Abstract
In vitro mimics of cellular machines have been recently engineered and utilized to investigate processes within cells. These devices can provide novel insights into biological mechanisms and have the potential to improve biotechnological processes such as separation. In particular, several devices have been designed to mimic translocation through nuclear pore complexes (NPCs). We describe here the fabrication of a biomimetic NPC using nanoporous filters lined with FG-repeats of proteins that create a selectivity barrier. We show the utility of this nanoselective filter as a testbed for the investigation of nucleocytoplasmic transport and demonstrate that this device closely reproduces key features of trafficking through the NPC.
INTRODUCTION
The transport of macromolecules across the nuclear envelope is facilitated by proteinaceous assemblies called nuclear pore complexes (NPCs). These eightfold symmetric molecular machines are selective for the passage of certain macromolecules, yet provide high throughput to maintain proper cellular order and function (Wente, 2000). The NPCs are highly conserved among species and they contain approximately 30 distinct proteins called nucleoporins or nups in multiple copies (Alber et al., 2007; Cronshaw, Krutchinsky, Zhang, Chait, & Matunis, 2002; Rout et al., 2000). In budding yeast, for example, the NPC has a molecular mass of ~50 MDa and is ~100 nm in diameter. In the classical case, translocation through this complex is a receptor-mediated process and transported cargo molecules have a nuclear localization signal for nuclear import or a nuclear export signal for nuclear export. These signal sequences are recognized by soluble transport factors (karyopherins, Kaps) that can overcome the NPC barrier by transient binding to certain nups that contain phenylalanine–glycine repeats called FG-nups (Rout, Aitchison, Magnasco, & Chait, 2003). The directionality of this transport is in large part determined by the GTP-bound state of the small GTPase Ran. In the nucleus, Ran-GTP predominates and both displaces cargo from importing Kaps and promotes cargo binding to exporting Kaps; in the cytoplasm, RanGAP ensures Ran is predominantly in the Ran-GDP form, promoting cargo binding to importing Kaps and displacing cargo from exporting Kaps.
Nanopores have been used to separate molecules based on various differential properties such as size, charge, hydrophobicity, and affinity (Iqbal, Akin, & Bashir, 2007; Jirage, Hulteen, & Martin, 1999; Lakshmi & Martin, 1997; Lee et al., 2002). We and others have built upon these approaches and have recently shown that nanobiological assays can be used for investigation of the detailed mechanism of nucleocytoplasmic transport in vitro by engineering and employing a minimalistic NPC mimic (Jovanovic-Talisman et al., 2009).
To construct a functional NPC mimic, we lined nanoscale pores in polycarbonate membranes (30–100 nm pore diameter and ~15–30 nm thickness of functionalized layer) with copies of a nuclear pore proteins involved in creating selective barrier in vivo: FG-nups. We used recombinant transport factor proteins (carriers) and their binding partners (cargo) to recapitulate features of receptor-mediated transport. Using confocal microscopy, we tested how efficiently various proteins and protein mixtures cross the functionalized membrane and we showed that this simple architecture is sufficient for selective transport. Concomitantly with our work, Elbaum lab published a chemical NPC mimic capable of conducting receptor-mediated transport (Caspi, Zbaida, Cohen, & Elbaum, 2008) using nanoporous filters (50 nm diameter and 6 μm length) functionalized with polyisopropylacrylamide (pNIPAM) chains. Their approach relies on weak interactions in pNIPAM; they used mobile pNIPAM molecules as carriers and single-stranded (ss) DNA as the cargo. Using fluorescence microscopy, they showed that pNIPAM/ssDNA complex crosses the pore more efficiently then ssDNA alone. Several other groups have published alternative approaches to NPC mimics in last few years, and here, we discuss just a few. Dekker and colleagues (Kowalczyk et al., 2011) utilized SiN solid-state nanopores (~20–40 nm diameter and ~20 nm length) lined with FG-nups and showed NPC-like selectivity on the single-molecule level using ionic current measurements (compared to bulk flux measurements utilized in previous work; Caspi et al., 2008; Jovanovic-Talisman et al., 2009). Maglia and coworkers (Soskine et al., 2012) also performed ionic current measurements, but instead of synthetic pores, they used the biological nanopore cytolysin A (~4–7 nm in diameter and 13 nm in length). The authors incorporated cytolysin A into planar lipid bilayers and functionalized it with aptamers evolved to bind to thrombin or lysozyme to show selective transport that resembles that of the NPC. These and similar NPC mimic devices provide quantitative testbeds for studying nucleocytoplasmic transport and afford tools for studying highly debated details of NPC selectivity mechanism. Here, we provide a detailed step-by-step protocol for our NPC mimic assembly and measurement of transport rates.
