Abstract
The LXCXE peptide motif facilitates interaction between the RB tumor suppressor and a large number of cellular proteins that are expected to impinge on diverse biological processes. In vitro and in vivo analyses demonstrated that LXCXE-binding function is dispensable for RB promoter association and control of basal gene expression. Dependence on this function of RB is unmasked after DNA damage, wherein LXCXE-binding is essential for exerting control over E2F3 and suppressing cell cycle progression in the presence of genotoxic stress. Gene expression profiling revealed that the transcriptional program coordinated by this specific aspect of RB is associated with progression of human hepatocellular carcinoma and poor disease outcome. Consistent with these findings, biological challenge revealed a requirement for LXCXE-binding in suppression of genotoxin-initiated hepatocellular carcinoma in vivo. Together, these studies establish an essential role of the LXCXE-binding motif for RB-mediated transcriptional control, response to genotoxic insult, and tumor suppression.
Keywords: RB, N750F, Hepatocellular Carcinoma, LXCXE, Tumor Suppressor
INTRODUCTION
The retinoblastoma tumor suppressor (RB) is functionally inactivated in a large fraction of human tumors (Burkhart and Sage, 2008; Knudsen and Knudsen, 2008). RB interacts with both transcription factors and transcriptional co-repressors (Blais and Dynlacht, 2007; Zhang and Dean, 2001) to effectively mediate transcriptional repression of target genes (Markey et al., 2002; Nevins, 2001). This RB-mediated transcriptional repression has been hypothesized to be critically dependent on recruitment of co-repressors via a highly conserved LXCXE binding domain (Lee et al., 1998).
In distinct models RB plays a potent role in actively suppressing proliferation, controlling DNA damage checkpoint responses, or promoting differentiation (Burkhart and Sage, 2008; Knudsen et al., 2006). Thus, specific functional deficits of RB, such as those arising in the LXCXE-binding motif, could be relevant to tumor suppression or regulation of disease progression through distinct mechanisms. Consistent with this concept, the LXCXE binding pocket is targeted by viral oncoproteins, coordinates interaction with multiple proteins of biological significance, and is mutated in cancer (Henley et al., 2010; Morris and Dyson, 2001).
The specific role of this domain for RB function has been interrogated using targeted mutations based on the RB crystal structure (Chen and Wang, 2000; Dick et al., 2000). Interestingly, despite yielding strong resistance to LXCXE-containing oncogenes (Balsitis et al., 2005), these mutations had relatively modest effects on transcriptional control and cell cycle inhibition in cell culture models (Chen and Wang, 2000; Isaac et al., 2006). However, in the paradigms of myogenic differentiation or oncogene-induced senescence, there were specific deficits in the establishment of gene silencing (Talluri et al., 2010). Parallel studies have suggested that LXCXE-binding function could also be important for coordinating chromatin structure, mitotic fidelity, and modulating the intrinsic tumor susceptibility of p53 deficiency (Coschi et al., 2010; Francis et al., 2011; Manning et al., 2010; Talluri et al., 2010). However, recent studies also indicate that LXCXE-function has no effect on ErbB2 mediated mammary tumor development (Francis et al., 2011). Together, these findings suggest distinct functions of the LXCXE-binding pocket in specific biological settings
Here, we sought to define the processes that are uniquely dependent on the LXCXE-binding surface of RB, and relevant to tumor suppression, in the absence of other genetic events. Using an integrative series of cell culture and animal models, we demonstrate specific dependence on this conserved functional domain of RB in response to DNA damage. While having limited impact on spontaneous tumorigenesis or basal gene expression, the LXCXE-binding surface is critical for controlling the response to DNA damage and suppression of tumorigenesis by genotoxic carcinogens. This mechanism could be particularly relevant in multiple cancers that are initiated as a consequence of genetic damage.
RESULTS
The LXCXE-binding motif: dispensable for RB chromatin association, important for recruitment of co-repressors
To dissect the specific importance of LXCXE-binding on overall RB function, the NF allele of RB was employed (Chen and Wang, 2000). This amino acid substitution (hN757F is analogous to mN750F) disrupts hydrogen-bonding surfaces and introduces a bulky hydrophobic residue to sterically preclude LXCXE-protein binding (Figure 1A). Despite specificity in protein-binding interactions, the impact of LXCXE-binding on RB-chromatin association is unknown. Chimeric GFP-RB and GFP-N757F genes were generated and the GFP-epitope was utilized to chromatin immunoprecipitate (ChIP) GFP-RB and GFP-N757F. These analyses revealed that both wild type and mutant proteins effectively interacted with E2F target gene promoters (Figure 1B). Subsequently, FRAP was performed to delineate changes in mobility associated with disruption of the LXCXE-binding function of RB. Consistent with prior studies, GFP was freely diffusible, while GFP-H2B was immobile due to chromatin association (Stengel et al., 2009). In contrast, both GFP-RB and GFP-N757F exhibited identical dynamic behavior that is consistent with the transient interaction of co-repressors with chromatin (Figure 1C and Supp. Figure S1A). Thus, the predominant determinant for RB promoter occupancy is independent of LXCXE-interactions.
Figure 1. The LXCXE binding motif plays a critical role in mediating repressor complex formation in vitro.
