Abstract
Context
Variations in the brain-derived neurotrophic factor (BDNF) gene have been associated with psychiatric disorders, such as schizophrenia, bipolar and major depressive (MDD) disorder, and with antidepressant action. Deep sequencing of the BDNF gene may identify new genetic variations and bring further insights into psychiatric genetics.
Objective
To better characterize sequence variability in the BDNF gene.
Design
A genomic DNA region of 72 kb that contained the entire BDNF coding sequence and 5kb of flanking regions was re-sequenced in more than 500 subjects.
Setting and Participants
Re-sequencing data was obtained in 264 controls and 272 MDD individuals collected from the Mexican-American community in Los Angeles; individuals were accessed by the same bilingual clinical research team.
Main Outcome Measures
Identification of novel genetic polymorphisms in the BDNF gene, assessment of their frequencies and associations with MDD risk or response to antidepressants.
Results
We identified 83 novel single nucleotide polymorphisms (SNP): 30 in untranslated regions, in coding sequences, 37 in introns, and 12 in upstream regions. 3 of 4 rare novel coding SNPs were non-synonymous. Association analyses of MDD and controls revealed that 6 SNPs were associated with MDD (rs12273539, rs11030103, rs6265, rs28722151, rs41282918, rs11030101) and two haplotypes in different blocks were significantly associated with MDD. One recently reported 5′ UTR (untranslated region) SNP, rs61888800, was associated with antidepressant response after adjusting for age, gender, medication and baseline HAM-D21 score.
Conclusions
We identified 83 new BDNF polymorphisms and found that genetic frequency variations across populations exist for this gene. One single SNP (rs12273539) and two haplotypes (one including Val66, another near exon VIIIh) remained significant after adjusting for multiple testing. One 5′ UTR SNP was associated with antidepressant response. Further studies using larger independent samples are needed to confirm this association and to understand the implications of these novel BDNF variations in psychiatric disorders.
INTRODUCTION
The neutrophins are secreted peptides that are critically involved in differentiation and survival of neuronal populations 1–3. Brain-derived neurotrophic factor (BDNF) 4–7 is a neurotrophin that is abundantly and widely expressed in the CNS 8, 9. During the past decade, BDNF has emerged as a key factor implicated in complex behavioral patterns in the developing CNS and in disease. BDNF modulates signaling pathways that rapidly affect local synaptic function but it also has long-term effects on gene transcription. It promotes neuronal survival in the peripheral and CNS via the transcription factor cAMP-response element (CREB), which influences the expression of Bcl-2, a pro-survival gene. It also has important roles in excitatory synaptic transmission and plasticity10–13, memory processing and storage 13–18, and kindling and temporal lobe epilepsy 19–22. This relevance to crucial CNS functions has raised interest in its role in neurodegenerative and psychiatric disorders.
Allelic variations of the BDNF gene have been implicated in several conditions. Specifically, the allelic variation Thr2Ile (substitution of isoleucine for threonine at aminoacid position 2 in the coding sequence) has been implicated in the congenital central hypoventilation syndrome 23. The variations in BDNF have been extensively studied and implicated in the susceptibility to memory and hippocampal function impairments 24, and several psychiatric disorders25, such as obsessive-compulsive 26, eating 27, 28, bipolar 29,30–34, schizophrenia35, major depression 36, 37, and Alzheimer’s disease 38–40. In spite of conflicting findings in replication studies have been noted for several of these associations, it is interesting to note that the less frequent variation Met66, which is associated with poorer episodic memory and abnormal hippocampal activation in functional magnetic resonance imaging, generally confers a protective effect to neuropsychiatric conditions.
The genetic factors that contribute to human disease show enormous variation in the allelic spectra in number and population frequency of disease-predisposing alleles. Common complex disorders are multi-factorial and probably composed of both common genetic variants (common disease/common allele model) with small effect and rare sequence variants (rare variant/common disease model) with larger effect41. Although, the common allele is the prevalent of these two competing models in the genetic influence in common complex conditions, it has been predicted that re-sequencing studies may identify many rarer variants (>5%) of intermediate effect associated with common disorders and they may also be able to identify structural variations in genomic DNA, such as duplication and deletions of DNA sequences 42, 43.
Given the functional importance of BDNF in the CNS, the understanding of its allelic variants may be relevant to understanding its role in neuropsychiatric conditions. In spite a number of studies conducted to examine the association of BDNF variants, most of them have been focused on genotyping tag single nucleotide polymorphisms (tagSNPs) or the functional coding SNP rs6265. To our knowledge, no study has comprehensive surveyed BDNF sequence variation through direct sequencing and correlated the identified genetic variants with disease susceptibility. To discover new BDNF genetic variants and detect rare variants, we sequenced the whole BDNF gene and 5 kb flanking region in a total of 536 DNA samples comprised of 264 control and 272 depressed Mexican-American individuals. We further investigated all the identified genetic variants for association with risk for major depression and for relation to efficacy of antidepressant treatment.
METHODS
Participants
Participants were 264 controls and 272 patients with major depressive disorder (MDD) aged 19–68 years old. All participants were Mexican-Americans and had at least 3 grandparents born in Mexico. MDD was defined as a DSM-IV (Diagnostic and Statistical Manual of Mental Disorders, 4th Ed) diagnosis of current, unipolar major depressive episode and a HAM-D21 (21-Item Hamilton Depression Rating Scale) score of 18 or greater with item number 1 (depressed mood) rated 2 or greater. All MDD patients were enrolled in a pharmacogenetic study of antidepressant treatment response as previously described, and registered at clinical trials.gov (ID number NCT00265291)44, 45. The demographic characteristics and the numbers of subjects in each subgroup were presented in Supplementary Table 1 and a flowchart (Supplementary Figure 1). Briefly, in their primary language, all MDD patients had a comprehensive psychiatric and medical assessment based on the diagnostic and ratings instruments that had been fully validated in English and in Spanish. Exclusion criteria included active medical illnesses that could be etiologically related to the ongoing depressive episode, current or active suicidal ideation with a plan and strong intent, pregnancy, lactation, current use of medications with significant central nervous system activity, which interfere with EEG activity (e.g. benzodiazepines) or any other antidepressant treatment within the 2 weeks prior to enrollment, illicit drug use and/or alcohol abuse in the last 3 months or current enrollment in psychotherapy. Control individuals for our genomic studies were in general good health but were not screened for medical or psychiatric illness, they were age- and gender- matched and recruited from the same Mexican-American community in Los Angeles by the same bilingual clinical research team.
Antidepressant Treatment
All patients had an initial comprehensive psychiatric and medical assessment and, if enrolled, had weekly structured follow-up assessments for 9 weeks. The study consisted of two phases: a 1-week single-blind placebo lead-in phase to minimize the impact of placebo responders followed, if subjects continued to meet the inclusion criteria after phase 1, by random assignment to one of the two treatment groups: fluoxetine 10–40 mg/day or desipramine 50–200 mg/day, administered in a double-blind manner for 8 weeks. Our primary clinical outcome measure was HAM-D21 score and clinical remission on antidepressants was defined as having a final (week 8) HAM-D21 score < 844. In addition, the relative response change was also computed as the difference in HAM-D21 score between pre- and post-treatment divided by the pre-treatment HAM-D21 score.
