Abstract
Schizophrenia is a highly heritable neuropsychiatric disorder of complex genetic etiology. Previous genome-wide surveys have revealed a greater burden of large, rare CNVs in schizophrenia cases and identified multiple rare recurrent CNVs that increase risk of schizophrenia although with incomplete penetrance and pleiotropic effects. Identification of additional recurrent CNVs and biological pathways enriched for schizophrenia CNVs requires greater sample sizes. We conducted a genome-wide survey for CNVs associated with schizophrenia using a Swedish national sample (4,719 cases and 5,917 controls). High-confidence CNV calls were generated using genotyping array intensity data and their effect on risk of schizophrenia was measured. Our data confirm increased burden of large, rare CNVs in schizophrenia cases as well as significant associations for recurrent 16p11.2 duplications, 22q11.2 deletions and 3q29 deletions. We report a novel association for 17q12 duplications (odds ratio=4.16, P=0.018), previously associated with autism and mental retardation but not schizophrenia. Intriguingly, gene set association analyses implicate biological pathways previously associated with schizophrenia through common variation and exome sequencing (calcium channel signaling and binding partners of the fragile X mental retardation protein). We found significantly increased burden of the largest CNVs (>500Kb) in genes present in the post-synaptic density, in genomic regions implicated via schizophrenia genome-wide association studies, and in gene products localized to mitochondria and cytoplasm. Our findings suggest that multiple lines of genomic inquiry – genome-wide screens for CNVs, common variation, and exonic variation – are converging on similar sets of pathways and/or genes.
Keywords: schizophrenia, genetics, genomics, copy number variation, structural variation
Introduction
Schizophrenia is an often devastating psychiatric disorder with substantial morbidity, mortality, and personal and societal costs.1–3 An important genetic component is indicated by a sibling recurrence risk of 8.6, high heritability estimates (0.64 in a national family study, 0.81 in a meta-analysis of twin studies, and 0.32 estimated directly from common SNPs), and prior genomic findings.4–7
Recent studies into the genetic architecture of this disease have identified both common and rare variation.7–13 GWAS have implicated 22 genome-wide significant loci plus biological pathways including genes regulated by miR-137, neuronal calcium channel signaling, and binding partners of fragile X mental retardation protein (FMRP). Exome sequencing of 2,536 schizophrenia cases and 2,543 controls implicated gene sets enriched for rare exonic variations, including genes involved in calcium channel signaling, FMRP interactors, and the neuronal activity-regulated cytoskeleton-associated (ARC) complex of the postsynaptic density (PSD).14 Genomic evaluation of copy number variation (CNV) has established a role for large rare CNVs (>100 kb, <1%) in risk for schizophrenia. Multiple studies have reported a greater burden of rare CNVs in schizophrenia cases versus controls.15–17 Eight rare CNVs of strong effect (odds ratio 4–20) increase risk for schizophrenia (e.g., 22q11del and 16p11dup), and are often recurrent mutations in genomic hotspots with incomplete penetrance and pleiotropy.10,11,13 Enrichment analyses of genes intersected by rare CNVs implicated functional categories related to synaptic activity and neurodevelopment, and components of the PSD (particularly the N-methyl-D-aspartate receptor, NMDAR, and ARC complexes).12,15,18
The cumulative results from genomic studies of common and rare variation strongly suggest that schizophrenia is notably polygenic (i.e., many genes of differing effect sizes confer the risk for schizophrenia). Besides the eight CNVs of strongest effects,13 it remains unclear whether additional CNVs contribute to the risk for schizophrenia. Larger samples are required to yield new insights and identify novel loci of lower frequencies or modest effects.
Of particular interest, several gene sets (e.g., calcium channel signaling and FMRP interactors) have convergent genomic results from GWAS and exome sequencing. This convergence from different methodologies minimizes risk of bias and suggests that different types of alleles can perturb the same biological pathways that contribute to the etiology of schizophrenia. However, it remains unclear whether CNVs similarly play a role in these pathways of high risk. Previous enrichment analyses of genes intersected by rare CNVs did not explicitly examine the relationship between rare CNVs and common SNPs.
The purpose of this study was to identify CNV alleles, genes, or gene sets that confer risk for schizophrenia in a well-powered sample and to examine the overlap and relative impact of common SNPs and rare CNVs. We conducted a genome-wide CNV survey in a Swedish sample (4,719 cases with schizophrenia and 5,917 controls)7. This sample is well suited for this study given its national sampling frame and relative homogeneity. While the primary data source were genome-wide SNP arrays, all samples were also genotyped with Illumina exome arrays enabling CNV validation with an independent technology.19
Materials and Methods
Additional information is in the Supplemental Methods. To advance knowledge of schizophrenia, CNV data on subsamples were included in prior reports.20,21 We present here the primary analyses for this project, and note that CNV data for 57% of the sample have never been reported before.
Subjects
Subject ascertainment, diagnosis, and validation are described elsewhere7 and summarized in the Supplemental Methods. Briefly, all procedures were approved by ethical committees in Sweden and the US, and all subjects provided written informed consent. Cases with schizophrenia were identified using the Swedish Hospital Discharge Register 22,23 which captures all public and private inpatient hospitalizations.24–27 Case inclusion criteria: ≥2 hospitalizations with a discharge diagnosis of schizophrenia, both parents born in Scandinavia, and age≥ 18 years. Case exclusion criteria: hospital register diagnosis of any medical or psychiatric disorder mitigating a confident diagnosis of schizophrenia. The validity of this case definition of schizophrenia is strongly supported as detailed in the Supplementary Note of reference 7. Controls were selected at random from Swedish population registers, with the goal of obtaining an appropriate control group and avoiding “super-normal” controls that can cause substantial bias in psychiatric research. 28,29 Control inclusion criteria: never hospitalized for schizophrenia or bipolar disorder, both parents born in Scandinavia, and age ≥18 years. Participation rates were lower for cases than for controls (53.3% versus 58.3%) but similar to participation rates in epidemiology (41% for cross-sectional and 56% for case-control studies),30,31 and a large Norwegian longitudinal study (42%).
Genotyping and quality control
DNA was extracted from peripheral venous blood for all subjects. Genotyping was done in six batches (Sw1–6) at the Broad Institute using Affymetrix 5.0 (3.9%, Sw1), Affymetrix 6.0 (38.6%, Sw2–4), and Illumina OmniExpress (57.4%, Sw5–6). Genotypes were called using Birdsuite (Affymetrix) or BeadStudio (Illumina). Quality control filters excluded SNPs for missingness ≥ 0.05 or minor allele frequency < 0.01 and subjects for missingness ≥ 0.02, autosomal heterozygosity deviation, and one of any pair of subjects with high relatedness (π̂ > 0.2). A total of 11,244 subjects (5,001 cases with schizophrenia and 6,243 controls) remained and were used for subsequent CNV calling and quality control. All genomic locations are given in NCBI build 37/UCSC hg19 coordinates.
CNV calling and quality control
We used Birdseye to detect CNVs.32 Birdseye applies a hidden Markov model to normalized probe intensities using model priors tuned to each GWAS array. Basic quality control included removal of low-confidence CNVs with confidence scores < 10, spanning < 10 probes, or < 10 kb in length32 followed by removal of CNVs with > 50% reciprocal overlap with large genomic gaps (e.g., centromeres) or regions subject to rearrangement in white blood cells. We annealed adjoining CNVs that appeared to be artificially split by Birdseye by recursively joining CNVs if the called region is ≥ 80% of the entire region to be joined. The largest CNVs (≥ 5 Mb) and chrX CNVs were visually inspected and those of low confidence were removed. We excluded subjects whose genotyping arrays had excessive noise (probe intensity variance or genomic “waviness” exceeding platform specific thresholds, Table S3, Figure S1) or excessive CNV calls scattered across many chromosomes (≥ 40 segments or total length ≥ 6 Mb). These procedures resulted in a final sample size of 10,636 subjects (4,719 cases and 5,917 controls, Table S4).
Because Birdseye was optimized to identify rare CNVs,32 we imposed a 0.01 frequency threshold by removing CNVs with > 50% of its length spanning a region with > 107 CNVs. Our main analyses were conducted using CNVs ≥ 100 kb and spanning ≥ 15 probes, which gave an estimated false positive rate of 3.3% based on NanoString validation of 212 CNVs detected from Affymetrix 6.0 SNP arrays (unpublished data).
CNV validation
The same DNA samples from all cases and controls were genotyped on Illumina exome arrays. We develop CNV calling procedures for these data (essentially, an exon-focused set of 250K probes), and have shown that the exome array has high sensitivity and specificity to identify CNVs ≥ 400 kb.19 Therefore, we used these additional data for large-scale validation. A CNV (≥400 kb) is considered validated if it is overlapped by an exome array CNV in the same sample by 50% of its length. Table S5 displays the validation results stratified by array type and for deletions and duplications separately and combined. The validation rates were 89%, 83%, and 92% for Affymetrix 5.0, Affymetrix 6.0, and Illumina Omni Express arrays. For all nominally associated CNVs that had sufficient probe coverage from the exome array, we manually inspected CNV calls and probe intensity plots in the same samples.
Statistical analysis
All analyses were conducted using PLINK33 and R.34 As confounders can create spurious associations, we tested a series of metrics (Table S7) for their impact on genome-wide CNV burden using multiple logistic regression with genotyping batch, subject ancestry, age, and sex included as covariates. As anticipated, genotyping batch (Sw1–6) could potentially influence association testing and was controlled for in all analyses. For each association test, 100,000 permutations were performed to evaluate statistical significance with the permutation procedure swapping case-control status within genotyping batches to control for batch effect.20 As shown in Table S4, each genotyping batch used one specific type of arrays. Thus, controlling for genotyping batch effect simultaneously controlled for the difference in genotyping method as permutation swapped case-control status within a specific array type.
