I Burkholderia, consisting of the BCC and related organisms, enabling clear distinction of this group from all other Burkholderia. This clade, which contains all well characterized pathogens within the genus, represents the most clinically relevant group within the Burkholderia. All species within this clade are potentially pathogenic to human, animals, or plants and most have been isolated from clinical human samples (Simpson et al., 1994; Mahenthiralingam et al., 2002, 2005; Biddick et al., 2003; O'Carroll et al., 2003). One example of a CSI that is specific to the Clade I Burkholderia is shown in Figure 3A. In this case, a one amino acid deletion is present in a highly conserved region of a periplasmic amino acid-binding protein. The indel is flanked on both sides by highly conserved regions indicating that it is not the result of alignment artifacts and that it is a reliable genetic characteristic. This CSI is present in all of the sequenced members of the Clade I Burkholderia, but absent in all other bacterial homologs of this protein. Our work has identified 5 additional CSIs in other widely distributed proteins that are specific for the Clade I Burkholderia and sequence alignments for these CSIs are shown in Supplemental Figures 1–5 and a summary of their characteristics is provided in Table 2.
Figure 3.
Partial sequence alignments of (A) a periplasmic amino acid-binding protein showing a 1 amino acid deletion identified in all members of Clade I of the genus Burkholderia (B) a dehydrogenase showing a 1 amino acid insertion (boxed) identified only in members of Clade II of the genus Burkholderia. These CSIs were not found in the sequence homologs of these proteins from any other sequenced bacteria. In each case, sequence information for a Burkholderia species and a limited number other bacteria are shown, but unless otherwise indicated, similar CSIs were detected in all members of the indicated group and not detected in any other bacterial species in the top 250 BLAST hits. The dashes (–) in the alignments indicate identity with the residue in the top sequence. GenBank identification (GI) numbers for each sequence are indicated in the second column. Sequence information for other CSIs specific to the members of Clade I and Clade II of the genus Burkholderia are presented in Supplemental Figures 1–5 and Supplemental Figure 6, respectively, and their characteristics are summarized in Table 2.
Table 2.
Conserved signature indels specific for the two major clades within the genus Burkholderia.
| Protein Name | GI Number | Figures | Indel size | Indel positiona | Specificity |
|---|---|---|---|---|---|
| Periplasmic amino acid-binding protein | 385357135 | Figure 3A | 1 aa del | 135–195 | Clade I |
| Putative lyase | 167724527 | Supplemental Figure 1 | 1 aa del | 70–121 | Clade I |
| 4-hydroxybenzoate 3-monooxygenase | 238023559 | Supplemental Figure 2 | 1 aa ins | 101–171 | Clade I |
| 6-phosphogluconate dehydrogenase, decarboxylating | 330820932 | Supplemental Figure 3 | 1 aa ins | 137–202 | Clade I |
| Putative lipoprotein | 121598811 | Supplemental Figure 4 | 1 aa del | 363–393 | Clade I |
| Sarcosine oxidase subunit alpha | 493818877 | Supplemental Figure 5 | 3 aa ins | 904–965 | Clade I |
| Dehydrogenase | 497456569 | Figure 3B | 1 aa ins | 279–333 | Clade II |
| LysR family transcriptional regulator | 187919777 | Supplemental Figure 6 | 2 aa del | 260–294 | Clade II |
The region of the specified protein that contains the indel.
Two additional CSIs identified in this work are specific for the Clade II Burkholderia species which is made up of mainly environmental organisms. One of these CSIs, shown in Figure 3B, consists of a one amino acid insertion in a dehydrogenase protein that is uniquely found in members of the Clade II Burkholderia and absent in all other Burkholderia species as well all other bacterial groups. A sequence alignment for another CSI that is specific for the Clade II Burkholderia (a 2 aa deletion in a LysR family of transcription regulator protein) is shown in Supplemental Figure 6 and its characteristics are summarized in Table 2.
CSIs distinguishing different main groups within the clade I Burkholderia
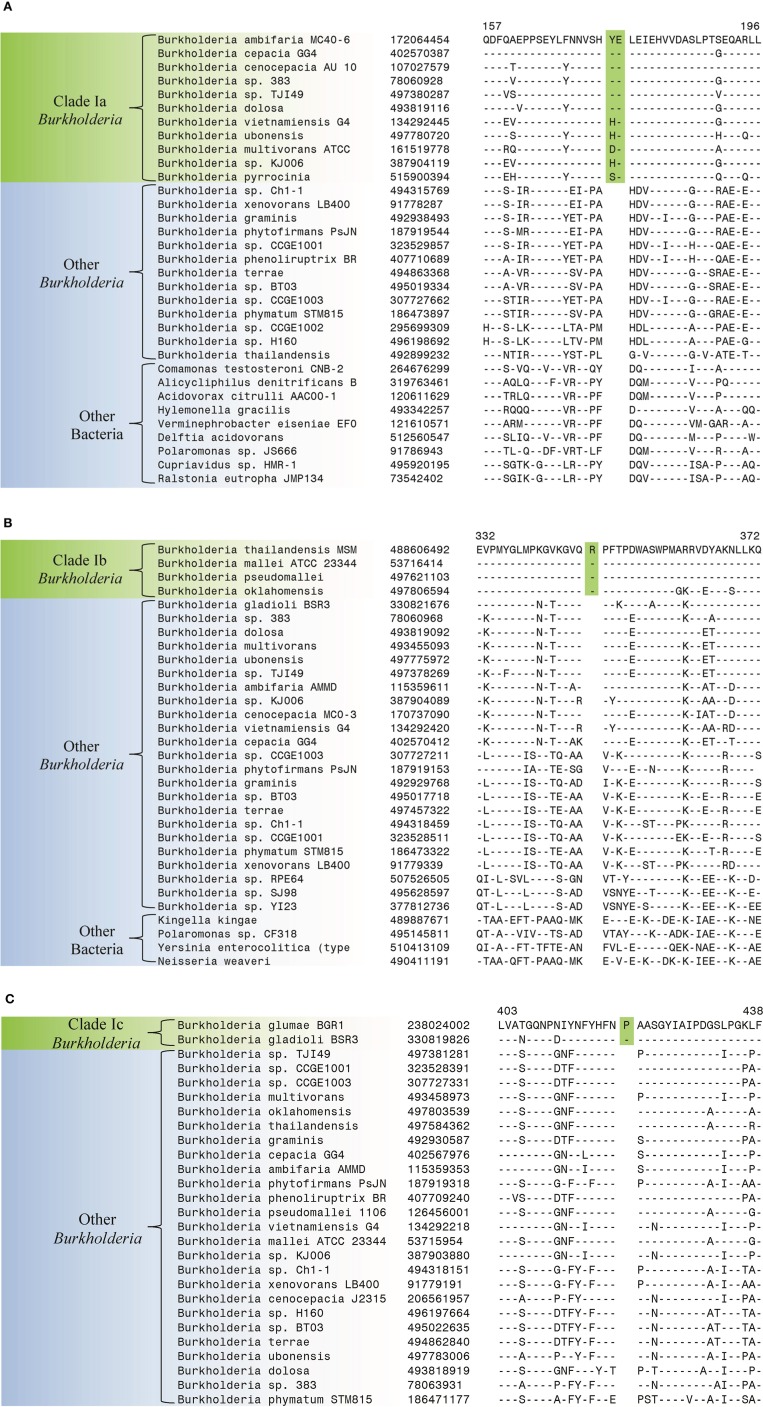
The species within Clade I of the genus Burkholderia are responsible for a range of human, animal, and plant diseases (Biddick et al., 2003; Mahenthiralingam et al., 2005). The members of Clade I (i.e., the BCC and related Burkholderia) are commonly separated into 3 main groups which correspond to clades identified in our phylogenetic trees. The first group, the members of the BCC (Clade 1a), are prevalent pathogens in cystic fibrosis patients, the second group, the B. pseudomallei group (Clade Ib), contains the causative agents of melioidosis and glanders, while the third group contains the plant pathogenic Burkholderia species (Clade Ic) (White, 2003; Mahenthiralingam et al., 2005; Whitlock et al., 2007; Nandakumar et al., 2009). Our analysis has identified 3 CSIs that are specific for all members of the BCC clade (Clade 1a). One example of a BCC clade specific CSI is shown in Figure 4A. This CSI consists of a 2 amino acid insertion in a conserved region of a histidine utilization repressor which is only found in members of the BCC. Sequence alignments for two other BCC clade specific CSIs are shown in Supplemental Figures 7, 8 and their characteristics are summarized in Table 3.
Figure 4.
Partial sequence alignments of (A) a histidine utilization repressor showing a 2 amino acid insertion (boxed) identified in all members of the Burkholderia cepacia complex (Clade Ia) within the genus Burkholderia (B) a periplasmic oligopeptide-binding protein showing a 1 amino acid insertion (boxed) identified in all members of the Burkholderia pseudomallei group (Clade Ib) within the genus Burkholderia (C) a SMP-30/gluconolaconase/LRE-like region-containing protein showing a 1 amino acid insertion (boxed) identified in all members of the phytopathogenic Burkholderia clade (Clade Ic). These CSIs were not found in the sequence homologs of these proteins from any other sequenced bacteria in the top 250 BLAST hits. Sequence information for other CSIs specific to subclades within Clade I of the genus Burkholderia are presented in Supplemental Figures 7–15 and their characteristics are summarized in Table 3.
Table 3.
