Abstract
Nanobodies are single domain antibodies derived from the variable regions of Camelidae atypical immunoglobulins. They show great promise as high affinity reagents for research, diagnostics and therapeutics due to their high specificity, small size (~15 kDa) and straightforward bacterial expression. However, identification of repertoires with sufficiently high affinity has proven time consuming and difficult, hampering nanobody implementation. Here, we present a rapid, straightforward approach that generates large repertoires of readily expressible recombinant nanobodies with high affinities and specificities against a given antigen. We demonstrate the efficacy of this approach through the production of large repertoires of nanobodies against two antigens, GFP and mCherry, with Kd values into the sub-nanomolar range. After mapping diverse epitopes on GFP, we were also able to design ultra-high affinity dimeric nanobodies with Kds down to ~30 pM. The approach presented is well-suited for the routine production of high affinity capture reagents for various biomedical applications.
Introduction
There is a continuing need in biomedicine for antibodies that recognize target molecules with high affinity and specificity. When high affinity antibodies are not available, common protein tags such as GFP, mCherry, and FLAG have been invaluable for many biological applications. However, most such studies still demand high quality antibodies against these protein tags, particularly when affinity isolation is required1–4. Although monoclonal or polyclonal antibodies remain the primary bait reagents available for these purposes, their large size, limited availability, and batch to batch variation have often proved problematic for biochemical or proteomic studies5.
Single domain antibodies, also referred to as “nanobodies”6, have emerged as an alternative to traditional antibodies. These are usually derived from camelids such as llamas, which make a unique subset of immunoglobulins consisting of heavy chain homodimers devoid of light chains7–9; their variable region (VHH) is the smallest antigen-binding single polypeptide chain naturally found in the antibody world8–12. Nanobodies are recombinant antigen-binding domains derived from these VHH regions. Unlike monoclonal antibodies, they can be readily produced in large amounts in bacterial expression systems9, 13. Moreover, nanobodies are usually extremely stable, can bind antigens with affinities in the nanomolar range, and are smaller in size (approximately 15 kDa) than other antibody constructs11, 14–18. However, rapid and robust techniques for the isolation of extensive repertoires of high affinity nanobodies have proven elusive – the labor-intensive nature and poor efficiency of current approaches have proven a major bottleneck for the widespread implementation of these reagents8, 12, 14, explaining why demand for these reagents greatly exceeds supply8.
Here, we present a highly optimized pipeline that allows the rapid production of large repertoires of high affinity nanobodies against selected proteins. The approach is based on high-throughput DNA sequencing of a marrow lymphocyte VHH cDNA library from an immunized llama combined with mass spectrometric (MS) identification of high affinity VHH regions derived from serum of the same animal.
Results
Strategy for nanobody identification
Our approach to nanobody discovery centers on MS identification of affinity-purified heavy-chain antibodies isolated from an individual llama, in correlation with a DNA sequence database generated from the same animal (Fig. 1). This concept is inspired by our previous efforts to identify circulating neutralizing HIV antibodies in humans by MS in conjunction with patient-specific antibody cDNA databases19. Our approach represents a novel pipeline for nanobody production where each stage has been highly optimized (Supplementary Protocol).
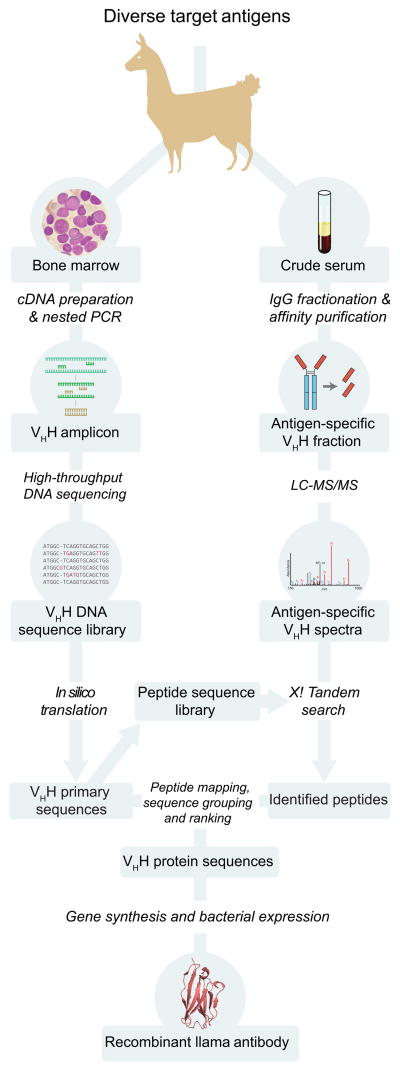
Figure 1.
Overview of nanobody identification and production pipeline. After llama immunization, cDNA from bone marrow aspirates is used for PCR amplification of the heavy-chain only variant’s variable region, which is then subjected to high-throughput DNA sequencing. Separately, the serum-derived VHH protein fraction from the same llama is affinity-purified against the antigen of interest, then analyzed by LC-MS/MS. The MS data is searched against a sequence database generated from the DNA sequencing reads, allowing identification of corresponding VHH sequences. These sequences are codon-optimized for gene synthesis, allowing efficient bacterial expression of recombinant protein. The example nanobody structure shown was obtained from PDB ID 3K1K29.
To generate nanobody repertoires of maximal utility, we chose the GFP and mCherry tags for our first target antigens, due to their central roles in cell biological studies. Further, while these fluorescent proteins have a broadly similar beta barrel structure, they are significantly evolutionarily divergent, making for very distinct immunogens20. After immunization of individual llamas with these antigens and confirmation of an immune response, we serially fractionated serum bleeds to obtain exclusively VHH-containing heavy chain antibodies (Supplementary Fig. 1a), taking advantage of the differential specificity of Protein A and Protein G for VHH versus conventional antibodies7. The VHH-containing fraction was affinity purified over antigen-coupled resin, washed with MgCl2 at various stringencies (Supplementary Fig. 1b), and digested with papain on-resin to cleave away the constant regions and leave behind the desired minimal VHH variable region fragments. Finally, the antigen-bound VHH fragments were eluted and separated by SDS-PAGE, allowing the purification of the ~15 kDa VHH fragments away from residual conventional Fab fragments and Fc fragments (both ~25 kDa), and undigested antibodies (~50 kDa) (Supplementary Fig. 1c). The gel-purified bands were trypsin-digested and analyzed by liquid chromatography-MS and MS/MS (Fig. 2a). We recovered the highest affinity VHH fragments by using the highest stringency washes, which also decreased the complexity of the eluted sample, aiding MS analysis.
