Abstract
Background
Intercellular adhesion molecule-1 (ICAM-1) K469E polymorphism has been implicated in susceptibility to coronary artery disease (CAD). Several studies investigated the association of this polymorphism with CAD in different populations but the results were contradictory. A meta-analysis was conducted to assess the association between ICAM-1 K469E polymorphism and CAD susceptibility.
Material/Methods
Databases including PubMed, EMBASE, China National Knowledge Infrastructure (CNKI), and Weipu Database were searched to find relevant studies. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the strength of associations. A random-effects model was used.
Results
Fifteen case-control studies including 3088 cases and 3466 controls were included. Overall, a significant association between ICAM-1 K469E polymorphism and CAD was observed in the dominant model (OR=1.80; 95% CI 1.62–2.01; P<0.00001; Pheterogeneity=0.40). In subgroup analysis by ethnicity, a significant association was found among Asians (OR=1.92; 95% CI 1.51–2.43; P<0.00001; Pheterogeneity=0.98) and among Caucasians (OR=1.64; 95% CI 1.30–2.08; P<0.0001; Pheterogeneity=0.04). In the subgroup analysis by age, a significant association was found among young patients (OR=1.46; 95% CI 1.10–1.93; P=0.008; Pheterogeneity=0.21) and old patients (OR=1.92; 95% CI 1.75–2.10; P<0.00001; Pheterogeneity=0.99).
Conclusions
Results of this meta-analysis suggest that ICAM-1 K469E polymorphism confers a risk factor of CAD.
MeSH Keywords: Coronary Artery Disease; Intercellular Adhesion Molecule-1; Meta-Analysis; Polymorphism, Single Nucleotide
Background
Coronary artery disease (CAD) is the leading cause of death worldwide. Genetic susceptibility to CAD may be determined by specific polymorphic variants that encode proteins involved in the atherosclerotic processes. Adhesion molecules are one of the main markers of endothelial dysfunction.
Intercellular adhesion molecule-1 (ICAM-1) is widely distributed and expressed constitutively at low levels on leukocytes, vascular endothelial cells, fibroblasts, and epithelial cells. ICAM-1 specifically participates in trafficking of inflammatory cells, in leukocyte effector functions, in adhesion of antigen-presenting cells to T lymphocytes, in microbial pathogenesis, and in signal transduction pathways through outside-in signaling events [1]. ICAM-1 plays an important role in the adhesion of circulating leukocytes to the blood vessel wall and transendothelial migration to the vascular intima [2]. An elevated level of soluble ICAM-1 (sICAM-1) was observed in patients with confirmed coronary or cerebral atherosclerosis [3]. Furthermore, sICAM-1 concentrations have been associated with future CAD risk [4].
The ICAM-1 gene is located in 19p13.2 and its K469E polymorphism (rs5498) has been suggested to have functional activity [5]. This polymorphism was suggested to affect mRNA splicing patterns that modify cell–cell interactions and influence inflammatory response [5]. In addition, this variant might have possible functional value in the etiology of atherosclerosis [6]. Studies of the association between ICAM-1 K469E polymorphism and CAD risk [7–21] have yielded conflicting and inconclusive results. Thus, we carried out a meta-analysis to ascertain whether there was a genetic effect of ICAM-1 K469E polymorphism on CAD susceptibility.
Material and Methods
Publication search
We conducted a systematic literature search using the databases: PubMed, EMBASE, and Weipu (last search was updated March 2014). The search terms were: (“coronary artery disease” or CAD or “coronary heart disease” or CHD) and (“intercellular adhesion molecule-1” or ICAM-1) and (polymorphism or mutation or variant). No publication date or language restrictions were imposed. All the searched studies were retrieved, and their references were checked for other relevant publications. Review articles were also searched to find additional eligible studies.
Inclusion and exclusion criteria
Case-control or cohort studies with sufficient published data for estimating an odds ratio (OR) and corresponding 95% confidence interval (CI) were included in this meta-analysis. Studies were excluded if any of the following criteria existed: (1) the studies were not relevant to ICAM-1 K469E polymorphism or CAD, (2) non-clinical studies or the design based on family or sibling pairs, (3) genotype frequencies or number were not reported, or (4) reviews, abstracts, or comments. For overlapping studies, only the 1 with the largest sample size was included.