Genome-wide burden analysis
We conducted burden analyses across a range of CNV frequencies, sizes, and types. The burden of rare CNVs was measured as the number of CNVs, the genomic length impacted by CNVs, and the number of genes impacted by CNVs (“gene count”) using one-sided statistical tests (assuming increased CNV burden in cases). Odds ratios measure the increase in the likelihood of having disease per unit increase in CNV burden, and were computed in R using logistic regression with batch as a covariate.
Known loci
Large CNVs with previously reported associations with schizophrenia, other psychiatric disorders, or developmental delay (Table 2)10,11,13,35,36 were examined using regional association testing in PLINK for deletions and duplications separately. For single genes, (i.e. NRXN1, VIPR2), we considered all >100kb CNV events that disrupted the gene (≥1bp overlap). For all other regions, we considered all >100kb CNV events that had >50% reciprocal overlap with a region (PLINK --cnv-union-overlap 0.5). Odds ratios and confidence intervals were computed in R. When zero events occurred, odds ratios were estimated by applying the standard continuity correction (i.e., adding 0.5 to each cell of the 2x2 table). 37
Table 2.
| Table 2a. Known pathogenic CNVs associated with psychiatric disorders (deletions) | ||||||
|---|---|---|---|---|---|---|
| CNV Region a | Location (mb) | Freqcases | Freqcontrol | OR (CI) b | EMP-1sided c (adjusted) | EMP-2sided d (adjusted) |
| 1q21.1 * | chr1:145.0–148.0 | 5 | 1 | 6.27 (0.7–296.41) | 0.048 (0.29) | |
| 2p16.3 (NRXN1 exons) * | chr2:50.1–51.2 | 2 | 0 | 6.27 (0.24-Inf) | 0.19 (0.57) | |
| 3q29 * | chr3:195.7–197.3 | 6 | 0 | 16.32 (1.48-Inf) | 0.009 (0.022) | |
| 7q11.23 | chr7:72.7–74.1 | 0 | 0 | |||
| 7q36.3 (VIPR2) | chr7:158.8–158.9 | 0 | 0 | |||
| 15q11.2 | chr15:23.6–28.4 | 0 | 0 | |||
| 15q13.3 * | chr15:30.9–33.5 | 7 | 2 | 4.39 (0.84–43.34) | 0.05 (0.1) | |
| 16p13.11 | chr16:15.4–16.3 | 3 | 4 | 0.94 (0.14–5.56) | 1 (1) | 1 (1) |
| 16p11.2 distal * | chr16:28.2–29.0 | 2 | 1 | 2.51 (0.13–147.88) | 0.43 (0.80) | |
| 16p11.2 | chr16:29.5–30.2 | 0 | 0 | |||
| 17q12 * | chr17:34.8–36.2 | 0 | 0 | |||
| 22q11.2 * | chr22:18.7–21.8 | 6 | 0 | 16.32 (1.48-Inf) | 0.0037 (0.022) | |
| Table 2b. Known pathogenic CNVs associated with psychiatric disorders (duplications) | ||||||
|---|---|---|---|---|---|---|
| CNV Region a | Location (mb) | Freqcases | Freqcontrol | OR (CI) b | EMP-1sided c (adjusted) | EMP-2sided d (adjusted) |
| 1q21.1 | chr1:145.0–148.0 | 2 | 1 | 2.51 (0.13–147.88) | 0.36 (0.98) | |
| 2p16.3 (NRXN1) | chr2:50.1–51.2 | 0 | 0 | |||
| 3q29 | chr3:195.7–197.3 | 1 | 0 | 3.76 (0.03-Inf) | 0.40 (0.88) | |
| 7q11.23 | chr7:72.7–74.1 | 2 | 0 | 6.27 (0.24-Inf) | 0.24 (0.57) | |
| 7q36.3 (VIPR2) * | chr7:158.8–158.9 | 0 | 2 | 0.25 (0–6.68) | 1 (1) | 1 (1) |
| 15q11.2 | chr15:23.6–28.4 | 0 | 0 | |||
| 15q13.3 | chr15:30.9–33.5 | 1 | 2 | 0.63 (0.01–12.05) | 1 (1) | 1 (1) |
| 16p13.11 | chr16:15.4–16.3 | 12 | 6 | 2.51 (0.87–8.16) | 0.168 (0.713) | |
| 16p11.2 distal | chr16:28.2–29.0 | 2 | 4 | 0.63 (0.06–4.38) | 1 (1) | 0.68 (1) |
| 16p11.2 * | chr16:29.5–30.2 | 10 | 2 | 6.28 (1.34–59) | 0.0031 (0.022) | |
| 17q12 | chr17:34.8–36.2 | 5 | 1 | 6.27 (0.7–296.41) | 0.074 (0.318) | |
| 22q11.2 | chr22:18.7–21.8 | 0 | 3 | 0.18 (0–3.03) | 1 (1) | 0.26 (0.83) |
For NRXN1 in Table (2a), we considered all >100kb deletions disrupting exons (≥1bp overlap). For VIPR2 and NRXN1 in Table (2b), we considered all >100Kb CNV events that disrupted the gene (≥1bp overlap). For all other regions, we considered all >100kb CNV events that had >50% reciprocal overlap with a region (PLINK --cnv-union-overlap 0.5).
When 0 events occurred, ORs were estimated by applying the standard continuity correction that added a value of 0.5 to each cell of the 2x2 table.
Test was one-sided assuming higher rate in cases and statistical significance was estimated empirically by 100,000 permutations within batches. The multiple-testing-adjusted P value was obtained in PLINK using the default max-T method.
For simplicity, results for 2-sided tests were only shown when Freqcotnrol > Freqcase.
The star indicates previously implicated loci in SCZ.
Single-site and gene-based association
We excluded known CNV loci in an attempt to identify novel deletions or duplications. The single-site analysis compared the number of CNV events between cases and controls per marker, where PLINK defines markers as to the start and stop sites of all CNV segments. For gene-based association, we used Ensembl gene models (20,007 protein coding genes)38 and, for each gene, we determined the counts of CNVs disrupting the gene (≥1bp overlap) in cases versus controls. To select loci for validation and replication, we extracted nominally associated loci (P < 0.01) and excluded spurious CNVs by manually inspecting probe intensities and overlap with segmental duplications.
Statistical power
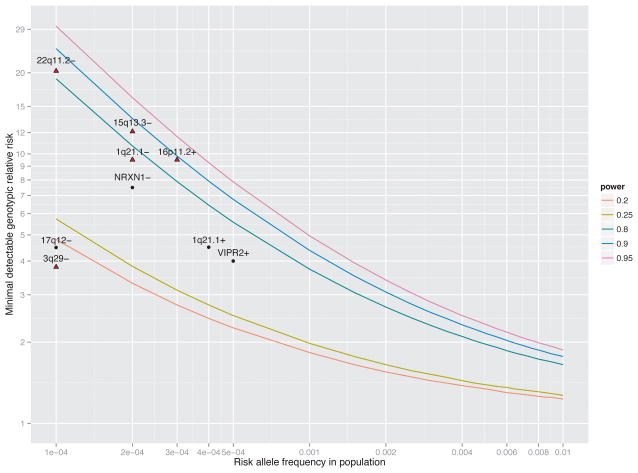
Power analyses to detect single CNV loci were conducted using the R/gap package (see URLs). Assuming a dominant model, lifetime risk of SCZ of 0.7%,39 and α =0.05, we computed the minimal detectable genotypic risk ratio to achieve 20%, 25%, 80%, 90%, or 95% power over a range of frequency of risk alleles in the population (Figure 1). An additive model produced nearly identical results.
Figure 1.
Power to detect novel and known risk loci. The three solid lines depict the minimal detectable genotypic relative risk (y-axis) as a function of the risk allele frequency in the population (x-axis) to achieve 20%, 25%, 80%, 90%, and 95% power. Assumptions were 4,719 cases/5,917 controls, a dominant model, lifetime risk of schizophrenia of 0.7%, and α = 0.05. An additive model produced nearly identical results. Nine known CNVs associated with schizophrenia are shown as red diamonds (P < 0.05 in this study) or black dots (not detected). For each risk locus, the estimated OR and allele frequency in controls were obtained from the literature. If not found in controls, allele frequencies were set to 0.0001. The four most replicated loci (22q11.2−, 15q13.3−, 1q21.1, and 16p11.2+) can be detected given our sample size. Loci 17q12−, 3q29−, 1q21.1+, and VIPR2+ likely have imprecise estimates of GRR or frequency as evidenced in large discrepancies between various reports.
Gene set analyses
We evaluated the collective effects of rare CNVs in predefined sets of genes using the “--cnv-enrichment-test” in PLINK relative to all genic CNVs (i.e. CNVs overlapping any gene by ≥1bp). This method explicitly compares the rate of CNVs impacting a specific gene set in cases versus controls while controlling for sources of bias.40 Restricting to only genic CNVs ensures specificity of the enrichment to the set of interests.40 Specifically, PLINK fits a logistic regression model: , where Pi is the probability that individual i is affected, ci is the number of genic-CNVs that an individual i has, si is the mean size of those events measured in kb, gi is the count of genes within a given gene set affected by genic-CNVs, and other terms are logistic regression coefficients. The “--cnv-enrichment-test” tests if γ, the coefficient associated with gene counts, is significantly different from 0 and evaluates its statistical significance via 100,000 permutations swapping case-control status within genotyping batches to control for batch effect.20 To estimate odds ratios for the increased risk of SCZ per affected gene, we fit the above logistic regression model with genotyping batch included as an additional covariate and computed eγ where γ is the estimated coefficient associated with gene counts after correcting for background difference in rate and size of genic-CNVs and genotyping batch effect.