(A) Modeled crystal structure of the LXCXE-binding domain of RB or N757F, as bound to HPV-E7. (B) Chromatin immunoprecipitation at the human Cyclin A, Plk1, RNR2 or human Albumin promoter in Saos-2 cells transfected with indicated constructs. (C) Fluorescence recovery after photobleaching (FRAP) in Saos-2 cells transfected with indicated constructs. [D–F] pMAFs were transduced with either Ad-Cre or Ad-LacZ. (D, upper) PCR was carried out to confirm recombination of exon 19 of the Rb gene. (D, lower) Immunoblot for total pRB. (E) qRT-PCR was performed for indicated transcripts. (F) ChIP was performed for indicated proteins at the Cyclin A promoter. Error bars represent SD.
To assess the function of the LXCXE-binding motif in the context of endogenous RB protein, an N750F knock-in allele was employed (Francis et al., 2009). In this model, a single point mutation was generated to produce the N750F protein from the endogenous murine Rb1 locus. Interestingly, while these mice were viable, RbN750F/− mice displayed less-than-expected Mendelian birth ratios (Supp. Figure S1B). RbN750F/− mice that were born were runted and ultimately died of wasting in the absence of any defined malignancy (Supp. Figure S1C, D). Since Rb heterozygous mice are prone to develop pituitary tumors at high penetrance, a cohort of mice were aged to explore a potential tumor suppressive role of LXCXE-binding pocket dysfunction. In this context, mice homozygous for the N750F allele remained tumor free and lived beyond 2 years of age (Supp. Figure S1D). Thus, unlike Rb heterozygous mice that experience LOH to develop pituitary tumors, RbN750F/N750F mice are competent for such tumor suppression.
To more rigorously explore the functionality of the LXCXE-binding pocket in the context of a defined, acute knockout, an allelic series was generated including Rbf/f, Rb+/f and RbN750F/f littermates. In these populations, the discrete impact of complete Rb loss, maintenance of only a single Rb+ allele, or the unique influence of a single RbN750F allele could be evaluated following delivery of cre-recombinase. Adenoviral transduction of primary adult fibroblasts derived from these mice resulted in efficient recombination of the Rbf allele (Figure 1D, upper), and loss of RB protein (Figure 1D, lower). Analysis via qRT-PCR demonstrated that RNA levels of endogenous RB/E2F target genes were significantly elevated with the deletion of Rb. However, cells harboring a single Rb+ allele or expressing the N750F mutant RB protein maintained similar levels of these RB target genes (Figure 1E). To determine whether this effect could be attributed to repressor complex formation, ChIP was performed at the CCNA2 promoter. Loss of RB or LXCXE binding activity increased promoter acetylation; however, this event was significantly suppressed by the presence of a single wild type RB allele (Figure 1F). In accordance with these findings, the Cyclin A2 promoter association with LXCXE-domain-dependent transcriptional co-repressors was reduced in RbN750F/f; Ad-Cre cells when compared to Rb+/f; Ad-Cre cells (Figure 1F and Supp. Figure S1E). Importantly, these data demonstrate that under unchallenged growth conditions, a single RbN750F allele is sufficient to suppress aberrant gene expression despite attenuated co-repressor recruitment.
Perturbation of the LXCXE-binding motif unmasks E2F3-mediated response to genotoxic stress
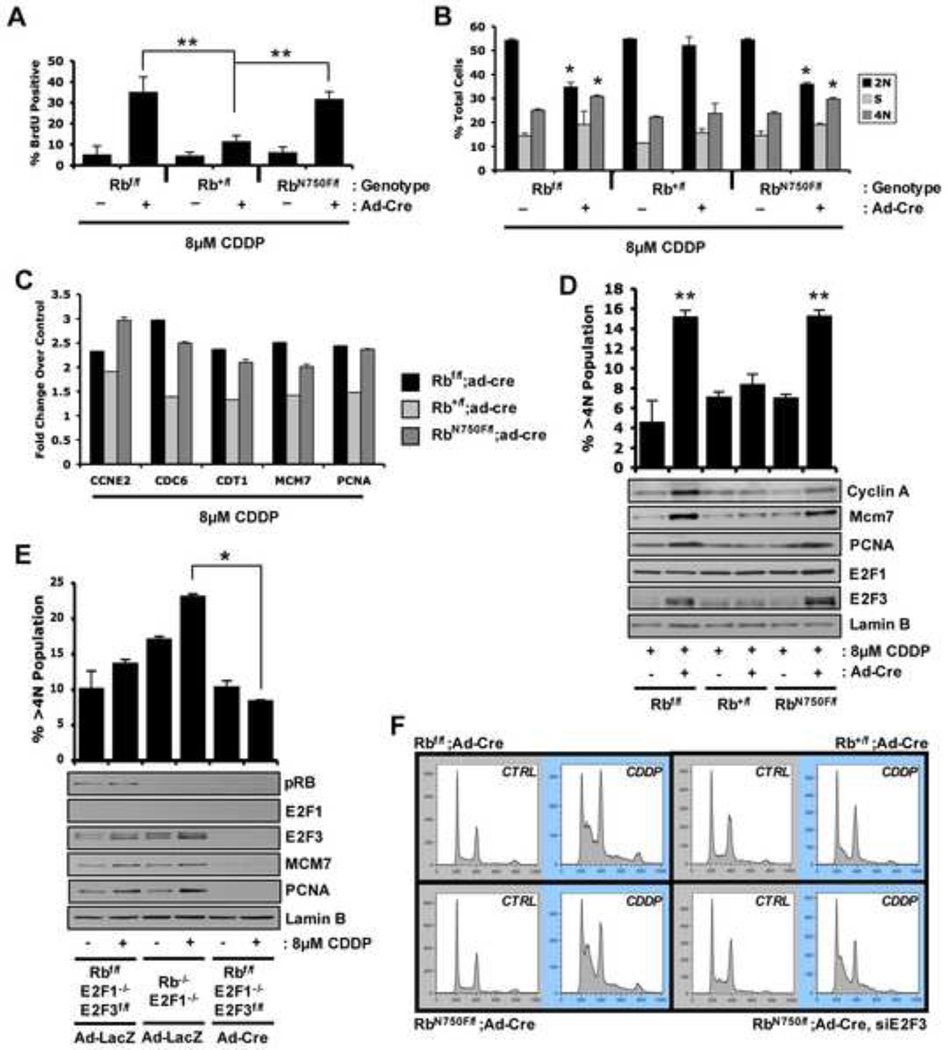