17.1 PROTEIN PRODUCTION AND PURIFICATION
17.1.1 FG-nups
FG-nups are essential components of the selectivity barrier in the NPCs. To create a nanoselective filter, we use two distinct S. cerevisiae FG-nups that contain two major classes of FG-repeat motifs: (1) Nsp1 with large blocks of FSFG-repeats and (2) Nup100 with large blocks of GLFG- and SLFG-repeats.
These FG-nup constructs are engineered to include a single C-terminal cysteine residue that facilitates proper orientation of the protein upon attachment to gold surfaces, and the result is an effective mimic of anchoring as it occurs in NPCs themselves. Due to their intrinsically disordered nature, FG-nups are extremely susceptible to proteolytic cleavage. We find that FG-nups obtained using codon-optimized sequences and co-transformed with transport factors give excellent yields. Because it can be challenging to obtain pure, full length FG-nups, we give a detailed protocol here.
Nsp1FG-Cys-His6 (residues 30–591 of Nsp1) in pET21b is transformed into BL21-Gold(DE3) cells and Nup100FG-Cys-His6 (residues 1–570 of Nup100) in pET21b is co-transformed with untagged Kap95 in pET24a into BL21-CodonPlus(DE3)-RIL cells.
-
Cells are grown in LB (Luria Bertani) medium with appropriate antibiotic selection, induced with 1 mM isopropyl-thio-β-D-galactopyranoside (IPTG) at optical density (OD600) ~ 0.8, and harvested after a 2-h incubation at 30 °C (Nsp1FG) or 4-h incubation at 22 °C (Nup100FG). Cell pellets are stored at −80 °C until next step is performed.
REMARKS
Starting from 1 l of culture, we generally obtain 1 mg of pure recombinant Nsp1FG-Cys-His6, and 2.5 mg of pure recombinant Nup100FG-Cys-His6.
To further reduce nup cleavage following modification can be done: cells are grown at 37 °C until OD600 reaches ~0.6, temperature is then reduced to 30 °C (Nsp1FG) or 22 °C (Nup100FG), and cells are induced at OD600 ~ 0.8. This ensures that cells reach incubation temperature at time of induction. Incubation times and temperatures have been extensively tested; lower temperatures generally produce better results. Generally 20 g of wet cell pellet are obtained per 6 l of culture.
for 1 g of wet cell paste, cells are resuspended in 4 ml of resuspension buffer: 50 mM sodium phosphate pH 8, 300 mM NaCl buffer supplemented with 8 M urea and protease inhibitors: (a) 1% (v/v) fresh solution P (18 mg/ml phenylmethylsulfonyl fluoride (PMSF), 0.3 mg/ml pepstatin A in ethanol), (b) 50 μM ethylenediaminetetraacetic acid (EDTA), (c) complete protease inhibitor cocktail tablets, EDTA-free, Roche (1 tablet per 50 ml of cell extract), (d) 5 mg/ml 6-amino-n-hexanoic acid (for Nup100FG). Resuspended cells are homogenized with a Polytron (90 s at setting 5) and lysed by passing six times through a microfluidizer.
Cell debris is removed with ultracentrifugation (90-min room temperature spin at 145,000 gav in a Ti50.2 rotor (Beckman)) followed by filtration through a 0.45 μm filter.
Supernatant is incubated with appropriate amount of His-affinity resin (we use approximately 5 ml of TALON resin (Clontech) or 1 ml HisTrapFF column (GE Healthcare) for 6 l of culture) preincubated with resuspension buffer for 30 min at room temperature. Resin is washed with ~10 bed volumes of buffer A: 50 mM sodium phosphate pH 8, 300 mM NaCl, 0.8 M urea, supplemented with protease inhibitors and imidazole; we use approximately 2 or 35 mM imidazole for Talon resin and HisTarpFF column, respectively. This value should be optimized for each resin and FG-nup construct; the use of automated HPLC/FPLC system and imidazole gradients is useful. Protein is eluted with five bed volumes of buffer A supplemented with protease inhibitors and 150 and 500 mM imidazole for Talon resin and HisTrapFF column, respectively.