Conserved signature indels specific for groups within Clades I and II.
| Protein Name | GI Number | Figures | Indel size | Indel positiona | Specificity |
|---|---|---|---|---|---|
| Histidine utilization repressor | 172064454 | Figure 4A | 2 aa ins | 157–196 | Clade Ia |
| Molybdate ABC transporter substrate-binding protein | 189352411 | Supplemental Figure 7 | 1 aa ins | 110–158 | Clade Ia |
| Acid phosphatase | 221203041 | Supplemental Figure 8 | 1 aa ins | 305–338 | Clade Ia |
| Periplasmic oligopeptide-binding protein | 488606492 | Figure 4B | 1 aa ins | 332–372 | Clade Ib |
| OpgC protein | 53716883 | Supplemental Figure 9 | 1 aa ins | 137–204 | Clade Ib |
| Polysaccharide deacetylase family protein | 167725414 | Supplemental Figure 10 | 1 aa ins | 29–63 | Clade Ib |
| Thioredoxin domain protein | 497613277 | Supplemental Figure 11 | 1 aa ins | 247–294 | Clade Ib |
| SMP-30/gluconolaconase/LRE-like region-containing protein | 238024002 | Figure 4C | 1 aa ins | 403–438 | Clade Ic |
| Cation efflux protein | 330820376 | Supplemental Figure 12 | 1 aa ins | 129–160 | Clade Ic |
| putative peptidoglycan-binding LysM/M23B peptidase | 238024763 | Supplemental Figure 13 | 1 aa ins | 155–198 | Clade Ic |
| SMP-30/gluconolaconase/LRE-like region-containing protein | 238024002 | Supplemental Figure 14 | 2 aa del | 80–130 | Clade Ic |
| hypothetical protein bgla_2g22890 | 330821370 | Supplemental Figure 15 | 1 aa ins | 322–358 | Clade Ic |
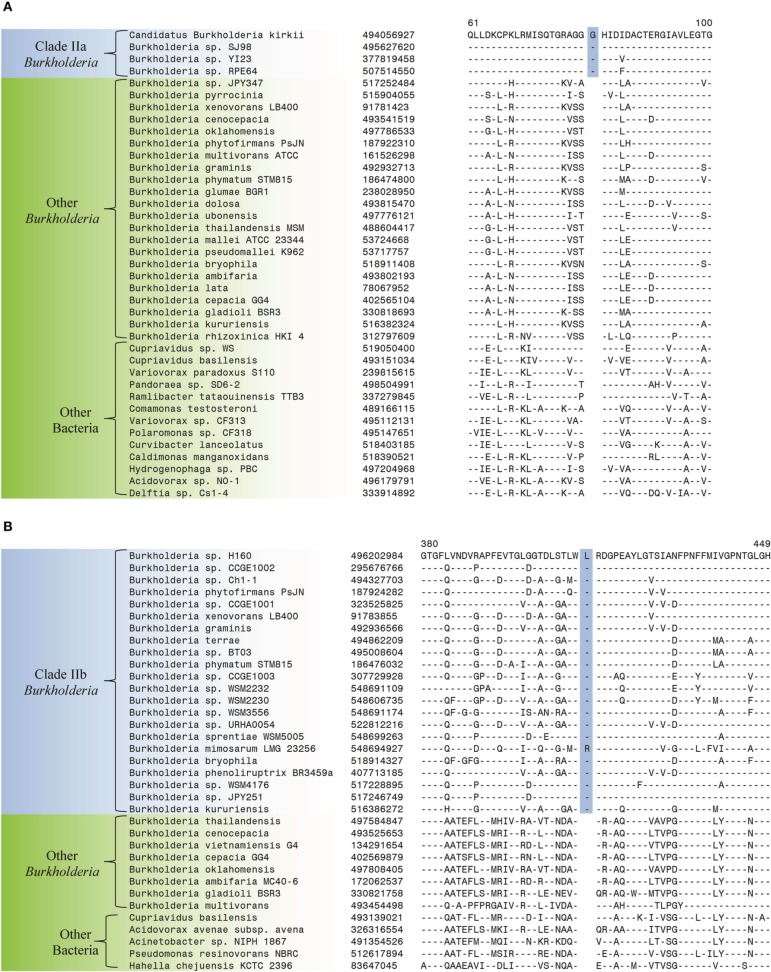
| 3-phosphoglycerate dehydrogenase | 494056927 | Figure 5A | 1 aa ins | 61–100 | Clade IIa |
| Hypothetical protein BYI23_A021470 | 377821591 | Supplemental Figure 16 | 1 aa del | 16–76 | Clade IIa |
| Prepilin peptidase | 377821714 | Supplemental Figure 17 | 1 aa ins | 179–230 | Clade IIa |
| Uracil-DNA glycosylase | 495619839 | Supplemental Figure 18 | 2 aa ins | 191–230 | Clade IIa |
| Hypothetical protein BYI23_A015260 | 377820970 | Supplemental Figure 19 | 2 aa ins | 221–270 | Clade IIa |
| Carboxylate-amine ligase | 377822128 | Supplemental Figure 20 | 1 aa del | 321–362 | Clade IIa |
| NADH:ubiquinone oxidoreductase subunit M | 494056355 | Supplemental Figure 21 | 3 aa ins | 303–348 | Clade IIa |
| NADH:ubiquinone oxidoreductase subunit L | 494056354 | Supplemental Figure 22 | 1 aa ins | 538–585 | Clade IIa |
| ABC transporter | 377821271 | Supplemental Figure 23 | 1 aa del | 59–99 | Clade IIa |
| Hypothetical protein BYI23_A002220 | 377819666 | Supplemental Figure 24 | 2 aa ins | 133–172 | Clade IIa |
| 16S rRNA-processing protein RimM | 494056031 | Supplemental Figure 25 | 1 aa ins | 147–201 | Clade IIa |
| FAD linked oxidase domain-containing protein | 377819737 | Supplemental Figure 26 | 1 aa ins | 106–144 | Clade IIa |
| Preprotein translocase subunit SecD | 495626933 | Supplemental Figure 27 | 1 aa del | 306–341 | Clade IIa |
| Mechanosensitive ion channel protein MscS | 494057445 | Supplemental Figure 28 | 3 aa ins | 101–143 | Clade IIa |
| Hypothetical protein BYI23_A006130 | 377820057 | Supplemental Figure 29 | 1 aa ins | 199–253 | Clade IIa |
| Uroporphyrinogen-III synthase | 494056428 | Supplemental Figure 30 | 7 aa ins | 37–79 | Clade IIa |
| 4-hydroxyacetophenone monooxygenase | 496202984 | Figure 5B | 1 aa ins | 380–449 | Clade IIb |
| Transposase A-like protein | 187923943 | Supplemental Figure 31 | 1 aa ins | 5–50 | Clade IIb |
| Group 1 glycosyl transferase | 186475830 | Supplemental Figure 32 | 1 aa ins | 153–194 | Clade IIb |
| 4-hydroxyacetophenone monooxygenase | 496202984 | Supplemental Figure 33 | 3 aa ins | 145–219 | Clade IIb |
| Undecaprenyl-phosphate glucose phosphotransferase | 209521823 | Supplemental Figure 34 | 1 aa ins | 208–275 | Clade IIb |
| putative flavin-binding monooxygenase-like protein | 186476032 | Supplemental Figure 35 | 3 aa ins | 102–148 | Clade IIb |
The region of the specified protein that contains the indel.
Our work has also identified 4 CSIs that are specific for the B. pseudomallei group (Clade Ib) which contains the most prevalent human pathogen within the genus, B. pseudomallei (Wiersinga et al., 2006). One example of a CSI specific to the B. pseudomallei group, which consists of a 1 amino acid insertion in a conserved region of a periplasmic oligopeptide-binding protein, is shown in Figure 4B. Sequence alignments for three other CSIs in three different proteins that are specific for the B. pseudomallei group are shown in Supplemental Figures 9–11 and their characteristics are summarized in Table 3.
We have also identified 5 CSIs that are specific for the major plant pathogenic group within the genus Burkholderia (Clade 1c) which contains the species B. glumae and B. gladioli. An example of a CSI representing this group is shown in Figure 4C. This CSI consists of a 1 amino acid insertion in a conserved region of a SMP-30/gluconolaconase/LRE-like region-containing protein that is found in the members of Clade 1c of the genus Burkholderia but absent in all other Burkholderia and all other bacterial groups. Sequence alignments for the other 4 CSIs are shown in Supplemental Figures 12–15 and their key features are highlighted in Table 3.
CSIs that are specific for two groups within the clade II Burkholderia
The species within Clade II of the genus Burkholderia inhabit a variety of environmental niches, but there is little evidence of their colonization of healthy or immunocompromised human patients (Coenye and Vandamme, 2003). The branching of different groups within Clade II is not well resolved in 16S rRNA trees and there is currently a lack of sequence data that can be used to generate trees based on concatenated gene sets that reliably resolve the interrelationships of the clade while sufficiently reflecting the total diversity of species within the clade (Figures 1, 2) (Cole et al., 2009; NCBI, 2014). Despite the limited sequence data, we have been able to identify two robust groups within Clade II that are supported by a number of CSIs. The first Clade, Clade IIa, primarily consists of unclassified members of the genus and candidatus Burkholderia species (Figure 1). Clade IIa is supported by 16 CSIs identified in this work. One example of a CSI specific for Clade IIa, consisting of a 1 amino acid insertion in 3-phosphoglycerate dehydrogenase, is shown in Figure 5A. This insertion is present in a highly conserved region of this protein in all sequenced members of Clade IIa and absent in all other Burkholderia and all other bacterial groups. Sequence alignments for the other 15 CSIs that are specific for Clade IIa Burkholderia spp. are shown in Supplemental Figures 16–30 and their characteristics are summarized in Table 3.
Figure 5.
Partial sequence alignments of (A) 3-phosphoglycerate dehydrogenase showing a 1 amino acid insertion (boxed) identified in all members of Clade IIa of the genus Burkholderia (B) 4-hydroxyacetophenone monooxygenase showing a 1 amino acid insertion (boxed) identified only in members of Clade IIb of the genus Burkholderia. These CSIs were not found in the sequence homologs of these proteins from any other sequenced bacteria in the top 250 BLAST hits. Sequence information for other CSIs specific to subclades within Clade II of the genus Burkholderia are presented in Supplemental Figures 16–35 and their characteristics are summarized in Table 3.