Generally, deletion of RB has little effect on proliferation under ideal growth conditions (Supp. Figure S2A and Herrera et al., 1996); however, RB plays a vital role in response to anti-proliferative stresses, such as DNA damage (Harrington et al., 1998; Knudsen and Knudsen, 2008). Therefore, primary fibroblast cultures were treated with 8µM cisplatin (CDDP) to induce DNA damage. RB ablation or the presence of a single RbN750F allele enabled ongoing DNA replication following CDDP treatment; in contrast, RB-proficient (Rb+) cells arrested similar to wild type (Figure 2A). Consistent with these findings, Rbf/f; Ad-Cre and RbN750F/f; Ad-Cre cells efficiently bypassed CDDP-mediated cell cycle arrest leading to an altered cell cycle profile (Figure 2B) and aberrant ploidy (>4N) (Figure 2D). Thus, LXCXE-binding is functionally required for the appropriate checkpoint response to DNA damage. Analyses of gene expression revealed that in the presence of DNA damage, a single wild type allele of Rb was able to suppress RB/E2F target genes; however, a single RbN750F mutant allele was deficient in target gene suppression (Figure 2C, D). Because E2F1 and E2F3 are induced as part of such DNA damage responses (Martinez et al., 2010), it was reasoned that the LXCXE-binding function of RB could be critical for constraining these proteins. While there was modest alteration in the accumulation of E2F1, the levels of E2F3 were specifically deregulated in the absence of a functional LXCXE-binding domain (Figure 2D). These findings imply a critical role for the LXCXE-binding function of RB in modulating E2F3 protein expression in the context of DNA damage stress to maintain cell cycle control.
Figure 2. LXCXE binding function modifies response to cellular stress in vitro.
Using 8µM CDDP treatment for 24 hours: (A) Coverslips were stained for BrdU incorporation. (B) Flow cytometry was performed; data indicate percentage of total cells with given DNA content. (C) qRT-PCR was performed for indicated transcripts. (D) Total proportion of cells with greater than 4N DNA content was quantified via flow cytometry and graphed. Total protein was resolved via SDS-PAGE and immunoblotted for indicated proteins. (E) Primary MEFs were transduced with Ad-LacZ or Ad-Cre for 48 hours prior to cisplatin treatment. Total proportion of cells with greater than 4N DNA content was quantified via flow cytometry and graphed. Total protein was resolved via SDS-PAGE and immunoblotted for indicated proteins. (F) Primary MAFs were transfected with siCTRL or siE2F3 prior to cisplatin treatment and flow cytometry. Error bars represent SD. *, p ≤ 0.05. **, p ≤ 0.01.
To assess the role of E2F3 in RB-mediated DNA damage response, cre-mediated knockout of E2F3 was performed in E2F1-deficient mouse embroyonic fibroblasts. Ablation of E2F3 in conjunction with RB deletion resulted in a downregulation of RB/E2F target genes and a corresponding decrease in aberrant ploidy after CDDP challenge (Figure 2E and Supp. Figure S2B). In order to more thoroughly interrogate this mechanism in the absence of LXCXE-binding function, siRNA mediated knockdown of E2F3 was performed in RbN750F/f; Ad-Cre adult fibroblasts (Supp. Figure S2C). Analyses of cell cycle profiles after induction of DNA damage revealed that knockdown of E2F3 was capable of reverting the aberrant DNA damage response of RbN750F/f; Ad-Cre cells to a physiological response that closely resembled that of Rb+/f; Ad-Cre cells (Figure 2F and Supp. Figure S2D). Combined, these results indicate that E2F3 plays a significant role in the aberrant response to DNA damage in an RB-compromised background.