-
Eluted proteins are reduced with fresh 10 mM tris(2-carboxyethyl)phosphine (TCEP) for 1 h (alternatively 5 mM dithiothreitol can be used), aggregates are removed by centrifugation (19,000 gav for 15 min at room temperature), and supernatant is loaded onto a Superose 6 10/300 column equilibrated with 20 mM HEPES pH 7.4, 110 mM potassium acetate, 2 mM MgCl2, 1 μM CaCl2, 1 μM ZnCl2, 0.1% Tween 20 buffer (TBT) supplemented with 50 μM TCEP. Pure protein fractions are stored at −20 °C.
REMARKS
Urea and TCEP are prepared fresh and added to solutions just before the use. TCEP is acidic and 1 M stock solution can be prepared in 0.5 M HEPES buffer pH 7.4.
17.1.2 Purification and fluorescent labeling of transport factors, cargo proteins, and control proteins
Transport factors recognize cargo substrates and mediate nuclear import and/or export. Human nuclear transport factor 2 (NTF2) (Moore & Blobel, 1994) is a dimeric protein that facilitates nuclear import of a GTPase Ran, ensuring efficient regulation of protein transport through the NPC. For our studies, we tag NTF2 with glutathione S-transferase (GST) or yellow fluorescent protein (YFP) to ensure the size above passive diffusion limit. Brief protocol for obtaining pure proteins is given:
Human NTF2–GST in pGEX-2T and NTF2–YFP-His6 in pET21b are transformed into BL21DE3 cells.
Cells are grown in LB medium supplemented with antibiotics, induced with 0.1 mM IPTG at OD600 ~ 0.8, and incubated for ~12 h at 30 °C (NTF2–GST) and 23 °C (NTF2–YFP).
Cells are harvested, lysed, and protein is purified from the clarified lysate using a glutathione Sepharose resin (NTF2–GST) or HisTrapFF and Superose 6 columns (NTF2–YFP).
To investigate the mechanism of nucleocytoplasmic transport using nanoselective filter, we also purified karyopherins Kap95 (using Kap95–GST plasmid in pGEX-2T-K vector) and Kap121 (using Kap121–GST plasmid in pGEX-4T-1 vector) as described before (Jovanovic-Talisman et al., 2009; Leslie et al., 2004). The GST tag is removed using a thrombin cleavage capture kit (Novagen) according to the manufacturer’s instructions. Cargo protein: Importin-β-binding site of importin alpha (Ibb) tagged with GFP, and control protein: GFP are obtained using Ibb-eGFP-His6 and eGFP-His6 plasmids in pET21b vector, respectively. Ibb-GFP forms a complex with Kap95, whereas GFP does not.
Pure control proteins (BSA, IgG, RNase A, bovine apo-transferrin) can be purchased from a variety of vendors.
REMARKS
It is essential to obtain the proper oligomerization state of transport factors (for instance, dimeric NTF2, monomeric Kaps); thus, the size exclusion chromatography step in the purification process is important. Formation of complexes (e.g., between transport factor and cargo) is always confirmed by size exclusion chromatography.
Either fluorescent proteins or fluorescent dyes can be used as reporters for confocal microscopy measurements. Fluorescent proteins are less bright, the tag can be bulky, or even interfere with protein function when engineered improperly; however, stoichiometric and site-specific labeling is achieved and proteins do not tend to aggregate. Alternatively, a variety of bright, fluorescent, commercially available dyes can be used (see below). Proteins are labeled per manufacturer’s recommendation with the goal of achieving close to stoichiometric labeling. Size exclusion chromatography is used to remove free dye and aggregated labeled proteins. Additionally, an ultracentrifugation step (112,000×gav for 45 min in a TLA55 rotor (Beckman) at 4 °C) can be used to remove microaggregates before each experiment. Size exclusion chromatography and native gels are used to confirm proper oligomerization state of the labeled proteins. The fluorescent proteins in native gels are detected using a Typhoon 9400 Variable Imager.
REMARKS
We found that for many transport factors, labeling sulfhydryl groups is preferable to primary amines as cysteine labeling generally appears to result in less aggregation.
17.2 PREPARATION OF NANOSELECTIVE FILTERS
We use commercially available polycarbonate membranes (6 μm thick) with pore sizes of 30, 50, or 100 nm as our templates.