Genomic DNA Collection and Sequencing
At the initial visit, blood samples were collected under informed consent from the participating individuals into EDTA (K2EDTA) BD Vacutainer EDTA tubes (Becton Dickinson, Franklin Lakes, NJ), and genomic DNA was isolated by using Gentra Puregene DNA purification kits (Gentra Systems, Indianapolis, IN). BDNF DNA sequencing was completed to identify genetic polymorphisms by the Wellcome Trust Sanger Institute following their ExoSeq protocol (http://www.sanger.ac.uk/humgen/exoseq/). A 72 kb genomic DNA region, containing the entire BDNF coding region and 5kb flanking region, was sequenced. Briefly, DNA sequences were extracted from the Vega database (http://vega.sanger.ac.uk/index.html). Primers were designed automatically using Primer3 to amplify DNA and primer pairs were checked for uniqueness prior to ordering and pre-screened to determine the optimum conditions for amplification. After amplification a sample of the products were visualized on an agarose gel, to confirm the size of the PCR product. The remaining PCR product was then cleaned-up using two enzymes, Exonuclease 1 and Shrimp Alkaline Phosphatase. Bi-directional sequencing of amplicons was carried out using Big DyeTM chemistry. SNPs were called using ExoTrace, a website algorithm developed for the detection of heterozygotes in sequence traces, which processes the sense and antisense sequence reads separately and subsequently and combines the results to allow SNP scoring.
Nucleotide Diversity (θ) and Population Differentiation (FST) Estimation
Nucleotide diversity (θ) and its standard deviation (S(θ)) were calculated by SNP class under the assumption of an infinite neutral allele model as follows46, 47: θ = K/aL, , and , where K = the number of observed SNPs among L base pairs of genomic sequence in a sample of n alleles. All calculations were based on n=990 for all the sites given that the average sample size was 495 individuals across all the polymorphisms. The pairwise FST values were estimated for the dbSNPs which were both detected in our Mexican American sample and reported in HapMap sample and were calculated as described by Weir48–50.
Hardy-Weinberg Equilibrium (HWE) Test and Population Stratification Analysis
Case-control study design is an efficient method to examine associations between candidate alleles and disease. But in order to compare allele frequencies and to be able to treat chromosomes as independent observations, the genotype frequencies must be in Hardy-Weinberg equilibrium51. Deviation from Hardy-Weinberg equilibrium was tested separately for healthy controls and patients by using PLINK program Version1.00 (http://pngu.mgh.harvard.edu/~purcell/plink/)52. SNPs that are not in HWE in the healthy control group were excluded from the allele-based association analyses of cases and controls.
Another confounding factor that may impact the internal validity of case-control studies is the presence of population stratification. We used two approaches to test for the hidden stratification in our data. Firstly, 54 unlinked SNPs across 22 autosomal chromosomes were employed to analyze a combined sample with the genotype data download from three HapMap ethnic samples using STRUCTURE program. Three distinct clusters were identified with an average proportion of at least 92% of individuals correctly assigned to the given ethnic populations (CEU, CHB+JPT, YRI) (Supplementary Figure 2A). We then used this panel of SNPs to test our sample and observed an almost equal proportion assigned to each clusters given K=2, 3, 4 in both cases and controls (Supplementary Figure 2B). Secondly, genotype frequencies from each of the 54 unlinked SNPs were compared between cases and controls using the method described by Pritchard et al53. No significant difference was found based on an overall test statistic (χ2=100.50, df =108, p=0.68).
Genetic Association Analyses of Case and Controls
For SNP-based association analysis, Fisher’s Exact test (2-tailed) was performed to compare allele frequencies and genotype distributions between depressed and healthy individuals by using PLINK program. In the allelic association analysis, each polymorphism was tested in controls to ensure the fitting with HWE; the odds ratio (OR) on the 2×2 contingency table of allele counts and its 95% confidence interval were also estimated for the polymorphism associated with the diagnosis of depression. In the genotypic association analysis, the SNP effects were tested under a codominant model on the 2×3 contingency table of genotype counts. In addition, logistic regression analyses were performed to test whether the observed SNP-depression association remained valid after controlling age and gender using SAS package (SAS Institute, Cary, NC).
For haplotype-based association analysis, haplotype blocks were identified by searching for “spine” of strong LD running from one marker to another along the legs of the triangle in the LD chart and haplotype population frequencies were estimated by using expectation maximization (EM) algorithm performed in computer program Haploview (Version 4, Broad Institute, http://www.broad.mit.edu/mpg/haploview/) 54. Haplotype frequencies were compared between depressed and control individuals to test whether a certain haplotype was associated with a diagnosis of depression.
To correct for multiple testing, 20,000 permutations were performed to estimate the adjusted p values for both single SNP-based analyses and haplotype-based analyses by using Haploview.
Genetic Association Analysis of Response to Antidepressants
Data analyses were performed using both intention-to-treat (ITT) and completed-treatment samples. ITT sample consisted of patients who were randomized to one arm and received at least one dose of antidepressant medication and completed-treatment sample consisted to patients who completed 8-week of antidepressant treatment. The last observation carried forward (LOCF) approach was used to imput missing outcome in the ITT analysis. For discrete outcome (remission vs. non-remission), we investigated the allelic and genotypic association with the response to antidepressant treatment using the similar approaches to those in the analysis of cases and controls. For the quantitative outcome (relative reduction % in HAM-D21 scores between pre- and post-treatment), we conducted the analyses based on three genetic models (additive, dominant and recessive) and first performed the analyses using the combined samples of patients treated with desipramine or fluoxetine. We then performed the analysis separately by antidepressant medication (desipramine only, fluoxetine only). We employed a multiple linear regression model to examine the association between genotype and relative HAM-D21 score reduction by controlling for age, gender, and baseline (pre-treatment) HAM-D21 score using PLINK program.
Power Calculation
Power to test the allelic association with depression was estimated with a range of effect size (odds ratio, OR) between 1.35 and 2.25 and minor allele frequency (MAF) between 0.1 and 0.25 using PAWE program 55. Power analyses showed that at a two-sided significance level of 0.05, sample sizes of 265 cases and 265 controls can achieve 80% power to detect an allelic OR of 1.68, 1.57, 1.50 and 1.46 with an MAF of 0.10, 0.15, 0.20 and 0.25, respectively. Power calculations for the association of BDNF variants with antidepressant treatment continuous outcome were given for a range of allele frequencies and Cohen’s effect size (mean difference in unit of standard deviation) based on dominant genetic model and using the Quanto (Version 1.2.3) program 56, 57. Sample size is assumed to be 200 for combined sample and 100 for each antidepressant treatment group based on an ITT design. Power analyses showed that at a two-sided significance level of 0.05 and when the allele frequency ≥ 0.15, the power is ≥ 89% to uncover a moderate effect size of 0.5 for a sample of 200 patients and ≥ 78% to detect a medium effect size of 0.6 for a sample of 100 patients.
RESULTS
Detection of Sequence Variation
Approximately 72 kb of DNA sequence containing 5 kb of flanking regions was systematically screened for novel nucleotide sequence variations in a sample of 536 Mexican American individuals, 264 controls and 272 depressed. A total of 130 nucleotide sequence variations were identified (Table 1). They included 83 novel SNPs and 47 dbSNPs: 40 in untranslated regions (UTRs), 6 in coding sequences, 62 in intronic sequences, and 22 in the flanking regions. Among 6 coding SNPs, 3 novel non-synonymous SNPs [NT_009237.17_26467094 (Ala/Thr), NT_009237.17_26467235 (His/Gly), NT_009237.17_26467246 (Gly/Asp)], and 1 synonymous SNP (NT_009237.17_26466714) were found and their minor allele frequencies were respectively 0.0019, 0.0019, 0.001, and 0.001 in the combined sample of cases and controls. Seventy-nine other novel polymorphisms included: 30 UTR SNPs, 37 intronic SNPs and 12 upstream SNPs (Supplementary Table 2). The minor allele frequencies for the novel polymorphisms ranged from 0.0009 to 0.2445 with an allele distribution of ≤0.001, 37.6%; >0.001 and <0.01, 50.5%; and >0.01, 11.9% in the combined sample of cases and controls.
Table 1.