Data extraction
Two authors independently reviewed full manuscripts of eligible studies. The following variables were extracted from each study, if available: first author’s surname, year of publication, ethnicity, age, sample size, and genotype numbers in cases and controls.
Methodological quality assessment
Two authors completed the quality assessment independently. The Newcastle–Ottawa Scale (NOS) was used to evaluate the methodological quality, which scored studies by the selection of the study groups, the comparability of the groups, and the ascertainment of the outcome of interest [22–24]. We considered a study that was awarded 0–3, 4–6, and 7–9 as a low-, moderate-, or high-quality study, respectively. Discrepancies were resolved by consensus and discussion.
Statistical analysis
Bielinski et al. suggested that ICAM-1 K469 caused an increase in sICAM-1 expression [25]. Thus, the strength of association between the ICAM-1 K469E polymorphism and CAD risk was measured by OR and 95% CI in the dominant model (KK + KE vs. EE). A random-effects model, using the inverse variance method, was used to calculate the pooled ORs. The statistical significance of an OR was determined with the Z test.
Hardy-Weinberg equilibrium (HWE) was tested using the chi-squared test and it was considered statistically significant when P<0.05. Heterogeneity was evaluated by Q statistic and was considered statistical significant at P value <0.10. Subgroup analyses were performed by ethnicity and age. Sensitivity analysis was performed through sequentially excluded individual studies to assess the stability of the results. In addition, sensitivity analysis was also conducted by omitting the studies not in HWE and the studies with small sample size (n<200). Publication bias was analyzed by several methods. Visual inspection of asymmetry in funnel plots was carried out. Egger’s test was also used to statistically assess publication bias [26].
All statistical tests were performed by using the Review Manager 5.1.2 (2011, The Cochrane Collaboration) and STATA 11.0 software (Stata Corporation, College Station, TX). A P value <0.05 was considered statistically significant.
Results
Study characteristics
A total of 15 case-control studies with 3088 cases and 3466 controls met our inclusion criteria [7–21]. There were 8 studies of Asians and 7 studies of Caucasians. Six studies were performed in young patients (age <60), and 9 studies were conducted in old patients (age >60). Six studies were not in HWE. The quality scores ranged from 5 to 9, suggesting that the methodological quality was generally acceptable. The characteristics of each study and genotype distribution are presented in Table 1.
Table 1.
Studies and data included in this meta-analysis.
| Study | Year | Ethnicity | Age of Case | No. of Case | No. of Control | Case | Control | HWE | Score | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KK | KE | EE | KK | KE | EE | ||||||||
| Jiang | 2002 | Caucasian | 62.2 | 349 | 213 | 139 | 148 | 62 | 60 | 66 | 87 | No | 8 |
| Zak | 2005 | Caucasian | 45.9 | 146 | 121 | 12 | 86 | 48 | 8 | 68 | 45 | No | 6 |
| Shang | 2005 | Asian | 65.9 | 89 | 117 | 33 | 39 | 17 | 32 | 45 | 109 | No | 6 |
| Wang | 2005 | Asian | 64.9 | 165 | 199 | 96 | 61 | 8 | 91 | 90 | 18 | Yes | 7 |
| Podgoreanu | 2006 | Caucasian | 63.7 | 52 | 382 | 14 | 26 | 12 | 50 | 177 | 155 | Yes | 8 |
| Wei | 2006 | Asian | 64.1 | 225 | 230 | 124 | 84 | 17 | 101 | 103 | 26 | Yes | 6 |
| Zhang | 2006 | Asian | 68.0 | 173 | 141 | 111 | 52 | 10 | 69 | 59 | 13 | Yes | 5 |
| Aminian | 2007 | Caucasian | 56.6 | 303 | 141 | 90 | 144 | 64 | 36 | 69 | 35 | Yes | 8 |
| Wen | 2008 | Asian | 64.0 | 71 | 164 | 28 | 30 | 13 | 40 | 65 | 59 | No | 5 |
| Zhou | 2008 | Asian | 59.2 | 103 | 197 | 20 | 45 | 38 | 33 | 62 | 102 | No | 7 |
| Sarecka-Hujar | 2009 | Caucasian | 43.8 | 191 | 203 | 12 | 118 | 61 | 8 | 122 | 73 | No | 8 |
| Sakowicz | 2010 | Caucasian | 41.0 | 160 | 131 | 106* | 54 | 14 | 69 | 48 | Yes | 6 | |
| Li | 2010 | Asian | 58.6 | 93 | 101 | 47 | 39 | 7 | 52 | 36 | 13 | Yes | 7 |
| Liu | 2011 | Asian | 64.7 | 312 | 302 | 178 | 112 | 22 | 130 | 138 | 34 | Yes | 8 |
| Buraczynska | 2012 | Caucasian | 62.8 | 656 | 824 | 288 | 340 | 28 | 272 | 379 | 173 | Yes | 9 |
The combined number of KK and KE genotypes.