RB-mediated DNA damage response is modified by LXCXE-binding function in vivo
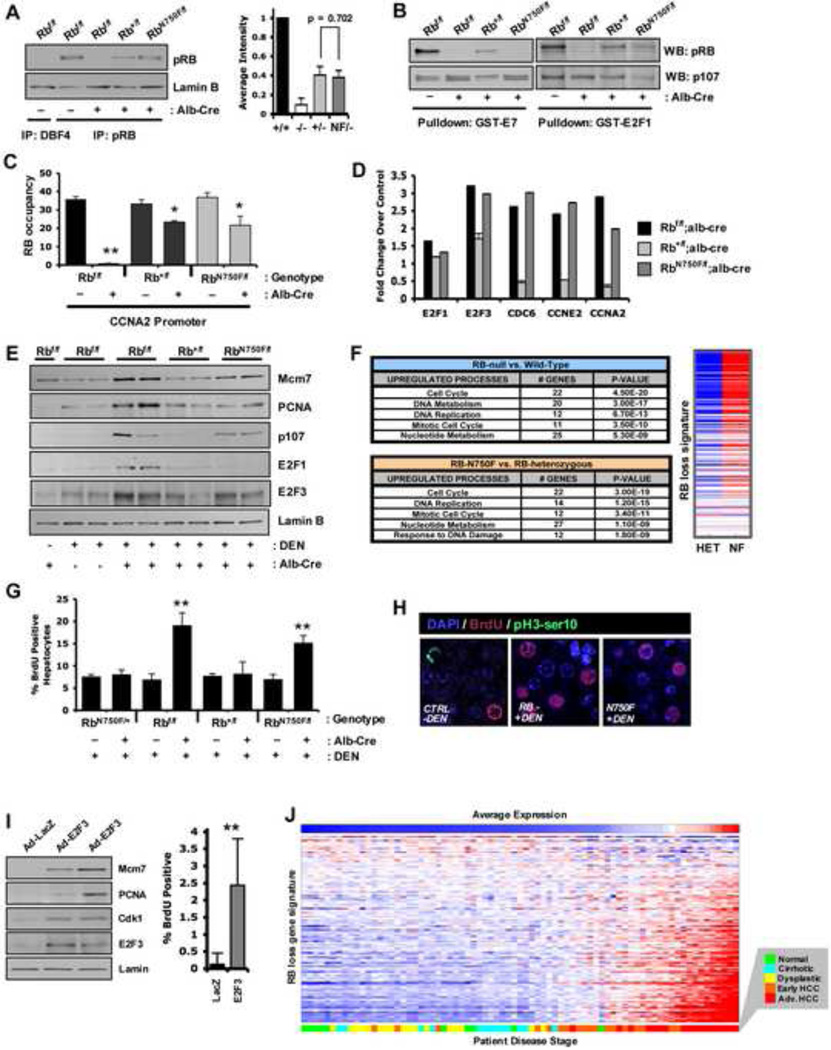
To functionally dissect the activity of the LXCXE-binding pocket in vivo, a mouse model was employed wherein an albumin-cre transgene drives the efficient recombination of the Rbf allele in the liver (Supp. Figure S3A and Mayhew et al., 2005). In this model, a single copy of either the mutant or wild type Rb1 expressed reduced, yet comparable levels of RB protein (Figure 3A). To confirm the specific deficiency of the N750F mutant, the ability of total liver RB protein to bind immobilized GST-E7 or GST-E2F1 was evaluated. These data demonstrated that the RB protein present in the RbN750F/f; Alb-Cre livers was incapable of binding to the LXCXE-containing protein E7, yet retained the ability to interact with E2F1 (Figure 3B and Supp. Figure S3B). To ensure specificity to RB, the interaction of endogenous p107 in the same lysates was confirmed. Furthermore, consistent with the studies in cell culture, both wild type and NF RB proteins associated with the CCNA2 promoter (Figure 3C), thus providing a system to study the impact of LXCXE-binding function in vivo.
Figure 3. The N750F mutant compromises RB-mediated transcription and cell cycle control and is associated with human HCC.
(A) Total protein lysates were immunoprecipitated and indicated proteins were detected via immunoblotting. (B) Purified GST-E7 and GST-E2F1 were incubated with total liver protein lysates and resulting complexes were resolved via SDS-PAGE. (C) Quantitative ChIP was performed for the presence of RB protein. (D) qRT-PCR was performed for indicated transcripts. (E) Total liver protein was immunoblotted for indicated proteins. (F) Ontological analysis of genes regulated by RB loss or LXCXE-binding disruption. Heatmap displays differential expression of RB loss signature genes between genotypes. (G) Percentage of BrdU positive hepatocytes were quantified. (H) Immunofluorescence for the presence of BrdU and pH3-ser10 in indicated liver sections. (I) Male mice were transduced with adenovirus encoding either LacZ or hE2F3 and analyzed for protein expression and BrdU. (J) Human tissue samples of varying disease states in the liver were sorted on expression of the RB loss signature. Error bars represent SD. *, p ≤ 0.05. **, p ≤ 0.01.
Since the RB LXCXE-interaction motif had a profound effect on DNA damage response in vitro, this response was interrogated in vivo. Neonatal mice were subjected to a single injection of diethylnitrosamine (DEN) and sacrificed 24 hours later. While this dose of DEN is not necrotic (Supp. Figure S3C), it does induce DNA damage sufficient to initiate liver tumor development (Mayhew et al., 2007; Rajewsky et al., 1966). In this model, DEN treatment resulted in increased expression of E2F3 protein in both Rbf/f; Alb-Cre and RbN750F/f; Alb-Cre livers (Figure 3D, E). These findings indicate that the LXCXE-binding function plays an important role in coordinating transcriptional stress responses in vivo.
Gene expression profiling of DEN-treated liver tissue from wild type versus Rbf/f; Alb-Cre or Rb+/f; Alb-Cre versus RbN750F/f; Alb-Cre revealed that deregulation of gene expression processes were largely synonymous across the two comparisons (Figure 3F and Supp. Table T1). In keeping with these findings, the LXCXE-binding deficient model was significantly enriched for deregulation of consensus RB loss signature genes (Figure 3F and Ertel et al., 2010). These findings indicate that the LXCXE-binding domain plays a significant role in coordinating general RB function in the response to DNA damage stress in vivo. To determine the subsequent impact of the deregulated transcription on liver biology, the cell cycle response to DNA damage stress was evaluated. Loss of RB lead to deregulated cell cycle entry following DEN treatment, as scored by BrdU incorporation; this event was specifically phenocopied with loss of the LXCXE-binding function (Figure 3G). Interestingly, while efficient DNA replication was observed, there were virtually no mitotic nuclei in liver tissue exposed to DEN treatment (Figure 3H), despite detectable levels of phosphorylated serine 10 on Histone H3 in interphase nuclei, indicating G2-phase arrest (Supp. Figure S3D). Thus, the impact of disabling RB, and more specifically, LXCXE-binding in the acute response to DNA damage in vivo is restricted to control over S-phase in this model. In keeping with an important role for E2F3 in mediating the observed effects on transcription, the ectopic expression of E2F3 in vivo was sufficient to drive cell cycle entry in a fashion analogous to DNA damage stress in the absence of LXCXE-binding function (Figure 3I).