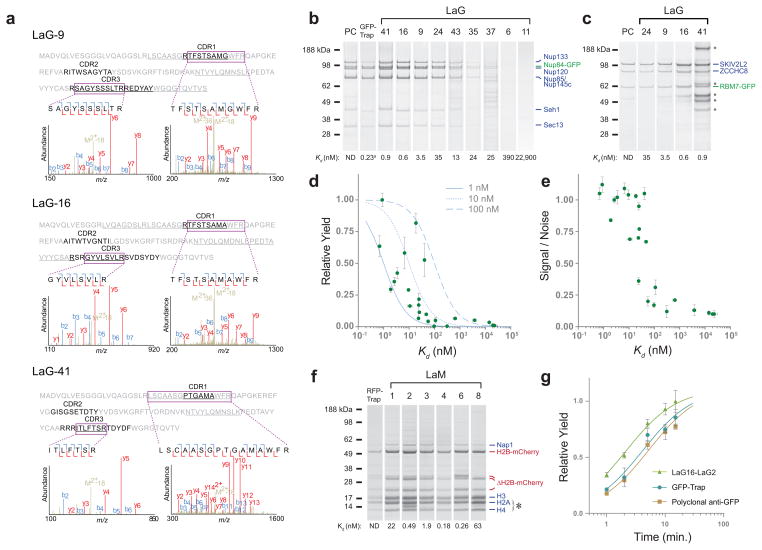
Figure 2.
MS analysis of GFP-binding VHH IgG and characterization of recombinant nanobodies. (a) Representative tandem mass spectra of identified peptides (shown boxed). Peptides were mapped to the informative CDR regions of three candidate VHH sequences, which were then chosen for production and characterization. The regions of these sequences covered by MS are underlined. (b) Indicated LaGs, commercial GFP-Trap®, or polyclonal anti-GFP llama antibody (PC) were conjugated to magnetic Dynabeads, and used for affinity isolations of S. cerevisiae Nup84-GFP or (c) RBM7-GFP from HeLa cells. Elutions were analyzed by SDS-PAGE, and duplicate Coomassie-stained bands identified by MS. Representative examples across a range of affinities are shown, and are labeled with the Kd for GFP as determined by SPR. (d) Relative yields of affinity isolated Nup84-GFP protein are plotted against the corresponding LaG’s in vitro affinity for GFP (green circles). Theoretical curves of the expected fraction of ligand bound to an immobilized binding partner at various Kds are also shown for three hypothetical ligand concentrations (grey lines). (e) The relative signal to noise ratio of three known Nup84 complex components to a known contaminant region was plotted against each LaG’s Kd. Experiments were done in duplicate, with error bars showing s.e.m. (f) S. cerevisiae mCherry-HTB2 (histone H2B) was affinity isolated by LaMs or RFP-Trap® conjugated to Dynabeads. Elutions were analyzed by SDS-PAGE, and Coomassie-stained bands identified by MS. The asterisk indicates the location of LaM nanobody leakage from the Dynabeads. LaM lanes are labeled with the Kd for mCherry as determined by SPR. (g) Affinity isolations of yeast Nup84-GFP were performed using a LaG16-LaG2 dimer with G4S, polyclonal anti-GFP, or commercial GFP- Trap®. The complex was isolated at various time points, and relative yield determined by quantification of Coomassie-stained bands of known Nup84 complex components. Experiments were done in duplicate, with error bars showing s.e.m.
To create an animal-specific antibody sequence database, lymphocyte mRNA samples from individual immunized llamas were obtained for high-throughput sequencing. Mononuclear cells were isolated from bone marrow aspirates, enriching for long-lived antibody secreting plasma cells, which transcribe elevated levels of immunoglobulin RNA19, 21, 22. Importantly, we do not create expression or display libraries, and thus remove the need for efficient exogenous expression, folding, and presentation of the clones.
Total RNA from these lymphocytes was reverse transcribed, and a nested PCR was performed to specifically amplify sequences encoding the VHH variable regions14. This PCR product was sequenced using high-throughput 454 (GFP) or MiSeq (mCherry), resulting in ~800,000 and ~3,000,000 unique reads, respectively. These reads were translated, filtered and trypsin-digested in silico to create a searchable peptide database for MS analysis (Fig. 1 and Supplementary Fig. 2).
The identification of specific VHH sequences is more challenging than typical proteins, as they consist in large part of highly conserved framework regions that are less easily distinguished by MS. Moreover, rather than searching well-established databases, a VHH cDNA database must be generated for each immunized animal. To deal with both challenges, we developed a bioinformatic pipeline that is able to identify the highest probability matches from a large pool of related VHH sequences (Llama Magic; http://www.llamamagic.org). In this pipeline, VHH sequences were ranked by a metric based on MS/MS sequence coverage of complementarity determining region 3 (CDR3, the most diverse VHH region) as well as CDR1 and CDR2 coverage, total VHH coverage, sequencing counts, mass spectral counts, and the expectation values of matched peptides (Supplementary Fig. 2 and 3). Preliminary attempts to identify VHH sequences solely by their CDR3 regions revealed that identical CDR3 sequences are frequently shared between multiple distinct VHH sequences, with diverse CDR1 and CDR2 sequences. It is likely that this is a result of somatic gene conversion, in which, after V(D)J recombination, secondary recombination occurs between upstream V gene segments and already rearranged V(D)J genes23, 24. Our automatic ranking pipeline, coupled with careful manual inspection, overcame these issues and provided us 44 high-probability hits against GFP, classified as LaG (Llama antibody against GFP) 1-44, which we subjected to further screening (Supplementary Fig. 4). A smaller subset of eight clones was chosen for follow up (LaM 1-8) for mCherry (Supplementary Fig. 5).
Codon optimized genes for these hits were synthesized and cloned into a bacterial expression vector. After expression, lysates were passed over antigen-coupled resin to identify nanobodies that displayed both robust expression as well as high and specific affinity (Supplementary Fig. 6). From these screens, we found 25 specific nanobodies against GFP (LaGs) and 6 against mCherry (LaMs). Phylogenetic analysis of the verified nanobodies revealed substantial sequence diversity among clones (Supplementary Fig. 7). While not directly analogous, the high success rate of this single screening step (57–75%) is favorable in comparison to the final panning and selection steps of phage display, in which up to 107 clones are screened to identify even a few positive clones12, 14, 25, 26. The affinity of these nanobodies was further assessed by either surface plasmon resonance (SPR) or in vitro binding assays with immobilized nanobodies (Supplementary Fig. 8–10). For the larger repertoire of LaGs, these experiments revealed a wide range of affinities, with Kds from 0.5 nM to over 20 μM (Table 1), and identified 16 nanobodies with high affinity binding (≤50 nM) (Supplementary Fig. 8). The Kds of the six LaMs were consistently strong, ranging from 0.18 nM to 63 nM (Table 1 and Supplementary Fig. 9).
Table 1.