The second group within Clade II of the Burkholderia (Clade IIb), is comprised of a large variety of environmental Burkholderia species (Coenye and Vandamme, 2003; Suarez-Moreno et al., 2012). Our analysis has identified 6 CSIs that are specific to this large group of Burkholderia species. One example of a CSI specific to the members of Clade IIb of the genus Burkholderia is shown in Figure 5B. The CSI consists of a one amino acid insertion in 4-hydroxyacetophenone monooxygenase, which is only present in members of Clade IIb of the genus Burkholderia and not in protein homologs from any other sequenced bacterial group. Information for other 5 CSIs which are specific to members of Clade IIb of the genus Burkholderia are shown in Supplemental Figures 31–35 and their characteristics are summarized in Table 3.
Discussion
The genus Burkholderia is one of the largest groups of species within the class Betaproteobacteria (Palleroni, 2005; Parte, 2013). The genus contains a variety of bacteria that inhabit a wide range of ecological niches including a number of bacteria that have pathogenic potential (Yabuuchi et al., 1992; Coenye and Vandamme, 2003; Mahenthiralingam et al., 2005; Palleroni, 2005; Compant et al., 2008). The phylogeny of the genus Burkholderia has been studied using a wide array of methodologies based on phenotypic, biochemical, genetic, and genomic characteristics (Stead, 1992; Gillis et al., 1995; Payne et al., 2005; Tayeb et al., 2008; Onofre-Lemus et al., 2009; Spilker et al., 2009; Ussery et al., 2009; Gyaneshwar et al., 2011; Vandamme and Dawyndt, 2011; Zhu et al., 2011; Estrada-de los Santos et al., 2013). These studies have provided novel insights into the evolutionary relationship of the species within the genus Burkholderia. However, no taxonomic changes have been made to date due to a lack of discrete, distinguishing characteristics identified for the different phylogenetic lineages within the genus (Estrada-de los Santos et al., 2013).
In the present work, we have outlined two major groups of species within the genus Burkholderia: Clade I, which contains all pathogenic members of the genus, and Clade II, which contains a large variety of environmental species. These two groups were found to branch distinctly in a highly resolved phylogenetic tree based on a large number of concatenated protein sequences produced in this work (Figure 1). Evidence for the distinctness of Clade I organisms from other Burkholderia species has been observed in a wide range of previous phylogenetic studies (Payne et al., 2005; Tayeb et al., 2008; Yarza et al., 2008; Spilker et al., 2009; Ussery et al., 2009; Gyaneshwar et al., 2011; Vandamme and Dawyndt, 2011; Zhu et al., 2011; Suarez-Moreno et al., 2012; Estrada-de los Santos et al., 2013; Segata et al., 2013). Importantly, we have also identified 6 and 2 CSIs that serve as discrete molecular characteristics of Clade I and Clade II, respectively (Figure 6 and Table 2). These CSIs are the first discrete features that have been identified that are unique to either Clade I or Clade II of the genus Burkholderia. These CSIs act as independent verification of the phylogenetic trends identified in this and other studies and provide clear evidence that the species from the Clade I are distinct from all other Burkholderia and that they are derived from a common ancestor exclusive of all other Burkholderia. Although sequence information for Clade II members is at present somewhat limited, based upon the shared presence of two CSIs by them, it is likely that they are also derived from a common ancestor exclusive of other bacteria.
Figure 6.
A summary diagram depicting the distribution of identified CSIs and the proposed names of the two major groups (Clade I and II) within Burkholderia. The major Burkholderia clades are indicated by brackets and highlighting.
Additionally, we have identified molecular evidence, in the form of large numbers of CSIs, which support the distinctiveness of several smaller groups within the genus Burkholderia. The most important of these groups, the B. cepacia complex (BCC; Clade Ia) and the B. pseudomallei group (Clade Ib), are supported by the 3 and 4 of the identified CSIs, respectively. The BCC are a group of opportunistic pathogens which colonize immunodificient human hosts and are among the most prevalent and lethal infections in cystic fibrosis patients (Mahenthiralingam et al., 2002, 2005; Biddick et al., 2003; Hauser et al., 2011). The 17 species that make up the BCC are closely related and form a tight monophyletic cluster within the genus Burkholderia (Vandamme and Dawyndt, 2011). The B. pseudomallei group consists of 4 closely related species: B. pseudomallei, the causative agent of the highly lethal septicemia melioidosis (White, 2003; Limmathurotsakul and Peacock, 2011), B. mallei, the causative agent of the equine disease glanders and occasional human infections (Whitlock et al., 2007), and the largely non-pathogenic organisms, Burkholderia thailandensis and Burkholderia oklahomensis (Deshazer, 2007). The identified CSIs are highly specific characteristics of these two important pathogenic groups and they provide novel and useful targets for the development of diagnostic assays for either the BCC or the B. pseudomallei group (Ahmod et al., 2011; Wong et al., 2014). We have identified CSIs for three other groups within the genus Burkholderia: A group of plant pathogenic Burkholderia related to the BCC and B. pseudomallei group (Clade Ic), a group containing unnamed and candidate Burkholderia species (Clade IIa), and a group consisting of environmental Burkholderia (Clade IIb). We have identified 6, 16, and 6 CSIs for these three groups, respectively. These CSIs provide important differentiating characteristics for these groups, particularly for Clades IIa and IIb which are related groups that have no other identified differentiating characteristics (Suarez-Moreno et al., 2012).
The phylogenetic analyses, identified CSIs, and the pathogenic characteristics of the different Burkholderia species presented in this work strongly suggest that the genus Burkholderia is made up of at least two distinct lineages. One lineage consisting of the BCC and related organisms (Clade I) and another consisting of a wide range of environmental organisms (Clade II). This latter clade is phylogenetically highly diverse and there is a paucity of sequence information available for its members. Thus, it is possible that in future this latter clade may be found to consist of more than one distinct bacterial lineage, however, it is currently clear that Clade I and Clade II represent distinct lineages. Evidence for the distinctness of the Clade I members from other Burkholderia species has been identified in a number of previous phylogenetic studies (Payne et al., 2005; Tayeb et al., 2008; Yarza et al., 2008; Spilker et al., 2009; Ussery et al., 2009; Gyaneshwar et al., 2011; Vandamme and Dawyndt, 2011; Zhu et al., 2011; Suarez-Moreno et al., 2012; Estrada-de los Santos et al., 2013; Segata et al., 2013). Estrada-de los Santos et al. (2013) recently completed a phylogenetic analysis of the genus Burkholderia utilizing the multilocus sequence analysis of atpD, gltB, lepA, and recA genes in combination with the 16S rRNA gene, which provides compelling evidence for the presence of two distinct evolutionary lineages within the genus Burkholderia. However, these authors have refrained from formally proposing a division of the genus into two genera due to a paucity of differentiating characteristics for the two groups. Our comparative analysis of Burkholderia genomes has identified a set of distinctive molecular characteristics that clearly differentiate the two evolutionary lineages within the genus Burkholderia in addition the phylogenetic evidence. In light of the abundance of phylogenetic and molecular evidence for the presence of two distinct evolutionary lineages within the genus Burkholderia, and the distinct pathogenicity profiles of the members of these two groups, we are proposing that genus Burkholderia should be divided into two separate genera. The first of these monophyletic genera, which comprises of all the clinically relevant species and clearly distinguished from all other Burkholderia species, will retain the name Burkholderia (Clade I). For the remainder of the Burkholderia species (Clade II), which include a wide range of environmental species, we propose the name Paraburkholderia gen. nov. An emended description of the genus Burkholderia and a description of Paraburkholderia gen. nov. are provided below. Brief descriptions of the new species combinations within Paraburkholderia gen. nov. are presented in Table 4.
Table 4.
Descriptions of the new combinations in the genus Paraburkholderia gen. nov.