To discover novel gene sets associated with schizophrenia, we used established pathways (KEGG, GO)41–45 and TargetScan (v6.2, predicted 3′ UTR targets of micro-RNAs).46 To assess gene sets previously implicated in schizophrenia and other psychiatric disorders, we analyzed neuronal calcium signaling genes, genes making RNAs that bind to FMRP (fragile X mental retardation protein, the product of FMR1),7,14,47 genes spanned by a de novo CNV in Kirov et al,12 genes making proteins found in the neuronal postsynaptic density,12 expert-curated lists of synaptic genes,48 the “genes2cognition” database,49 genes implicated in autism50 or mental retardation,45,51–53 genes whose knock-out in mouse yields a neurological or behavioral phenotype,54,55 and human nuclear-encoded mitochondrial genes in MitoCarta.56 All gene sets were established a priori and independently of this work. Kirov et al12 reported that eight de novo CNVs overlapped four known SCZ-associated CNVs (3q29, 15q11.2, 15q13.3 and 16p11.2).
To correct for multiple comparisons in the discovery analysis (i.e. KEGG, GO, TargetScan), we used the false discovery rate (FDR) control as implemented in R/qvalue. 57 Given the discovery nature of the analysis, FDR is less conservative than Bonferroni approach and has greater power to find truly significant results while effectively reducing false positives. To correct for multiple comparisons in the analysis of gene sets previously implicated in SCZ, we applied the Holm-Bonferroni (HB) 58 method and acknowledge the conservativeness of this approach. The HB adjusted P-values (adj_P) were computed in R considering all 126 association tests performed to establish statistical significance. For each significant enrichment identified, we examined specific genes affected by CNVs driving the enrichment and their potential overlap with known CNVs/genes associated with schizophrenia and other psychiatric disorders.
Rare CNVs and significant GWAS loci
We evaluated whether CNVs in schizophrenia cases were enriched for smaller GWAS P-values. The GWAS P-values were obtained from a meta-analysis of the Swedish samples (5,001 cases with SCZ and 6,243 controls) and independent Psychiatric Genomic Consortium (PGC) samples (8,832 cases with SCZ and 12,067 controls). 7 There were 2,691 genes with P < 10−3. We applied a logistic regression model40 for the likelihood of schizophrenia as a function of having a CNV overlapping any of these genes while accounting for background differences in the rate and size of genic CNVs. The extended MHC region (chr6:25–34 mb) was excluded and the “--cnv-enrichment-test” in PLINK relative to all genic CNVs was used with permutation to control for batch effects.
Rare CNV and risk profile score burden
We evaluated the relative impact of rare CNV burden and common variant allelic burden (i.e., risk profile scores, RPS)7,20,59 on risk of schizophrenia. RPS were used the 2011 PGC GWAS mega-analysis9 as the discovery set and then computed in the independent Swedish subjects using the -score function in PLINK. We selected high-quality, relatively independent PGC SNPs with unambiguous directions of effect using a threshold of PT < 0.01 (described in reference 7). For rare CNVs, we use the data from the Swedish sample and focused on the number of CNV events as burden metrics. We first evaluated the burden of known SCZ-associated CNVs that were replicated at the nominal level in the Swedish sample (1q21.1del, 3q29del, 15q13.3del, 22q11.2del, 16p11.2dup). We evaluated the burden of all >100kb CNVs stratified by type, size, and frequency categories. We fit an additive logistic regression model of disease status on RPS and rare CNV burden as predictors and genotyping batch as covariate. An additive model is justified because the Spearman correlations between CNV burden and RPS were all near zero (P > 0.05) and the interaction term, when included in the logistic regression model, was not significant. To estimate the proportion of variance in case-control status accounted for by RPS and CNV burden, we computed the difference in the Nagelkerke pseudo R2 score contrasting a full model with a reduced model in a series of logistic regression models: (1) logit(Pr(case)) ~ RPS burden + CNV burden + genotyping batch. (2) logit(Pr(case)) ~ RPS burden + genotyping batch. (3) logit(Pr(case)) ~ genotyping batch. For RPS, the pseudo R2 contrast model (2) with (3). For CNV burden, the pseudo R2 contrast models (1) with (2). CNV burden could also be obtained by contrasting a logistic regression model containing CNV burden plus batch covariate with (3), which gave the same results because of the independence of RPS and CNV burden.
Exploratory analyses
Bloom syndrome (OMIM #210900) is characterized by excessive homologous recombination in affected individuals, and is caused by mutations in BLM. Using the subsample with exome sequencing genotypes,14 we compared CNV burden between individuals with and without deleterious mutations in BLM by fitting a linear model that had CNV burden as dependent variable, BLM mutation status as a predictor and genotyping batch as covariate.
Replication
We obtained replication association results from 6,882 schizophrenia cases and 11,255 controls. Cases were from the United Kingdom CLOZUK and CardiffCOGS samples.8 Cases were genotyped at the Broad Institute using Illumina OmniExpress or OmniCombo arrays. Controls were from four external studies of non-psychiatric disorders. Details of the replication samples and CNV calling and quality control are documented in Supplementary Methods. Replication was attempted for all novel association regions with P < 0.01 in the Swedish case-control comparisons. For each region, we applied matching procedures to count the number of CNV events in the UK samples. Specifically, for single genes, we computed the counts of CNV events disrupting the gene (≥1bp overlap). For all other region, we computed the counts of CNV events that overlapped the region by >50% of its length. Statistical testing for replication was using Fisher’s exact test and the stratified Cochran-Mantel-Hänszel exact test to account for the case-control ratio difference between the Swedish and the UK samples.
Results
Large chromosomal anomalies can be identified using genotyping arrays. 60 We identified four schizophrenia cases and three controls as genomic outliers (defined as > 40 CNVs or > 6 Mb, thresholds determined empirically as > 3 SD above the sample means). Six of seven abnormalities were confirmed using a second technology (Table S6 and Figure S2). One case and one control had trisomy of chr8 or chr3 (Figure S2a–b), both confirmed using qPCR.60 Three individuals had deletion (one case) or duplication events (two controls) at 15q11.2 (20.5Mb – 22.5 Mb, Figure S2f), which overlapped a CNV associated with mental retardation. One case (Figure S2g) had multiple deletions that appeared to be consistent with mosaicism. Due to their large size, all seven individuals with genomic outliers were excluded from all subsequent analyses.
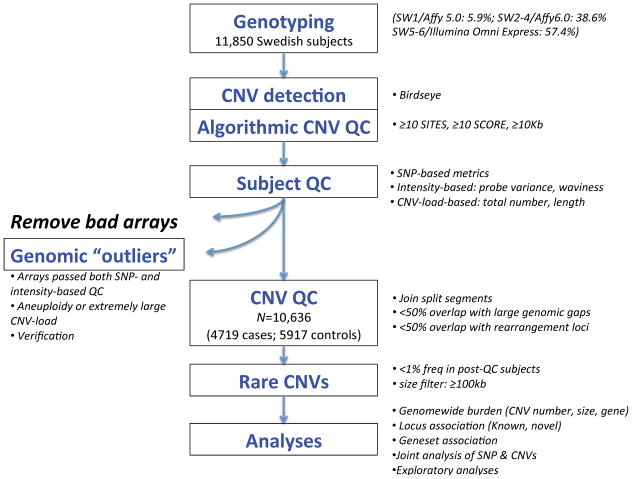
Extensive quality control procedures (Figure 2) were used to establish a stringent CNV dataset in 4,719 schizophrenia cases and 5,917 controls (Table 1, Table S8). There was no case-control or chip bias in the proportions of CNV deletions or median CNV size. However, chip effects were observed in the mean numbers of CNVs per subject. We observed greater mean numbers of CNVs in cases than in controls with ratios that varied by chip type, most notably in the 3.9% samples that were genotyped on the older platform Affymetrix 5.0. Therefore, we controlled for chip effect in all analyses as detailed in Methods so that our findings are robust against this confounder.
Figure 2.
Experimental workflow and CNV datasets.
Table 1.
Subject and CNV characteristics.
| Sample characteristics | Cases | Controls |
|---|---|---|
|
| ||
| Subjects (after quality control) | ||
| Sw1 (Affymetrix 5.0) | 207 | 206 |
| Sw2–4 (Affymetrix 6.0) | 1,847 | 2,137 |
| Sw5–6 (Illumina Omni Express) | 2,665 | 3,574 |
| Total sample | 4,719 | 5,917 |
|
| ||
| Male sex | 0.599 | 0.513 |
|
| ||
| Median age at sampling | 54 (45–62) | 57 (48–65) |
|
| ||
| Median hospital admissions for schizophrenia | 6 (3–13) | n/a |
|
| ||
| Median total inpatient days | 239 (79–694) | n/a |
|
| ||
| Median years from first to last admission | 9.7 (2.9–19.5) | n/a |
|
| ||
| Proportion of all CNVs that are deletions | ||
| Sw1 | 0.40 | 0.47 |
| Sw2–4 | 0.37 | 0.36 |
| Sw5–6 | 0.40 | 0.40 |
| Total sample | 0.38 | 0.38 |
|
| ||
| Median CNV size | ||
| Sw1 | 181.3 | 178.0 |
| Sw2–4 | 182.2 | 188.2 |
| Sw5–6 | 183.1 | 190.7 |
| Total sample | 182.7 | 188.8 |
|
| ||
| Mean number of CNVs per subject | ||
| Sw1 | 1.058 | 0.845 |
| Sw2–4 | 1.236 | 1.209 |
| Sw5–6 | 0.758 | 0.684 |
| Total sample | 0.958 | 0.879 |
After quality control, the combined sample size is 10,636, where Sw1=413 (3.9%, Affymetrix 5.0), Sw2–4=3,984 (37.5%, Affymetrix 6.0), and Sw5–6=6,239 (58.7%, Illumina OmniExpress). Values in parentheses are inter-quartile ranges. Cases had significantly more males (p < 0.0001) and were significantly younger (p < 0.0001) than controls although these differences were not of large magnitude. The higher median age in controls is in the direction of greater confidence in control classification (i.e., controls had greater time at risk for psychiatric hospitalization). The mean number of >500Kb CNVs and of singleton CNVs are reported in Table S8.