Characteristics, affinities, and specificities for LaG, LaM, and LaG dimer proteins. Kds for GFP and mCherry binding were determined by SPR unless otherwise noted. Kds are also shown for LaG dimers fused using a glycine-rich peptide linker (3 repeats of GGGGS, or G4S), or a 3xFLAG linker. For yeast Nup84-GFP and mammalian RBM7-GFP affinity isolations using LaG-conjugated Dynabeads, Coomassie-stained bands from elutions separated by SDS-PAGE were quantified, and known specific and nonspecific bands were used to calculate signal to noise (S:N) ratios. Bead binding assays were used to determine affinity for variant fluorescent proteins, and divergent AmCFP (for LaGs) or DsRed (for LaMs) binding abilities are shown. GFP epitopes were determined by NMR for LaGs, and grouped into three broad classes (I–III). The number of residues identified in the binding site, and its calculated accessible surface area (ASA), are also shown.
| Clone ID | Mol. Wt (Da) | Kd (nM) | Nup84-GFP S:N | RBM7-GFP S:N | Binds AmCFP (LaG)/DsRed (LaM) | GFP Epitope | No. of binding site residues | ASA of binding site residues (Å2) |
|---|---|---|---|---|---|---|---|---|
| LaG-2 | 15,919 | 191, 16 | 1.03 | 0.42 | − | III | 55 | 2,204 |
| LaG-3 | 15,329 | 25 | 0.77 | 1.13 | + | nd | nd | nd |
| LaG-6 | 15,700 | 310 | 0.12 | nd | + | nd | nd | nd |
| LaG-9 | 16,062 | 3.5 | 1.02 | 1.04 | + | I | 62 | 2,551 |
| LaG-10 | 15,748 | 97 | 0.17 | nd | + | nd | nd | nd |
| LaG-12 | 16,090 | 56 | 0.20 | nd | + | nd | nd | nd |
| LaG-14 | 16,002 | 1.9 | 0.84 | 0.58 | + | I | 66 | 2,519 |
| LaG-16 | 16,306 | 0.7 | 1.05 | 0.92 | + | I | 60 | 2,605 |
| LaG-17 | 15,823 | 50 | 0.67 | nd | + | I | 60 | 2,543 |
| LaG-19 | 15,528 | 24.61 | 0.95 | 1.06 | + | II | 54 | 2,404 |
| LaG-21 | 15,452 | 7 | 1.09 | nd | + | II | 56 | 2,340 |
| LaG-24 | 14,763 | 41 | 1.05 | 1.09 | − | III2 | nd | nd |
| LaG-26 | 16,221 | 2.6 | 1.00 | nd | + | II | 53 | 2,070 |
| LaG-27 | 15,565 | 9.5 | 1.04 | nd | + | II | 57 | 2,216 |
| LaG-29 | 15,449 | 110 | 0.31 | nd | + | nd | nd | nd |
| LaG-30 | 16,159 | 0.5 | 1.04 | nd | + | nd | nd | nd |
| LaG-35 | 16,010 | 23.51 | 0.70 | nd | + | nd | nd | nd |
| LaG-37 | 16,329 | 24 | 0.36 | nd | + | nd | nd | nd |
| LaG-41 | 15,471 | 0.9 | 1.12 | 0.41 | + | II | 53 | 2,091 |
| LaG-42 | 15,490 | 600 | 0.21 | nd | + | nd | nd | nd |
| LaG-43 | 16,167 | 11 | 0.69 | nd | + | I | 55 | 2,381 |
| LaG-5 | 15,589 | 14,2001 | 0.11 | nd | nd | nd | nd | nd |
| LaG-8 | 15,953 | 20,0001 | 0.10 | nd | nd | nd | nd | nd |
| LaG-11 | 16,221 | 22,9001 | 0.10 | nd | nd | nd | nd | nd |
| LaG-18 | 16,459 | 3,8001 | 0.13 | nd | nd | nd | nd | nd |
|
| ||||||||
| LaG16-G4S-2 | 30,791 | 0.036 | nd | nd | nd | nd | nd | nd |
| LaG16-3xFLAG-2 | 32,972 | 0.268 | nd | nd | nd | nd | nd | nd |
| LaG41-G4S-2 | 29,956 | 0.150 | nd | nd | nd | nd | nd | nd |
|
| ||||||||
| LaM-1 | 15,380 | 22 | n/a | n/a | − | n/a | n/a | n/a |
| LaM-2 | 15,151 | 0.49 | n/a | n/a | − | n/a | n/a | n/a |
| LaM-3 | 15,196 | 1.9 | n/a | n/a | + | n/a | n/a | n/a |
| LaM-4 | 14,866 | 0.18 | n/a | n/a | + | n/a | n/a | n/a |
| LaM-6 | 14,428 | 0.26 | n/a | n/a | − | n/a | n/a | n/a |
| LaM-8 | 14,666 | 63 | n/a | n/a | − | n/a | n/a | n/a |
Kd determined by bead binding assay.
Determined by binding assays and mutagenesis.
Specificity and efficacy of recombinant nanobodies
We performed a variety of experiments to assess our nanobodies. Affinity isolations were performed on endogenous GFP- and mCherry-tagged proteins in yeast and human cells. All 25 positive LaGs were used for the isolation of GFP-tagged Nup84, a structural nuclear pore complex component, in budding yeast (Fig. 2b)27, 28. We plotted each LaG’s observed Kd against a quantification of either signal-to-background or yield from a Nup84-GFP affinity capture (Fig. 2d, e and Table 1). Almost all LaGs were able to pull down detectable amounts of Nup84-GFP and its associated proteins, and many performed as well or better than either our best affinity-purified polyclonal antibodies1, or than the single commercially available GFP-Trap® anti-GFP nanobody (ChromoTek GmbH), which has a Kd of 0.59 nM (Fig. 2b, g)29. When determining depletion of Nup84-GFP by Western blot, LaG-16, for instance, displays slightly higher yields than GFP-Trap® (Supplementary Fig. 11). Generally speaking, a strong correlation is seen between low Kd and both high signal to background and high yield. This correlation is consistent with the relationship theoretically predicted for the percentage of the low abundance yeast target proteins bound in solution30 (Fig. 2d). Our ability to compare structurally similar nanobodies raised against a single antigen provides a unique opportunity to demonstrate the importance of very low Kd to high quality antibody performance in this type of application. Even nanobodies with Kds around 10 nM, typically considered high affinity for an antibody, start displaying a precipitous decline in affinity purification performance. These findings highlight the importance of ultra-high affinity reagents, such as the nanobodies described here, for proteomic and interactomic studies.
Affinity capture experiments were also performed on GFP-tagged RBM7, a component of the human nuclear exosome, from HeLa cells (Fig. 2c)4, yielding performances comparable to those seen with Nup84-GFP. However, differences in the amount of contaminants were seen for certain LaGs, notably LaG-41, from purifications in yeast versus HeLa cells (Fig. 2b, c). These results underscore how even high affinity reagents can give unpredictable background in certain cell types, demonstrating the utility of obtaining and testing large repertoires of such affinity reagents to improve the chances that at least one is likely to be optimal for any particular application. Similarly, Dynabead-conjugated LaMs were used to isolate mCherry-tagged histone H2B from yeast (Fig. 2f). For all six LaMs tested, the core nucleosome complex was efficiently isolated, demonstrating the affinity and specificity of this second group of nanobodies. Consistent with the low Kds of all the identified LaMs, the yield and specificity of all affinity isolations were high. Commercial RFP-Trap® nanobody (ChromoTek GmbH) was tested in parallel, giving consistently lower yields.