| New Combination | Basonym | Type Strain | References |
|---|---|---|---|
| Paraburkholderia acidipaludis comb. nov. | Burkholderia acidipaludis | SA33 | Aizawa et al., 2010b |
| NBRC 101816 | |||
| VTCC-D6-6 | |||
| Candidatus Paraburkholderia andongensis comb. nov. | Candidatus Burkholderia andongensis | — | Lemaire et al., 2011 |
| Paraburkholderia andropogonis comb. nov. | Burkholderia andropogonis | ATCC 23061 | Gillis et al., 1995 |
| CCUG 32772 | |||
| CFBP 2421 | |||
| CIP 105771 | |||
| DSM 9511 | |||
| ICMP 2807 | |||
| JCM 10487 | |||
| LMG 2129 | |||
| NCPPB 934 | |||
| NRRL B-14296 | |||
| Paraburkholderia aspalathi comb. nov. | Burkholderia aspalathi | VG1C | Mavengere et al., 2014 |
| DSM 27239 | |||
| LMG 27731 | |||
| Paraburkholderia bannensis comb. nov. | Burkholderia bannensis | E25 | Aizawa et al., 2011 |
| BCC 36998 | |||
| NBRC 103871 | |||
| Paraburkholderia bryophila comb. nov. | Burkholderia bryophila | 1S18 | Vandamme et al., 2007 |
| CCUG 52993 | |||
| LMG 23644 | |||
| Paraburkholderia caballeronis comb. nov. | Burkholderia caballeronis | TNe-841 | Martínez-Aguilar et al., 2013 |
| CIP 110324 | |||
| LMG 26416 | |||
| Paraburkholderia caledonica comb. nov. | Burkholderia caledonica | W50D | Coenye et al., 2001a |
| CCUG 42236 | |||
| CIP 107098 | |||
| JCM 21561 | |||
| LMG 19076 | |||
| NBRC 102488 | |||
| Candidatus Paraburkholderia calva comb. nov. | Candidatus Burkholderia calva | — | Van Oevelen et al., 2004 |
| Paraburkholderia caribensis comb. nov. | Burkholderia caribensis | MWAP64 | Achouak et al., 1999 |
| CCUG 42847 | |||
| CIP 106784 | |||
| DSM 13236 | |||
| LMG 18531 | |||
| Paraburkholderia caryophylli comb. nov. | Burkholderia caryophylli | ATCC 25418 | Yabuuchi et al., 1992 |
| CCUG 20834 | |||
| CFBP 2429 | |||
| CFBP 3818 | |||
| CIP 105770 | |||
| DSM 50341 | |||
| HAMBI 2159 | |||
| ICMP 512 | |||
| JCM 9310 | |||
| JCM 10488 | |||
| LMG 2155 | |||
| NCPPB 2151 | |||
| Paraburkholderia choica comb. nov. | Burkholderia choica | LMG 22940 | Vandamme et al., 2013 |
| CCUG 63063 | |||
| Paraburkholderia denitrificans comb. nov. | Burkholderia denitrificans | KIS30-44 | Lee et al., 2012 |
| DSM 24336 | |||
| KACC 12733 | |||
| Paraburkholderia diazotrophica comb. nov. | Burkholderia diazotrophica | JPY461 | Sheu et al., 2013 |
| NKMU-JPY461 | |||
| BCRC 80259 | |||
| KCTC 23308 | |||
| LMG 26031 | |||
| Paraburkholderia dilworthii comb. nov. | Burkholderia dilworthii | WSM3556 | De Meyer et al., 2014 |
| LMG 27173 | |||
| HAMBI 3353 | |||
| Paraburkholderia eburne comb. nov. | Burkholderia eburne | RR11 | Kang et al., 2014 |
| KEMC 7302-065 | |||
| JCM 18070 | |||
| Paraburkholderia endofungorum comb. nov. | Burkholderia endofungorum | HKI 456 | Partida-Martinez et al., 2007 |
| CIP 109454 | |||
| DSM 19003 | |||
| Paraburkholderia ferrariae comb. nov. | Burkholderia ferrariae | FeGl01 | Valverde et al., 2006 |
| CECT 7171 | |||
| DSM 18251 | |||
| LMG 23612 | |||
| Paraburkholderia fungorum comb. nov. | Burkholderia fungorum | Croize P763-2 | Coenye et al., 2001a |
| CCUG 31961 | |||
| CIP 107096 | |||
| JCM 21562 | |||
| LMG 16225 | |||
| NBRC 102489 | |||
| Paraburkholderia ginsengisoli comb. nov. | Burkholderia ginsengisoli | KMY03 | Kim et al., 2006 |
| KCTC 12389 | |||
| NBRC 100965 | |||
| Paraburkholderia glathei comb. nov. | Burkholderia glathei | ATCC 29195 | Vandamme et al., 1997 |
| CFBP 4791 | |||
| CIP 105421 | |||
| DSM 50014 | |||
| JCM 10563 | |||
| LMG 14190 | |||
| Paraburkholderia graminis comb. nov. | Burkholderia graminis | C4D1M | Viallard et al., 1998 |
| ATCC 700544 | |||
| CCUG 42231 | |||
| CIP 106649 | |||
| LMG 18924 | |||
| Paraburkholderia grimmiae comb. nov. | Burkholderia grimmiae | R27 | Tian et al., 2013 |
| CGMCC 1.11013 | |||
| DSM 25160 | |||
| Paraburkholderia heleia comb. nov. | Burkholderia heleia | SA41 | Aizawa et al., 2010a |
| NBRC 101817 | |||
| VTCC-D6-7 | |||
| Candidatus Paraburkholderia hispidae comb. nov. | Candidatus Burkholderia hispidae | — | Lemaire et al., 2012 |
| Paraburkholderia hospita comb. nov. | Burkholderia hospita | LMG 20598 | Goris et al., 2002 |
| CCUG 43658 | |||
| Paraburkholderia humi comb. nov. | Burkholderia humi | LMG 22934 | Vandamme et al., 2013 |
| CCUG 63059 | |||
| Candidatus Paraburkholderia kirkii comb. nov. | Candidatus Burkholderia kirkii | — | Van Oevelen et al., 2002a |
| Paraburkholderia kururiensis comb. nov. | Burkholderia kururiensis | KP23 | Zhang et al., 2000 |
| ATCC 700977 | |||
| CCUG 43663 | |||
| CIP 106643 | |||
| DSM 13646 | |||
| JCM 10599 | |||
| LMG 19447 | |||
| Paraburkholderia megapolitana comb. nov. | Burkholderia megapolitana | A3 | Vandamme et al., 2007 |
| CCUG 53006 | |||
| LMG 23650 | |||
| Paraburkholderia mimosarum comb. nov. | Burkholderia mimosarum | PAS44 | Chen et al., 2006 |
| BCRC 17516 | |||
| LMG 23256 | |||
| Candidatus Paraburkholderia nigropunctata comb. nov. | Candidatus Burkholderia nigropunctata | — | Van Oevelen et al., 2004 |
| Paraburkholderia nodosa comb. nov. | Burkholderia nodosa | Br3437 | Chen et al., 2007 |
| BCRC 17575 | |||
| LMG 23741 | |||
| Paraburkholderia oxyphila comb. nov. | Burkholderia oxyphila | OX-01 | Otsuka et al., 2011 |
| DSM 22550 | |||
| NBRC 105797 | |||
| Candidatus Paraurkholderia petitii comb. nov. | Candidatus Burkholderia petitii | — | Lemaire et al., 2011 |
| Paraburkholderia phenazinium comb. nov. | Burkholderia phenazinium | ATCC 33666 | Viallard et al., 1998 |
| CCUG 20836 | |||
| CFBP 4793 | |||
| CIP 106502 | |||
| DSM 10684 | |||
| JCM 10564 | |||
| LMG 2247 | |||
| NCIMB 11027 | |||
| Paraburkholderia phenoliruptrix comb. nov. | Burkholderia phenoliruptrix | AC1100 | Coenye et al., 2004 |
| CCUG 48558 | |||
| LMG 22037 | |||
| Paraburkholderia phymatum comb. nov. | Burkholderia phymatum | STM815 | Vandamme et al., 2002a |
| LMG 21445 | |||
| CCUG 47179 | |||
| Paraburkholderia phytofirmans comb. nov. | Burkholderia phytofirmans | PsJN | Sessitsch et al., 2005 |
| CCUG 49060 | |||
| LMG 22146 | |||
| Paraburkholderia rhizoxinica comb. nov. | Burkholderia rhizoxinica | HKI 454 | Partida-Martinez et al., 2007 |
| CIP 109453 | |||
| DSM 19002 | |||
| Paraburkholderia rhynchosiae comb. nov. | Burkholderia rhynchosiae | WSM3937 | De Meyer et al., 2013b |
| LMG 27174 | |||
| HAMBI 3354 | |||
| Candidatus Paraburkholderia rigidae comb. nov. | Candidatus Burkholderia rigidae | — | Lemaire et al., 2012 |
| Paraburkholderia sabiae comb. nov. | Burkholderia sabiae | Br3407 | Chen et al., 2008 |
| BCRC 17587 | |||
| LMG 24235 | |||
| Paraburkholderia sacchari comb. nov. | Burkholderia sacchari | CCT 6771 | Brämer et al., 2001 |
| CCUG 46043 | |||
| CIP 107211 | |||
| IPT 101 | |||
| LMG 19450 | |||
| Paraburkholderia sartisoli comb. nov. | Burkholderia sartisoli | RP007 | Vanlaere et al., 2008 |
| CCUG 53604 | |||
| ICMP 13529 | |||
| LMG 24000 | |||
| Candidatus Paraburkholderia schumannianae comb. nov. | Candidatus Burkholderia schumannianae | — | Lemaire et al., 2012 |
| Paraburkholderia sediminicola comb. nov. | Burkholderia sediminicola | HU2-65W | Lim et al., 2008 |
| KCTC 22086 | |||
| LMG 24238 | |||
| Paraburkholderia silvatlantica comb. nov. | Burkholderia silvatlantica | SRMrh-20 | Perin et al., 2006 |
| ATCC BAA-1244 | |||
| LMG 23149 | |||
| Paraburkholderia soli comb. nov. | Burkholderia soli | GP25-8 | Yoo et al., 2007 |
| DSM 18235 | |||
| KACC 11589 | |||
| Paraburkholderia sordidicola comb. nov. | Burkholderia sordidicola | CCUG 49583 | Lim et al., 2003 |
| JCM 11778 | |||
| KCTC 12081 | |||
| Paraburkholderia sprentiae comb. nov. | Burkholderia sprentiae | WSM5005 | De Meyer et al., 2013a |
| LMG 27175 | |||
| HAMBI 3357 | |||
| Paraburkholderia symbiotica comb. nov. | Burkholderia symbiotica | JPY-345 | Sheu et al., 2012 |
| NKMU-JPY-345 | |||
| BCRC 80258 | |||
| KCTC 23309 | |||
| LMG 26032 | |||
| Paraburkholderia telluris comb. nov. | Burkholderia telluris | LMG 22936 | Vandamme et al., 2013 |
| CCUG 63060 | |||
| Paraburkholderia terrae comb. nov. | Burkholderia terrae | KMY02 | Yang et al., 2006 |
| KCTC 12388 | |||
| NBRC 100964 | |||
| Paraburkholderia terrestris comb. nov. | Burkholderia terrestris | LMG 22937 | Vandamme et al., 2013 |
| CCUG 63062 | |||
| Paraburkholderia terricola comb. nov. | Burkholderia terricola | CCUG 44527 | Goris et al., 2002 |
| LMG 20594 | |||
| Paraburkholderia tropica comb. nov. | Burkholderia tropica | Ppe8 | Reis et al., 2004 |
| ATCC BAA-831 | |||
| DSM 15359 | |||
| LMG 22274 | |||
| Paraburkholderia tuberum comb. nov. | Burkholderia tuberum | STM678 | Vandamme et al., 2002a |
| CCUG 47178 | |||
| LMG 21444 | |||
| Paraburkholderia udeis comb. nov. | Burkholderia udeis | LMG 27134 | Vandamme et al., 2013 |
| CCUG 63061 | |||
| Paraburkholderia unamae comb. nov. | Burkholderia unamae | MTl-641 | Caballero-Mellado et al., 2004 |
| ATCC BAA-744 | |||
| CIP 107921 | |||
| Paraburkholderia xenovorans comb. nov. | Burkholderia xenovorans | LB400 | Goris et al., 2004 |
| CCUG 46959 | |||
| LMG 21463 | |||
| NRRL B-18064 | |||
| Paraburkholderia zhejiangensis comb. nov. | Burkholderia zhejiangensis | OP-1 | Lu et al., 2012 |
| KCTC 23300 |
Emended description of the genus Burkholderia (Yabuuchi et al., 1993 emend. Gillis et al., 1995)
The genus contains the type species B. cepacia (Yabuuchi et al., 1993). The species from this genus are gram-negative, straight or slightly curved rods, which exhibit motility mediated by one or more polar flagella. Only, B. mallei lacks flagella and is non-motile. The species do not produce sheaths or prosthecae and do not go through any resting stages. Most species are able to accumulate and utilize poly-β-hydroxybutyrate (PHB) for growth. The species are mostly aerobic chemoorganotrophs, but some species are capable of anaerobic respiration using nitrate as the terminal electron acceptor. The G+C content for the members of the genus ranges from 65.7 to 68.5%. The members of the genus form a distinct monophyletic clade in phylogenetic trees, and they are distinguished from all other bacteria by the conserved sequence indels reported in this work in the following proteins: Periplasmic amino acid-binding protein, 4-hydroxybenzoate 3-monooxygenase, 6-phosphogluconate dehydrogenase, Sarcosine oxidase subunit alpha, a putative lipoprotein, and a putative lyase (Table 2).