Genome-wide CNV burden
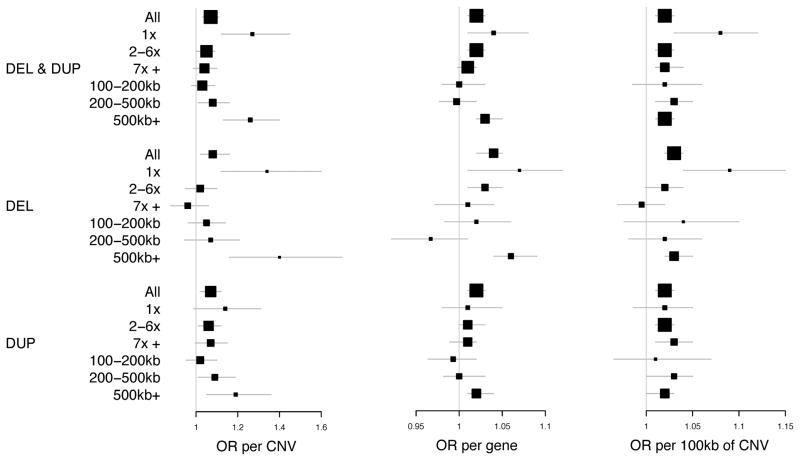
We assessed the role of rare CNVs >100kb. CNVs were stratified by type, frequency, and size. We computed odds ratios (ORs) using logistic regression with genotyping batch as a covariate. Complete results are in Tables S9–10 and summarized in Figure 3. We confirmed the literature finding that schizophrenia cases had a significantly greater genome-wide burden of CNVs than controls: ORs of 1.07 per CNV (95% CI 1.03–1.11, Pemp = 3x10−4), 1.02 (95% CI 1.01–1.03, Pemp = 1x10−5) per gene affected by CNVs, and 1.02 (95% CI 1.01–1.03, Pemp = 3x10−5) per 100Kb affected by CNVs. Deletions were enriched in cases to a greater extent than duplications. The rarest CNVs (single occurrence) and the largest CNVs (> 500 kb) were enriched in cases to a greater extent than other frequency or size categories. As expected, the rarest deletions (OR 1.34 per single-occurrence deletion, 95% CI 1.12–1.6, Pemp = 6x10−4) and the largest deletions (OR 1.4 per >500 kb deletion, 95% CI 1.16–1.7, Pemp = 4x10−4) showed the strongest effects albeit with broad CIs. Next, we examined the distribution of event size in the rarest deletions (Table S11); and for all size categories (100–200kb, 200–500kb, >500kb), we observed greater OR estimates using single-occurrence deletions compared to those obtained from corresponding categories using all deletions. Similarly, we examined the distribution of allele frequency in the largest deletions (Table S12); and for single- and 2–6 occurrence categories, we observed greater OR estimates using >500kb deletions compared to those obtained from corresponding categories using all deletions. The highest OR was obtained in >500kb single-occurrence deletions. These results (Tables S11–12), although based on a smaller sample size, imply that size and rarity could be independent predictors of case-controls status in the Swedish sample.
Figure 3.
Genome-wide CNV burden. These forest plots show OR estimates and 95% confidence intervals for each burden test. The size of the square is proportional to precision. Allele frequency categories: 1x means single-occurrence CNVs observed once in a case or control (MAF < 0.0001). These were conservatively defined as having no overlap with any other CNVs. 2–6x means 2 to 6 occurrences (MAF 0.0001–0.0005). 7x+ means ≥7 occurrences (MAF 0.0005–0.01).
Finally, the largest size category (>500 kb) included CNVs in known schizophrenia-associated regions that were enriched in cases (Table 2). After excluding 42 CNVs in these regions, the OR was 1.17 per >500 kb CNV (95% CI 1.01–1.31, Pemp =0.033), slightly smaller than in the full data (OR=1.26), and was mostly due to single-occurrence deletions (OR = 1.77, 95% CI 1.11–2.82, Pemp =0.011).
CNV loci
We examined regions known to increase risk for schizophrenia and other psychiatric disorders.10,11,13 Complete results are shown in Table 2 and are briefly summarized below. We previously verified these CNVs using exome genotyping arrays. 19 We confirmed literature findings of associations between risk for schizophrenia and 22q11.2 deletions (OR=15.06, 95% CI: 1.48-Inf, Pemp =0.0037), 16p11.2 duplications (OR=6.28, 95% CI: 1.34–59, Pemp =0.0031), and 3q29 deletions (OR=15.06, 95% CI: 1.48-Inf, Pemp =0.009). Weaker evidence was observed for 15q13.3 deletions (OR=4.39, 95% CI: 0.84–43.34, Pemp =0.05) and 1q21.1 deletions (OR=6.27, 95% CI: 0.7–296.4, Pemp =0.048). We observed more deletions disrupting NRXN1 exons in SCZ cases (2 deletions in cases disrupting one exon of NRXN1 but 0 exonic deletion in controls (Figure S5)). We observed more duplications at 22q11.2 in controls (0 cases and 5 controls), of which three spanned the full length of the known 22q11.2 deletion locus and two spanned ~30%. The full report of 22q11.2 duplications in a careful CNV analysis of 47,005 individuals (21,138 SCZ cases and 25,867 controls) has recently been published,61 where the discovery sample included controls across studies of both psychiatric and non-psychiatric phenotypes and the replication samples were obtained from multiple sources including our Swedish sample. Rees et al 61 reported 22q11.2 duplications as significantly less common in SCZ cases than in the general population (0.014% vs 0.0085%, OR=0.17, P=0.00086) and suggested 22q11.2 duplications as the first putative protective mutation for SCZ.
Duplications at 17q12 have been implicated in autism and mental retardation but their association with schizophrenia has not been previously reported. We observed more 17q12 duplications in schizophrenia cases (5 cases and 1 controls, OR=6.27, 95% CI: 0.7–296.4, Pemp =0.07). In the replication data, 5 cases and 2 controls had 17q12 duplications (OR=4.16, 95% CI: 1.28-Inf, P=0.018 in a one-sided Fisher’s exact test; OR=4.05, 95% CI: 1.35-Inf, P=0.024 in a one-sided Cochran-Mantel-Hänszel exact test). Table S13 and Figures S3–4 depict the 22q11.2 and 17q12 duplications from GWAS and exome arrays, confirming these events.
We tested regions known to increase risk for developmental delay.36 Of 173 loci, 21 deletions and 16 duplications overlapped (>20%) the Swedish schizophrenia results (Table 2). Among the loci unique to developmental delay, no significant association with schizophrenia (empirical P > 0.05) was observed.
After excluding previously implicated loci (Table 2), 10,11,13,35,36 we identified 15 regions nominally associated with schizophrenia (empirical P < 0.01). Five regions had substantial overlap with segmental duplications (>50%) and were excluded. None of the remaining 10 regions had strong evidence for association in the replication dataset (Table S14).
Gene set testing
Given that the rarest (single-occurrence) and the largest CNVs (>500 kb) had the strongest risks for schizophrenia, these subclasses were examined further. Gene set testing results obtained from single-occurrence CNVs did not reveal additional gene sets with significant enrichment (data not shown). Gene set testing results obtained from CNVs >500kb showed higher effect sizes compared to the corresponding tests based on CNVs >100Kb (Table 3, Table S16).
Table 3.