To determine how such functional processes may relate to human disease, deregulation of transcriptional activity, as occurs with loss of RB/LXCXE-binding function, was evaluated in reference to human HCC development. These data showed that disruption of RB/LXCXE function was associated with progression to HCC (Figure 3J). Utilizing analysis of an independent data set, genetic deletion encompassing the Rb1 gene (13q14) statistically correlated with upregulation of the published RB loss signature in cases of hepatocellular carcinoma (Supp. Figure S3E). In total, these findings suggest that deregulated transcriptional responses could be important in liver tumor development.
The LXCXE-binding cleft of RB is critical for liver tumor suppression
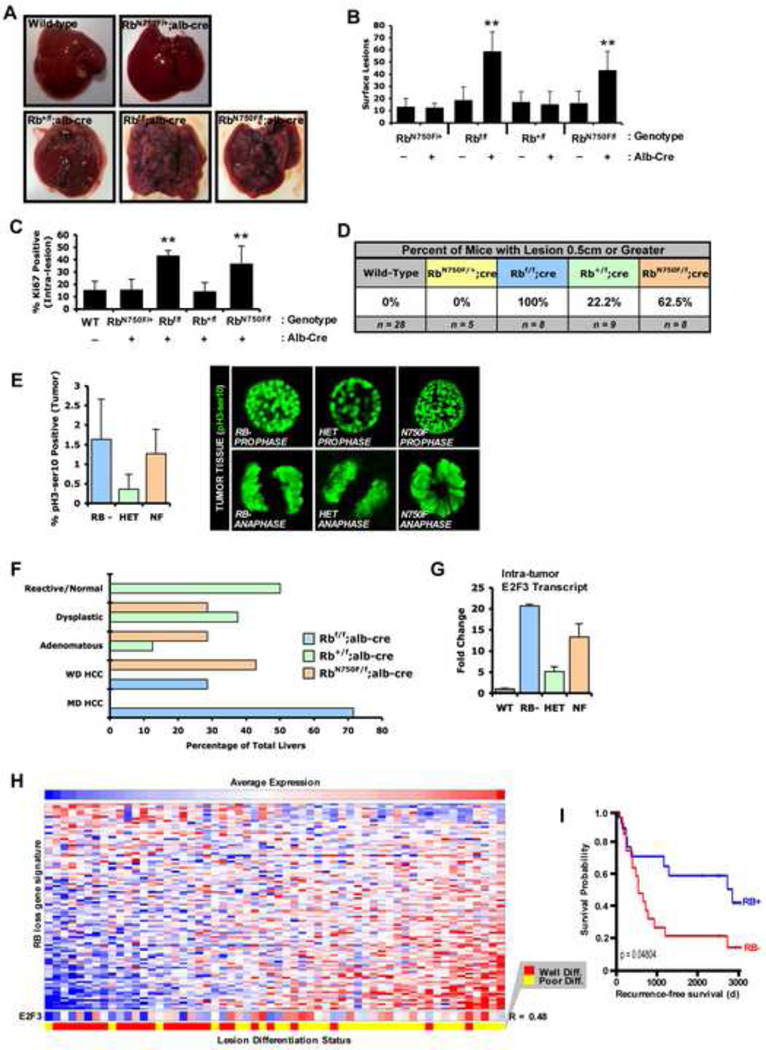
Aberrant response to DNA damage signaling can contribute to tumor development, and germline Rb mutation is associated with radiation induced secondary malignancies (Kleinerman, 2009). To interrogate the impact of heterozygosity and LXCXE-inactivation on tumor suppression, an established model of liver tumor initiation-progression was employed (Huang et al., 2005). With this protocol, the majority of Rb+/f; Alb-Cre livers harbored small lesions and were visually similar to livers with two functional copies of Rb. In contrast, both Rbf/f; Alb-Cre and RbN750F/f; Alb-Cre livers were permeated with lesions that covered the entirety of the organ (Figure 4A). Thus, these data demonstrate that the LXCXE-binding motif of RB plays a significant role in suppression of genotoxin initiated liver tumorigenesis. Quantifiably, Rb+/f; Alb-Cre livers expressed no more visible lesions than wild type livers, indicating that a single allele of wild type Rb is largely capable of suppressing tumor initiation. In contrast, disruption of the RB/LXCXE interaction was sufficient to abrogate this tumor suppressive capability (Figure 4B). Lesions arising in RB-null or RB-mutant livers were highly proliferative (Figure 4C and Supp. Figure S4A, B) and reach larger size (Figure 4D). Whereas previous studies have suggested that defective chromosomal condensation is a contributor to tumor development in the presence of LXCXE-deficient RB alleles (Coschi et al., 2010), there was no obvious evidence of such phenomena in this model (Figure 4E). Interestingly, a subset of hepatocytes within RB-null or RB-mutant lesions harbored dual positive BrdU/pH3-ser10 staining (Supp. Figure S4D) that has been recently shown to be indicative of an aberrant DNA re-replication phenomenon that occurs specifically in hepatocytes (Bourgo et al., 2011). Together, these findings indicate that LXCXE-binding integrity is essential for suppressing genotoxin-initiated tumorigenesis in vivo.