HWE – Hardy-Weinberg equilibrium; NA – not available.
Quantitative data synthesis
All studies
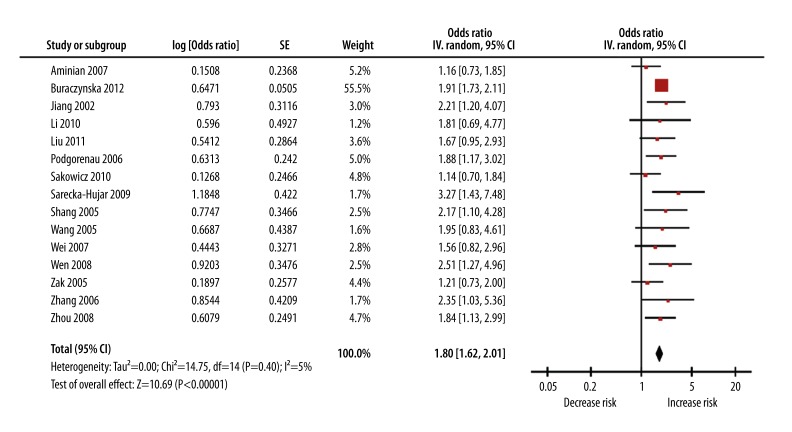
The pooled OR was 1.80 (95% CI 1.62–2.01) and the Z test for overall effect was 10.69 (P<0.00001) (Figure 1). There was small heterogeneity (I2=5% and Pheterogeneity=0.40).
Figure 1.
Meta-analysis with a random-effects model for the association between ICAM-1 K469E polymorphism and CAD risk.
Subgroup analyses
In the subgroup analysis by ethnicity, a significant association was found among Asians (OR=1.92; 95% CI 1.51–2.43; P<0.00001; Pheterogeneity=0.98) and among Caucasians (OR=1.64; 95% CI 1.30–2.08; P<0.0001; Pheterogeneity=0.04) in the dominant genetic model. In the subgroup analysis by age, a significant association was found among young patients (OR=1.46; 95% CI 1.10–1.93; P=0.008; Pheterogeneity=0.21). There was a significant association between ICAM-1 K469E polymorphism and CAD risk among old patients (OR=1.92; 95% CI 1.75–2.10; P<0.00001; Pheterogeneity=0.99).
Sensitivity analysis
To assess the stability of the results of the meta-analysis, we performed a sensitivity analysis through sequentially excluding individual studies. Statistically similar results were obtained after sequentially excluding each study (data not shown). Omitting the studies deviating from HWE also did not change the result (OR=1.75; 95% CI 1.52–2.00; P<0.00001; Pheterogeneity=0.35). Additionally, when the studies with small sample sizes were excluded, the result was still significant (OR=1.86; 95% CI 1.70–2.05; P<0.00001; Pheterogeneity=0.42).
Publication bias
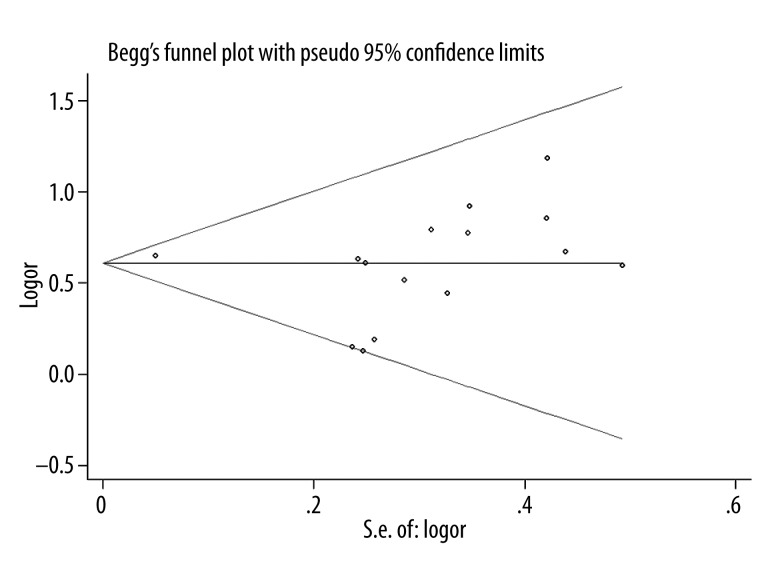
Funnel plot was performed to assess the publication bias in this meta-analysis; it showed a symmetrical inverse funnel shape (Figure 2). Egger’s test also indicated no significant publication bias (P=0.584).