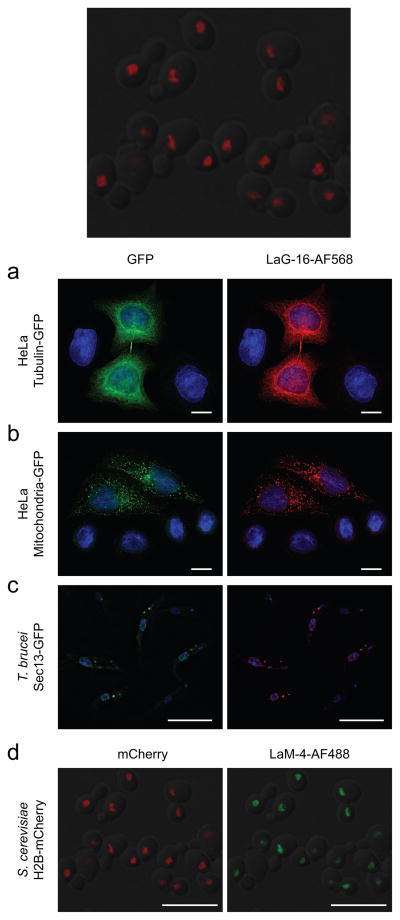
Nanobodies are powerful new tools for fluorescence microscopy, both standard and super-resolution31. We therefore tested the effectiveness of a selection of the LaG and LaM repertoire for immunofluorescence microscopy (Fig. 3 and Supplementary Fig. 12). As target proteins, we first made use of emGFP-tagged tubulin and mitochondria-targeted emGFP, transiently transfected into HeLa cells32. Fixed cells were stained with LaG-16 conjugated to Alexa Fluor® 568, giving specific and strong staining of GFP-tagged microtubule or mitochondrial structures respectively, with negligible non-specific staining of untransfected cells (Fig. 3a, b). To demonstrate the versatility of these reagents, we also used them for immunofluorescence in a Trypanosoma brucei strain with eGFP-tagged Sec13. This protein localizes to both the nuclear pore complex and COPII-coated vesicles, and indeed the AF568-nanobody signal colocalized with GFP to give the expected nuclear rim and endoplasmic reticulum staining33 (Fig. 3c). To determine if our anti-mCherry nanobodies were similarly well-suited for immunofluorescence microscopy, we conjugated LaM-4 to Alexa Fluor® 488 and stained S. cerevisiae expressing mCherry-tagged histone H2B; this also showed specific, colocalized nuclear staining (Fig. 3d).
Figure 3.
Efficacy of LaG and LaM nanobodies in immunofluorescence microscopy. HeLa cells transiently transfected with (a) tubulin-emGFP or (b) an emGFP-tagged mitochondrial marker were fixed and immunostained with LaG-16 conjugated to Alexa Fluor® 568 (AF568). Cells were visualized in the green (left) and red (right) channels, with DAPI counter-staining of nuclei (blue). (c) T. brucei cells expressing GFP-tagged Sec13 were mixed 1:1 with wild-type cells, fixed, and stained with LaG-16-AF568, with DAPI counterstaining. (d) An S. cerevisiae strain with mCherry-tagged histone H2B was fixed and permeabilized, then directly stained with LaM-4 conjugated to Alexa Fluor® 488. Yeast were visualized in the red (left) and green (right) channels. All scale bars are 10 μm.
We also compared the fluorescence spectra of GFP in the presence or absence of various LaGs to look for spectral shifts upon binding, as have previously been reported, and observed moderate increases in fluorescence for several LaGs, with a maximum increase in fluorescence intensity of approximately 60% (Supplementary Fig. 13)29.
One additional question of specificity we sought to address was the ability of our nanobodies to recognize other fluorescent homologs of Aequorea victoria GFP and Discosoma mCherry. We tested the 13 highest affinity LaGs against a variety of fluorescent proteins: eGFP, two YFP variants, two CFP variants, BFP, mCherry, and DsRed (Fig. 4a). None of these nanobodies bound DsRed or mCherry, two Discosoma sp.-derived proteins with low sequence identity to eGFP (<30%), or TurboYFP, derived from Phialidium sp., which has 53% sequence identity to eGFP20, 34, 35. All bound standard Aequorea victoria-derived CFP, YFP, and BFP variants (>96% eGFP identity). Two LaGs did not bind a moderately divergent (78% eGFP identity) CFP sequence from Aequorea macrodactyla, while all others did36. These results indicate that while identified LaGs bind specifically to fluorescent proteins with high identity to eGFP, differential binding activities can be obtained through selection of variants from other species. Our anti-mCherry LaM nanobodies bound to mCherry, but not to any form of GFP, YFP, or CFP tested (Fig. 4b). Interestingly, two LaMs (LaM-3 and LaM-4) bound to standard DsRed, which has approximately 80% sequence identity to mCherry. Given the different fluorescent protein affinities observed with the LaG and LaM nanobodies, including specificity for AmCFP and DsRed, these reagents have diverse potential uses in differential labeling and affinity capture experiments from cells simultaneously expressing different fluorescently-tagged proteins.
Figure 4.

Nanobody fluorescent protein binding. (a) Thirteen high-affinity LaGs were conjugated to magnetic beads and incubated with various recombinant fluorescent proteins. All LaGs bound A. victoria (Av) GFP variants, while none bound mCherry or DsRed from Discosoma (Ds), or Phialidium (Phi) YFP. Mixed binding was observed for A. macrodactyla (Am) CFP. (b) Immobilized LaMs were similarly incubated with fluorescent proteins. All LaMs bound Discosoma mCherry, while none bound A. victoria GFP, A. macrodactyla CFP, or Phialidium YFP. Mixed binding was observed for DsRed. Example structural models were obtained from PDB IDs 1EMA52 (Av), 4HE453 (Phi), and 1GGX54 (Ds); the AmCFP model is a Phyre server prediction36, 55.
Mapping of the nanobody epitopes on GFP
We identified the epitopes on GFP recognized by the twelve highest affinity LaGs using chemical shift perturbation, a well-established nuclear magnetic resonance (NMR) technique. This method allows the mapping of binding sites on a protein by following changes in its characteristic “fingerprint” spectrum (typically the 15N-1H HSQC) occurring as a result of adding an unlabeled ligand into a 15N-labeled protein sample37.
Because previous studies have already made backbone 15N-1H chemical shift assignments of the GFPuv variant38, 39 (closely related to standard eGFP with 97% sequence identity), we prepared 15N-labeled GFPuv, measured its 15N-1H HSQC spectrum and obtained chemical shift assignments based on those published38, 39 (Supplementary Fig. 14a). We then tested complexes between 12 high affinity LaGs and 15N-labeled GFPuv and measured their 15N-1H HSQC spectra. For 11 out of the 12 cases, we observed clear and specific changes in chemical shifts of a large percentage of cross-peaks compared to the 15N-1H HSQC spectrum of GFPuv alone (Supplementary Figs. 14b, c and 15). In the 12th case, LaG-24, the nanobody did not bind the GFPuv variant.