Description of the genus paraburkholderia gen. nov.
The genus contains the type species Paraburkholderia graminis comb. nov. (Basonym: Burkholderia graminis, Viallard et al., 1998) The species from this genus are gram-negative straight or slightly curved rods with one or more polar flagella. Other morphological and metabolic characteristics are similar to genus Burkholderia. The G+C content for the members of the genus ranges from 61.4 to 65.0%. The species are not associated with humans. The members of this genus generally form a distinct clade in the neighborhood of genus Burkholderia in phylogenetic trees, and they lack the molecular signatures which are specific for Burkholderia. Most of the sequenced members from this genus contain the conserved sequence indels reported in this work in the protein sequences of an unnamed dehydrogenase and a LysR family transcriptional regulator (Table 2).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fgene.2014.00429/abstract
References
- Achouak W., Christen R., Barakat M., Martel M.-H., Heulin T. (1999). Burkholderia caribensis sp. nov., an exopolysaccharide-producing bacterium isolated from vertisol microaggregates in Martinique. Int. J. Syst. Bacteriol. 49, 787–794. 10.1099/00207713-49-2-787 [DOI] [PubMed] [Google Scholar]
- Ahmod N. Z., Gupta R. S., Shah H. N. (2011). Identification of a Bacillus anthracis specific indel in the yeaC gene and development of a rapid pyrosequencing assay for distinguishing B. anthracis from the B. cereus group. J. Microbiol. Methods 87, 278–285. 10.1016/j.mimet.2011.08.015 [DOI] [PubMed] [Google Scholar]
- Aizawa T., Ve N. B., Nakajima M., Sunairi M. (2010a). Burkholderia heleia sp. nov., a nitrogen-fixing bacterium isolated from an aquatic plant, Eleocharis dulcis, that grows in highly acidic swamps in actual acid sulfate soil areas of Vietnam. Int. J. Syst. Evol. Microbiol. 60, 1152–1157. 10.1099/ijs.0.015198-0 [DOI] [PubMed] [Google Scholar]
- Aizawa T., Ve N. B., Vijarnsorn P., Nakajima M., Sunairi M. (2010b). Burkholderia acidipaludis sp. nov., aluminium-tolerant bacteria isolated from Chinese water chestnut (Eleocharis dulcis) growing in highly acidic swamps in South-East Asia. Int. J. Syst. Evol. Microbiol. 60, 2036–2041. 10.1099/ijs.0.018283-0 [DOI] [PubMed] [Google Scholar]
- Aizawa T., Vijarnsorn P., Nakajima M., Sunairi M. (2011). Burkholderia bannensis sp. nov., an acid-neutralizing bacterium isolated from torpedo grass (Panicum repens) growing in highly acidic swamps. Int. J. Syst. Evol. Microbiol. 61, 1645–1650. 10.1099/ijs.0.026278-0 [DOI] [PubMed] [Google Scholar]
- Biddick R., Spilker T., Martin A., LiPuma J. J. (2003). Evidence of transmission of Burkholderia cepacia, Burkholderia multivorans and Burkholderia dolosa among persons with cystic fibrosis. FEMS Microbiol. Lett. 228, 57–62. 10.1016/S0378-1097(03)00724-9 [DOI] [PubMed] [Google Scholar]
- Brämer C. O., Vandamme P., da Silva L. F., Gomez J., Steinbüchel A. (2001). Polyhydroxyalkanoate-accumulating bacterium isolated from soil of a sugar-cane plantation in Brazil. Int. J. Syst. Evol. Microbiol. 51, 1709–1713. 10.1099/00207713-51-5-1709 [DOI] [PubMed] [Google Scholar]
- Caballero-Mellado J., Martínez-Aguilar L., Paredes-Valdez G., Estrada-de los Santos P. (2004). Burkholderia unamae sp. nov., an N2-fixing rhizospheric and endophytic species. Int. J. Syst. Evol. Microbiol. 54, 1165–1172. 10.1099/ijs.0.02951-0 [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Chain P. S., Denef V. J., Konstantinidis K. T., Vergez L. M., Agullo L., Reyes V. L., et al. (2006). Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. U.S.A. 103, 15280–15287. 10.1073/pnas.0606924103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlebois R. L., Doolittle W. F. (2004). Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 14, 2469–2477. 10.1101/gr.3024704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-M., de Faria S. M., Chou J.-H., James E. K., Elliott G. N., Sprent J. I., et al. (2008). Burkholderia sabiae sp. nov., isolated from root nodules of Mimosa caesalpiniifolia. Int. J. Syst. Evol. Microbiol. 58, 2174–2179. 10.1099/ijs.0.65816-0 [DOI] [PubMed] [Google Scholar]
- Chen W.-M., De Faria S. M., James E. K., Elliott G. N., Lin K.-Y., Chou J.-H., et al. (2007). Burkholderia nodosa sp. nov., isolated from root nodules of the woody Brazilian legumes Mimosa bimucronata and Mimosa scabrella. Int. J. Syst. Evol. Microbiol. 57, 1055–1059. 10.1099/ijs.0.64873-0 [DOI] [PubMed] [Google Scholar]
- Chen W.-M., James E. K., Coenye T., Chou J.-H., Barrios E., De Faria S. M., et al. (2006). Burkholderia mimosarum sp. nov., isolated from root nodules of Mimosa spp. from Taiwan and South America. Int. J. Syst. Evol. Microbiol. 56, 1847–1851. 10.1099/ijs.0.64325-0 [DOI] [PubMed] [Google Scholar]
- Ciccarelli F. D., Doerks T., Von Mering C., Creevey C. J., Snel B., Bork P. (2006). Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283–1287. 10.1126/science.1123061 [DOI] [PubMed] [Google Scholar]
- Coenye T., Henry D., Speert D. P., Vandamme P. (2004). Burkholderia phenoliruptrix sp. nov., to accommodate the 2, 4, 5-trichlorophenoxyacetic acid and halophenol-degrading strain AC1100. Syst. Appl. Microbiol. 27, 623–627. 10.1078/0723202042369992 [DOI] [PubMed] [Google Scholar]
- Coenye T., Laevens S., Willems A., Ohlén M., Hannant W., Govan J., et al. (2001a). Burkholderia fungorum sp. nov. and Burkholderia caledonica sp. nov.,two new species isolated from the environment, animals and human clinical samples. Int. J. Syst. Evol. Microbiol. 51, 1099–1107. 10.1099/00207713-51-3-1099 [DOI] [PubMed] [Google Scholar]
- Coenye T., Mahenthiralingam E., Henry D., LiPuma J. J., Laevens S., Gillis M., et al. (2001b). Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 51, 1481–1490. 10.1099/00207713-51-4-1481 [DOI] [PubMed] [Google Scholar]
- Coenye T., Vandamme P. (2003). Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5, 719–729. 10.1046/j.1462-2920.2003.00471.x [DOI] [PubMed] [Google Scholar]
- Cole J. R., Wang Q., Cardenas E., Fish J., Chai B., Farris R. J., et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S., Nowak J., Coenye T., Clement C., Ait Barka E. (2008). Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 32, 607–626. 10.1111/j.1574-6976.2008.00113.x [DOI] [PubMed] [Google Scholar]
- Coutinho B. G., Passos da Silva D., Previato J. O., Mendonca-Previato L., Venturi V. (2013). Draft genome sequence of the rice endophyte Burkholderia kururiensis M130. Genome Announc. 1, e0022512–e0022512. 10.1128/genomeA.00225-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer S. E., Cnockaert M., Ardley J. K., Maker G., Yates R., Howieson J. G., et al. (2013a). Burkholderia sprentiae sp. nov., isolated from Lebeckia ambigua root nodules. Int. J. Syst. Evol. Microbiol. 63(Pt 11), 3950–3957. 10.1099/ijs.0.048777-0 [DOI] [PubMed] [Google Scholar]
- De Meyer S. E., Cnockaert M., Ardley J. K., Trengove R. D., Garau G., Howieson J. G., et al. (2013b). Burkholderia rhynchosiae sp. nov., isolated from Rhynchosia ferulifolia root nodules. Int. J. Syst. Evol. Microbiol. 63(Pt 11), 3944–3949. 10.1099/ijs.0.048751-0 [DOI] [PubMed] [Google Scholar]
- De Meyer S. E., Cnockaert M., Ardley J. K., Van Wyk B.-E., Vandamme P. A., Howieson J. G. (2014). Burkholderia dilworthii sp. nov., isolated fromLebeckia ambigua root nodules. Int. J. Syst. Evol. Microbiol. 64(Pt 4), 1090–1095. 10.1099/ijs.0.058602-0 [DOI] [PubMed] [Google Scholar]
- Deshazer D. (2007). Virulence of clinical and environmental isolates of Burkholderia oklahomensis and Burkholderia thailandensis in hamsters and mice. FEMS Microbiol. Lett. 277, 64–69. 10.1111/j.1574-6968.2007.00946.x [DOI] [PubMed] [Google Scholar]
- Estrada-de los Santos P., Vinuesa P., Martínez-Aguilar L., Hirsch A. M., Caballero-Mellado J. (2013). Phylogenetic analysis of Burkholderia species by multilocus sequence analysis. Curr. Microbiol. 67, 51–60. 10.1007/s00284-013-0330-9 [DOI] [PubMed] [Google Scholar]
- Gao B., Gupta R. S. (2012). Microbial systematics in the post-genomics era. Antonie Van Leeuwenhoek 101, 45–54. 10.1007/s10482-011-9663-1 [DOI] [PubMed] [Google Scholar]