Gene set association, previously implicated in schizophrenia
| Gene Set | CNVs >100Kb | CNVs >500Kb | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name (source) | Genes | Type | CNVs | Genes | OR | 95% CI | Pemp | Adj_P | CNVs | Genes | OR | 95% CI | Pemp | Adj_P |
| mir137 targets (TargetScan) | 1089 | DEL & DUP | 799 | 274 | 1.04 | (0.909, 1.19) | 0.3303 | 1 | 259 | 106 | 1.24 | (0.969, 1.59) | 0.03377 | 1 |
| DEL | 228 | 96 | 1.03 | (0.797, 1.32) | 0.4005 | 1 | 101 | 48 | 1.43 | (0.936, 2.17) | 0.04573 | 1 | ||
| DUP | 571 | 218 | 1.04 | (0.888, 1.22) | 0.382 | 1 | 158 | 72 | 1.13 | (0.819, 1.55) | 0.2149 | 1 | ||
| Calcium signaling (KEGG:hsa04020) | 178 | DEL & DUP | 238 | 45 | 2.48 | (1.36, 4.51) | 0.0004 | 0.041 | 87 | 19 | 4.39 | (1.46, 13.2) | 0.00042 | 0.047 |
| DEL | 55 | 19 | 0.87 | (0.656, 1.16) | 0.856 | 1 | 26 | 8 | 1.18 | (0.73, 1.91) | 0.2473 | 1 | ||
| DUP | 183 | 34 | 1.07 | (0.834, 1.37) | 0.2788 | 1 | 61 | 14 | 1.53 | (0.992, 2.36) | 0.01746 | 1 | ||
| FRMP targets (Darnell et al.) | 810 | DEL & DUP | 913 | 225 | 1.14 | (1.02, 1.27) | 0.0079 | 0.66 | 286 | 86 | 1.27 | (1.07, 1.51) | 9.00E-04 | 0.095 |
| DEL | 209 | 79 | 1.09 | (0.903, 1.33) | 0.1683 | 1 | 89 | 31 | 1.7 | (1.08, 2.67) | 0.00038 | 0.043 | ||
| DUP | 704 | 185 | 1.16 | (1.01, 1.32) | 0.0162 | 1 | 197 | 69 | 1.17 | (0.956, 1.43) | 0.04766 | 13 | ||
| PSD (Kirov et al) | 668 | DEL & DUP | 655 | 161 | 1.13 | (0.982, 1.3) | 0.0358 | 1 | 295 | 64 | 1.4 | (1.12, 1.74) | 0.00062 | 0.067 |
| DEL | 174 | 58 | 1.17 | (0.875, 1.57) | 0.1604 | 1 | 93 | 26 | 1.57 | (1, 2.45) | 0.01749 | 1 | ||
| DUP | 481 | 132 | 1.12 | (0.958, 1.32) | 0.0614 | 1 | 202 | 47 | 1.35 | (1.05, 1.73) | 0.00648 | 0.55 | ||
| PSD/ARC (Kirov et al) | 28 | DEL & DUP | 103 | 5 | 1.12 | (0.757, 1.66) | 0.307 | 1 | 79 | 3 | 1.2 | (0.759, 1.91) | 0.2138 | 1 |
| DEL | 47 | 4 | 1.19 | (0.662, 2.12) | 0.2981 | 1 | 38 | 3 | 1.1 | (0.557, 2.16) | 0.3793 | 1 | ||
| DUP | 56 | 4 | 1.09 | (0.644, 1.86) | 0.4031 | 1 | 41 | 2 | 1.22 | (0.648, 2.29) | 0.2741 | 1 | ||
| PSD/mGluR5 (Kirov et al) | 38 | DEL & DUP | 35 | 11 | 1.7 | (0.854, 3.4) | 0.0485 | 1 | 24 | 3 | 4.59 | (1.56, 13.5) | 0.00022 | 0.025 |
| DEL | 15 | 6 | 1.77 | (0.594, 5.3) | 0.1635 | 1 | 10 | 2 | 16.7 | (0.817, 341) | 5.00E-04 | 0.056 | ||
| DUP | 20 | 7 | 1.61 | (0.658, 3.95) | 0.1009 | 1 | 14 | 2 | 2.62 | (0.817, 8.41) | 0.02174 | 1 | ||
| PSD/NMDAR (Kirov et al) | 61 | DEL & DUP | 52 | 22 | 2.37 | (1.31, 4.29) | 0.0010 | 0.11 | 28 | 11 | 5.8 | (2, 16.8) | 3.00E-05 | 0.0038 |
| DEL | 19 | 9 | 2.04 | (0.763, 5.46) | 0.0774 | 1 | 10 | 5 | 17.27 | (0.849, 351) | 0.00058 | 0.063 | ||
| DUP | 33 | 16 | 2.56 | (1.22, 5.39) | 0.0038 | 0.34 | 18 | 7 | 3.79 | (1.24, 11.6) | 0.00235 | 0.22 | ||
| PSD/PSD-95 (Kirov et al) | 65 | DEL & DUP | 80 | 17 | 1.81 | (1.15, 2.86) | 0.0021 | 0.20 | 23 | 6 | 4 | (1.34, 11.9) | 0.00118 | 0.12 |
| DEL | 35 | 9 | 2.87 | (1.32, 6.22) | 0.0011 | 0.11 | 18 | 5 | 32.18 | (1.71, 604) | 2.00E-05 | 0.0025 | ||
| DUP | 45 | 11 | 1.31 | (0.725, 2.35) | 0.141 | 1 | 5 | 3 | 0.23 | (0.026, 2.1) | 0.9425 | 1 | ||
| GWAS P<0.001 (Ripke et al.) | 2691 | DEL & DUP | 3543 | 819 | 1.04 | (0.981, 1.1) | 0.119 | 1 | 937 | 355 | 1.21 | (1.1, 1.33) | 4.00E-05 | 0.0049 |
| DEL | 1210 | 299 | 1.04 | (0.923, 1.16) | 0.2869 | 1 | 279 | 110 | 1.31 | (1.07, 1.61) | 0.0041 | 0.3649 | ||
| DUP | 2333 | 679 | 1.04 | (0.977, 1.12) | 0.1464 | 1 | 658 | 295 | 1.2 | (1.08, 1.33) | 0.00021 | 0.025 | ||
| Mitochondrion (Kirov et al) | 193 | DEL & DUP | 143 | 52 | 1.14 | (0.844, 1.55) | 0.204 | 1 | 67 | 25 | 1.63 | (1, 2.64) | 0.01944 | 1 |
| DEL | 60 | 21 | 2.12 | (1.22, 3.67) | 0.0015 | 0.15 | 40 | 13 | 4.31 | (1.71, 10.9) | 4.00E-05 | 0.0049 | ||
| DUP | 83 | 42 | 0.79 | (0.528, 1.17) | 0.8908 | 1 | 27 | 17 | 0.61 | (0.289, 1.29) | 0.9201 | 1 | ||
| MitoCarta (Pagliarini et al.) | 892 | DEL & DUP | 756 | 232 | 1.06 | (0.948, 1.19) | 0.1439 | 1 | 229 | 99 | 1.28 | (1.04, 1.56) | 0.00295 | 0.27 |
| DEL | 311 | 86 | 1.18 | (0.98, 1.42) | 0.0277 | 1 | 105 | 39 | 1.74 | (1.12, 2.71) | 0.00019 | 0.022 | ||
| DUP | 445 | 190 | 0.98 | (0.835, 1.15) | 0.6469 | 1 | 124 | 79 | 1.07 | (0.824, 1.38) | 0.3387 | 1 | ||
DEL: deletions. DUP: duplications. All tests were one-sided assuming enrichment in cases using genic CNVs. #CNV = the number of events that overlapped any gene in the geneset by ≥1bp. #genes = the number of unique genes in the geneset that had at least 1 genic CNV hit (≥1bp overlap). OR=odds ratio, indicating the increase in risk for schizophrenia correcting for rate and size of genic CNVs and genotyping batch effect (a continuity correction applied if necessary). CI: confidence interval. Pemp, empirical P values were obtained in PLINK by 100,000 permutations and permuting phenotype labels within genotyping batches. Adj_P: Holm-Bonferroni multiple-testing adjusted P values considering all 126 tests performed in Table 3 and Table S16.
We hypothesized that different types of genetic variation can perturb the same biological pathways critical to the etiology of schizophrenia. To test this, we compared CNV rates in cases versus controls in gene sets previously implicated in schizophrenia (Table 3). The strongest enrichment was observed for CNVs (deletions and duplications combined) in neuronal calcium channel signaling (OR=2.48, 95% CI: 1.36–4.51, adj_Pemp =0.041 for events >100Kb; OR=4.39, 95% CI: 1.46–13.2, adj_Pemp =0.047 for events >500Kb). For CNVs >500Kb in SCZ cases, 56 events overlapped 11 genes of the calcium signaling pathway, among which 17 (30%) events overlapped (by >50%) known risk CNV loci (Table S17). A significant enrichment in FMRP targets44 (OR=1.7, 95% CI: 1.08–2.67, adj_Pemp =0.043) was observed for deletions >500Kb, of which 76 case deletions (36 in known loci) overlapped 28 genes (Table S17). Previously, the calcium channel and FMPR targets were enriched for rare exonic mutations in exome sequencing of half of the Swedish samples 7,14,47 and were enriched for common variants in a meta-analysis of the Swedish samples and independent PGC samples (a total of 13,833 cases with SCZ and 18,310 controls). 7,14,47 We then examined the overlap between genes affected by common variants and by rare CNVs that are driving the enrichment signals in these two shared pathways. Table S15 shows that common variants affected more genes than rare CNVs did, and that common variants and rare CNVs had no overlap for calcium channel and a small overlap for FMRP targets. These results suggest that different types of risk alleles were independently enriched in SCZ cases than in controls.
For genes found at the post-synaptic density (PSD), we observed increased burden for the gene set used by Kirov et al.12 (OR=1.4, 95% CI: 1.12–1.74, adj_Pemp =0.067 for >500Kb deletions and duplications combined), and a stronger enrichment using the GO category “postsynaptic density” (OR=6.74, 95% CI: 1.66–27.3 for >500kb deletions, Table 4). We next tested gene sets comprising components of the PSD as in Kirov et al.12, and observed significant enrichment (adj_Pemp <0.05) in the NMDAR gene set and in genes encoding mGluR5 for >500Kb deletions and duplications combined, and in the PSD-95 (post-synaptic density protein 95) complex for >500Kb deletions only (Table 3). Most of the case CNVs (~73%) overlapped three known risk CNV loci (3q29, 16p11.2, 22q11.2) and overlapped genes previously enriched for case de novo CNVs12 (Table S17). Overlap between de novo CNVs with 3q29 and 16p11.2 was previously reported.12
Table 4.