A chemical shift difference was calculated for all spectra, and residues exhibiting a difference higher than 0.03 ppm were judged to be in the binding interface (Supplementary Fig. 14b, c)37, 40. All the identified epitopes corresponded to large interfaces comprising more than 50 amino acids, consistent with the high affinity binding observed (Fig. 5 and Table 1). The binding epitopes of the nanobodies can be divided into 3 distinct groups. The binding site of group I, containing 5 nanobodies (LaG-16, LaG-9, LaG-14, LaG-43 and LaG-17) overlaps with the binding site of group II, also containing 5 nanobodies (LaG-19, LaG-21, LaG-26, LaG-27 and LaG-41), whereas the two group III nanobodies (LaG-2 and LaG-24) exhibit a binding epitope on the opposite side of the GFP molecule compared to groups I and II. As a control, we also used this NMR approach to determine the GFPuv binding site of the commercial GFP-Trap® nanobody, the structure of whose complex with GFP has been crystallographically determined (PDB ID 3K1K)29, and showed that the NMR-mapped epitope matched the published results (Supplementary Fig. 16)29, 41. Comparing the binding epitopes of our nanobodies with that of GFP-Trap®, groups I and II show little or no overlap with the GFP-Trap® binding site, while group III, which binds on the same face of GFP, shows significant overlap (Fig. 5).
Figure 5.
Mapping of nanobody binding epitopes on GFP by NMR. Binding epitopes of the 11 strongest binding nanobodies on GFPuv, shown in their respective epitope group type (groups I – III). For each nanobody, two opposite sides (via a 180° rotation along a vertical axis) of the GFPuv are shown, with the binding site of the respective nanobody colored green. All GFPuv molecules are presented in space-filling mode and have the same orientation in all panels. The 3 panels on the lowest right show the GFP-Trap® nanobody (top) binding epitope and dimerization site (middle) on GFPuv as well as its ribbon diagram depicting the secondary structure elements (bottom).
Dimerized nanobodies as ultra-high affinity reagents
Because NMR identified multiple epitopes for these 12 LaGs, we engineered heterodimers of LaGs with non-overlapping binding sites on GFP that could potentially bind with higher affinity, an approach that has been successfully used in various applications to develop high avidity reagents42, 43. A LaG16-LaG2 fusion with a flexible glycine-rich peptide linker (encoding three repeats of GGGGS) showed the highest affinity by SPR, with a Kd of 36 pM. Dimers of other LaGs or with a different linker (a 3xFLAG tag), displayed Kds in the range of 100–200 pM. We also determined whether the higher affinity of these dimers yielded faster affinity isolations after conjugation to magnetic beads, compared to single nanobodies or polyclonal anti-GFP. We therefore performed time courses of yeast Nup84-GFP isolations and compared the relative yields of known Nup84 complex components. The LaG16-LaG2 dimer showed higher yields at earlier time points, reaching approximately 80% of maximum yield after only 5 minutes and 90% after 10 minutes (Fig. 2g and Supplementary Fig. 17). These picomolar affinity reagents open the door for increasingly rapid affinity isolations, potentially allowing the capture of weakly or transiently associated complex components for interactome studies. In addition, their high avidity would allow for the detection of low abundance antigens, as is required for many diagnostic applications.
Discussion
Our optimized pipeline for the production and generation of nanobodies allows for the rapid generation of a large antibody repertoire against multiple epitopes in a chosen antigen. Notably, this approach identifies high affinity nanobody sequences directly from animal serum, taking advantage of the complex selection and maturation processes occurring in the animal’s immune system, avoiding intermediary expression systems. The pipeline allows for the rapid production of a comprehensive repertoire of specific high affinity nanobodies for use in the characterization of target macromolecules, such as the GFP- and mCherry-tagged proteins shown here. The laboratory effort required after the collection of samples from llamas (50–70 days after initial immunization, once an immune response is generated) is modest. The direct work required, including IgG purification (2 days), MS (2 days), cDNA generation and PCR (2 days), and final cloning and screening (3–6 days), can be performed over approximately 10 days. Animal handling, high-throughput sequencing, MS, and gene synthesis can be readily outsourced, and depending on turnaround times, each can typically be carried out in 1–2 weeks. The entire process can take as little as 4–6 weeks after an immune response is generated, with only standard techniques required in the primary laboratory. This is faster and more direct than other approaches available, which often require specialized high-throughput capability. Our approach puts the generation of large repertoires and quantities of high affinity single chain antibodies into the hands of the average researcher. Our LaM and LaG reagents, generated against the widely used GFP and mCherry tags, will be of immediate general use for the affinity isolation and enhanced visualization of these tags.
Our approach is well-suited to the development of nanobody reagents against various types of protein targets, including proteins that are difficult to tag. The versatility and potential of nanobodies is huge, as reflected by the interest of the research community8, 10, 44, 45. Nanobodies are much smaller than antibodies, resistant to aggregation, and can be readily humanized8, 46, 47. They have great potential in drug development, as they can bind with great specificity and efficacy to disease targets such as tumor cells, either independently (as a monomer or an ultra-high affinity nanobody dimer), or as a fusion with other protein domains, molecules, or drugs48–51. As demonstrated here, the ability of our method to quickly and easily identify large repertoires of high affinity bacterially-expressed nanobodies against a chosen target antigen has the potential to significantly advance a field that otherwise can take years to generate such reagents.
Online Methods
Isolation of VHH antibodies
For detailed information on the entire nanobody identification procedure, please see the Supplementary Protocol. In short, a 5 year old female llama, Barbie, was immunized with recombinant GFP-His6, and a 4 year old male llama, Marley, with recombinant mCherry-His6 through subcutaneous injections of 5 mg of protein with CFA. Three additional injections of 5 mg protein, with IFA, were performed at three week intervals. Serum bleeds were obtained 10 days after the final injection. 2.5 ml of serum was diluted ten-fold in 20 mM sodium phosphate, pH 7.0, and incubated with Protein G-agarose resin for 30 min. The flow-through was then incubated for 30 min with Protein A-agarose resin. Both resins were washed with 20 mM sodium phosphate, pH 7.0, and bound VHH IgG was eluted with 100 mM acetic acid, pH 4.0 and 500 mM NaCl (Protein G resin) or 100 mM acetic acid, pH 3.5 and 150 mM NaCl (Protein A resin). These elutions were pooled and dialyzed into PBS. 3 mg of this VHH fraction was then incubated with Sepharose-conjugated GFP. This resin was washed with 10 mM sodium phosphate, pH 7.4 and 500 mM NaCl, followed by 1–4.5 M MgCl2 in 20 mM Tris, pH 7.5, and then equilibrated in PBS. The resin was then digested with 0.3 mg/ml papain in PBS plus 10 mM cysteine, for 4 hours at 37°C. The resin was then washed with 1) 10 mM sodium phosphate, pH 7.4 and 500 mM NaCl 2) PBS plus 0.1% Tween-20 3) PBS 4) 0.1 M NH4OAc, 0.1 mM MgCl2, 0.02% Tween-20. Bound protein was then eluted for 20 min with 0.1 M NH4OH and 0.5 mM EDTA, pH 8.0. These elutions were dried down in a SpeedVac and resuspended in LDS plus 25 mM DTT. The samples were alkylated with iodoacetamide and run on a 4–12% Bis-Tris gel. The ~15 kDa band corresponding to the digested VHH region was then cut out and prepared for MS.