- Gillis M., Van Van T., Bardin R., Goor M., Hebbar P., Willems A., et al. (1995). Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int. J. Syst. Bacteriol. 45, 274–289 10.1099/00207713-45-2-274 [DOI] [Google Scholar]
- Gogarten J. P., Doolittle W. F., Lawrence J. G. (2002). Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 19, 2226–2238. 10.1093/oxfordjournals.molbev.a004046 [DOI] [PubMed] [Google Scholar]
- Goris J., Dejonghe W., Falsen E., De Clerck E., Geeraerts B., Willems A., et al. (2002). Diversity of transconjugants that acquired plasmid pJP4 or pEMT1 after inoculation of a donor strain in the A-and B-horizon of an agricultural soil and description of Burkholderia hospita sp. nov. andBurkholderia terricola sp. nov. Syst. Appl. Microbiol. 25, 340–352. 10.1078/0723-2020-00134 [DOI] [PubMed] [Google Scholar]
- Goris J., De Vos P., Caballero-Mellado J., Park J., Falsen E., Quensen J. F., et al. (2004). Classification of the biphenyl-and polychlorinated biphenyl-degrading strain LB400T and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol. 54, 1677–1681. 10.1099/ijs.0.63101-0 [DOI] [PubMed] [Google Scholar]
- Gupta R. S. (1998). Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62, 1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. S. (2001). The branching order and phylogenetic placement of species from completed bacterial genomes, based on conserved indels found in various proteins. Int. Microbiol. 4, 187–202. 10.1007/s10123-001-0037-9 [DOI] [PubMed] [Google Scholar]
- Gupta R. S. (2009). Protein signatures (molecular synapomorphies) that are distinctive characteristics of the major cyanobacterial clades. Int. J. Syst. Evol. Microbiol. 59, 2510. 10.1099/ijs.0.005678-0 [DOI] [PubMed] [Google Scholar]
- Gupta R. S. (2014). Identification of Conserved Indels that are Useful for Classification and Evolutionary Studies Methods in Microbiology, Vol. 41 Oxford: Academic Press. [Google Scholar]
- Gupta R. S., Griffiths E. (2002). Critical issues in bacterial phylogeny. Theor. Popul. Biol. 61, 423–434. 10.1006/tpbi.2002.1589 [DOI] [PubMed] [Google Scholar]
- Gyaneshwar P., Hirsch A. M., Moulin L., Chen W.-M., Elliott G. N., Bontemps C., et al. (2011). Legume-nodulating betaproteobacteria: diversity, host range, and future prospects. Mol. Plant Microbe Interact. 24, 1276–1288. 10.1094/MPMI-06-11-0172 [DOI] [PubMed] [Google Scholar]
- Harris J. K., Kelley S. T., Spiegelman G. B., Pace N. R. (2003). The genetic core of the universal ancestor. Genome Res. 13, 407–412. 10.1101/gr.652803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser A. R., Jain M., Bar-Meir M., McColley S. A. (2011). Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin. Microbiol. Rev. 24, 29–70. 10.1128/CMR.00036-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M. T., Seth-Smith H. M., Crossman L. C., Sebaihia M., Bentley S. D., Cerdeno-Tarraga A. M., et al. (2009). The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 191, 261–277. 10.1128/JB.01230-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M. T., Titball R. W., Peacock S. J., Cerdeno-Tarraga A. M., Atkins T., Crossman L. C., et al. (2004). Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U.S.A. 101, 14240–14245. 10.1073/pnas.0403302101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K. W., Koh C. L., Sam C. K., Yin W. F., Chan K. G. (2012). Complete genome sequence of Burkholderia sp. Strain GG4, a betaproteobacterium that reduces 3-oxo-N-acylhomoserine lactones and produces different N-acylhomoserine lactones. J. Bacteriol. 194, 6317–6312. 10.1128/JB.01578-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin F., Thompson J. D., Gouy M., Higgins D. G., Gibson T. J. (1998). Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23, 403. 10.1016/S0968-0004(98)01285-7 [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. CABIOS, 8, 275–282. [DOI] [PubMed] [Google Scholar]
- Kang S. R., Srinivasan S., Lee S. S. (2014). Burkholderia eburnea sp. nov., isolated from peat soil. Int. J. Syst. Evol. Microbiol. 64(Pt 4), 1108–1115. 10.1099/ijs.0.051078-0 [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Asif H., Studholme D. J., Khan I. A., Azim M. K. (2013). Genome characterization of a novel Burkholderia cepacia complex genomovar isolated from dieback affected mango orchards. World J. Microbiol. Biotechnol. 29, 2033–2044. 10.1007/s11274-013-1366-5 [DOI] [PubMed] [Google Scholar]
- Kim H.-B., Park M.-J., Yang H.-C., An D.-S., Jin H.-Z., Yang D.-C. (2006). Burkholderia ginsengisoli sp. nov., a β-glucosidase-producing bacterium isolated from soil of a ginseng field. Int. J. Syst. Evol. Microbiol. 56, 2529–2533. 10.1099/ijs.0.64387-0 [DOI] [PubMed] [Google Scholar]
- Kim H. S., Schell M. A., Yu Y., Ulrich R. L., Sarria S. H., Nierman W. C., et al. (2005). Bacterial genome adaptation to niches: divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC Genomics 6:174. 10.1186/1471-2164-6-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Vikram S., Raghava G. P. (2012). Genome sequence of the nitroaromatic compound-degrading Bacterium Burkholderia sp. strain SJ98. J. Bacteriol. 194, 3286–3212. 10.1128/JB.00497-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak M. J., Song J. Y., Kim S. Y., Jeong H., Kang S. G., Kim B. K., et al. (2012). Complete genome sequence of the endophytic bacterium Burkholderia sp. strain KJ006. J. Bacteriol. 194, 4432–4433. 10.1128/JB.00821-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides N., Overbeek R., Ouzounis C. (1999). Universal protein families and the functional content of the last universal common ancestor. J. Mol. Evol. 49, 413–423. 10.1007/PL00006564 [DOI] [PubMed] [Google Scholar]
- Lackner G., Moebius N., Partida-Martinez L., Hertweck C. (2011). Complete genome sequence of Burkholderia rhizoxinica, an Endosymbiont of Rhizopus microsporus. J. Bacteriol. 193, 783–784. 10.1128/JB.01318-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-M., Weon H.-Y., Yoon S.-H., Kim S.-J., Koo B.-S., Kwon S.-W. (2012). Burkholderia denitrificans sp. nov., isolated from the soil of Dokdo Island, Korea. J. Microbiol. 50, 855–859. 10.1007/s12275-012-1554-2 [DOI] [PubMed] [Google Scholar]
- Lemaire B., Robbrecht E., van Wyk B., Van Oevelen S., Verstraete B., Prinsen E., et al. (2011). Identification, origin, and evolution of leaf nodulating symbionts of Sericanthe (Rubiaceae). J. Microbiol. 49, 935–941. 10.1007/s12275-011-1163-5 [DOI] [PubMed] [Google Scholar]
- Lemaire B., Van Oevelen S., De Block P., Verstraete B., Smets E., Prinsen E., et al. (2012). Identification of the bacterial endosymbionts in leaf nodules of Pavetta (Rubiaceae). Int. J. Syst. Evol. Microbiol. 62, 202–209. 10.1099/ijs.0.028019-0 [DOI] [PubMed] [Google Scholar]
- Lim J. H., Baek S.-H., Lee S.-T. (2008). Burkholderia sediminicola sp. nov., isolated from freshwater sediment. Int. J. Syst. Evol. Microbiol. 58, 565–569. 10.1099/ijs.0.65502-0 [DOI] [PubMed] [Google Scholar]
- Lim J., Lee T. H., Nahm B. H., Choi Y. D., Kim M., Hwang I. (2009). Complete genome sequence of Burkholderia glumae BGR1. J. Bacteriol. 191, 3758–3759. 10.1128/JB.00349-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. S., Choi B. S., Choi A. Y., Kim K. D., Kim D. I., Choi I. Y., et al. (2012). Complete genome sequence of the fenitrothion-degrading Burkholderia sp. strain YI23. J. Bacteriol. 194, 896–811. 10.1128/JB.06479-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y. W., Baik K. S., Han S. K., Kim S. B., Bae K. S. (2003). Burkholderia sordidicola sp. nov., isolated from the white-rot fungus Phanerochaete sordida. Int. J. Syst. Evol. Microbiol. 53, 1631–1636. 10.1099/ijs.0.02456-0 [DOI] [PubMed] [Google Scholar]
- Limmathurotsakul D., Peacock S. J. (2011). Melioidosis: a clinical overview. Br. Med. Bull. 99, 125–139. 10.1093/bmb/ldr007 [DOI] [PubMed] [Google Scholar]