Figure 4. N750F mutant is impaired for tumor suppression and dictates tumor grade and proliferation rate.
(A) Representative, whole livers are pictured. (B) At the time of sacrifice all visible lesions were counted and graphed. (C) Paraffin embedded sections of liver tissue were stained for the presence of Ki-67. (D) Percentage of mice with or without a liver harboring dissectable lesions (diameter ≥ 0.5cm) were scored positive or negative based on such criteria. Comparative statistical evaluation is in Supp. Figure S4F. (E) Individual dissected tumors were stained for phosphorylated serine 10 of Histone H3. (F) Pathologist-scored H&E sections were analyzed for the most predominant histological grade of lesions. (G) qRT-PCR was performed for indicated transcripts on excised lesions. (H) Human liver tissue samples independently scored as either well- or poorly-differentiated, sorted based on the RB loss signature. E2F3 expression was correlated to level of differentiation (R=0.48). (I) Kaplan-Meier analyses comparing samples with high or low Rb loss signature expression. Error bars represent SD. *, p ≤ 0.05. **, p ≤ 0.01.
Histological analyses revealed that RB-null or RB-mutant tumor reached higher stage than those harboring either wild type or heterozygous lesions (Figure 4F and Supp. Figure S4C). Importantly, lesions in Rbf/f; Alb-Cre and RbN750F/f; Alb-Cre livers frequently progressed to HCC, in contrast to normal tissue or dysplastic lesions observed in the Rb+/f; Alb-Cre livers. To determine if this finding reflected loss of heterozygosity in the larger tumors, expression of the Rb1 transcript was examined. These large tumors retained Rb1 mRNA both with a single wild type or single LXCXE-binding deficient allele (Supp. Figure S4E). The advanced progression and increased proliferation of lesions in RB-null or RB-mutant livers suggested differential E2F activity in these lesions. Consistent with this concept, E2F3 transcript was differentially expressed among the varying genotypes (Figure 4G). RB-null lesions expressed the highest levels of E2F3 mRNA and correspondingly progressed to HCC most frequently, while RB-heterozygous lesions expressed contrastingly low levels of E2F3 transcript and never progressed to HCC. To determine if this deregulated gene expression program was relevant to human disease, gene expression patterns were analyzed. Strikingly, the deregulation of the RB loss signature was associated with poorly differentiated forms of disease (Figure 4H). Further strengthening the role of E2F3 in RB-mediated tumor suppression, E2F3 transcript expression was correlated (R = 0.48) to de-differentiated lesions (Figure 4H), and this coincided with disease states that harbored poor prognosis in human patients (Figure 4I).
DISCUSSION
As the target of viral oncoproteins and a highly conserved peptide motif necessary for multiple protein interactions, the LXCXE binding pocket of RB would be expected to harbor critical function necessary for RB activity. However, the disruption of LXCXE protein binding has little impact on basal transcriptional control attributed to RB (Chen and Wang, 2000; Isaac et al., 2006). Presumably, the basis for this finding is that LXCXE-deficient RB mutants retain the capacity to bind to E2F proteins sufficiently to mask their functional transactivation. This is consistent with the concept that LXCXE-binding defective proteins are compromised for stable repressive processes associated with cell cycle exit, such as in the context of induced senescence or myogenic differentiation (Chen and Wang, 2000; Talluri et al., 2010). In contrast, the work herein demonstrates a novel function for the LXCXE-binding domain in coordinating acute transcriptional responses to genotoxic stress that underlie RB-mediated tumor suppression.
While the mechanism behind RB mediated DNA damage response has remained elusive, the present findings demonstrate a role for the LXCXE-binding domain of RB in control of E2F activity following DNA damage stress. In particular, E2F3 levels are induced following DNA damage in a fashion that is controlled through RB. More specifically, loss of RB or LXCXE-deficiency enables significantly increased levels of E2F3 following DNA damage both in fibroblasts and liver tissue in vivo. The functional significance of E2F3 is supported by knockdown and overexpression studies: E2F3 deficiency precludes the checkpoint bypass that occurs with loss of LXCXE activity, while E2F3 overexpression mimics LXCXE-deficiency. Thus, in response to DNA damage, RB uses a functional domain that is independent of that involved in direct E2F-binding to control gene expression through E2F3. Importantly, such gene expression changes are associated with aggressive liver cancers that harbor poor disease outcome in human patients.