-
Thin gold layer can be deposited on one side of polycarbonate membrane in various manners; we chose gold sputtering and used a Edwards S150 Sputter Coater following the manufacturer’s instructions. The thickness of the gold layer can be monitored by observing the blue–green color of the gold layer and measured using transmission electron microscopy. Reproducible results can be obtained by maintaining identical sputtering conditions from one experiment to the next. 15–30 nm thicknesses are optimal for our experiments as this range mimics the approximate length of the central pore of the NPC in vivo.
REMARKS
Pore diameters are somewhat batch dependent and can be accurately measured using scanning electron microscopy after gold sputtering (or after coating with PEG or FG-nups, see below). To examine the coated membranes by scanning electron microscopy, membranes are treated with 1% glutaraldehyde in TBT buffer, 0.2% aqueous tannic acid, and 1% OsO4 in sodium cacodylate buffer pH 7.4 with water washing steps in between. The membranes are kept in methanol overnight and dried with hexamethyldisilazane. Scanning EM is done after ~30 s gold coating. Images can be taken with a LEO 1550 field emission scanning electron microscope (Zeiss).
We observed that a ~2-fold increase in gold layer thickness (that should allow more FG-nups to bind to the internal gold surface at the entrance to the pores without changingthe effectiveporesize) yielded a ~2-foldincreasein selectivity. The following steps are used to create nanoselective filters:
Before chemisorption, gold sputtered membranes are cleaned with 25% HNO3 for an hour, rinsed in water, and air-dried (gold side up).
Approximately 20 μl of 0.4 mg/ml FG-nups in TBT, or as negative control, 100 μl of 2 mM methoxy polyethylene glycol (mPEG)-thiol in 8 M urea are reduced for 30 min with fresh 5 mM TCEP and placed on a glass slide in a humid chamber (glass slide is placed in the middle of the Petri dish lined with wet wipes).
The membrane is placed gold side down on top of the protein or PEG solution for 1 h. Occasional short (several seconds) incubations of the chamber in a room temperature sonication bath help distribute the coating more evenly.
The FG-nup-coated membranes are subsequently put in ~100 μl of 2 mM reduced 356 Da mPEG-thiol solution in 8 M urea for 1 h to coat any remaining exposed gold area.
-
then, membranes are washed in TBT buffer; pieces of appropriate size (approximately 2 × 2 mm) are cut and maintained in TBT at room temperature. Only freshly prepared membranes are used for experiments.
REMARKS
After sulfhydryl chemisorption, a color change is evident on the membrane.
The number of proteins around the pore can be calculated using amino acid analysis. Our samples were analyzed at the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University (New Haven, CT, USA). In typical experiments, ~0.04–0.05 FG-nup molecules per nm2 were found. Taking pore diameter (33 nm) and thickness of the gold layer (15 nm) into account, this corresponds to approximately 60–80 molecules of FG-nups present around each pore.
17.3 DEVICE SETUP
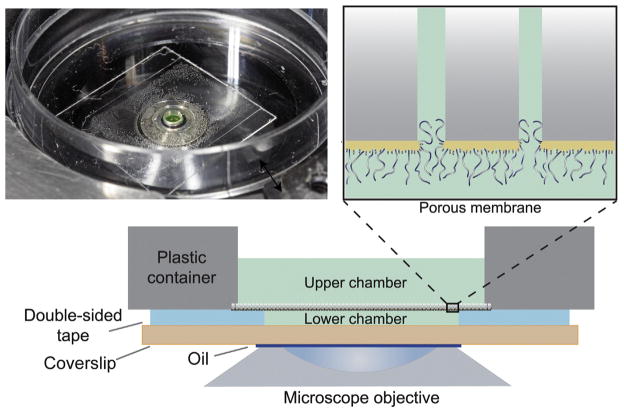
The assembly with the nanoselective-functionalized membrane used for flux measurements is shown in Fig. 17.1.
FIGURE 17.1.
Device setup. Photograph (top left), porous membrane diagram (top right), and schematic representation (bottom) of biomimetic NPC device.
Using a hole-puncher, a ~1.6-mm diameter hole is made in a square piece of ~ 25 μm thick optically clear double-sided tape.
-
The double-sided tape is bonded to a clean glass coverslip of appropriate thickness for microscopy.