Top-ranking significant gene sets from the discovery analysis
| Source | Gene set | Type | CNVs >100Kb | CNVs >500Kb | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Genes | # CNVs | OR | 95% CI | Pemp | Genes within CNVs (genes) | CNVs | OR | 95% CI | Pemp | Genes within CNVs (genes) | ||
| KEGG | hsa04660 T cell receptor signaling pathway | 108 | DEL | 29 | 4.42 | (1.61, 12.2) | 1e-04 | CBLB,DLG1,LAT,MAP3K14,MAP3K7,MAPK3,NFAT5,NRAS,PAK2,PDCD1,PPP3CA,RELA,VAV3 (13) | 19 | 15.72 | (0.87, 285) | 4e-05 | CBLB,DLG1,LAT,MAP3K7,NFAT5,PAK2,PPP3CA (7) |
| KEGG | hsa04640 Hematopoietic cell lineage | 85 | DEL | - | - | - | 17 | 14.4 | (1.9, 109) | 2e-05 | CD19,CD36,GP1BB,IL7R,TFRC (5) | ||
| KEGG | hsa04621 NOD-like receptor signaling pathway | 58 | DUP | 22 | 9.27 | (2.17, 39.7) | 1e-05 | CCL5,CXCL1,CXCL2,ERBB2IP,HSP90AA1,IL8,MAPK3,MAPK8,NAIP,NLRC4,RIPK2 (11) | 15 | 7.05 | (1.6, 31) | 2e-04 | CXCL1,CXCL2,IL8,MAPK3,MAPK8,NAIP,RIPK2 (7) |
| TargetScan | hsa-miR-10a | 144 | DEL& DUP | - | - | - | - | - | 50 | 2.86 | (1.54, 5.32) | 3e-05 | AP002478.1,BCL2L11,BCR,C10orf68,C16orf54,CEP350,CLEC18A, CNST,COL4A4,CSMD1,ELOVL2, EPHA2,GJA5,LCA5,PVRL3,SDPR,SLCO4C1,TFRC,TIAM1,ZNF254 (20) |
| GO | GO:16458 gene silencing | 71 | DEL | 14 | 31.52 | (1.67, 595) | 1e-05 | DGCR8,ERI1,NCBP2 (3) | 14 | 28.21 | (1.48, 537) | 1e-05 | DGCR8,ERI1,NCBP2 (3) |
| GO | GO:2000602 regulation of interphase of mitotic cell cycle | 162 | DEL | 17 | 17.34 | (2.31, 130) | 1e-05 | ATM,CDC45,DLG1,PRMT2,RPL24,USP17L2 (6) | 13 | 26.17 | (1.36, 504) | 1e-05 | CDC45,DLG1,RPL24 (3) |
| GO | GO:42471 ear morphogenesis | 99 | DEL | 12 | 28.67 | (1.5, 548) | 5e-05 | CDH23,GBX2,PTPRQ,SOBP,TBX1 (5) | - | - | - | - | - |
| GO | GO:14069 postsynaptic density | 100 | DEL | - | - | - | 25 | 6.74 | (1.66, 27.3) | 1e-05 | DLG1,DLGAP1,FBXO45,LIN7A,MAGI2,P2RX6,SEMA4C (7) | ||
| GO | GO:16327 apicolateral | 109 | DEL | - | - | - | 24 | 9.61 | (2.23, 41.4) | 1e-05 | CLDN23,CLDN5,DLG1,FRMPD2, JAM2,LIN7A,MAGI2,MICALL2,PMP22,PVRL3 (10) | ||
| GO | GO:287 magnesium ion binding | 179 | DEL | - | - | - | 24 | 6.71 | (1.66, 27.1) | 2e-05 | ABL2,COMT,FAN1,MAP3K7,NT5C3,STK38L,TSSK2 (7) | ||
| GO | GO:32526 response to retinoic acid | 101 | DEL | - | - | - | 14 | 28.67 | (1.51, 546) | 1e-05 | ABL2,PDGFA,TBX1,TFRC (4) | ||
| GO | GO:10498 proteasomal protein catabolic process | 190 | DUP | - | - | - | 23 | 8.39 | (2.51, 28.1) | 2e-05 | ANAPC1,CRBN,DERL3,FBXO45, KCTD13,MAD2L1,PSMA7,STUB1,UBB,WWP2 (10) | ||
| GO | GO:16052 carbohydrate catabolic process | 131 | DUP | - | - | - | 14 | 6.88 | (2.04, 23.3) | 2e-05 | AGL,ALDOA,ALDOB,HK2,OGDHL,PGM1 (6) | ||
| GO | GO:16585 chromatin remodeling complex | 114 | DUP | - | - | - | 22 | 26.31 | (3.58, 194) | 1e-05 | BAZ1B,DPY30,HDAC9,INO80B,INO80E,NCOR1,SMARCA2,SMARCB1,USP22 (9) | ||
| GO | GO:2755 MyD88-dependent toll-like receptor signaling pathway | 75 | DUP | - | - | - | 14 | 17.55 | (2.35, 131) | 1e-05 | MAP2K3,MAPK3,MAPK8,RIPK2, UBB (5) | ||
The number of genesets (N) tested within each database: KEGG: N=228, TargetScan: N=87, and GO: N=1499. All the gene sets listed here were significantly enriched in cases than in controls based on p-values. For KEGG and TargetScan, all have q-value <0.01 implying that <1% of significant tests or <0.27 test will result in false discoveries; and for GO, all have q-value <0.004, implying that <0.4% of significant tests or <0.59 test will result in false discoveries. DEL: deletions. DUP: duplications. All tests were one-sided assuming enrichment in cases using genic CNVs. #CNV = the number of events that overlapped any gene in the geneset by ≥1bp. #genes = the number of unique genes in the geneset that had at least 1 genic CNV hit (≥1bp overlap). OR=odds ratio, indicating the increase in risk for schizophrenia correcting for rate and size of genic CNVs and genotyping batch effect (a continuity correction applied if necessary). CI: confidence interval. Pemp, empirical P values were obtained in PLINK by 100,000 permutations and permuting phenotype labels within genotyping batches.
Gene products often localize to particular sub-cellular compartments. Identifying compartments enriched for genes impacted by CNVs in schizophrenia patients could provide greater insight into pathophysiological mechanisms related to the disorder. Besides the PSD genes, we tested additional gene sets enriched for de novo CNVs in Kirov et al.12 (Table S16) and observed significant enrichment for gene products localized to cytoplasm (OR=3.38, 95% CI: 1.49–7.68, adj_Pemp=0.012 for duplications >500Kb) and mitochondria (OR=4.31, 95% CI: 1.17–10.9, adj_Pemp=0.0049 for deletions >500Kb). Given the potential role of mitochondrial dysfunction in psychiatric disorders, 62,63 we further tested human nuclear-encoded mitochondrial genes in MitoCarta and found significant enrichment of case deletions (1.74, 95% CI: 1.12–2.71, adj_Pemp=0.022 for deletions >500Kb, Table 3). Half of the case CNVs (~51%) overlapped known CNV risk loci (Table S17).
Since many CNVs associated with schizophrenia also show pleiotropic effects for autism and mental retardation, we tested the genes implicated in these disorders in the Swedish sample (Table S16). Large CNVs (>500Kb duplications and deletions combined) in schizophrenia cases were enriched in genes implicated in mental retardation (OR=2.71, 95% CI:1.41–3.34, adj_Pemp = 0.0049). Approximately 33% case CNVs overlapped known CNV risk loci (Table S17), which is not surprising as many of the MR genes have been identified by genomic re-arrangements.
Schizophrenia is often associated with cognitive deficits. Therefore, we tested whether CNVs in schizophrenia patients were enriched for genes associated with cognitive ability in humans and neurological or behavioral phenotypes in mice (Table S16). We observed significant enrichment of large deletions (>500Kb) in synaptic genes based on functional group analysis48 (OR=1.67, 95% CI: 1.15–2.43, adj_Pemp=0.036). Although not significant based on multiple-testing adjustment, increased burden of deletions was observed in genes for which knockout in mouse yields a neurological or behavioral phenotype (OR=1.26, 95% CI: 1.06–1.49, Pemp=0.0021 for deletions >100Kb; OR=1.53, 95% CI: 1.11–2.1, Pemp=0.00099 for deletions>500kb).
The markedly increased sample size of this study relative to others provides the opportunity to identify novel biological pathways associated with schizophrenia. Therefore, we performed a series of exploratory analyses using gene sets based on KEGG, GO, and TargetScan (targets of microRNAs other than miR-137). Table 4 lists the top ranking gene sets that were significantly enriched in cases than in controls (all had p < 0.0002). For KEGG and TargetScan, all had q-value <0.01, implying that <1% of significant tests or <0.27 test will result in false discoveries; and for GO, all had q-value <0.004, implying that <0.4% of significant tests or <0.59 test will result in false discoveries. These gene sets included multiple elements of the immune system (T cell receptor signaling, hematopoietic cell lineage, positive regulation of monocyte proliferation), members of chromatin remodeling complexes (e.g. HDAC9, NCOR1), and a novel association with target genes of microRNA miR-10a as well as basic elements of microRNA processing machinery necessary for gene silencing (i.e. DGCR8, ERI1, NCBP2). We investigated the potential role of miR-10a using the Swedish GWAS data.7 However, unlike miR-137, no significant GWAS hit was found in the vicinity of mirR-10a and the targets of miR-10a were not enriched for smaller GWAS P-values (data not shown).
Rare CNVs and significant GWAS loci
We asked whether CNVs were enriched in cases compared to controls in the genes implicated by GWAS meta-analysis of the Swedish samples and independent PGC samples (a total of 13,833 cases with SCZ and 18,310 controls).7 For 2,619 genes with GWAS P < 10−3, we observed significant enrichment for large CNVs (OR=1.21, 95% CI: 1.1–1.33, adj_Pemp=0.0049 for >500kb deletions and duplications combined, Table 3).
Rare CNV and common variant burden
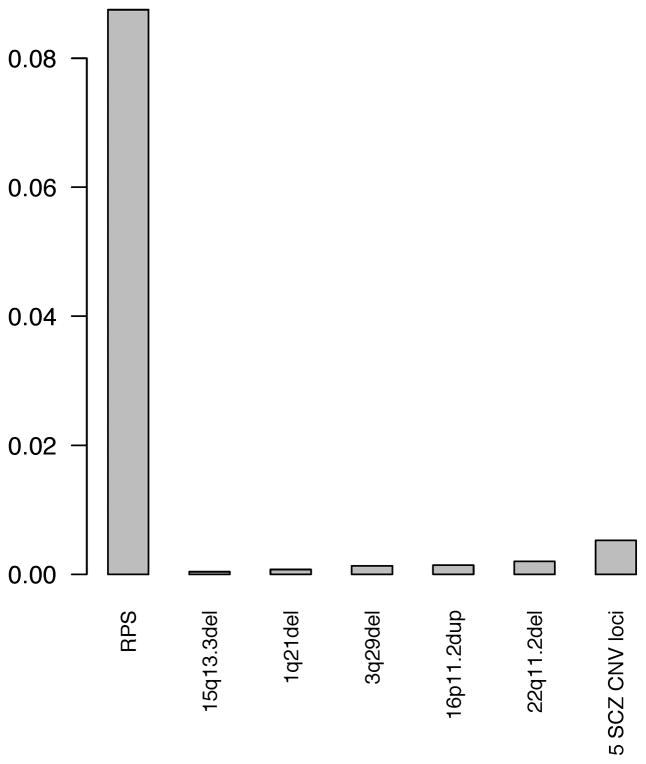
We computed the Spearman correlations between rare CNV burden and common variant burden (i.e., RPS), which were all near zero and not statistically significant (P > 0.05). Thus, the two types of genetic burden are uncorrelated. The relative impact of rare CNV burden and RPS burden was quantified by fitting multiple logistic regression models and estimating the effect size (R2). The two classes of variation were significantly, independently, and additively enriched in schizophrenia cases compared to controls (Table S18). Figure 4 and Tables S19 show the R2 values using CNV burden at known SCZ-associated loci (1q21.1del, 3q29del, 15q13.3del, 22q11.2del, 16p11.2dup, and all 5 loci combined). Figure S6 and Table S20 show the R2 values using burden of >100kb single-occurrence deletions, >500kb deletions, and CNVs in other categories. In summary, RPS accounted for at least an order of magnitude more variance than rare CNVs of strongest effects in this sample. Similar results were obtained using gene count and total length as CNV burden metrics (data not shown).