The mechanisms through which the LXCXE-binding domain of RB contributes to tumor suppression reflects the complexity of RB function in distinct tissue settings. Prior studies have shown that mice deficient in RB-mediated LXCXE protein binding are not necessarily tumor prone (Francis et al., 2009). As such, deficiency of LXCXE-binding function in RbN750F/N750F animals has not been associated with increased pituitary tumorigenesis as is observed in Rb1 heterozygous mice (Jacks et al., 1992). Due to the premature mortality of RbN750F/− animals described here, it is possible that RbN750F/− mice die too young to observe compromised tumor suppression and that gene dosage effects are relevant for the suppression of pituitary tumorigenesis. However, recent studies have demonstrated that LXCXE-binding deficiency of RB increases susceptibility to lymphomagenesis in p53-deficient mice (Coschi et al., 2010). In similar fashion, LXCXE-deficiency alters susceptibility to p53-mutant-driven mammary tumorigenesis, while having no effect on ErbB2 mediated tumorigenesis in the same tissue (Francis et al., 2011). These studies in particular suggest that disruption of the LXCXE-binding pocket can cooperate with p53 loss, and is associated with gross defects in mitotic condensation (Coschi et al., 2010). In the liver tumorigenesis model used here, initiated cells that harbor oncogenic mutations are responsible for tumorigenesis. In such models, the LXCXE-binding function of RB is critically important for limiting S-phase progression following DNA damage, while G2/M checkpoints remain intact. Although we fail to observe major deficiencies in chromosomal condensation in these tumors, potential latent deficits in mitotic condensation could still contribute to tumorigenesis. It is important to note that, LXCXE-binding deficiency is associated with an intermediate tumor phenotype between wild type and RB-deficient backgrounds in the liver. These findings suggest that even following initial genotoxic challenge, LXCXE-deficiency only partially compromises the tumor suppressive functions of RB. However, such tumors are highly proliferative compared to the rare tumors that emerge in Rb heterozygous mice, suggesting that in the context of established disease, the LXCXE-binding domain of RB could play an important role in limiting progression. To this end, tumorigenic proliferation is associated with excessive DNA damage, implying that the LXCXE-mediated transcriptional control of E2F3 could play an important role in modulating tumor progression. This assertion is supported by the analyses of gene expression and disease outcome in clinical specimens.
In total, these findings demonstrate compromised RB-mediated tumor suppression after alteration of a single protein-binding domain. While RB promoter association and unchallenged function remained intact in the absence of LXCXE-binding activity, the dormant impact of this event was unmasked during DNA damage response and ultimately played a vital role in suppression of tumor development. In sum, these findings define key activities through which RB functions to control cell cycle transitions and suppress tumor development.
MATERIALS AND METHODS
For complete details, please see Supp. Materials and Methods.
Carcinogen treatments
Diethylnitrosamine
Neonatal mice (14d) received a single IP-injection of 20mg/kg body weight of DEN dissolved in 0.9% saline. Mice were sacrificed 24 hours later.
Tumorigenesis Protocol
Neonatal mice (14d) received a single IP-injection of 20mg/kg body weight of DEN. Every two weeks thereafter, mice received an IP-injection of 3mg/kg body weight of TCPOBOP dissolved in corn oil.
Real-Time PCR
In all cases, RNA was isolated using the Trizol reagent (Gibco). Total RNA was quantified via Nanodrop and converted into cDNA.
Adenoviral transduction and plasmid transfection in vitro
Adenoviral Transduction
Cells were transduced using Ad-LacZ, Ad-GFP, Ad-Cre or Ad-GFP-Cre adenoviruses at a multiplicity of infection of 10 for 48 hours.
Transfection
Transfection of plasmids was carried out using Fugene 6 transfection reagent (Roche, Palo Alto, CA) following manufacturer specifications.
Adenoviral transduction in vivo
Adenoviral delivery was performed on male mice, anesthetized with isoflurane prior to intravenous injection of 1×109 plaque-forming units as previously described (Mayhew et al., 2005).
Chromatin immunoprecipitation (ChIP)
Nuclei from pMAFs were cross-linked, isolated, and lysed and the chromatin was sonicated to shear. Equal amounts of chromatin were immunoprecipitated with specific antibodies. The resulting protein slurry was added to the IP and mixed for overnight at 4°C.
Protein complex immunoprecipitation (Pulldown)
GST-E7 and GST-E2F1 were expressed and purified from bacteria using glutathione-conjugated beads as previously described (Knudsen and Wang, 1996).
Microarray and Gene Ontology
Independent samples for RB-heterozygous mice (4), RB-N750F mice (5), Wild type RB mice (3) and RB null mice (3) were used in analyses. Ontology was done using upregulated genes (1.2-fold or higher) via DAVID (http://david.abcc.ncifcrf.gov/).
Comparative gene expression analyses (GSE4180 and GSE6764)
Gene expression and lesion differentiation data in hepatocellular carcinoma was obtained from the Gene Expression Omnibus. A subset of genes corresponding with those previously identified as the Rb loss signature (Ertel et al., 2010) was used to evaluate the samples in this dataset.
Supplementary Material
HIGHLIGHTS.
A specific RB functional domain regulates transcriptional response to stress.
E2F3 is a key effector through which RB modulates DNA damage checkpoints.
LXCXE-binding is essential for suppression of tumorigenesis after genotoxic stress.
A single RB binding surface coordinates gene expression deregulated in human disease.
ACKNOWLEDGEMENTS
The authors thank Dr. Gustavo Leone for generously providing mouse embryonic fibroblasts for study of E2F3 function. Dr. Karen Knudsen and Ms. Elizabeth Schade graciously contributed to the editing of the manuscript.