REMARKS
If nonspecific adsorption of fluorescent protein occurs, PEG-coated coverslips can be used. They can be purchased (e.g., MicroSurfaces, Inc., Minneapolis), or PEG can be deposited using standard methods such as Schlapak et al. (2006).
-
Approximately 0.2 μl of fluorescently labelled protein(s) (~0.5–10 μM in TBT buffer) is applied to the opening of the double-sided tape, directly on the coverslip. This forms the lower chamber with volume of approximately 0.05 μl.
REMARKS
For our experiments, we use the following protein and protein combinations: (a) two transport factors, (b) a transport factor (with or without cargo) and a control protein, (c) transport factor alone, and (d) control protein alone. For two-component systems, we use labelling with two different fluorophores that spectrally show minimal overlap (e.g., Alexa Fluor 488 and Alexa Fluor 633) (see details Section 17.1.2).
For the two-color experiments, we usually use twofold excess of nonbinding proteins (e.g., BSA) over binding proteins (e.g., NTF2–GST).
A piece of the coated membrane is gently placed on top of the tape, gold side down. It is important to remove excess TBT from the membrane without drying out the surface.
A ~2-mm diameter hole is created with a drill in the middle of a 35 mm plastic dish (1 mm thickness of the bottom layer). This dish is positioned on top of the assembly, sealing the functionalized membrane via the double-sided tape.
2.0 μl of fluorescent protein mix (same as in the lower chamber described in point 3) is put in the hole of the plastic dish, creating the upper chamber; this solution is identical for both the top and bottom chambers. For measurements, the assembly is placed on the oil-covered objective of an inverted confocal microscope.
17.4 FLUX MEASUREMENTS
-
After assembly, the functionalized membrane device is placed on the stage of a Leica TCS SP spectral confocal inverted microscope equipped with a PL 100×/1.4 oil-immersion objective lens. The lower chamber, nanoselective filter, and a small part of the upper chamber are imaged using x-z scanning. For fluorophore excitation, we use the 488 nm line of an argon ion laser (to excite Alexa Fluor 488, GFP, FITC, YFP, or Cy3 fluorescence) and/or the 633 nm line of a HeNe laser (to excite Cy5 and Alexa Fluor 633 fluorescence). We observe fluorescence in either one channel (one-component system) or two channels (two-component system).
REMARKS
While other confocal microscopes can certainly be used, we found that Leica SP confocal microscopes provided excellent x-z scanning capabilities.
The high voltage and off-set of the photomultiplier tube should be adjusted to guarantee that a linear relationship is established between fluorophore concentration and recorded signal as well as to ensure that no significant bleaching occurs during the measurement.
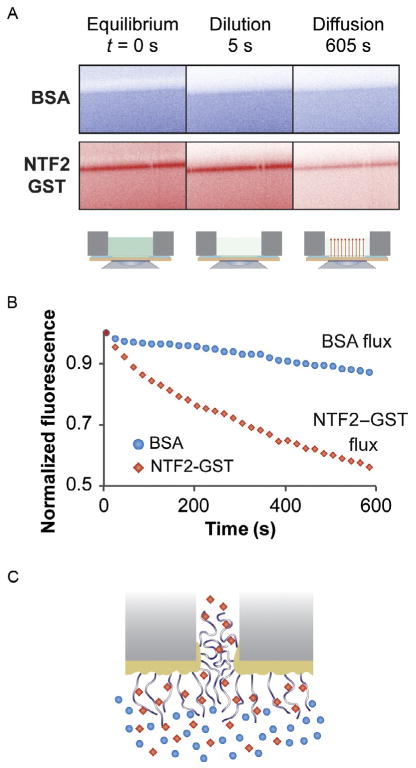
Flux measurements are performed by first obtaining an x-z scan before dilution of the upper chamber (Fig. 17.2A, left panel). At that stage, control proteins such as BSA (Fig. 17.2A, top) do not bind to FG-nup membranes, whereas transport factors such as NTF2–GST (Fig. 17.2A, bottom) bind to FG-nup membrane (band with increased fluorescent density). Note that in this case, the control protein (BSA) has approximately the same Stokes radius as the transport factor construct (NTF2–GST, which is a dimer in solution).
-
Next, proteins are rapidly diluted in the upper chamber with ~100 μl of TBT buffer (Fig. 17.2A, middle panel). Finally, to measure the diffusion from the lower chamber, fluorescence scans are acquired every 20 s for a total measuring period of ~10 min (Fig. 17.2A, right panel). After 10 min, BSA is largely retained in the lower chamber, whereas NTF2–GST diffuses out.