Figure 4.
Relative impact of rare CNV burden and common variant allelic burden. We computed the difference in the Nagelkerke pseudo R2 score to estimate the proportion of variance of case-control status in the Swedish samples accounted for by the common variant allelic burden (risk profile scores, RPS) and by the rare CNV burden (as measured by the number of CNV at known SCZ-associated loci). We examined CNV burden of 1q21.1del, 3q29del, 15p13.3del, 22q11.2del, 16p11.2dup, individually and combined. The Y-axis of the barplot shows the estimates of effect size (i.e. Nagelkerke pseudo R2). RPS accounted for at least an order of magnitude more variance than rare CNVs in this sample.
Exploratory analyses
We investigated the impact of deleterious mutations in BLM. Bloom syndrome is a rare Mendelian disorder characterized by genomic instability and excessive homologous recombination in affected individuals and is caused by recessive mutations in BLM. From exome sequencing in a subset of this sample (2,222 schizophrenia cases and 2,278 controls post-QC), there were 30 cases and 33 controls with a disruptive exonic mutation in BLM.14 The presence of a BLM mutation was not associated with case-control status (P = 0.88 chi-squared test), but individuals with BLM mutations had greater CNV burden compared to individuals without mutations in BLM (Table S21). For genic deletions, we observed a significantly (P = 0.0019) greater burden (as measured by the number of impacted genes) in individuals with deleterious BLM mutations (with the presence of a deleterious BLM mutation increasing CNV genic deletion burden by a mean of 0.96).
Discussion
This sample from Sweden provided an ideal CNV screening set since it is one of the largest schizophrenia samples yet collected in a single country and is relatively homogenous and genomically well-characterized. Furthermore, we had the opportunity to validate CNVs using exome array data from the same samples as well as test for replication using large scale samples from the UK. 61
Consistent with previous reports, our results confirm increases in CNV burden in cases and recapitulate known specific CNV risk loci. We also identified an association of duplications at 17q12 with schizophrenia that is reciprocal of the known 17q12 deletion. We did not identify any novel CNV loci as enriched among schizophrenia cases. Our power analyses suggest that most rare and recurrent CNVs carrying moderate to high risk have been discovered with GWAS arrays (Figure 1). The CNV burden analyses imply that additional recurrent CNVs do exist; as these were among the rarest events, far larger sample sizes will be required for their confident identification.
Our primary analyses applied a stringent size filter (≥100kb and spanning ≥15 probes) because these events can be most reliably detected from GWAS arrays. 11,13 For secondary CNV analyses, we used more liberal thresholds (≥ 20 kb and spanning ≥10 probes) because of tradeoffs between sensitivity and specificity. However, we did not observe additional novel findings that exceeded genome-wide significance.
Gene set enrichment testing replicated a number of findings from the literature and identified novel biological processes potentially related to schizophrenia pathophysiology. Consistent with previous GWAS, exome sequencing, and CNV findings, we found enrichment for neuronal calcium channel signaling, FMRP binding partners, glutamate receptors (NMDA and mGluR5). The FMRP targets were also linked to synaptic function and autism.44 Novel pathways identified included: signaling components within the immune system not encoded by the MHC region previously associated with schizophrenia (T cell receptor signaling, hematopoietic cell lineage, positive regulation of monocyte proliferation), new microRNA pathway (miR-10a) along with basic microRNA processing elements (e.g. DGCR8, ERI1, and NCBP2), and members of chromatin remodeling complexes (e.g. HDAC9, NCOR1). These novel pathways underscore the importance of the immune system and microRNA-based regulation of gene expression for schizophrenia etiology and suggest that MHC and miR-137 may be just the first components of larger biological networks yet to be identified. Furthermore, the detection of multiple pathways involved in transcriptional regulation is consistent with the schizophrenia GWAS results, where the vast majority of associated loci are thought to tag regulatory variation.
On the phenotype level, we found that CNVs in schizophrenia patients are enriched for genes associated with mental retardation, cognitive ability in humans as well as neurological or behavioral phenotypes in knockout mice. These results are consistent with the pleiotropic effects of large, rare CNVs previously described in the human genetics literature. Furthermore, the mouse finding suggests that murine models provide a biologically relevant system to examine the pathological effects of gene dosage (via knockout or overexpression) on brain and behavior. On the subcellular compartment level, we identified enrichment for gene products localized to three regions: the post-synaptic density, mitochondria, and cytoplasm. The synaptic enrichments are consistent with pathways mentioned above (neuronal calcium, glutamate, and FMRP signaling) as well as a growing body of literature implicating genes related to synaptic transmission in schizophrenia and bipolar disorder. 7,64,65
Mitochondria generate ATP by oxidative phosphorylation and play important roles in the regulation of cellular calcium levels, steroid synthesis, production of free radicals, and regulation of apoptosis. Brain is highly dependent on ATP generated by mitochondria. Mitochondrial dysfunction and a disturbance of energy metabolism have been observed in schizophrenia patients and, likewise, mitochondrial disorders can present with psychotic, affective, and cognitive symptoms. 62,63 Our data reveal that nuclear encoded genes that localize to the mitochondria are over-represented among large, rare CNVs in schizophrenia patients, lending credence to the mitochondrial dysfunction hypothesis.
By exome sequencing of over 5,000 of these subjects, Purcell et al.14 found that three classes of variation, as measure by rare CNV burden, common variant/RPS burden, and rare exonic variant burden, were uncorrelated and independently and additively enriched in SCZ cases compared to controls. Consistent with this report,14 we found that rare CNV burden and common variant/RPS burden are independently and additively enriched in SCZ cases. We note that it is currently unknown how environmental factors 5,6,66 interact with genetic burden. Consistent with Purcell et al.14, RPS burden accounted for at least an order of magnitude more variance than rare CNVs in this sample. This suggests that while the SCZ-associated rare and large CNVs are much more penetrant individually than SCZ-associated common SNPs, they contribute much less to the overall population risk of schizophrenia than do the SNPs.
In the exome sequencing study, Purcell et al.14 found increased burden of rare exonic mutations in genes implicated by GWAS and in a known CNV risk locus 3q29/DLG1. Here we found increased burden of rare CNVs in genes implicated by GWAS. GWAS results are robust as they were based on a meta-analysis of the Swedish samples and a much larger independent PGC samples. The associated common variants were far more distributed both in more subjects and from more sequences in the genome than rare exonic variants or large rare CNVs. The burden of rare exonic variants, common variants, and rare CNVs were uncorrelated and independent. Together, these findings suggest that multiple lines of genomic inquiry – genome-wide screens for CNVs, common variation, and exonic variation – are converging on similar sets of pathways and/or genes. This convergence helps to increase confidence in the robustness of the findings and provides results relevant to resolving the rare “versus” common variation debate. Further work will be required to characterize the potentially mechanisms of these pathways.
We observed increased genic deletions in individuals with exonic BLM mutation, which is consistent with the fact that BLM mutations increase homologous recombination in the affected individuals. To identify genomic loci influencing individual differences in CNV burden, future investigation using large-scale trio data would be informative in order to assess how parental genotypes associated with meiotic CNVs in offspring.
We believe it is possible that CNV discovery for schizophrenia has entered a difficult phase where almost all low-hanging fruit have been collected (i.e., large, recurrent CNVs measurable by SNP arrays). Increasing CNV discovery could well await dramatic improvements in sample size, technology, and cost. The detection of large, very rare recurrent CNVs will require marked increases in sample sizes. The application of whole genome sequencing to large case-control samples remains expensive but offers the possibility of confident identification of CNVs as small as 1 kb, particularly those that impact exons in single genes. 67 CNV technologies based on genotyping arrays do not perform well for copy number polymorphisms (frequency > 0.01), 68 and is it certainly possible that this class of variation has explanatory power in schizophrenia but is currently not well measured.
Supplementary Material
Acknowledgments
We are deeply grateful for the participation of all subjects contributing to this research, and to the team that conducted the fieldwork (Emma Flordal-Thelander, Ann-Britt Holmgren, Marie Hallin, Marie Lundin, Ann-Kristin Sundberg, Christina Pettersson, Radja Satgunanthan-Dawoud, Sonja Hassellund, Malin Rådstrom, Birgitta Ohlander, Leila Nyrén, and Isabelle Kizling). Funding support was provided by NIMH R01 MH077139 (Sullivan), NIMH R01 MH095034 (Sklar), NIMH K01 MH093517 (Szatkiewicz), the Stanley Center for Psychiatric Research, the Sylvan Herman Foundation, the Karolinska Institutet, Karolinska University Hospital, the Swedish Research Council, the Swedish County Council, the Söderström Königska Foundation, and the Netherlands Scientific Organization (NWO 645-000-003). The work at UK was funded by Medical Research Council (MRC) Centre (G0800509) and Program Grants (G0801418), the European Community’s Seventh Framework Programme (HEALTH-F2-2010-241909 (Project EU-GEI), an MRC PhD Studentship to ER, a clinical research fellowship to JTRW from the MRC/Welsh Assembly Government and the Margaret Temple Award from the British Medical Association. The UK samples were genotyped at the Broad Institute, USA, funded by a philanthropic gift to the Stanley Center for Psychiatric Research. The funders had no role in study design, execution, analysis, and manuscript preparation.