Detected BDNF SNPs in Mexican Americans
| Location* | Sequence Screened (bp) | No. of Novel SNP | No. of dbSNP | No. of Total SNP | Nucleotide Diversity ± SD (x10−4) | Transition % |
|---|---|---|---|---|---|---|
| Coding | 792 | 4 | 2 | 6 | 10.1 ± 4.5 | 100 |
| 5′ UTR | 2434 | 16 | 2 | 18 | 9.9 ± 2.9 | 72.2 |
| Intron | 60703 | 37 | 25 | 62 | 1.4 ± 0.3 | 71.0 |
| 3′ UTR | 2928 | 14 | 8 | 22 | 10.5 ± 2.8 | 69.6 |
| Upstream | 4989 | 12 | 10 | 22 | 5.9 ± 1.6 | 68.2 |
| Total | 71846 | 83 | 47 | 130 | 2.4 ± 0.5 | 71.8 |
Intron-exon boundaries were based on multiple alternative 5′ exons in NCBI AceView Database.
Nucleotide Diversity
The nucleotide diversity was estimated in each class of sites (coding, 3′ UTR, 5′ UTR and intronic) by correcting for both sample size and the length of the screened site (Table 1). The nucleotide diversities were comparable for coding (0.0010 ± 0.0005), 3′UTR (0.0011 ± 0.0003) and 5′ UTR (0.0010 ± 0.0003) regions, but the estimate showed much lower nucleotide diversity in intronic region (0.00014 ± 0.00003). SNPs in UTRs or coding regions showed a 6-fold more diversity compared to those in intronic region. For the type of substitution, all the identified coding polymorphisms were transition, whereas the transition rates were 71.0%, 69.6%, 72.2% and 68.2% for intronic, 3′ UTR, 5′ UTR, and upstream regions, respectively.
Population Differentiation
Among the 47 dbSNPs detected, 18 were reported in three HapMap ethnic groups: White (CEU), Black (YRI), and Asian (CHB+JPT) in the NCBI database as of 06/25/2008. Pairwise FST values between Mexican Americans (MA) and each HapMap ethnic sample were computed for the shared 18 SNPs and showed in Table 2. Overall, the greater similarity in allele frequencies was found between Mexican Americans and Caucasians with a lower mean FST in MA vs CEU of 0.03, compared to that in MA vs YRI of 0.1 and in MA vs CHB+JPT of 0.09. For the single-locus estimates of FST values, large FST values (> 0.1) were observed at 4 SNPs (rs7124442, rs11819808, rs4923468, and rs7931755) in MA vs YRI (22.2%) and at 5 SNPs (rs6265, rs11030102, rs11030104, rs988748, and rs10767664) in MA vs CHB+JPT (27.8%), but less often (5.5%) in MA vs CEU (1 SNP: rs12273539).
Table 2.
Allele Frequencies and Fst Values for BDNF dbSNPs Shared by Mexican Americans and HapMap Samples
| SNP | Major/ Minor Allele | Minor Allele Frequency (MAF) | Fst | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mexican American (MA) | HapMap Sample | |||||||||
| Cases | Controls | All | CEU | YRI | HCB+JPT | MA vs CEU | MA vs YRI | MA vs HCB+JPT | ||
| rs7124442 | T/C | 0.23 | 0.26 | 0.25 | 0.37 | 0.54 | 0.07 | 0.03 | 0.18 | 0.09 |
| rs6265 | G/A | 0.10 | 0.15 | 0.12 | 0.18 | 0.00 | 0.48 | 0.01 | 0.08 | 0.31 |
| rs11030101 | A/T | 0.26 | 0.33 | 0.29 | 0.40 | 0.12 | 0.31 | 0.02 | 0.07 | 0.00 |
| rs11819808 | C/T | 0.01 | 0.00 | 0.01 | 0.00 | 0.28 | 0.00 | 0.00 | 0.45 | 0.00 |
| rs11030102 | C/G | 0.17 | 0.18 | 0.18 | 0.29 | 0.07 | 0.01 | 0.04 | 0.04 | 0.11 |
| rs12273539 | C/T | 0.35 | 0.23 | 0.29 | 0.00 | 0.34 | 0.13 | 0.20 | 0.00 | 0.06 |
| rs11030104 | A/G | 0.12 | 0.15 | 0.14 | 0.20 | 0.00 | 0.49 | 0.01 | 0.08 | 0.29 |
| rs11030109 | G/A | 0.02 | 0.03 | 0.03 | 0.04 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 |
| rs988748 | C/G | 0.15 | 0.16 | 0.15 | 0.22 | 0.05 | 0.49 | 0.01 | 0.04 | 0.25 |
| rs4923468 | C/A | 0.01 | 0.02 | 0.02 | 0.02 | 0.19 | 0.02 | 0.00 | 0.26 | 0.00 |
| rs10767664 | A/T | 0.14 | 0.16 | 0.15 | 0.20 | 0.04 | 0.49 | 0.00 | 0.04 | 0.26 |
| rs7931755 | A/G | 0.01 | 0.01 | 0.01 | 0.00 | 0.25 | 0.00 | 0.00 | 0.41 | 0.00 |
| rs2030324 | G/A | 0.37 | 0.41 | 0.39 | 0.57 | 0.55 | 0.56 | 0.06 | 0.05 | 0.05 |
| rs12273363 | T/C | 0.15 | 0.16 | 0.16 | 0.19 | 0.07 | 0.01 | 0.00 | 0.03 | 0.09 |
| rs908867 | C/T | 0.04 | 0.05 | 0.04 | 0.12 | 0.10 | 0.04 | 0.04 | 0.03 | 0.00 |
| rs7931247 | C/T | 0.37 | 0.42 | 0.39 | 0.57 | 0.55 | 0.56 | 0.06 | 0.05 | 0.05 |
| rs12288512 | G/A | 0.14 | 0.15 | 0.15 | 0.19 | 0.07 | 0.01 | 0.00 | 0.02 | 0.09 |
| rs11030123 | G/A | 0.04 | 0.05 | 0.04 | 0.12 | 0.10 | 0.04 | 0.04 | 0.02 | 0.00 |
Single SNP-Based Association Analyses of Cases and Controls
SNP-based allelic association analyses revealed that 6 polymorphisms were associated with MDD (rs12273539, p=0.00009; rs11030103, p=0.008; rs6265, p=0.009; rs28722151, p=0.01; rs41282918, p=0.01; rs11030101, p=0.02) (Table 3). All these 6 SNPs had a minor allele frequency of ≥ 0.14 and their genotypes were in Hardy-Weinberg equilibrium in controls. Genotyped-based analyses also showed that the 6 polymorphisms were associated with depression phenotype with a p ≤ 0.04 (Table 3). Among the 6 associated SNPs, 4 were intronic variants with OR ranging from 1.37 to 1.80; 1 SNP was 3′ UTR variant (rs41282918) with an effect of OR=2.13 (95% CI: 1.18–3.86); and 1 SNP was non-synonymous variant (rs6265) with an effect of OR=1.66 (95% CI: 1.14–2.41). Logistic regression analyses did not reveal a significant difference in age or gender between cases and controls and the associations of the 6 SNPs with depression remained similar after adjusting for age and gender. Permutation analysis showed that only SNP rs12273539 remained significant after adjusting for multiple tests with a corrected p value of 0.002.
Table 3.