REMARKS
The height of the lower chamber should be close to constant over the time course of the experiment. If the height changes significantly (more then 10%), the membrane is likely not bonded well and the device is discarded.
-
Image J software is used to obtain the time-dependent mean fluorescence intensity of the lower chamber from a confocal scan series. Next, fluorescence intensity values are corrected for background fluorescence, normalized, and plotted versus time after dilution. The experimental values are fitted by a simple exponential using SigmaPlot:
(17.1) where c(t) is the time-dependent mean fluorescence of the lower chamber, k is the transport rate constant, Pn is the permeability coefficient of a membrane with n nanopores, and L is the height of the lower chamber.
FIGURE 17.2.
Flux measurements. (A) Scheme of the experiment: control protein (bovine serum albumin, BSA), labeled with fluorescein isothiocyanate (FITC) top, and transport factor (nuclear transport factor 2, NTF2) tagged with glutathione S-transferase (GST) and labeled with Cy5, bottom, are mixed together and their transport through FG-nup-functionalized membrane is measured in two channels. (B) Measurements for BSA (circles) and NTF2–GST (diamonds) diffusion through this nanoselective filter. (C) Schematic representation of FG-nup-functionalized membrane with transport factor (diamonds) and control protein (circles).
The flux through a single nanopore is:
| (17.2) |
where σ is the area density of nanopores and Δc is concentration difference. Pore size and density can be obtained from scanning EM measurements or manufacturers’ specifications. Examples of flux measurements for the transport factor NTF2–GST (diamonds) and a control protein BSA (circles) are given in Fig. 17.2B, and a schematic representation of transport is given in Fig. 17.2C. Each measurement is repeated multiple times and reported with standard error.
We use a flux ratio as a measure of transport efficiency. This value is obtained for each protein by calculating the ratio of its flux through a FG-nup-functionalized membrane relative to its flux through a control membrane coated with 356 Da PEG; a value of 1 indicates no change in flux between two membranes, while a value of 0 indicates no flux through the FG-nup-functionalized membrane. Detailed description of the evaluation and quantification of transport measurements has been given in detail previously (Jovanovic-Talisman et al., 2009; Peters, 2003).
We can measure fluxes of various proteins and protein mixtures across FG-nup-and PEG-functionalized membranes. We find that control proteins have significantly reduced fluxes across FG-nup-functionalized membranes compared with transport factor constructs having the same Stokes radius when transport factors are present. This indicates that transient binding of transport factors is an important feature of the NPC selectivity mechanism.
17.5 MATERIALS AND REAGENTS (LISTED BY ALPHABETICAL ORDER)
17.5.1 For proteins and protein preparation
17.5.1.1 Equipment
HPLC, Agilent Technologies, Santa Clara, CA
Microfluidizer, Microfluidics, Westwood, MA
Polytron, Kinematica, Luzern, Switzerland
Superose 6 10/300 column, #17-5172-01 GE Healthcare, Piscataway, NJ
Typhoon 9400 Variable Imager, Amersham Biosciences, Piscataway, NJ
17.5.1.2 Bacterial strains and plasmids
BL21-CodonPlus(DE3)-RIL cells, #230240, Agilent Technologies, Inc
BL21-Gold(DE3) cells, #230130, Agilent Technologies, Inc.