- Lu P., Zheng L.-Q., Sun J.-J., Liu H.-M., Li S.-P., Hong Q., et al. (2012). Burkholderia zhejiangensis sp. nov., a methyl-parathion-degrading bacterium isolated from a wastewater-treatment system. Int. J. Syst. Evol. Microbiol. 62(Pt 6), 1337–1341 10.1099/ijs.0.035428-0 [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E., Baldwin A., Vandamme P. (2002). Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51, 533–538. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E., Urban T. A., Goldberg J. B. (2005). The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3, 144–156. 10.1038/nrmicro1085 [DOI] [PubMed] [Google Scholar]
- Mardis E. R. (2008). The impact of next-generation sequencing technology on genetics. Trends Genet. 24, 133–141. 10.1016/j.tig.2007.12.007 [DOI] [PubMed] [Google Scholar]
- Martínez-Aguilar L., Salazar-Salazar C., Méndez R. D., Caballero-Mellado J., Hirsch A. M., Vásquez-Murrieta M. S., et al. (2013). Burkholderia caballeronis sp. nov., a nitrogen fixing species isolated from tomato (Lycopersicon esculentum) with the ability to effectively nodulate Phaseolus vulgaris. Antonie Van Leeuwenhoek 104, 1063–1071. 10.1007/s10482-013-0028-9 [DOI] [PubMed] [Google Scholar]
- Mavengere N. R., Ellis A. G., Le Roux J. J. (2014). Burkholderia aspalathi sp. nov., isolated from root nodules of the South African legume Aspalathus abietina Thunb. Int. J. Syst. Evol. Microbiol. 64, 1906–1912. 10.1099/ijs.0.057067-0 [DOI] [PubMed] [Google Scholar]
- Nandakumar R., Shahjahan A., Yuan X., Dickstein E., Groth D., Clark C., et al. (2009). Burkholderia glumae and B. gladioli cause bacterial panicle blight in rice in the southern United States. Plant Dis. 93, 896–905 10.1094/PDIS-93-9-0896 [DOI] [PubMed] [Google Scholar]
- Nazir R., Hansen M. A., Sorensen S., van Elsas J. D. (2012). Draft genome sequence of the soil bacterium Burkholderia terrae strain BS001, which interacts with fungal surface structures. J. Bacteriol. 194, 4480–4481. 10.1128/J.B.00725-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI (2014). NCBI Genome Database. http://www.ncbi.nlm.nih.gov/genome/
- Nierman W. C., DeShazer D., Kim H. S., Tettelin H., Nelson K. E., Feldblyum T., et al. (2004). Structural flexibility in theBurkholderia mallei genome. Proc. Natl. Acad. Sci. U.S.A. 101, 14246–14251. 10.1073/pnas.0403306101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll M. R., Kidd T. J., Coulter C., Smith H. V., Rose B. R., Harbour C., et al. (2003). Burkholderia pseudomallei: another emerging pathogen in cystic fibrosis. Thorax 58, 1087–1091. 10.1136/thorax.58.12.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira Cunha C., Goda Zuleta L. F., Paula de Almeida L. G., Prioli Ciapina L., Lustrino Borges W., Pitard R. M., et al. (2012). Complete genome sequence of Burkholderia phenoliruptrix BR3459a (CLA1), a heat-tolerant, nitrogen-fixing symbiont of Mimosa flocculosa. J. Bacteriol. 194, 6675–6676. 10.1128/JB.01821-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onofre-Lemus J., Hernández-Lucas I., Girard L., Caballero-Mellado J. (2009). ACC (1-aminocyclopropane-1-carboxylate) deaminase activity, a widespread trait in Burkholderia species, and its growth-promoting effect on tomato plants. Appl. Environ. Microbiol. 75, 6581–6590. 10.1128/AEM.01240-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormeno-Orrillo E., Rogel M. A., Chueire L. M., Tiedje J. M., Martinez-Romero E., Hungria M. (2012). Genome sequences of Burkholderia sp. strains CCGE1002 and H160, isolated from legume nodules in Mexico and Brazil. J. Bacteriol. 194, 6927–6912. 10.1128/JB.01756-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y., Muramatsu Y., Nakagawa Y., Matsuda M., Nakamura M., Murata H. (2011). Burkholderia oxyphila sp. nov., a bacterium isolated from acidic forest soil that catabolizes (+)-catechin and its putative aromatic derivatives. Int. J. Syst. Evol. Microbiol. 61, 249–254. 10.1099/ijs.0.017368-0 [DOI] [PubMed] [Google Scholar]
- Palleroni N. J. (2005). Genus I. Burkholderia Yabuuchi et al. 1993, 398VP (Effective publication: Yabuuchi et al. 1992, 1268) emend. Gillis et al. 1995, 286*, in Bergey's Manual of Systematic Bacteriology, 2 Edn, Vol. 2, eds Brenner D. J., Krieg N. R., Garrity G. M., Staley J. T. (New York, NY: Springer; ), 575–600. [Google Scholar]
- Parte A. C. (2013). LPSN–list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 42, D613–D616. 10.1093/nar/gkt1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partida-Martinez L. P., Groth I., Schmitt I., Richter W., Roth M., Hertweck C. (2007). Burkholderia rhizoxinica sp. nov. and Burkholderia endofungorum sp. nov., bacterial endosymbionts of the plant-pathogenic fungus Rhizopus microsporus. Int. J. Syst. Evol. Microbiol. 57, 2583–2590. 10.1099/ijs.0.64660-0 [DOI] [PubMed] [Google Scholar]
- Payne G. W., Vandamme P., Morgan S. H., LiPuma J. J., Coenye T., Weightman A. J., et al. (2005). Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl. Environ. Microbiol. 71, 3917–3927. 10.1128/AEM.71.7.3917-3927.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perin L., Martínez-Aguilar L., Paredes-Valdez G., Baldani J., Estrada-de Los Santos P., Reis V., et al. (2006). Burkholderia silvatlantica sp. nov., a diazotrophic bacterium associated with sugar cane and maize. Int. J. Syst. Evol. Microbiol. 56, 1931–1937. 10.1099/ijs.0.64362-0 [DOI] [PubMed] [Google Scholar]
- Reis V., Estrada-De los Santos P., Tenorio-Salgado S., Vogel J., Stoffels M., Guyon S., et al. (2004). Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant-associated bacterium. Int. J. Syst. Evol. Microbiol. 54, 2155–2162. 10.1099/ijs.0.02879-0 [DOI] [PubMed] [Google Scholar]
- Rokas A., Holland P. W. H. (2000). Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 15, 454–459. 10.1016/S0169-5347(00)01967-4 [DOI] [PubMed] [Google Scholar]
- Segata N., Bornigen D., Morgan X. C., Huttenhower C. (2013). PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 4, 2304. 10.1038/ncomms3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y. S., Lim J., Choi B. S., Kim H., Goo E., Lee B., et al. (2011). Complete genome sequence ofBurkholderia gladioli BSR3. J. Bacteriol. 193, 3149–3111. 10.1128/JB.00420-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessitsch A., Coenye T., Sturz A., Vandamme P., Barka E. A., Salles J., et al. (2005). Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int. J. Syst. Evol. Microbiol. 55, 1187–1192. 10.1099/ijs.0.63149-0 [DOI] [PubMed] [Google Scholar]
- Sheu S.-Y., Chou J.-H., Bontemps C., Elliott G. N., Gross E., James E. K., et al. (2012). Burkholderia symbiotica sp. nov., isolated from root nodules of Mimosa spp. native to north-east Brazil. Int. J. Syst. Evol. Microbiol. 62(Pt 9), 2272–2278. 10.1099/ijs.0.037408-0 [DOI] [PubMed] [Google Scholar]
- Sheu S.-Y., Chou J.-H., Bontemps C., Elliott G. N., Gross E., dos Reis Junior F. B., et al. (2013). Burkholderia diazotrophica sp. nov., isolated from root nodules of Mimosa spp. Int. J. Syst. Evol. Microbiol. 63(Pt 2), 435–441. 10.1099/ijs.0.039859-0 [DOI] [PubMed] [Google Scholar]
- Shibata T. F., Maeda T., Nikoh N., Yamaguchi K., Oshima K., Hattori M., et al. (2013). Complete genome sequence of Burkholderia sp. Strain RPE64, bacterial symbiont of the bean bug Riptortus pedestris. Genome Announc. 1, 10–13. 10.1128/genomeA.00441-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson I. N., Finlay J., Winstanley D. J., Dewhurst N., Nelson J. W., Butler S. L., et al. (1994). Multi-resistance isolates possessing characteristics of both Burkholderia (Pseudomonas) cepacia and Burkholderia gladioli from patients with cystic fibrosis. J. Antimicrob. Chemother. 34, 353–361. 10.1093/jac/34.3.353 [DOI] [PubMed] [Google Scholar]
- Song J. Y., Kwak M. J., Lee K. Y., Kong H. G., Kim B. K., Kwon S. K., et al. (2012). Draft genome sequence of the antifungal-producing plant-benefiting bacterium Burkholderia pyrrocinia CH-67. J. Bacteriol. 194, 6649–6650. 10.1128/JB.01779-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilker T., Baldwin A., Bumford A., Dowson C. G., Mahenthiralingam E., LiPuma J. J. (2009). Expanded multilocus sequence typing for Burkholderia species. J. Clin. Microbiol. 47, 2607–2610. 10.1128/JCM.00770-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead D. (1992). Grouping of plant-pathogenic and some other Pseudomonas spp. by using cellular fatty acid profiles. Int. J. Syst. Bacteriol. 42, 281–295 10.1099/00207713-42-2-281 [DOI] [Google Scholar]