BDNF Polymorphisms Associated with Depression
| SNP | Position | SNP Type | Risk/Non-risk Allele | Control Risk Allele Freq | OR (95%CI) | p* |
|---|---|---|---|---|---|---|
| rs41282918 | 27635356 | 3′ UTR | A/C | 0.84 | 2.13(1.18, 3.86) | 0.01 (0.02) |
| rs6265 | 27636492 | Non-synonymous | G/A | 0.85 | 1.66(1.14, .41) | 0.009 (0.008) |
| rs11030101 | 27637320 | Intronic | A/T | 0.67 | 1.37(1.05, 1.78) | 0.02 (0.04) |
| rs28722151 | 27637752 | Intronic | C/G | 0.68 | 1.48(1.10, 1.99) | 0.01 (0.009) |
| rs11030103 | 27638909 | Intronic | G/A | 0.19 | 1.80(1.18, 2.74) | 0.008 (0.03) |
| rs12273539 | 27639887 | Intronic | T/C | 0.23 | 1.75(1.32, 2.31) | 0.00009 (0.0008) |
OR: Odd Ratio.
Results are based on Fisher’s exact test for comparisons of allele and genotype (in parenthesis) frequencies between depressed patients and controls.
Haplotype-Based Association Analysis of Cases and Controls
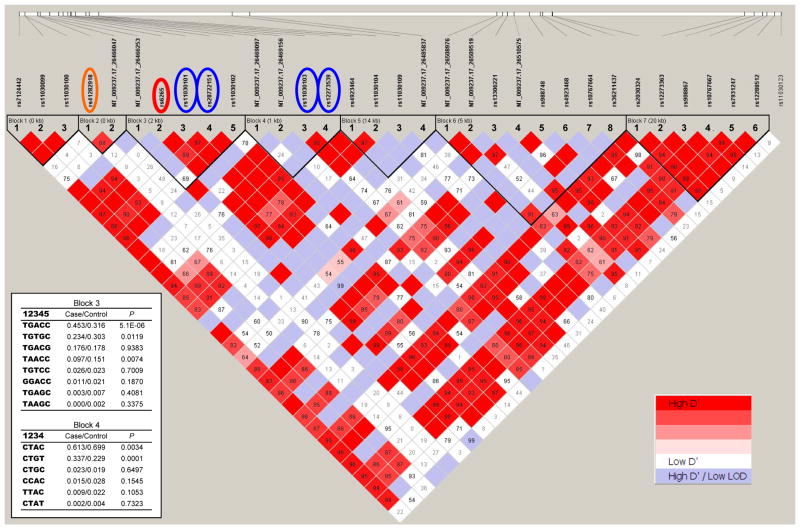
Figure 1 shows that 7 haplotype blocks were identified by searching for the solid spine of strong LD, Among the 130 detected polymorphisms, 33 SNPs with a minor allele frequency ≥1.5% were included in the haplotype analyses. Several haplotypes were found to be associated with the diagnosis of depression in block 3 (5 SNPs: rs56820186, rs6265, rs11030101, rs28722151, and rs11030102) and block 4 (4 SNPs: rs57083135, NT_009237.17_26469156, rs110303103, and rs12273539). Block 3 included three SNPs associated with depression (Table 3). The most significant association in block 3 was found for a common haplotype TGACC, and the haplotype frequency was 0.453 in cases and 0.316 in controls (χ2=20.80, p=0.000005; permutation adjusted p=0.0002). In block 4, the most significant association was found for haplotype CTGT, and the haplotype frequency was 0.337 in cases and 0.229 in controls (χ2=15.06, p=0.0001; permutation adjusted p=0.002). No other haplotypes were associated with depression after adjusting for multiple testing in the permutation tests.
Figure 1. Linkage disequilibrium (LD) pattern in BDNF.
Standard color scheme in Haploview program is used to display the level of logarithm of odds (LOD) and the D′ (right inserted key). Estimated statistics of the D′ are shown in each box. They indicate the LD relationship between each pair of SNPs and they are not labeled if D′=1.00. The BDNF gene structure is illustrated by a long horizontal white bar with vertical lines indicating the relative positions of SNPs and black box representing alternative exons named by Pruunsild et al7. SNPs associated with depression are marked in orange (UTR), red (coding) and blue (intronic) circles. Left inset shows haplotype frequencies in cases and controls and the p values for the association analysis between haplotype and diagnosis of depression in blocks 3 and 4.
Genetic Association Analysis of Response to Antidepressants
In the present study, there were 200 MDD patients who received at least one dose of antidepressant treatment (ITT sample of 103 received desipramine and 97 received fluoxetine) and 142 MDD patients who completed 8-week antidepressant treatment (completed-treatment sample of 68 with desipramine and 74 with fluoxetine). For the discrete outcome (remission vs non-remission), no detected polymorphisms were found to be significantly associated with the remission status in allelic and genotype-based analyses using ITT or completed-treatment samples. For the quantitative outcome (relative reduction in HAM-D21 score), one newly reported 5′ UTR SNP, rs61888800, was found to be associated with the better response to antidepressant treatment (p=0.02) after adjusting for age, gender, medication and baseline HAM-D21 score in the combined sample of patients treated with desipramine or fluoxetine in completed-treatment sample analysis. Patients who had GG genotype showed a larger average reduction of HAM-D21 score of 66.3% (95CI: 62.0–70.7%) compared to those who had non-GG genotype and had an average relative reduction of HAM-D21 score of 56.5% (95CI: 48.6–64.57%). For the medication-specific analyses, eight BDNF polymorphisms were found to be associated with the HAM-D score reduction among the patients treated with desipramine in both ITT and completed-treatment analyses with a p ≤ 0.05 after controlling for age, gender and baseline HAM-D score (Table 4), but no remained significant after adjusting for multiple testing. Among the 8 SNPs associated with response to desipramine treatment, all showed 14% larger reduction of HAM-D score in the patients homozygous for major allele except rs12273539, which showed 14% smaller reduction in patients homozygous for major allele in completed-treatment analysis and showed similar pattern but with a smaller reduction in ITT analysis. No associated polymorphism remained significant after adjusting for multiple testing through permutation and no detected SNPs were found significantly associated with the reduction of HAM-D scores in fluoxetine-treated group.
Table 4.
BDNF Polymorphisms Associated with Response to Antidepressant Treatment with Desipramine
| SNP (Type*) | Chr Position | Genotype | Intent-to-treat analysis^ | Complete-case analysis^ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Mean ± SD | β (95% CI) | p | N | Mean ± SD | β (95% CI) | p | |||
| rs7124442 (3′ UTR) | 27633617 | CC/CT | 43 | 37.97 ± 30.35 | 14.88 (2.99, 26.77) | 0.02 | 28 | 50.17 ± 25.58 | 14.6 (3.09, 26.11) | 0.02 |
| TT | 58 | 48.17 ± 30.77 | 40 | 60.88 ± 22.58 | ||||||
| rs11030102 (Intronic) | 27638172 | GG/GC | 33 | 35.64 ± 30.60 | 15.72 (3.14, 28.31) | 0.02 | 21 | 46.64 ± 27.50 | 15.09 (3.08, 27.11) | 0.02 |
| CC | 65 | 49.45 ± 30.04 | 47 | 60.86 ± 21.56 | ||||||
| rs12273539 (Intronic) | 27639887 | TT/TC | 53 | 47.30 ± 30.89 | −13.70 (−25.95, −1.45) | 0.03 | 35 | 61.13 ± 21.34 | −14.16 (−26.1, −2.23) | 0.02 |
| CC | 44 | 36.35 ± 31.42 | 28 | 49.66 ± 27.49 | ||||||
| rs61888800 (5′ UTR, Intronic) | 27678854 | TT/TG | 33 | 36.05 ± 30.83 | 17.12 (4.19, 30.05) | 0.01 | 21 | 47.28 ± 27.64 | 18.41 (6.24, 30.57) | 0.004 |
| GG | 60 | 49.84 ± 31.14 | 41 | 63.84 ± 20.96 | ||||||
| rs56133711 (Intronic) | 27679910 | AA/AG | 28 | 36.54 ± 29.80 | 15.52 (2.64, 28.40) | 0.02 | 18 | 48.57 ± 28.44 | 14.12 (1.20, 27.04) | 0.04 |
| GG | 63 | 50.16 ± 29.58 | 45 | 61.64 ± 21.44 | ||||||
| rs2030324 (Intronic) | 27683491 | AA/AG | 61 | 40.35 ± 30.63 | 12.81 (0.34, 25.29) | 0.05 | 37 | 52.44 ± 24.80 | 14.2 (2.78, 25.62) | 0.02 |
| GG | 39 | 48.79 ± 32.27 | 29 | 63.70 ± 21.84 | ||||||
| rs12273363 (Upstream) | 27701435 | CC/CT | 29 | 34.59 ± 28.61 | 15.81 (2.54, 29.09) | 0.02 | 19 | 48.29 ± 24.16 | 14.28 (1.47, 27.09) | 0.03 |
| TT | 71 | 46.39 ± 31.99 | 48 | 59.44 ± 24.00 | ||||||
| rs7931247 (Upstream) | 27703567 | TT/TC | 63 | 39.90 ± 30.23 | 13.2 (0.92, 25.49) | 0.04 | 38 | 51.72 ± 24.86 | 14.94 (3.53, 26.35) | 0.01 |
| CC | 39 | 48.79 ± 32.27 | 29 | 63.70 ± 21.84 | ||||||
Intron-exon boundaries were based on multiple alternative 5′ exons in NCBI AceView Database.