RT-PCR and DNA sequencing
Bone marrow aspirates were obtained from immunized llamas concurrent with serum bleeds. Bone marrow plasma cells were isolated on a Ficoll gradient using Ficoll-Paque (GE Healthcare). RNA was isolated from approximately 1–6 × 107 cells using Trizol LS reagent (Life Technologies), according to the manufacturer’s instructions. cDNA was reverse-transcribed using Ambion RETROscript (Life Technologies). A nested PCR was then performed with IgG specific primers. In the first step, CALL001 (5′-GTCCTGGCTGCTCTTCTACAAGG-3′) and CALL002 (5′-GGTACGTGCTGTTGA ACTGTTCC-3′) primers were used to amplify the IgG variable domain into the CH2 domain25. The approximately 600–750 bp band from VHH variants lacking a CH1 domain was purified on an agarose gel. Next, for 454 sequencing, VHH regions were specifically reamplified using framework 1- and 4-specific primers with 5′ 454 adaptor sequences: 454-VHH-forward (5′-CGTATCGCCTCCCTCGCGCCATCAGATGGCT[C/G]A[G/T]GTGCAGCTGGTGGAGTCTGG-3′) and 454-VHH-reverse (5′-CTATGCGCCTTGCCAGCCCGCTCAG GGAGACGGTGACCTGGGT-3′) (adaptor sequences are underlined)25. The approximately 400 bp product of this reaction was gel purified, then sequenced on a 454 GS FLX system after emPCR amplification, on one Pico Titer Plate. For Illumina MiSeq sequencing, the second PCR was instead performed with random 12-mers replacing adaptor sequences, to aid in cluster identification: MiSeq-VHH-forward (5′-NNNNNNNNNNNN ATGGCT[C/G]A[G/T]GTGCAGCTGGTGGAGTCTGG-3′) and MiSeq-VHH-reverse (5′-NNNNNNNNNNNN GGAGACGGTGACCTGGGT-3′). The product of this PCR was gel purified, ligated to MiSeq adaptors before library preparation using Illumina kits, and run on a MiSeq sequencer with 2 × 300 bp paired end reads.
Database preparation
The protein sequence databases used for identification were prepared by translating sequencing reads in all 6 reading frames, and for each read the longest Open Reading Frame (ORF) was selected. The selected ORF was digested with trypsin in silico and a list of unique tryptic peptides of 7 amino acids or longer was constructed and saved in a FASTA file. It is important to construct a FASTA file only containing unique peptides because even though most search engines can handle some sequence redundancy, they are not well equipped to handle the extreme redundancy that is provided by next generation sequencing of the single chain antibody locus and search engines either become very slow or crash if presented with such an extreme redundancy.
Mass spectrometry
Gel sections containing VHH domains were excised, destained, and dehydrated. The dehydrated gel slices were then subjected to in-gel digestion with proteomic-grade trypsin (80 μL; 25 ng trypsin, 25 mM ammonium bicarbonate) (Promega) at 37 °C overnight. The gel was extracted once with extraction solution (140 μL; 67% acetonitrile, 1.7 % formic acid). The resulting proteolytic digest was cleaned with a STAGE tip56 and loaded onto a home-packed reverse phase C18 column (75 μm I.D., 15 μm tip) (New Objective) with a pressurized bomb. The loaded peptides were subsequently separated with a linear gradient (0 % to 42 % acetonitrile, 0.5 % acetic acid, 120 min, 150 nL/min after flow splitting) generated by an Agilent 1260 HPLC and directly sprayed into an LTQ-Velos-Orbitrap mass spectrometer (Thermo Scientific) for analysis. In the mass spectrometer, a survey scan was carried out in the orbitrap (resolution = 30,000, AGC target = 1E6) followed by tandem MS in the ion trap (AGC target = 5E3) of the top twenty most intense peaks. Tandem MS was carried out with collision induced dissociation (isolation width = 2 Th, CE = 35 %, activation time = 5 ms). Internal calibration was used for improved mass accuracy (lock mass m/z = 371.1012). In order to scan more peptides, both predictive AGC and dynamic exclusion were enabled (Repeat counts: 2, repeat duration: 12 s, exclusion duration: 60 s). Single and unassigned charge species were excluded from tandem MS scans. The raw files were converted into mzXML format with ReAdW (version 4.3.1).
MS-based identification of VHH sequences
The MS search was performed on the custom database of tryptic peptides using the X! Tandem search engine. Then, the identified peptides filtered by expectation value were mapped to the sequences translated from 454 reads (longest ORF only, as described above). The CDR regions were located within the sequence based on approximate position in the sequence and the presence of specific leading and trailing amino acids. For example, to locate the CDR3 region, the algorithm searched for the left anchor YXC (X representing any amino acid) between position 93 and 103 of the sequence, and the right anchor WG between position n-14 and n-4 of the sequence, where n is the length of the sequence. Once the peptides were mapped to the sequences and their CDR regions, a metric was calculated to rank each sequence as a potential candidate based on the bioinformatics evidence available. The factors included in the metric were: MS coverage and length of individual CDR regions with CDR3 carrying highest weight, overall coverage including framework region, and a count of the 454 reads producing the sequence. Finally, sequences with similar CDR3 regions were grouped together, allowing for the identification of the highest confidence sequence corresponding to a particular CDR3. A sequence was assigned to a group where its hamming distance to an existing member was 1, i.e. there was one amino acid difference in the sequence, and different groups that have one shared sequence were further combined. By choosing sequence hits from different groups for production, we maximized the overall sequence diversity of the candidate pool. The candidate list was displayed for manual inspection as an interactive HTML page with CDR regions annotated, peptide mapping information and the ranking metrics shown for each sequence. All algorithms described above were implemented in Perl. An example of a candidate list view is shown in Supplementary Figure 3.
Web-based application for nanobody sequence identification: “Llama-Magic”
The pipeline that was used for identification of the Nanobody sequences has been automated and can be accessed through a web-based interface at http://www.llamamagic.org. Llama-Magic allows upload of FASTA files containing reads from High-throughput DNA sequencing. Once uploaded, the reads will be automatically translated and digested to create an MS searchable database of tryptic peptides, as described above. Next, the MS (mgf) files can be uploaded for a selected tryptic peptide sequence database, and the parent and fragment error can be chosen for the X! Tandem search. Once the mgf files are uploaded, the X! Tandem search will be executed and the matching peptides saved. Then (1) annotation of CDR regions, (2) mapping of the identified peptides and (3) ranking and grouping of candidates are performed automatically, producing an interactive display of the candidate list showing detailed information regarding each sequence and its corresponding rank. Llama-Magic is implemented in Perl, HTML and JavaScript. Manual inspection was performed to make sure a) long CDR3 peptides, which embrace both variable regions and framework regions, have fragmentation pattern within the variable regions; b) CDR3 peptides are unique enough (uniqueness score < 100);
Cloning
Nanobody sequences were codon-optimized for expression in E. coli and cloned into pCR2.1 after gene synthesis (Eurofins MWG Operon), incorporating BamHI and XhoI restriction sites at 5′ and 3′ ends, respectively. A pelB leader sequence was cloned into pET21b at NdeI and BamHI restriction sites using complementary primers: 5′-TATGAAATACTTATTGCCTACGGCAGCCGCTGGATTGTTATTACTCGCGGCCCAGCCGGCCATGGCTG-3′ and 5′-GATCCAGCCATGGCCGGCTGGGCCGCGAGTAATAACAATCCAGCGGCTGCCGTAGGCAA-TAAGTATTTCA-3′. Nanobody sequences were then subcloned into pET21b-pelB using BamHI and XhoI restriction sites, with primers also encoding a PreScission Protease cleavage site just before the C-terminal 6xHis tag.