Figure 2.
Funnel plot for ICAM-1 K469E polymorphism and CAD risk.
Discussion
This meta-analysis of 15 case-control studies including 3088 cases and 3466 controls evaluated the association between ICAM-1 K469E polymorphism and CAD risk. The result indicate that ICAM-1 K469E polymorphism might be a risk factor for developing CAD. Our results suggest that individuals who carry the KK or KE genotype may have an 80% increased CAD risk compared with EE genotype carriers. In the subgroup analysis by ethnicity, both significant associations were found in Asians and Caucasians, suggesting that both Asians and Caucasians who carry the KK or KE genotype might have an increased CAD risk. In the subgroup analysis by age, we found ICAM-1 K469E polymorphism showed increased early-onset CAD risk and late-onset CAD risk. This result indicates that the role of ICAM-1 K469E polymorphism is not selective by age.
Many recent studies suggest that sICAM-1 plays an important in the development of CAD. For example, Hulthe et al. showed that levels of sICAM-1 were associated with subclinical atherosclerosis and inflammatory variables [27]. Bongard et al. [28] reported that sICAM-1 was independently associated with the risk of having at least 1 carotid plaque and with the risk of having at least 1 femoral plaque. Furthermore, ICAM-1 K469E polymorphism could influence the levels of sICAM-1. Genome-wide association analysis indicated that ICAM-1 K469E polymorphism had a role in the genetic regulation of sICAM-1 levels [29]. In addition, Reilly et al. found that ICAM-1 K469E EE genotype was associated with lower coronary artery calcification (CAC) scores in men [30]. Taken together, these data indicate that ICAM-1 K469E polymorphism can influence the risk of CAD.
The role of the IL-33/ST2 signaling pathway in ischemic heart disease has been reported [31]. Choi et al. found that IL-33 mediated the expression of ICAM-1 and vascular cell adhesion molecule (VCAM)-1 in endothelial cells [32]. sST2 seems to act as a decoy-receptor for IL-33: it binds IL-33, thus, subtracting such a molecule from the interaction with ST2L [31]. Therefore, it is interesting to investigate whether sST2 can influence the expression of ICAM-1.
Heterogeneity is an important issue when interpreting the results of meta-analyses. In our meta-analysis, there was no significant heterogeneity in most of the comparisons and no publication bias was found. Therefore, heterogeneity and publication bias did not influence the results. We also conducted sensitivity analyses and no individual study was found to affect the overall result. Excluding the studies not in HWE or with small sample size also did not influence the overall result. These results indicate that our results are stable and robust. However, several limitations of this study should be addressed. First, the sample size was still relatively small for some stratified analyses. Second, only published studies were included in the meta-analysis; therefore, publication bias may have occurred, even though the use of a statistical test did not show it. Third, we were unable to obtain information from most studies on the presence or absence of a history of smoking, because of lack of the investigation of gene-environment interactions [33]. Finally, our meta-analysis was based on unadjusted estimates, whereas a more precise analysis could be performed if individual data were available and would allow for an adjustment estimate. Despite the limitations, our meta-analysis significantly increased the statistical power of the analysis based on the large number of cases and controls from different studies.
Conclusions
Results of this meta-analysis suggest that the ICAM-1 K469E polymorphism is a risk factor for CAD. Future large-scale studies are needed to validate our findings.
Footnotes
Conflicts of interest
None.