Mean: average relative reduction in HAM-D21 score; SD: standard deviation; β: regression coefficient for allele effect based on dominant model after adjusting for gender, age and baseline HAM-D21 score.
COMMENTS
We surveyed BDNF sequence variation by studying a 72kb genomic DNA region, which contained the entire BDNF coding and 5 kb of flanking sequences by direct sequencing. Our results provide a detailed description of BDNF sequence variations in Mexican Americans. Among the 130 SNPs that we detected in this study, 83 are novel and only 47 have been reported in NCBI dbSNP database, which has collected 254 BNDF SNPs to date(http://www.ncbi.nlm.nih.gov/projects/SNP). Most of these new polymorphisms (89%) are rare variants with a minor allele below 1% (Supplementary Table 2). This is not surprising because our study was conducted in large sample of 537 subjects of a specific ethnic group that has not been investigated extensively. The nucleotide diversity in that genomic region is 0.00024. Unexpectedly, we observed 6-fold more genetic diversity in coding regions (0.00101) compared to the intronic region (0.00014) although this estimate is very close to that (0.000238) observed in 6.8 kb BDNF non-coding region58. Some studies revealed that nucleotide diversity varies across both genes and functional classes, and gene-to-gene differences in SNP diversity are the most important factors that contribute to the variation46. To date, 254 BDNF SNPs (after excluding duplicates) have been reported in NCBI dbSNP database, including 7 in CDS (coding domain sequence) of 792 bp, 224 in intron of 60721 bp and 23 in UTR of 5362 bp. This also yields a higher frequency in coding regions (1 SNP per 113 bp) compared to intronic region (1 SNP per 271 bp). In addition, the sample size and population may also contribute to this frequency difference. Pairwise FST values revealed a substantial population differentiation in 18 dbSNPs using frequency data available from the NCBI database of 3 ethnic groups (CEU, YRI, CHB+JPT). For example, a high divergence of allele frequency was noted for non-synonymous SNP rs6265 across ethnic populations; minor allele (A allele) frequencies of 0.12, 0.18, 0.00 and 0.48 were found in Mexican American, Caucasian, African, and Asian, respectively. Our findings suggest that the genetic variation in the BDNF gene across different populations may be large and this heterogeneity may contribute to explain controversial findings in association of BNDF with depression from different populations.
It is noteworthy that rare variants in relevant genes in neurodevelopmental pathways have been associated with schizophrenia59, further supporting the rare variant/common disease model. The discovery of 83 mostly rare variants in BDNF, a gene that is found to be relevant to several psychiatric disorders, may therefore be of widespread interest.
We report here that 5 SNPs in the BDNF gene were significantly associated with depression, in addition to the non-synonymous SNP rs6265 which we reported previously36. Among the 6 SNPs, rs12273539, an intronic variant located 3.4 kb away from rs6265 and near alternative 5′exon VIIIh (Figure 1), showed the most significant association with depression and remained significant after adjustment for multiple testing. Unlike rs6265, rs12273539 showed much less similarity in allele frequency between Mexican Americans and Caucasians with a large Fst value of 0.20. Haplotype analyses revealed a strong LD (D′=1.00) between rs6265 and rs12273539 but they mapped to two LD blocks (blocks 3 and 4 in Figure 1). Two common haplotypes: TGACC that includes BDNF Val66 allele (G) in exon IX in block 3 and CTGT in block 4 near exon VIIIh, were found significantly associated with the increased risk for depression after correcting for multiple testing.
We also found that 8 SNPs were associated with drug response to desipramine treatment in both ITT and completed-treatment samples although the association did not remained significant after adjustment for multiple testing. Among the 8 SNPs, there were one 3′ UTR SNP (rs7124442) in block 1, two newly reported SNPs (5′ UTR SNP rs61888800 in exon Vh and intronic SNP56133711) in block 6, three SNPs (rs2030324 in intron; rs12273363 and rs7931247 in upstream region) in block 7, and one in each of block 3 (rs11030102) and block 4 (rs12273539) (Figure 1). Interestingly, SNP rs12273539, which showed the most significant association with depression status, was also associated with the drug response to desipramine treatment (β=−14.16%, p=0.024) in 8-week completers.
There are several implications to our findings. Firstly, they support the concept that BNDF genetic variants may differ in frequency and/or effect among different ethnic groups. For instance, our data support that in the variant rs6265 (Val66Met), the Val (G allele) carriers are at increased risk for depression, which is consistent with the data of several Caucasian studies 60–62. However, several studies in Asians have reported no association between depression and Val66Met63–65, or the association of the Met (A allele) variant with susceptibility to depression66, 67. Our population differentiation analysis also revealed that Mexican-Americans and Caucasians have a comparable Val66Met allele frequency (Fst=0.01), but they have substantial allele difference when compared to Asians (Fst=0.31). The observed results across ethnic groups may suggest heterogeneity in the BDNF allele frequencies and genetic polymorphisms among populations. Secondly, they suggest that other BDNF genetic variants besides Val66Met may contribute to susceptibility to depression. In this survey, we found 6 BDNF polymorphisms that were associated with depression risk. The strongest association was found to an intronic variant rs12273539. We also identified two haplotypes in different haplotype blocks, one containing rs6265 and the other containing rs12273539, that are significantly associated with depression after multiple testing adjustment (p ≤ 0.002). Thirdly, they suggest that the association of BDNF genetic variants with drug response to antidepressant treatment may be medication-specific and do not support a major role of Val66Met variant in antidepressant action. Among the 6 polymorphisms associated with depression in this study, only SNP rs12273539 was found to be associated with HAM-D21 score reduction in desipramine treatment in our sample. However, 7 other SNPs were found to be associated with desipramine treatment by showing ≥14% more average reduction in patients who are homozygous for major allele.
Three studies have recently assessed the association between Val66Met polymorphism and antidepressant response in MDD patients, but only one reported that Met carriers had a better response to 8-week citalopram treatment63. Gratacos et al reported a SNP rs908867 and a haplotype (TAT at rs12273363, rs908867 and rs1491850) in 5′ upstream region associated with antidepressant response68. Interestingly, in this region, we found 3 SNPs (rs2030324, rs12273363, and rs7931247 in block 7) associated with desipramine treatment although the association of rs908867 with response to antidepressant treatment was not significant in our study. The differential findings could be due to a number of factors such as medication type, outcome assessment, sample size, population substructure, and very importantly, the complexity and rich diversity in the regulation of BDNF multiple transcripts, in the coding and noncoding sequences,, and in the proBDNF and mature BDNF translation product sequences 7, 69.