Purification of nanobodies
pelB-fused nanobodies were expressed under a T7 promoter in Arctic Express (DE3) cells (Agilent), induced with IPTG at a final concentration of 0.1 mM. Cells were induced for 18–20 hours at 12°C, then pelleted by a 10 min spin at 5000 × g. The periplasmic fraction was then isolated by osmotic shock17. This fraction was bound to His-Select nickel affinity resin (Sigma), washed with His wash buffer (20 mM sodium phosphate pH 8.0, 1 M NaCl, 20 mM imidazole), and eluted with His elution buffer (20 mM sodium phosphate pH 8.0, 0.5 M NaCl, 0.3 M imidazole). The elution was then dialyzed into PBS.
Fluorescent protein binding assays
2 μg of fluorescent protein was added to 50 μl of 2 mg/ml E. coli lysate diluted in binding buffer (20 mM HEPES, pH 7.4, 350 mM NaCl, 0.01% Tween-20, 0.1 M PMSF, 3 μg/ml pepstatin A). This was incubated with 25 μl of nanobody-Dynabead slurry. After a 30 minute incubation at 4°C, beads were washed with binding buffer and bound protein was eluted with 15 μl LDS. Elutions were run on a 4–12% Bis-Tris gel.
Kd determinations
SPR measurements were obtained on a Proteon XPR36 Protein Interaction Array System (Bio-Rad). Recombinant GFP or mCherry was immobilized on a ProteOn GLC sensor chip: the chip surface was first activated with 50 mM sulfo-NHS and 50 mM EDC, run at a flow-rate of 30 μl/min for 300 sec. The ligand was then diluted to 5 μg/ml in 10 mM sodium acetate, pH 5.0, and injected at 25 μl/min for 180 sec. Finally, the surface was deactivated by running 1 M ethanolamine-HCl (pH 8.5) at 30 μl/min for 300 sec. This led to immobilization of approximately 600–800 response units (RU) of ligand.
Kds of recombinant nanobodies were determined by injecting 4 or 5 concentrations of each protein, in triplicate, with a running buffer of 20 mM HEPES, pH 8.0/150 mM NaCl/0.01% Tween. Proteins were injected at 50 μl/min for 120 sec, or 100 μl/min for 90 sec, followed by a dissociation time of 600 sec. Between injections, residual bound protein was eliminated by regeneration with 4.5 M MgCl2 in 10 mM Tris, pH 7.5, run at 100 μl/min for 36 sec. Binding sensorgrams from these injections were processed and analyzed using the ProteOn Manager software. Binding curves were fit to the data with a Langmuir model, using grouped ka, kd, and Rmax values.
Immunofluorescence microscopy
HeLa cells were cultured on coverslips in DMEM media with 10% FBS and penicillin/streptomycin at 37°C with 8% CO2 in a humidified environment. Cells tested negative for mycoplasma. Cells were transfected with CellLight Tubulin-GFP or Mitochondria-GFP BacMam 2.0 reagents (Life Technologies) using 4 μL of reagent per 5,000 cells, and processed after 18–20 hrs. Cells were fixed in ice-cold methanol for 10 minutes (for Tubulin-GFP) or in 2% paraformaldehyde for 10 minutes (for Mitochondria-GFP). Cells were permeabilized with 0.5% Triton for 10 min. and blocked for 1 hr with 10% goat serum/1% BSA in PBS. They were then incubated for 1 hr at room temperature with recombinant nanobody conjugated to Alexa Fluor® 568 succinimidyl ester (Life Technologies), diluted to 100 ng/ml in 1% BSA in PBS. Cells were washed four times with PBS/0.01% BSA, with 300 nM DAPI included in the final wash, then mounted with ProLong Diamond (Life Technologies).
Wild-type and Sec13-GFP tagged T. brucei strains were cultured to a cell density of 1×107 as previously described33. Cells from each strain were mixed 1:1, and fixed for 10 minutes with cold 4% formaldehyde. Approximately 1×106 cells were spotted onto coverslips, allowed to settle for 30 min., permeabilized with 0.1% Triton for 5 min., and blocked with 10% goat serum/1% BSA in PBS for 30 minutes. Cells were then stained, washed, and mounted identically to HeLa cells.
A S. cerevisiae W303 strain with Htb2 genomically tagged at the C-terminus with mCherry was grown to mid-log phase, and allowed to settle on Concanavalin A-coated coverslips. Yeast were fixed in 4% paraformaldehyde/2% sucrose/PBS, and blocked and permeabilized for 30 minutes in 0.25% Triton/2% milk/PBS31. Cells were stained overnight at 4°C with nanobody diluted to 3.3 μg/ml in 0.25% Triton/1% BSA/PBS. They were then washed 5 times with 0.01% BSA in PBS, the final two washes for 5 min. Cells were mounted in 70% glycerol/PBS.
All images were obtained on a Deltavision Image Restoration Microscrope (Applied Precision/Olympus), with an Olympus 100x/1.40 numerical aperture objective, or 60x/1.42 objective in the case of HeLa cells. Raw images were processed by a deconvolution algorithm using softWorX software (Applied Precision/GE Healthcare).
Affinity isolations of tagged protein complexes
Recombinant nanobodies were conjugated to epoxy-activated magnetic Dynabeads (Life Technologies), with minor modifications to published IgG coupling conditions57. 10 μg recombinant protein was used per 1 mg of Dynabeads, with conjugations carried out in 0.1 M sodium phosphate, pH 8.0 and 1 M ammonium sulfate, with an 18–20 hour incubation at 30°C. Affinity isolations of yeast Nup84-GFP were carried out as previously described, using binding buffer consisting of 20 mM HEPES, pH 7.4, 500 mM NaCl, 2 mM MgCl2, 0.1% CHAPS, 0.1 M PMSF, and 3 μg/ml pepstatin A57. For each experiment, 50 μl of bead slurry was used with 0.5 g of yeast cells. Similar conditions were used for HTB2-mCherry isolations (from yeast with HTB2 genomically tagged at the C-terminus with mCherry58), except lysate was sonicated 4 times for 10 s before centrifugation, and the binding buffer consisted of 20 mM HEPES, pH 8.0, 300 mM NaCl, 110 mM KOAc, 0.1% Tween-20, 0.1% Triton X-100, 0.1 M PMSF, and 3 μg/ml pepstatin A. Isolations of RBM7-GFP from HeLa cells were performed as previously described4. 10 μl of bead slurry was used with 100 mg of cells, using a binding buffer of 20 mM HEPES, pH 7.4, 300 mM NaCl, 0.5% Triton X-100, with cOmplete Protease Inhibitor, EDTA-free (Roche).
To determine affinity isolation yields, samples of resuspended lysate were taken before and after Dynabead binding. These were run on a 4–12% Novex Bis-Tris gel in MES running buffer (Life Technologies), and probed by Western blotting using mouse anti-GFP antibody (Roche, cat. no. 11 814 460 001) diluted 1:1,000 in TBST/2% dry milk and an anti-mouse, HRP-conjugated secondary (GE Healthcare, cat. no. NA931V) diluted 1:3,000 in TBST/2% dry milk. Signals were quantified using ImageJ software.