Source of support: Self financing
References
- 1.Hubbard AK, Rothlein R. Intercellular adhesion molecule-1 (ICAM-1) expression and cell signaling cascades. Free Radic Biol Med. 2000;28:1379–86. doi: 10.1016/s0891-5849(00)00223-9. [DOI] [PubMed] [Google Scholar]
- 2.Hayflick JS, Kilgannon P, Gallatin WM. The intercellular adhesion molecule (ICAM) family of proteins. New members and novel functions. Immunol Res. 1998;17:313–27. doi: 10.1007/BF02786454. [DOI] [PubMed] [Google Scholar]
- 3.Languino LR, Plescia J, Duperray A, et al. Fibrinogen mediates leukocyte adhesion to vascular endothelium through an ICAM-1-dependent pathway. Cell. 1993;73:1423–34. doi: 10.1016/0092-8674(93)90367-y. [DOI] [PubMed] [Google Scholar]
- 4.Hwang SJ, Ballantyne CM, Sharrett AR, et al. Circulating adhesion molecules VCAM-1, ICAM-1, and E-selectin in carotid atherosclerosis and incident coronary heart disease cases: the Atherosclerosis Risk In Communities (ARIC) study. Circulation. 1997;96:4219–25. doi: 10.1161/01.cir.96.12.4219. [DOI] [PubMed] [Google Scholar]
- 5.Iwao M, Morisaki H, Morisaki T. Single-nucleotide polymorphism g.1548G > A (E469K) in human ICAM-1 gene affects mRNA splicing pattern and TPA-induced apoptosis. Biochem Biophys Res Commun. 2004;317:729–35. doi: 10.1016/j.bbrc.2004.03.101. [DOI] [PubMed] [Google Scholar]
- 6.Gaetani E, Flex A, Pola R, et al. The K469E polymorphism of the ICAM-1 gene is a risk factor for peripheral arterial occlusive disease. Blood Coagul Fibrinolysis. 2002;13:483–88. doi: 10.1097/00001721-200209000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Jiang H, Klein RM, Niederacher D, et al. C/T polymorphism of the intercellular adhesion molecule-1 gene (exon 6, codon 469). A risk factor for coronary heart disease and myocardial infarction. Int J Cardiol. 2002;84:171–77. doi: 10.1016/s0167-5273(02)00138-9. [DOI] [PubMed] [Google Scholar]
- 8.Zak I, Balcerzyk A, Sarecka B, et al. Contemporaneous carrier-state of two or three “proatherosclerotic” variants of APOE, ICAM1, PPARA and PAI-1 genes differentiate CAD patients from healthy individuals. Clin Chim Acta. 2005;362:110–18. doi: 10.1016/j.cccn.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Shang Q, Lu FH, Wen PE, et al. The study of intercellular adhesion molecule-1 gene polymorphisms C469T in elderly patients with coronary heart disease. Chin J Geriatr. 2005;24:444–45. [Google Scholar]
- 10.Wang M, Li Y, Zhang PA, et al. Study on the intercellular adhesion molecule-1 gene polymorphisms in a Chinese population with myocardial infarction. Zhonghua Liu Xing Bing Xue Za Zhi. 2005;26:702–6. [PubMed] [Google Scholar]
- 11.Podgoreanu MV, White WD, Morris RW, et al. Inflammatory gene polymorphisms and risk of postoperative myocardial infarction after cardiac surgery. Circulation. 2006;114:I275–81. doi: 10.1161/CIRCULATIONAHA.105.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei YS, Tang RG, Yuan XH, et al. Association between polymorphism of intercellular adhesion molecule-1 gene K469E and coronary heart disease. Zhongguo Mian Yi Xue Za Zhi. 2006;22:1056–59. [Google Scholar]
- 13.Zhang SR, Xu LX, Gao QQ, et al. The correlation betweenICAM-1 gene K469E polymorphism and coronary heart disease. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2006;23:205–7. [PubMed] [Google Scholar]
- 14.Aminian B, Abdi Ardekani AR, Arandi N. ICAM-1 polymorphisms (G241R, K469E), in coronary artery disease and myocardial infarction. Iran J Immunol. 2007;4:227–35. [PubMed] [Google Scholar]
- 15.Wen PE, Lu FH, Zhou W, et al. Study on relationship between K/E polymorphism and angina. Zhongguo Gong Gong Wei Sheng. 2008;24:808–9. [Google Scholar]
- 16.Zhou YL, Zhu MA, Ding Y. Association of intercellular adhesion molecule-1 gene polymorphism and coronary heart disease. J Pract Diagn Ther. 2008;22:581–84. [Google Scholar]