Limitations of this study are related to the sample size is relative small, particularly for analyses of antidepressant treatment response. Although power analyses showed that at a single two-sided significance of 0.05 and allele frequency ≥ 0.15, a sample size of 200 patients can achieve 89% power to detect a moderate effect size of 0.5 that is close to what we observed in desipramine group, the power should be much lower if the genetic effect is medication-specific as our results suggest. Given the small sample size and that the lack of replication sample, the association with antidepressant treatment response should be interpreted with much caution and considered exploratory.
In conclusion, we have identified 83 novel BDNF genetic variants. Our data support the implication of BDNF in the susceptibility to major depressive disorder and in the therapeutic response to antidepressants. To our knowledge, this work is the most comprehensive genetic association study to date to have examined the association between BDNF sequence variation with both depression and antidepressant response. Given that a number of alternative BDNF tanscripts have been found to display complex splicing and expression patterns and the findings in different studies remain inconsistent, further comprehensive studies in larger independent samples are clearly warranted for conclusive results. Moreover, we suggest that deep sequencing of relevant genes in large numbers of patients can reveal substantial numbers of novel variants that may be useful targets for association studies.
Supplementary Material
Supplementary Figure 1. Study flow chart
The flow diagram shows the numbers of participants through each stage of the study. Brown boxes represent the sample in case-control study and blue boxes include the MDD patients enrolled in 1 week placebo lead in, 8-week double blind and randomized pharmacogenetic study of antidepressant treatment.
Supplementary Figure 2. Estimated population structure using 54 unlinked SNPs
In the rectangles on the left, each individual is represented by a single vertical line, which is broken into K colored segments, with lengths proportional to each of the K clusters inferred from STRUCTURE program using the genotype download from HapMap database (A) and Mexican-American MDD patients and healthy controls in sample (B) separately. In the triangle plots on the right, each individual is represented by a colored point corresponding to the predefined population. For a given point, the estimate is given by the distance to one edge of the triangle.
Acknowledgments
Funding sources: NIH grants GM61394, RR017365, MH062777, RR000865, RR16996, HG002500, and DK063240, and Institutional Funds from the University of Miami, Department of Psychiatry & Behavioral Sciences
This research was supported in part by National Institutes of Health (NIH) grants GM61394, RR017365, MH062777, RR000865, RR16996, HG002500, and DK063240, and Institutional Funds from the University of Miami, Department of Psychiatry & Behavioral Sciences. No sponsors or funders had any role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript. The Principal Investigator (J.L.) had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All the DNA sequencing was completed by the Wellcome Trust Sanger Institute, UK. The authors gratefully acknowledge the cooperation of participants. The authors also thank all the staff from University of California in Los Angels and University of Miami for sample collection, clinical assessments and technical assistance.
References
- 1.Barbacid M. Neurotrophic factors and their receptors. Curr Opin Cell Biol. 1995;7(2):148–155. doi: 10.1016/0955-0674(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 2.Levi-Montalcini R. The nerve growth factor 35 years later. Science. 1987;237(4819):1154–1162. doi: 10.1126/science.3306916. [DOI] [PubMed] [Google Scholar]
- 3.Segal RA, Greenberg ME. Intracellular signaling pathways activated by neurotrophic factors. Annu Rev Neurosci. 1996;19:463–489. doi: 10.1146/annurev.ne.19.030196.002335. [DOI] [PubMed] [Google Scholar]
- 4.Barde YA, Edgar D, Thoenen H. Purification of a new neurotrophic factor from mammalian brain. Embo J. 1982;1(5):549–553. doi: 10.1002/j.1460-2075.1982.tb01207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofer MM, Barde YA. Brain-derived neurotrophic factor prevents neuronal death in vivo. Nature. 1988;331(6153):261–262. doi: 10.1038/331261a0. [DOI] [PubMed] [Google Scholar]
- 6.Maisonpierre PC, Le Beau MM, Espinosa R, 3rd, Ip NY, Belluscio L, de la Monte SM, Squinto S, Furth ME, Yancopoulos GD. Human and rat brain-derived neurotrophic factor and neurotrophin-3: gene structures, distributions, and chromosomal localizations. Genomics. 1991;10(3):558–568. doi: 10.1016/0888-7543(91)90436-i. [DOI] [PubMed] [Google Scholar]
- 7.Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90(3):397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connor B, Dragunow M. The role of neuronal growth factors in neurodegenerative disorders of the human brain. Brain Res Brain Res Rev. 1998;27(1):1–39. doi: 10.1016/s0165-0173(98)00004-6. [DOI] [PubMed] [Google Scholar]
- 9.Murer MG, Yan Q, Raisman-Vozari R. Brain-derived neurotrophic factor in the control human brain, and in Alzheimer’s disease and Parkinson’s disease. Prog Neurobiol. 2001;63(1):71–124. doi: 10.1016/s0301-0082(00)00014-9. [DOI] [PubMed] [Google Scholar]
- 10.Bramham CR, Messaoudi E. BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Prog Neurobiol. 2005;76(2):99–125. doi: 10.1016/j.pneurobio.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Poo MM. Neurotrophins as synaptic modulators. Nat Rev Neurosci. 2001;2(1):24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- 12.Schinder AF, Poo M. The neurotrophin hypothesis for synaptic plasticity. Trends Neurosci. 2000;23(12):639–645. doi: 10.1016/s0166-2236(00)01672-6. [DOI] [PubMed] [Google Scholar]
- 13.Tyler WJ, Alonso M, Bramham CR, Pozzo-Miller LD. From acquisition to consolidation: on the role of brain-derived neurotrophic factor signaling in hippocampal-dependent learning. Learn Mem. 2002;9(5):224–237. doi: 10.1101/lm.51202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pang PT, Lu B. Regulation of late-phase LTP and long-term memory in normal and aging hippocampus: role of secreted proteins tPA and BDNF. Ageing Res Rev. 2004;3(4):407–430. doi: 10.1016/j.arr.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Rattiner LM, Davis M, Ressler KJ. Brain-derived neurotrophic factor in amygdala-dependent learning. Neuroscientist. 2005;11(4):323–333. doi: 10.1177/1073858404272255. [DOI] [PubMed] [Google Scholar]
- 16.Bekinschtein P, Cammarota M, Izquierdo I, Medina JH. Reviews: BDNF and memory formation and storage. Neuroscientist. 2008;14(2):147–156. doi: 10.1177/1073858407305850. [DOI] [PubMed] [Google Scholar]
- 17.Bekinschtein P, Cammarota M, Katche C, Slipczuk L, Rossato JI, Goldin A, Izquierdo I, Medina JH. BDNF is essential to promote persistence of long-term memory storage. Proc Natl Acad Sci U S A. 2008;105(7):2711–2716. doi: 10.1073/pnas.0711863105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada K, Nabeshima T. Brain-derived neurotrophic factor/TrkB signaling in memory processes. J Pharmacol Sci. 2003;91(4):267–270. doi: 10.1254/jphs.91.267. [DOI] [PubMed] [Google Scholar]