The authors thank two anonymous reviewers for their helpful comments. All authors reviewed and approved the final version of the manuscript. The corresponding authors had access to the full dataset.
Footnotes
URLs
R project “gap” package, http://cran.r-project.org/web/packages/gap/index.html. SHAPEIT software, http://www.shapeit.fr/pages/m02_formats/hapssample.html.
Conflicts of Interest
Dr Sullivan was on the SAB of Expression Analysis (Durham, NC, USA). Dr Sklar is on the Board of Directors of Catalytic, Inc. The other authors report no conflicts.
References
- 1.Saha S, Chant D, McGrath J. A systematic review of mortality in schizophrenia: is the differential mortality gap worsening over time? Archives of general psychiatry. 2007;64:1123–31. doi: 10.1001/archpsyc.64.10.1123. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. The Global Burden of Disease: 2004 Update. WHO Press; Geneva: 2008. [Google Scholar]
- 3.Knapp M, Mangalore R, Simon J. The global costs of schizophrenia. Schizophrenia bulletin. 2004;30:279–93. doi: 10.1093/oxfordjournals.schbul.a007078. [DOI] [PubMed] [Google Scholar]
- 4.Lichtenstein P, et al. The Swedish Twin Registry in the Third Millennium – an update. Twin Res Hum Genet. 2006;9:875–882. doi: 10.1375/183242706779462444. [DOI] [PubMed] [Google Scholar]
- 5.Lichtenstein P, et al. Common genetic influences for schizophrenia and bipolar disorder: A population-based study of 2 million nuclear families. Lancet. 2009;373:234–9. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Archives of General Psychiatry. 2003;60:1187–92. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 7.Ripke S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature Genetics. 2013;45:1150–9. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamshere ML, et al. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Molecular psychiatry. 2012 doi: 10.1038/mp.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Genome-wide association study identifies five new schizophrenia loci. Nature Genetics. 2011;43:969–76. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levinson DF, et al. Copy number variants in schizophrenia: Confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–16. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malhotra D, Sebat J. CNVs: Harbingers of a Rare Variant Revolution in Psychiatric Genetics. Cell. 2012;148:1223–41. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirov G, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Molecular psychiatry. 2012;17:142–53. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nature Reviews Genetics. 2012;13:537–51. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell SM, et al. A burden of ultra-rare disruptive mutations concentrated in synaptic gene networks increases risk of schizophrenia. Nature. 2014 [Google Scholar]
- 15.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–43. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 16.Kirov G, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buizer-Voskamp JE, et al. Genome-wide analysis shows increased frequency of copy number variation deletions in Dutch schizophrenia patients. Biological psychiatry. 2011;70:655–62. doi: 10.1016/j.biopsych.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malhotra D, et al. High Frequencies of De Novo CNVs in Bipolar Disorder and Schizophrenia. Neuron. 2011;72:951–63. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szatkiewicz JP, et al. Detecting large copy number variants using exome genotyping arrays. Molecular Psychiatry. 2013 doi: 10.1038/mp.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergen SE, et al. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared to bipolar disorder. Molecular Psychiatry. 2012;17:880–6. doi: 10.1038/mp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristjansson E, Allebeck P, Wistedt B. Validity of the diagnosis of schizophrenia in a psychiatric inpatient register. Nordisk Psykiatrik Tidsskrift. 1987;41:229–34. [Google Scholar]
- 23.Dalman C, Broms J, Cullberg J, Allebeck P. Young cases of schizophrenia identified in a national inpatient register--are the diagnoses valid? Social Psychiatry and Psychiatric Epidemiology. 2002;37:527–31. doi: 10.1007/s00127-002-0582-3. [DOI] [PubMed] [Google Scholar]
- 24.Hultman CM, Sparen P, Takei N, Murray RM, Cnattingius S. Prenatal and perinatal risk factors for schizophrenia, affective psychosis, and reactive psychosis of early onset: case-control study. Bmj. 1999;318:421–6. doi: 10.1136/bmj.318.7181.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zammit S, et al. Investigating the association between cigarette smoking and schizophrenia in a cohort study. Am J Psychiatry. 2003;160:2216–21. doi: 10.1176/appi.ajp.160.12.2216. [DOI] [PubMed] [Google Scholar]
- 26.Andersson RE, Olaison G, Tysk C, Ekbom A. Appendectomy and protection against ulcerative colitis. N Engl J Med. 2001;344:808–14. doi: 10.1056/NEJM200103153441104. [DOI] [PubMed] [Google Scholar]
- 27.Hansson LE, et al. The risk of stomach cancer in patients with gastric or duodenal ulcer disease. N Engl J Med. 1996;335:242–9. doi: 10.1056/NEJM199607253350404. [DOI] [PubMed] [Google Scholar]
- 28.Kendler KS. The super-normal control group in psychiatric genetics: possible artifactual evidence for coaggregation. Psychiatric Genetics. 1990;1:45–53. [Google Scholar]
- 29.Schwartz S, Susser E. The use of well controls: an unhealthy practice in psychiatric research. Psychol Med. 2010:1–6. doi: 10.1017/S0033291710001595. [DOI] [PubMed] [Google Scholar]
- 30.Hartge P. Participation in population studies. Epidemiology. 2006;17:252–4. doi: 10.1097/01.ede.0000209441.24307.92. [DOI] [PubMed] [Google Scholar]
- 31.Morton LM, Cahill J, Hartge P. Reporting participation in epidemiologic studies: a survey of practice. Am J Epidemiol. 2006;163:197–203. doi: 10.1093/aje/kwj036. [DOI] [PubMed] [Google Scholar]
- 32.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–60. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2011. [Google Scholar]
- 35.Sanders SJ, et al. Multiple Recurrent De Novo CNVs, Including Duplications of the 7q11.23 Williams Syndrome Region, Are Strongly Associated with Autism. Neuron. 2011;70:863–85. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper GM, et al. A copy number variation morbidity map of developmental delay. Nature genetics. 2011;43:838–46. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox D. The continuity correction. Biometrika. 1970;57:217–9. [Google Scholar]
- 38.Flicek P, et al. Ensembl 2013. Nucleic acids research. 2013;41:D48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha S, Chant D, Welham J, McGrath J. A systematic review of the prevalence of schizophrenia. PLoS Medicine. 2005;2:e141. doi: 10.1371/journal.pmed.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raychaudhuri S, et al. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic acids research. 2012;40:D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.GO Project. The Gene Ontology: enhancements for 2011. Nucleic acids research. 2012;40:D559–64. doi: 10.1093/nar/gkr1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mi H, et al. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic acids research. 2010;38:D204–10. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jupe S, Akkerman JW, Soranzo N, Ouwehand WH. Reactome - a curated knowledgebase of biological pathways: megakaryocytes and platelets. Journal of thrombosis and haemostasis: JTH. 2012 doi: 10.1111/j.1538-7836.2012.04930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am J Hum Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 47.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–61. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruano D, et al. Functional gene group analysis reveals a role of synaptic heterotrimeric G proteins in cognitive ability. American journal of human genetics. 2010;86:113–25. doi: 10.1016/j.ajhg.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Croning MD, Marshall MC, McLaren P, Armstrong JD, Grant SG. G2Cdb: the Genes to Cognition database. Nucleic acids research. 2009;37:D846–51. doi: 10.1093/nar/gkn700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- 51.Chiurazzi P, Schwartz CE, Gecz J, Neri G. XLMR genes: update 2007. European journal of human genetics: EJHG. 2008;16:422–34. doi: 10.1038/sj.ejhg.5201994. [DOI] [PubMed] [Google Scholar]
- 52.Inlow JK, Restifo LL. Molecular and comparative genetics of mental retardation. Genetics. 2004;166:835–81. doi: 10.1534/genetics.166.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Najmabadi H, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 54.Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2011;39:D842–8. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maddatu TP, Grubb SC, Bult CJ, Bogue MA. Mouse Phenome Database (MPD) Nucleic acids research. 2011 doi: 10.1093/nar/gkl1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–23. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holm S. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics. 1979;6:65–70. [Google Scholar]
- 59.International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruderfer DM, et al. Mosaic copy number variation in schizophrenia. European journal of human genetics: EJHG. 2013 doi: 10.1038/ejhg.2012.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rees E, et al. Evidence that duplications of 22q11.2 protect against schizophrenia. Molecular Psychiatry. 2013 doi: 10.1038/mp.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manji H, et al. Impaired mitochondrial function in psychiatric disorders. Nature reviews Neuroscience. 2012;13:293–307. doi: 10.1038/nrn3229. [DOI] [PubMed] [Google Scholar]
- 63.Robicsek O, et al. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Molecular psychiatry. 2013;18:1067–76. doi: 10.1038/mp.2013.67. [DOI] [PubMed] [Google Scholar]
- 64.Ferreira M, et al. Collaborative genome-wide association analysis of 10,596 individuals supports a role for Ankyrin-G (ANK3) and the alpha-1C subunit of the L-type voltage-gated calcium channel (CACNA1C) in bipolar disorder. Nature Genetics. 2008;40:1056–8. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Craddock N, Sklar P. Genetics of bipolar disorder. Lancet. 2013;381:1654–62. doi: 10.1016/S0140-6736(13)60855-7. [DOI] [PubMed] [Google Scholar]
- 66.Murray RM, Jones PB, Susser E, van Os J, Cannon M. The Epidemiology of Schizophrenia. Cambridge University Press; Cambridge, UK: 2003. [Google Scholar]
- 67.Handsaker RE, Korn JM, Nemesh J, McCarroll SA. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet. 2011;43:269–76. doi: 10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–74. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.