- Suarez-Moreno Z. R., Caballero-Mellado J., Coutinho B. G., Mendonca-Previato L., James E. K., Venturi V. (2012). Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 63, 249–266. 10.1007/s00248-011-9929-1 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S. (1986). Some probabilistic and statistical problems in the analysis of DNA sequences, in Lectures on Mathematics in the Life Sciences, 17 Edn, ed Miura R. M. (Providence (RI): American Mathematical Society; ), 57–86. [Google Scholar]
- Tayeb L. A., Lefevre M., Passet V., Diancourt L., Brisse S., Grimont P. A. (2008). Comparative phylogenies of Burkholderia, Ralstonia, Comamonas, Brevundimonas and related organisms derived from rpoB, gyrB and rrs gene sequences. Res. Microbiol. 159, 169–177. 10.1016/j.resmic.2007.12.005 [DOI] [PubMed] [Google Scholar]
- Tian Y., Kong B. H., Liu S. L., Li C. L., Yu R., Liu L., et al. (2013). Burkholderia grimmiae sp. nov., isolated from a xerophilous moss (Grimmia montana). Int. J. Syst. Evol. Microbiol. 63(Pt 6), 2108–2113. 10.1099/ijs.0.045492-0 [DOI] [PubMed] [Google Scholar]
- Ussery D. W., Kiil K., Lagesen K., Sicheritz-Ponten T., Bohlin J., Wassenaar T. M. (2009). The genus Burkholderia: analysis of 56 genomic sequences. Genome Dyn. 6, 140–457. 10.1159/000235768 [DOI] [PubMed] [Google Scholar]
- Valverde A., Delvasto P., Peix A., Velázquez E., Santa-Regina I., Ballester A., et al. (2006). Burkholderia ferrariae sp. nov., isolated from an iron ore in Brazil. Int. J. Syst. Evol. Microbiol. 56, 2421–2425. 10.1099/ijs.0.64498-0 [DOI] [PubMed] [Google Scholar]
- Vandamme P., Dawyndt P. (2011). Classification and identification of the Burkholderia cepacia complex: past, present and future. Syst. Appl. Microbiol. 34, 87–95. 10.1016/j.syapm.2010.10.002 [DOI] [PubMed] [Google Scholar]
- Vandamme P., De Brandt E., Houf K., Salles J. F., van Elsas J. D., Spilker T., et al. (2013). Burkholderia humi sp. nov., Burkholderia choica sp. nov., Burkholderia telluris sp. nov., Burkholderia terrestris sp. nov. and Burkholderia udeis sp. nov.: Burkholderia glathei-like bacteria from soil and rhizosphere soil. Int. J. Syst. Evol. Microbiol. 63(Pt 12), 4707–4718. 10.1099/ijs.0.048900-0 [DOI] [PubMed] [Google Scholar]
- Vandamme P., Goris J., Chen W. M., de Vos P., Willems A. (2002b). Burkholderia tuberum sp. nov. andBurkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25, 507–512. 10.1078/07232020260517634 [DOI] [PubMed] [Google Scholar]
- Vandamme P., Goris J., Chen W.-M., De Vos P., Willems A. (2002a). Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25, 507–512. 10.1078/07232020260517634 [DOI] [PubMed] [Google Scholar]
- Vandamme P., Holmes B., Vancanneyt M., Coenye T., Hoste B., Coopman R., et al. (1997). Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47, 1188–1200. 10.1099/00207713-47-4-1188 [DOI] [PubMed] [Google Scholar]
- Vandamme P., Opelt K., Knöchel N., Berg C., Schönmann S., De Brandt E., et al. (2007). Burkholderia bryophila sp. nov. andBurkholderia megapolitana sp. nov., moss-associated species with antifungal and plant-growth-promoting properties. Int. J. Syst. Evol. Microbiol. 57, 2228–2235. 10.1099/ijs.0.65142-0 [DOI] [PubMed] [Google Scholar]
- Vanlaere E., van der Meer J. R., Falsen E., Salles J. F., De Brandt E., Vandamme P. (2008). Burkholderia sartisoli sp. nov., isolated from a polycyclic aromatic hydrocarbon-contaminated soil. Int. J. Syst. Evol. Microbiol. 58, 420–423. 10.1099/ijs.0.65451-0 [DOI] [PubMed] [Google Scholar]
- Van Oevelen S., De Wachter R., Vandamme P., Robbrecht E., Prinsen E. (2002a). Identification of the bacterial endosymbionts in leaf galls of Psychotria (Rubiaceae, angiosperms) and proposal of'Candidatus Burkholderia kirkii'sp. nov. Int. J. Syst. Evol. Microbiol. 52, 2023–2027. 10.1099/ijs.0.02103-0 [DOI] [PubMed] [Google Scholar]
- Van Oevelen S., De Wachter R., Vandamme P., Robbrecht E., Prinsen E. (2002b). Identification of the bacterial endosymbionts in leaf galls of Psychotria (Rubiaceae, angiosperms) and proposal of ‘Candidatus Burkholderia kirkii’ sp. nov. Int. J. Syst. Evol. Microbiol. 52(Pt 6), 2023–2027. 10.1099/ijs.0.02103-0 [DOI] [PubMed] [Google Scholar]
- Van Oevelen S., De Wachter R., Vandamme P., Robbrecht E., Prinsen E. (2004). ‘Candidatus Burkholderia calva’ and ‘Candidatus Burkholderia nigropunctata’ as leaf gall endosymbionts of African Psychotria. Int. J. Syst. Evol. Microbiol. 54, 2237–2239. 10.1099/ijs.0.63188-0 [DOI] [PubMed] [Google Scholar]
- Viallard V., Poirier I., Cournoyer B., Haurat J., Wiebkin S., Ophel-Keller K., et al. (1998). Burkholderia graminis sp. nov., a rhizospheric Burkholderia species, and reassessment of [Pseudomonas] phenazinium,[Pseudomonas] pyrrocinia and [Pseudomonas] glathei as Burkholderia. Int. J. Syst. Bacteriol. 48, 549–563. 10.1099/00207713-48-2-549 [DOI] [PubMed] [Google Scholar]
- Weilharter A., Mitter B., Shin M. V., Chain P. S., Nowak J., Sessitsch A. (2011). Complete genome sequence of the plant growth-promoting endophyteBurkholderia phytofirmans strain PsJN. J. Bacteriol. 193, 3383–3384. 10.1128/JB.05055-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N. J. (2003). Melioidosis. Lancet 361, 1715. 10.1016/S0140-6736(03)13374-0 [DOI] [PubMed] [Google Scholar]
- Whitlock G. C., Estes D. M., Torres A. G. (2007). Glanders: off to the races with Burkholderia mallei. FEMS Microbiol. Lett. 277, 115–122. 10.1111/j.1574-6968.2007.00949.x [DOI] [PubMed] [Google Scholar]
- Wiersinga W. J., van der Poll T., White N. J., Day N. P., Peacock S. J. (2006). Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 4, 272–282. 10.1038/nrmicro1385 [DOI] [PubMed] [Google Scholar]
- Wong S. Y., Paschos A., Gupta R. S., Schellhorn H. E. (2014). Insertion/deletion-based approach for the detection of Escherichia coli O157:H7 in freshwater environments. Environ. Sci. Technol. 48, 11462–11470. 10.1021/es502794h [DOI] [PubMed] [Google Scholar]
- Wu D., Hugenholtz P., Mavromatis K., Pukall R., Dalin E., Ivanova N. N., et al. (2009). A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462, 1056–1060. 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuuchi E., Kosako Y., Oyaizu H., Yano I., Hotta H., Hashimoto Y., et al. (1992). Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36, 1251–1275. 10.1111/j.1348-0421.1992.tb02129.x [DOI] [PubMed] [Google Scholar]
- Yabuuchi E., Kosako Y., Oyaizu H., Yano I., Hotta H., Hashimoto Y., et al. (1993). Burkholderia gen. nov. validation of the publication of new names and new combinations previously effectively published outside the IJSB, List no 43. Int. J. Syst. Bacteriol. 43, 398–399 10.1099/00207713-43-2-398 [DOI] [Google Scholar]
- Yang H.-C., Im W.-T., Kim K. K., An D.-S., Lee S.-T. (2006). Burkholderia terrae sp. nov., isolated from a forest soil. Int. J. Syst. Evol. Microbiol. 56, 453–457. 10.1099/ijs.0.63968-0 [DOI] [PubMed] [Google Scholar]
- Yarza P., Richter M., Peplies J., Euzeby J., Amann R., Schleifer K. H., et al. (2008). The All-Species Living Tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 31, 241–250. 10.1016/j.syapm.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Yoo S.-H., Kim B.-Y., Weon H.-Y., Kwon S.-W., Go S.-J., Stackebrandt E. (2007). Burkholderia soli sp. nov., isolated from soil cultivated with Korean ginseng. Int. J. Syst. Evol. Microbiol. 57, 122–125. 10.1099/ijs.0.64471-0 [DOI] [PubMed] [Google Scholar]
- Zhang H., Hanada S., Shigematsu T., Shibuya K., Kamagata Y., Kanagawa T., et al. (2000). Burkholderia kururiensis sp. nov., a trichloroethylene (TCE)-degrading bacterium isolated from an aquifer polluted with TCE. Int. J. Syst. Evol. Microbiol. 50, 743–749. 10.1099/00207713-50-2-743 [DOI] [PubMed] [Google Scholar]
- Zhu B., Zhou S., Lou M., Zhu J., Li B., Xie G., et al. (2011). Characterization and inference of gene gain/loss along burkholderia evolutionary history. Evol. Bioinform. Online 7, 191. 10.4137/EBO.S7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.