- 19.He XP, Kotloski R, Nef S, Luikart BW, Parada LF, McNamara JO. Conditional deletion of TrkB but not BDNF prevents epileptogenesis in the kindling model. Neuron. 2004;43(1):31–42. doi: 10.1016/j.neuron.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 20.Croll SD, Suri C, Compton DL, Simmons MV, Yancopoulos GD, Lindsay RM, Wiegand SJ, Rudge JS, Scharfman HE. Brain-derived neurotrophic factor transgenic mice exhibit passive avoidance deficits, increased seizure severity and in vitro hyperexcitability in the hippocampus and entorhinal cortex. Neuroscience. 1999;93(4):1491–1506. doi: 10.1016/s0306-4522(99)00296-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kokaia M, Ernfors P, Kokaia Z, Elmer E, Jaenisch R, Lindvall O. Suppressed epileptogenesis in BDNF mutant mice. Exp Neurol. 1995;133(2):215–224. doi: 10.1006/exnr.1995.1024. [DOI] [PubMed] [Google Scholar]
- 22.Lahteinen S, Pitkanen A, Koponen E, Saarelainen T, Castren E. Exacerbated status epilepticus and acute cell loss, but no changes in epileptogenesis, in mice with increased brain-derived neurotrophic factor signaling. Neuroscience. 2003;122(4):1081–1092. doi: 10.1016/j.neuroscience.2003.08.037. [DOI] [PubMed] [Google Scholar]
- 23.Weese-Mayer DE, Bolk S, Silvestri JM, Chakravarti A. Idiopathic congenital central hypoventilation syndrome: evaluation of brain-derived neurotrophic factor genomic DNA sequence variation. Am J Med Genet. 2002;107(4):306–310. doi: 10.1002/ajmg.10133. [DOI] [PubMed] [Google Scholar]
- 24.Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112(2):257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 25.Licinio J, Wong ML. Brain-derived neurotrophic factor (BDNF) in stress and affective disorders. Molecular psychiatry. 2002;7(6):519. doi: 10.1038/sj.mp.4001211. [DOI] [PubMed] [Google Scholar]
- 26.Hall D, Dhilla A, Charalambous A, Gogos JA, Karayiorgou M. Sequence variants of the brain-derived neurotrophic factor (BDNF) gene are strongly associated with obsessive-compulsive disorder. American journal of human genetics. 2003;73(2):370–376. doi: 10.1086/377003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribases M, Gratacos M, Armengol L, de Cid R, Badia A, Jimenez L, Solano R, Vallejo J, Fernandez F, Estivill X. Met66 in the brain-derived neurotrophic factor (BDNF) precursor is associated with anorexia nervosa restrictive type. Molecular psychiatry. 2003;8(8):745–751. doi: 10.1038/sj.mp.4001281. [DOI] [PubMed] [Google Scholar]
- 28.Ribases M, Gratacos M, Fernandez-Aranda F, Bellodi L, Boni C, Anderluh M, Cavallini MC, Cellini E, Di Bella D, Erzegovesi S, Foulon C, Gabrovsek M, Gorwood P, Hebebrand J, Hinney A, Holliday J, Hu X, Karwautz A, Kipman A, Komel R, Nacmias B, Remschmidt H, Ricca V, Sorbi S, Wagner G, Treasure J, Collier DA, Estivill X. Association of BDNF with anorexia, bulimia and age of onset of weight loss in six European populations. Hum Mol Genet. 2004;13(12):1205–1212. doi: 10.1093/hmg/ddh137. [DOI] [PubMed] [Google Scholar]
- 29.Sklar P, Gabriel SB, McInnis MG, Bennett P, Lim YM, Tsan G, Schaffner S, Kirov G, Jones I, Owen M, Craddock N, DePaulo JR, Lander ES. Family-based association study of 76 candidate genes in bipolar disorder: BDNF is a potential risk locus. Brain-derived neutrophic factor. Molecular psychiatry. 2002;7(6):579–593. doi: 10.1038/sj.mp.4001058. [DOI] [PubMed] [Google Scholar]
- 30.Geller B, Badner JA, Tillman R, Christian SL, Bolhofner K, Cook EH., Jr Linkage disequilibrium of the brain-derived neurotrophic factor Val66Met polymorphism in children with a prepubertal and early adolescent bipolar disorder phenotype. Am J Psychiatry. 2004;161(9):1698–1700. doi: 10.1176/appi.ajp.161.9.1698. [DOI] [PubMed] [Google Scholar]
- 31.Lohoff FW, Sander T, Ferraro TN, Dahl JP, Gallinat J, Berrettini WH. Confirmation of association between the Val66Met polymorphism in the brain-derived neurotrophic factor (BDNF) gene and bipolar I disorder. Am J Med Genet B Neuropsychiatr Genet. 2005;139(1):51–53. doi: 10.1002/ajmg.b.30215. [DOI] [PubMed] [Google Scholar]
- 32.Neves-Pereira M, Mundo E, Muglia P, King N, Macciardi F, Kennedy JL. The brain-derived neurotrophic factor gene confers susceptibility to bipolar disorder: evidence from a family-based association study. American journal of human genetics. 2002;71(3):651–655. doi: 10.1086/342288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada T, Hashimoto R, Numakawa T, Iijima Y, Kosuga A, Tatsumi M, Kamijima K, Kato T, Kunugi H. A complex polymorphic region in the brain-derived neurotrophic factor (BDNF) gene confers susceptibility to bipolar disorder and affects transcriptional activity. Molecular psychiatry. 2006;11(7):695–703. doi: 10.1038/sj.mp.4001822. [DOI] [PubMed] [Google Scholar]
- 34.Tramontina J, Frey BN, Andreazza AC, Zandona M, Santin A, Kapczinski F. Val66met polymorphism and serum brain-derived neurotrophic factor levels in bipolar disorder. Molecular psychiatry. 2007;12(3):230–231. doi: 10.1038/sj.mp.4001941. [DOI] [PubMed] [Google Scholar]
- 35.Neves-Pereira M, Cheung JK, Pasdar A, Zhang F, Breen G, Yates P, Sinclair M, Crombie C, Walker N, St Clair DM. BDNF gene is a risk factor for schizophrenia in a Scottish population. Molecular psychiatry. 2005;10(2):208–212. doi: 10.1038/sj.mp.4001575. [DOI] [PubMed] [Google Scholar]
- 36.Ribeiro L, Busnello JV, Cantor RM, Whelan F, Whittaker P, Deloukas P, Wong ML, Licinio J. The brain-derived neurotrophic factor rs6265 (Val66Met) polymorphism and depression in Mexican-Americans. Neuroreport. 2007;18(12):1291–1293. doi: 10.1097/WNR.0b013e328273bcb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumacher J, Jamra RA, Becker T, Ohlraun S, Klopp N, Binder EB, Schulze TG, Deschner M, Schmal C, Hofels S, Zobel A, Illig T, Propping P, Holsboer F, Rietschel M, Nothen MM, Cichon S. Evidence for a relationship between genetic variants at the brain-derived neurotrophic factor (BDNF) locus and major depression. Biol Psychiatry. 2005;58(4):307–314. doi: 10.1016/j.biopsych.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Bagnoli S, Nacmias B, Tedde A, Guarnieri BM, Cellini E, Petruzzi C, Bartoli A, Ortenzi L, Sorbi S. Brain-derived neurotrophic factor genetic variants are not susceptibility factors to Alzheimer’s disease in Italy. Ann Neurol. 2004;55(3):447–448. doi: 10.1002/ana.10842. [DOI] [PubMed] [Google Scholar]
- 39.Riemenschneider M, Schwarz S, Wagenpfeil S, Diehl J, Muller U, Forstl H, Kurz A. A polymorphism of the brain-derived neurotrophic factor (BDNF) is associated with Alzheimer’s disease in patients lacking the Apolipoprotein E epsilon4 allele. Molecular psychiatry. 2002;7(7):782–785. doi: 10.1038/sj.mp.4001073. [DOI] [PubMed] [Google Scholar]
- 40.Kunugi H, Ueki A, Otsuka M, Isse K, Hirasawa H, Kato N, Nabika T, Kobayashi S, Nanko S. A novel polymorphism of the brain-derived neurotrophic factor (BDNF) gene associated with late-onset Alzheimer’s disease. Molecular psychiatry. 2001;6(1):83–86. doi: 10.1038/sj.mp.4000792. [DOI] [PubMed] [Google Scholar]
- 41.Campbell H, Manolio T. Commentary: rare alleles, modest genetic effects and the need for collaboration. Int J Epidemiol. 2007;36(2):445–448. doi: 10.1093/ije/dym055. [DOI] [PubMed] [Google Scholar]