eGFP-His6 in pET21b (Rout lab)
Ibb-eGFP-His6 in pET21b (Rout lab)
Kap121–GST in pGEX-4T-1 (J. Aitchison lab, Seattle Biomedical Research Institute)
Kap95 in pET24a (Rout lab)
Kap95–GST in pGEX-2T-K (J. Aitchison lab, Seattle Biomedical Research Institute)
Nsp1FG-Cys-His6 in pET21b (Chait/Rout lab)
NTF2(W7A)–GST in pGEX-2T (R. Peters lab; Jovanovic-Talisman et al., 2009)
NTF2(WT)–GST in pGEX-2T (R. Peters lab; Jovanovic-Talisman et al., 2009)
NTF2(WT)–YFP-His6 in pET21b (Chait/Rout lab)
Nup100FG-Cys-His6 in pET21b (Chait/Rout lab)
17.5.1.3 Reagents
6-amino-n-hexanoic acid, # A7824-100G, Sigma-Aldrich, St. Louis, MO
Complete protease inhibitor cocktail tablets, EDTA-free, #11873580001, Roche, Indianapolis, IN
Control proteins: bovine apo-transferrin, bovine serum albumin, and RNase A from bovine pancreas, Sigma-Aldrich, St. Louis, MO; Rabbit IgG, MP Biomedicals, Solon, OH
Fluorescent dyes: Alexa Fluor 488 C5 Maleimide, #A-10254; Alexa Fluor 633 C5 Maleimide, #A-20342; FITC, #F-1906, Life Technologies, Grand Island, NY, and Cy3, Cy5 Maleimide mono-Reactive Dye 5-packs, #PA23031 and #PA25031, GE Healthcare, Piscataway, NJ
Glutathione Sepharose 4 Fast Flow, 17-5132-01, GE Healthcare
HisTrapFF column, 17-5247-01 GE Healthcare, Piscataway, NJ
Imidazole, #I5513-25G, Sigma-Aldrich, St. Louis, MO
IPTG, #I56000-5.0, RPI Corp, Mount Prospect, IL
Pepstatin A, # P5318-5MG, Sigma-Aldrich, St. Louis, MO
PMSF, # 78830-1G, Sigma-Aldrich, St. Louis, MO
TALON resin # 635506, Clontech Laboratories, Inc., Mountain View, CA
TCEP, #20490 Thermo Scientific-Pierce, Rockford, IL
Thrombin Cleavage Capture Kit, #69022-3, Novagen, San Diego, CA
Urea, #U6504, Sigma-Aldrich, St. Louis, MO
17.5.2 For device assembly and flux measurements
17.5.2.1 Equipment/material
~25 μm thick optically clear double-sided tape (3 M)
35 × 10 mm Petri Dishes, # 351008, Corning Life Sciences, Tewksbury, MA
6-μm thick polycarbonate membranes with pore sizes of 30, 50, or 100 nm, #KN3CP02500, #KN5CP02500, and #K01CP02500, respectively, GE Osmonics, Piscataway, NJ
Edwards S150 Sputter Coater
Hole-puncher 1/16 in., # 3424A57, McMaster-Carr, Santa Fe Springs, CA
Leica TCS SP spectral confocal inverted microscope equipped with a PL
100×/1.4 oil-immersion objective lens, Leica Microsystems Buffalo Grove, IL
17.5.2.2 Reagents/buffer
Buffer A: 50 mM sodium phosphate pH 8, 300 mM NaCl, 0.8 M urea mPEG-thiol (356 Da, #11156-0695) Polypure, Norway or (30 kDa, #2M4F0R01) Nektar Therapeutics, CA
TBT buffer: 20 mM HEPES, pH 7.4, 110 mM KOAc, 2 mM MgCl2, 10 μM ZnCl2, 10 μM CaCl2, 0.1% Tween-20
CONCLUSIONS
Using an interplay between biochemistry, cell biology, nanotechnology, and microscopy we devised a mechanochemical mimic of the NPC, a 2 billion-year-old molecular machine. This device has key properties in common with the NPC in vivo and can be used to investigate the mechanism of nucleocytoplasmic transport. In addition to the approach presented here, several groups have developed mimetic devices that can be used to study nucleocytoplasmic shuttling (e.g., Bird & Baker, 2011; Caspi et al., 2008; Kowalczyk et al., 2011; Pagliara, Schwall, & Keyser, 2013; Soskine et al., 2012). These novel nanobiological approaches are excellent test devices for gaining a detailed understanding of the mechanism of transport between the nucleus and the cytoplasm.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang WZ, Kipper J, Devos D, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450(7170):695–701. doi: 10.1038/nature06405. http://dx.doi.org/10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Bird SP, Baker LA. An abiotic analogue of the nuclear pore complex hydrogel. Biomacromolecules. 2011;12(9):3119–3123. doi: 10.1021/bm200820x. http://dx.doi.org/10.1021/bm200820x. [DOI] [PubMed] [Google Scholar]
- Caspi Y, Zbaida D, Cohen H, Elbaum M. Synthetic mimic of selective transport through the nuclear pore complex. Nano Letters. 2008;8(11):3728–3734. doi: 10.1021/nl801975q. http://dx.doi.org/10.1021/nl801975q. [DOI] [PubMed] [Google Scholar]