Grant Support: ESK is supported by grant CA127387. JYJW is supported by grant CA058320.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Balsitis S, Dick F, Lee D, Farrell L, Hyde RK, Griep AE, Dyson N, Lambert PF. Examination of the pRb-dependent and pRb-independent functions of E7 in vivo. J Virol. 2005;79:11392–11402. doi: 10.1128/JVI.79.17.11392-11402.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD. E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol. 2007;19:658–662. doi: 10.1016/j.ceb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgo RJ, Ehmer U, Sage J, Knudsen ES. RB deletion disrupts coordination between DNA replication licensing and mitotic entry in vivo. Mol Biol Cell. doi: 10.1091/mbc.E10-11-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TT, Wang JY. Establishment of irreversible growth arrest in myogenic differentiation requires the RB LXCXE-binding function. Mol Cell Biol. 2000;20:5571–5580. doi: 10.1128/mcb.20.15.5571-5580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschi CH, Martens AL, Ritchie K, Francis SM, Chakrabarti S, Berube NG, Dick FA. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev. 24:1351–1363. doi: 10.1101/gad.1917610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick FA, Sailhamer E, Dyson NJ. Mutagenesis of the pRB pocket reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol Cell Biol. 2000;20:3715–3727. doi: 10.1128/mcb.20.10.3715-3727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertel A, Dean JL, Rui H, Liu C, Witkiewicz AK, Knudsen KE, Knudsen ES. RB-pathway disruption in breast cancer: Differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle. 9:4153–4163. doi: 10.4161/cc.9.20.13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis SM, Bergsied J, Isaac CE, Coschi CH, Martens AL, Hojilla CV, Chakrabarti S, Dimattia GE, Khoka R, Wang JY, Dick FA. A functional connection between pRB and transforming growth factor beta in growth inhibition and mammary gland development. Mol Cell Biol. 2009;29:4455–4466. doi: 10.1128/MCB.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis SM, Chakrabarti S, Dick FA. A context-specific role for retinoblastoma protein-dependent negative growth control in suppressing mammary tumorigenesis. PLoS One. 6:e16434. doi: 10.1371/journal.pone.0016434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley SA, Francis SM, Demone J, Ainsworth P, Dick FA. A cancer derived mutation in the Retinoblastoma gene with a distinct defect for LXCXE dependent interactions. Cancer Cell Int. 10:8. doi: 10.1186/1475-2867-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera RE, Sah VP, Williams BO, Makela TP, Weinberg RA, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac CE, Francis SM, Martens AL, Julian LM, Seifried LA, Erdmann N, Binne UK, Harrington L, Sicinski P, Berube NG, et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol Cell Biol. 2006;26:3659–3671. doi: 10.1128/MCB.26.9.3659-3671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008;8:714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen ES, Sexton CR, Mayhew CN. Role of the retinoblastoma tumor suppressor in the maintenance of genome integrity. Curr Mol Med. 2006;6:749–757. doi: 10.2174/1566524010606070749. [DOI] [PubMed] [Google Scholar]
- Lee JO, Russo AA, Pavletich NP. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- Manning AL, Longworth MS, Dyson NJ. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 24:1364–1376. doi: 10.1101/gad.1917310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markey MP, Angus SP, Strobeck MW, Williams SL, Gunawardena RW, Aronow BJ, Knudsen ES. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 2002;62:6587–6597. [PubMed] [Google Scholar]
- Markey MP, Bergseid J, Bosco EE, Stengel K, Xu H, Mayhew CN, Schwemberger SJ, Braden WA, Jiang Y, Babcock GF, et al. Loss of the retinoblastoma tumor suppressor: differential action on transcriptional programs related to cell cycle control and immune function. Oncogene. 2007;26:6307–6318. doi: 10.1038/sj.onc.1210450. [DOI] [PubMed] [Google Scholar]
- Martinez LA, Goluszko E, Chen HZ, Leone G, Post S, Lozano G, Chen Z, Chauchereau A. E2F3 is a mediator of DNA damage-induced apoptosis. Mol Cell Biol. 30:524–536. doi: 10.1128/MCB.00938-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew CN, Bosco EE, Fox SR, Okaya T, Tarapore P, Schwemberger SJ, Babcock GF, Lentsch AB, Fukasawa K, Knudsen ES. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 2005;65:4568–4577. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- Mayhew CN, Bosco EE, Solomon DA, Knudsen ES, Angus SP. Analysis of RB action in DNA damage checkpoint response. Methods Mol Biol. 2004;281:3–16. doi: 10.1385/1-59259-811-0:003. [DOI] [PubMed] [Google Scholar]
- Mayhew CN, Carter SL, Fox SR, Sexton CR, Reed CA, Srinivasan SV, Liu X, Wikenheiser-Brokamp K, Boivin GP, Lee JS, et al. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology. 2007;133:976–984. doi: 10.1053/j.gastro.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Morris EJ, Dyson NJ. Retinoblastoma protein partners. Adv Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- Rajewsky MF, Dauber W, Frankenberg H. Liver carcinogenesis by diethylnitrosamine in the rat. Science. 1966;152:83–85. doi: 10.1126/science.152.3718.83. [DOI] [PubMed] [Google Scholar]
- Stengel KR, Thangavel C, Solomon DA, Angus SP, Zheng Y, Knudsen ES. Retinoblastoma/p107/p130 pocket proteins: protein dynamics and interactions with target gene promoters. J Biol Chem. 2009;284:19265–19271. doi: 10.1074/jbc.M808740200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talluri S, Isaac CE, Ahmad M, Henley SA, Francis SM, Martens AL, Bremner R, Dick FA. A G1 checkpoint mediated by the retinoblastoma protein that is dispensable in terminal differentiation but essential for senescence. Mol Cell Biol. 30:948–960. doi: 10.1128/MCB.01168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HS, Dean DC. Rb-mediated chromatin structure regulation and transcriptional repression. Oncogene. 2001;20:3134–3138. doi: 10.1038/sj.onc.1204338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.