- 42.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, Bras JM, Schymick JC, Hernandez DG, Traynor BJ, Simon-Sanchez J, Matarin M, Britton A, van de Leemput J, Rafferty I, Bucan M, Cann HM, Hardy JA, Rosenberg NA, Singleton AB. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451(7181):998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 43.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong ML, Whelan F, Deloukas P, Whittaker P, Delgado M, Cantor RM, McCann SM, Licinio J. Phosphodiesterase genes are associated with susceptibility to major depression and antidepressant treatment response. Proc Natl Acad Sci U S A. 2006;103(41):15124–15129. doi: 10.1073/pnas.0602795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Licinio J, O’Kirwan F, Irizarry K, Merriman B, Thakur S, Jepson R, Lake S, Tantisira KG, Weiss ST, Wong ML. Association of a corticotropin-releasing hormone receptor 1 haplotype and antidepressant treatment response in Mexican-Americans. Molecular psychiatry. 2004;9(12):1075–1082. doi: 10.1038/sj.mp.4001587. [DOI] [PubMed] [Google Scholar]
- 46.Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nature genetics. 1999;22(3):239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- 47.Li WH. Molecular Evolution. Sunderland, MA: Sinauer Associates; 1997. [Google Scholar]
- 48.Weir BS. Genetic Data Analysis II. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 49.Weir BS, Cockerham CC. Estinmating F-statistics for the analysis of population structure. Evoluation. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 50.Weir BS, Hill WG. Estimating F-statistics. Annu Rev Genet. 2002;36:721–750. doi: 10.1146/annurev.genet.36.050802.093940. [DOI] [PubMed] [Google Scholar]
- 51.Balding DJ, Martin Bishop M, Cannings C, editors. Handbook of Statistical Genetics. 3. New York: JOHN WILEY & SONS, LTD; 2001. [Google Scholar]
- 52.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. American journal of human genetics. 1999;65(1):220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 55.Gordon D, Finch SJ, Nothnagel M, Ott J. Power and sample size calculations for case-control genetic association tests when errors are present: application to single nucleotide polymorphisms. Human heredity. 2002;54(1):22–33. doi: 10.1159/000066696. [DOI] [PubMed] [Google Scholar]
- 56.Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. American journal of epidemiology. 2002;155(5):478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 57.Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Statistics in medicine. 2002;21(1):35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- 58.Winantea J, Hoang MN, Ohlraun S, Rietschel M, Cichon S, Propping P, Nothen MM, Freudenberg J, Freudenberg-Hua Y. A summary statistic approach to sequence variation in noncoding regions of six schizophrenia-associated gene loci. Eur J Hum Genet. 2006;14(9):1037–1043. doi: 10.1038/sj.ejhg.5201664. [DOI] [PubMed] [Google Scholar]
- 59.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare Structural Variants Disrupt Multiple Genes in Neurodevelopmental Pathways in Schizophrenia. Science. 2008 doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 60.Lang UE, Hellweg R, Kalus P, Bajbouj M, Lenzen KP, Sander T, Kunz D, Gallinat J. Association of a functional BDNF polymorphism and anxiety-related personality traits. Psychopharmacology (Berl) 2005;180(1):95–99. doi: 10.1007/s00213-004-2137-7. [DOI] [PubMed] [Google Scholar]
- 61.Sen S, Nesse RM, Stoltenberg SF, Li S, Gleiberman L, Chakravarti A, Weder AB, Burmeister M. A BDNF coding variant is associated with the NEO personality inventory domain neuroticism, a risk factor for depression. Neuropsychopharmacology. 2003;28(2):397–401. doi: 10.1038/sj.npp.1300053. [DOI] [PubMed] [Google Scholar]
- 62.Strauss J, Barr CL, George CJ, Devlin B, Vetro A, Kiss E, Baji I, King N, Shaikh S, Lanktree M, Kovacs M, Kennedy JL. Brain-derived neurotrophic factor variants are associated with childhood-onset mood disorder: confirmation in a Hungarian sample. Molecular psychiatry. 2005;10(9):861–867. doi: 10.1038/sj.mp.4001685. [DOI] [PubMed] [Google Scholar]
- 63.Choi MJ, Kang RH, Lim SW, Oh KS, Lee MS. Brain-derived neurotrophic factor gene polymorphism (Val66Met) and citalopram response in major depressive disorder. Brain research. 2006;1118(1):176–182. doi: 10.1016/j.brainres.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 64.Hong CJ, Huo SJ, Yen FC, Tung CL, Pan GM, Tsai SJ. Association study of a brain-derived neurotrophic-factor genetic polymorphism and mood disorders, age of onset and suicidal behavior. Neuropsychobiology. 2003;48(4):186–189. doi: 10.1159/000074636. [DOI] [PubMed] [Google Scholar]
- 65.Tsai SJ, Cheng CY, Yu YW, Chen TJ, Hong CJ. Association study of a brain-derived neurotrophic-factor genetic polymorphism and major depressive disorders, symptomatology, and antidepressant response. Am J Med Genet B Neuropsychiatr Genet. 2003;123(1):19–22. doi: 10.1002/ajmg.b.20026. [DOI] [PubMed] [Google Scholar]
- 66.Hwang JP, Tsai SJ, Hong CJ, Yang CH, Lirng JF, Yang YM. The Val66Met polymorphism of the brain-derived neurotrophic-factor gene is associated with geriatric depression. Neurobiol Aging. 2006;27(12):1834–1837. doi: 10.1016/j.neurobiolaging.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 67.Iga J, Ueno S, Yamauchi K, Numata S, Tayoshi-Shibuya S, Kinouchi S, Nakataki M, Song H, Hokoishi K, Tanabe H, Sano A, Ohmori T. The Val66Met polymorphism of the brain-derived neurotrophic factor gene is associated with psychotic feature and suicidal behavior in Japanese major depressive patients. Am J Med Genet B Neuropsychiatr Genet. 2007;144(8):1003–1006. doi: 10.1002/ajmg.b.30520. [DOI] [PubMed] [Google Scholar]
- 68.Gratacos M, Soria V, Urretavizcaya M, Gonzalez JR, Crespo JM, Bayes M, de Cid R, Menchon JM, Vallejo J, Estivill X. A brain-derived neurotrophic factor (BDNF) haplotype is associated with antidepressant treatment outcome in mood disorders. Pharmacogenomics J. 2007 doi: 10.1038/sj.tpj.6500460. [DOI] [PubMed] [Google Scholar]
- 69.Liu QR, Lu L, Zhu XG, Gong JP, Shaham Y, Uhl GR. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain research. 2006;1067(1):1–12. doi: 10.1016/j.brainres.2005.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Study flow chart
The flow diagram shows the numbers of participants through each stage of the study. Brown boxes represent the sample in case-control study and blue boxes include the MDD patients enrolled in 1 week placebo lead in, 8-week double blind and randomized pharmacogenetic study of antidepressant treatment.
Supplementary Figure 2. Estimated population structure using 54 unlinked SNPs
In the rectangles on the left, each individual is represented by a single vertical line, which is broken into K colored segments, with lengths proportional to each of the K clusters inferred from STRUCTURE program using the genotype download from HapMap database (A) and Mexican-American MDD patients and healthy controls in sample (B) separately. In the triangle plots on the right, each individual is represented by a colored point corresponding to the predefined population. For a given point, the estimate is given by the distance to one edge of the triangle.