- Cronshaw JA, Krutchinsky AN, Zhang WZ, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. Journal of Cell Biology. 2002;158(5):915–927. doi: 10.1083/jcb.200206106. http://dx.doi.org/10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal SM, Akin D, Bashir R. Solid-state nanopore channels with DNA selectivity. Nature Nanotechnology. 2007;2(4):243–248. doi: 10.1038/nnano.2007.78. http://dx.doi.org/10.1038/nnano.2007.78. [DOI] [PubMed] [Google Scholar]
- Jirage KB, Hulteen JC, Martin CR. Effect of thiol chemisorption on the transport properties of gold nanotubule membranes. Analytical Chemistry. 1999;71(21):4913–4918. doi: 10.1021/ac990615i. http://dx.doi.org/10.1021/ac990615i. [DOI] [PubMed] [Google Scholar]
- Jovanovic-Talisman T, Tetenbaum-Novatt J, McKenney AS, Zilman A, Peters R, Rout MP, et al. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature. 2009;457(7232):1023–1027. doi: 10.1038/nature07600. http://dx.doi.org/10.1038/nature07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk SW, Kapinos L, Blosser TR, Magalhaes T, van Nies P, Lim RYH, et al. Single-molecule transport across an individual biomimetic nuclear pore complex. Nature Nanotechnology. 2011;6(7):433–438. doi: 10.1038/nnano.2011.88. http://dx.doi.org/10.1038/nnano.2011.88. [DOI] [PubMed] [Google Scholar]
- Lakshmi BB, Martin CR. Enantioseparation using apoenzymes immobilized in a porous polymeric membrane. Nature. 1997;388(6644):758–760. doi: 10.1038/41978. http://dx.doi.org/10.1038/41978. [DOI] [PubMed] [Google Scholar]
- Lee SB, Mitchell DT, Trofin L, Nevanen TK, Soderlund H, Martin CR. Antibody-based bio-nanotube membranes for enantiomeric drug separations. Science. 2002;296(5576):2198–2200. doi: 10.1126/science.1071396. http://dx.doi.org/10.1126/science.1071396. [DOI] [PubMed] [Google Scholar]
- Leslie DM, Zhang WZ, Timney BL, Chait BT, Rout MP, Wozniak RW, et al. Characterization of karyopherin cargoes reveals unique mechanisms of kap121p-mediated nuclear import. Molecular and Cellular Biology. 2004;24(19):8487–8503. doi: 10.1128/MCB.24.19.8487-8503.2004. http://dx.doi.org/10.1128/mcb.24.19.8487-8503.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(21):10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliara S, Schwall C, Keyser UF. Optimizing diffusive transport through a synthetic membrane Channel. Advanced Materials. 2013;25(6):844–849. doi: 10.1002/adma.201203500. http://dx.doi.org/10.1002/adma.201203500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R. Optical single transporter recording: Transport kinetics in microarrays of membrane patches. Annual Review of Biophysics and Biomolecular Structure. 2003;32:47–67. doi: 10.1146/annurev.biophys.32.110601.142429. http://dx.doi.org/10.1146/annurev.biophys.32.110601.142429. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Magnasco MO, Chait BT. Virtual gating and nuclear transport: The hole picture. Trends in Cell Biology. 2003;13(12):622–628. doi: 10.1016/j.tcb.2003.10.007. http://dx.doi.org/10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao YM, Chait BT. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. Journal of Cell Biology. 2000;148(4):635–651. doi: 10.1083/jcb.148.4.635. http://dx.doi.org/10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlapak R, Pammer P, Armitage D, Zhu R, Hinterdorfer P, Vaupel M, et al. Glass surfaces grafted with high-density poly(ethylene glycol) as substrates for DNA oligonucleotide microarrays. Langmuir. 2006;22(1):277–285. doi: 10.1021/la0521793. http://dx.doi.org/10.1021/la0521793. [DOI] [PubMed] [Google Scholar]
- Soskine M, Biesemans A, Moeyaert B, Cheley S, Bayley H, Maglia G. An engineered ClyA nanopore detects folded target proteins by selective external association and pore entry. Nano Letters. 2012;12(9):4895–4900. doi: 10.1021/nl3024438. http://dx.doi.org/10.1021/nl3024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR. Gatekeepers of the nucleus. Science. 2000;288(5470):1374–1377. doi: 10.1126/science.288.5470.1374. [DOI] [PubMed] [Google Scholar]