- 17.Sarecka-Hujar B, Zak I, Krauze J. Interactions between rs5498 polymorphism in the ICAM1 gene and traditional risk factors influence susceptibility to coronary artery disease. Clin Exp Med. 2009;9:117–24. doi: 10.1007/s10238-008-0022-0. [DOI] [PubMed] [Google Scholar]
- 18.Sakowicz A, Fendler W, Lelonek M, Pietrucha T. Genetic variability and the risk of myocardial infarction in Poles under 45 years of age. Arch Med Sci. 2010;6:160–67. doi: 10.5114/aoms.2010.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li YJ, Han M, Zheng B, et al. Relationship between intercellular adhesion molecule-1 K469E gene polymorphism and coronary heart disease. Chin J Gerontol. 2010;30:3494–95. [Google Scholar]
- 20.Liu W, Wei Y, Tan Z. Relationship between intercellular adhesion molecule-1 K469E gene polymorphism and coronary heart disease in Zhuang. Chin J Gerontol. 2011;30:581–82. [Google Scholar]
- 21.Buraczynska M, Zaluska W, Baranowicz-Gaszczyk I, et al. The intercellular adhesion molecule-1 (ICAM-1) gene polymorphism K469E in end-stage renal disease patients with cardiovascular disease. Hum Immunol. 2012;73:824–28. doi: 10.1016/j.humimm.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. 2011. URL: http://www.ohri.ca/programs/clinical_epidemiology/oxford.Asp.
- 23.Guan HB, Wu L, Wu QJ, et al. Parity and pancreatic cancer risk: a dose-response meta-analysis of epidemiologic studies. PLoS One. 2014;9:e92738. doi: 10.1371/journal.pone.0092738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L, Jiang Z, Li C, Shu M. Prediction of heart rate variability on cardiac sudden death in heart failure patients: A systematic review. Int J Cardiol. 2014;174:857–60. doi: 10.1016/j.ijcard.2014.04.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bielinski SJ, Reiner AP, Nickerson D, et al. Polymorphisms in the ICAM1 gene predict circulating soluble intercellular adhesion molecule-1(sICAM-1) Atherosclerosis. 2011;216:390–94. doi: 10.1016/j.atherosclerosis.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hulthe J, Wikstrand J, Mattsson-Hultén L, Fagerberg B. Circulating ICAM-1 (intercellular cell-adhesion molecule 1) is associated with early stages of atherosclerosis development and with inflammatory cytokines in healthy 58-year-old men: the Atherosclerosis and Insulin Resistance (AIR) study. Clin Sci (Lond) 2002;103:123–29. doi: 10.1042/cs1030123. [DOI] [PubMed] [Google Scholar]
- 28.Bongard V, Elias A, Bal dit Sollier C, et al. Soluble intercellular adhesion molecule-1 is associated with carotid and femoral atherosclerosis but not with intima-media thickness in a population-based sample. Atherosclerosis. 2002;164:297–304. doi: 10.1016/s0021-9150(02)00071-0. [DOI] [PubMed] [Google Scholar]
- 29.Paré G, Ridker PM, Rose L, et al. Genome-wide association analysis of soluble ICAM-1 concentration reveals novel associations at the NFKBIK, PNPLA3, RELA, and SH2B3 loci. PLoS Genet. 2011;7:e1001374. doi: 10.1371/journal.pgen.1001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reilly MP, Wolfe ML, Dykhouse J, et al. Intercellular adhesion molecule 1 (ICAM-1) gene variant is associated with coronary artery calcification independent of soluble ICAM-1 levels. J Investig Med. 2004;52:515–22. doi: 10.1136/jim-52-08-23. [DOI] [PubMed] [Google Scholar]
- 31.Ciccone MM, Cortese F, Gesualdo M, et al. A novel cardiac bio-marker: ST2: a review. Molecules. 2013;18:15314–28. doi: 10.3390/molecules181215314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi YS, Park JA, Kim J, et al. Nuclear IL-33 is a transcriptional regulator of NF-κB p65 and induces endothelial cell activation. Biochem Biophys Res Commun. 2012;421:305–11. doi: 10.1016/j.bbrc.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Gloria-Bottini F, Banci M, Saccucci P, et al. p53 codon 72 polymorphism and coronary artery disease: evidence of interaction with ACP1. Med Sci Monit. 2012;18(12):CR712–15. doi: 10.12659/MSM.883597. [DOI] [PMC free article] [PubMed] [Google Scholar]