Fluorescence spectra
Samples of recombinant GFP at 0.5 μM in PBS were mixed with either buffer or 10 μM of a LaG protein. Fluorescence spectra were obtained on a Synergy Neo (BioTek) microplate reader. Excitation spectra from 300 nm to 530 nm were taken at an emission wavelength of 560 nm, and emission spectra were measured from 450 nm to 600 nm at an excitation wavelength of 425 nm.
Phylogenetic analysis
Phylogenetic trees and alignments were generated from LaG amino acid sequences using the Phylogeny.fr web service59, 60.
Mapping of nanobody binding epitopes on GFP by NMR
Three variants of GFP were used in the preparation of NMR samples. GFP-His6 (eGFP), the variant used for immunization; GFPuv, the variant for which backbone 15N-1H chemical shift assignments were available from BMRB file 566639 and a crystal structure was available from PBD ID 1B9C41; GFPuv_A206K (GFPuv_M), a monomeric version of GFPuv61. Supplementary Table 1 summarizes the amino acid sequences of the three GFP variants.
All NMR samples contained between 500 and 20 μM 15N-GFP either alone or in the presence of a 1–1.2 molar excess of LaG, 10 mM sodium phosphate buffer, pH 7.4, 150 mM NaCl and 90% H2O/10% D2O. All NMR spectra (2D 1H-15N HSQC) were measured at 310 K on a Bruker Avance DPX-600 MHz spectrometer equipped with a TCI cryoprobe.
Backbone 1H-15N assignments of GFPuv were obtained from a comparison between a 1H-15N HSQC spectrum of GFPuv alone and a simulated 1H-15N HSQC based on BMRB 566639 (Supplementary Fig. 14a). Due to a very high similarity between the two, 1H-15N backbone assignment of GFPuv was obtained for 97% of 1H-15N backbone resonances for which assignment was available in BMRB5666. The accuracy of the GFPuv assignment was verified by mapping the binding site of a previously identified nanobody, GFP-Trap®29, on GFPuv. The crystal structure of the GFP/GFP-Trap complex is available in the PDB (PDB ID 3K1K)29 and a comparison between the X-ray crystallography-derived binding site (obtained by analysis of 3K1K by PISA - ‘Protein interfaces, surfaces and assemblies’ service at the European Bioinformatics Institute (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html)62) and the one determined by the chemical shift perturbation method, reveals they overlap, thereby confirming our assignment of GFPuv residues (Supplementary Fig. 16).
Backbone 1H-15N assignments of GFPuv_M were obtained from a comparison between a 1H-15N HSQC spectrum of GFPuv and that of GFPuv_M (Supplementary Fig. 14a). Assignment was verified by mapping the dimerization site of GFPuv and comparing it to the crystal structure of PDB ID 1B9C41 (analyzed for interacting residues using PISA62).
All chemical shift differences were calculated using equation (1) where CSD is the total
| (1) |
chemical shift difference and ΔδN and ΔδH are the chemical shift differences in the free and bound states between the amide nitrogens and protons, respectively. The CSD cutoff for binding site residues was 0.05ppm for GFP-Trap binding site and for GFPuv dimerization site and 0.03ppm for all LaG binding sites.
All LaG binding site residues are listed in Supplementary Table 2 and their respective 1H-15N HSQC spectra are shown in Supplementary Figure 15 overlaid with the 1H-15N HSQC spectrum of the free GFPuv_M.
Supplementary Material
Acknowledgments
We acknowledge support from US National Institutes of Health grants U54 GM103511 (MPR and BTC), P41 GM103314 (BTC), AI072529-08 and AI037526-20A1 (MCN). MCN is an HHMI Investigator. We wish to thank A. North from the Rockefeller University Bio-Imaging Resource Center for assistance with immunofluorescence microscopy. We also thank A. Viale from the Memorial Sloan Kettering Cancer Center Genomics Core Laboratory, and A. Luz from the Rockefeller University High-Throughput Screening Resource Center. We thank S. Reed-Paske and the other members of Capralogics, Inc. for their advice and animal husbandry. Finally, we thank members of the Rout and Chait labs, past and present, for helpful discussions and technical assistance, particularly A. Ferguson, K. Wei, H. Jiang, and S. Obado.
Footnotes
Author Contributions:
The project was conceived by M.P.R., B.T.C., P.C.F., Y.L., D.F. and M.C.N. Experiments relating to immunization, sample collection and processing were performed by P.C.F., M.K.T and M.O.; J.F.S. assisted with bone marrow processing. Mass spectrometry was carried out by Y.L. Experimental work was supervised by M.P.R. and B.T.C. Bioinformatic analysis was performed by Y.L., S.K., and D.F. Production and characterization of recombinant nanobodies was performed by P.C.F. and M.K.T. NMR analyses were performed by I.N. The manuscript was co-written by P.C.F., Y.L., I.N., M.P.R., and B.T.C., with contributions from all authors. M.P.R. communed with llamas in the Atacama desert.
Competing Financial Interests:
B.T.C. and M.P.R. are inventors on a US patent application encompassing the method described in this manuscript.
References
- 1.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Molecular & cellular proteomics: MCP. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nature biotechnology. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 3.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 4.Domanski M, et al. Improved methodology for the affinity isolation of human protein complexes expressed at near endogenous levels. Biotechniques. 2012;0:1–6. doi: 10.2144/000113864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gingras AC, Aebersold R, Raught B. Advances in protein complex analysis using mass spectrometry. J Physiol. 2005;563:11–21. doi: 10.1113/jphysiol.2004.080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortez-Retamozo V, et al. Efficient cancer therapy with a nanobody-based conjugate. Cancer Res. 2004;64:2853–2857. doi: 10.1158/0008-5472.can-03-3935. [DOI] [PubMed] [Google Scholar]
- 7.Hamers-Casterman C, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 8.Muyldermans S. Nanobodies: Natural Single-Domain Antibodies. Annual Review of Biochemistry. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 9.Harmsen MM, De Haard HJ. Properties, production, and applications of camelid single-domain antibody fragments. Appl Microbiol Biotechnol. 2007;77:13–22. doi: 10.1007/s00253-007-1142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romer T, Leonhardt H, Rothbauer U. Engineering antibodies and proteins for molecular in vivo imaging. Curr Opin Biotechnol. 2011;22:882–887. doi: 10.1016/j.copbio.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Dumoulin M, et al. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002;11:500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arbabi Ghahroudi M, Desmyter A, Wyns L, Hamers R, Muyldermans S. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS letters. 1997;414:521–526. doi: 10.1016/s0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- 13.Arbabi-Ghahroudi M, Tanha J, MacKenzie R. Prokaryotic expression of antibodies. Cancer Metastasis Rev. 2005;24:501–519. doi: 10.1007/s10555-005-6193-1. [DOI] [PubMed] [Google Scholar]
- 14.Rothbauer U, et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods. 2006;3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 15.Muyldermans S, et al. Camelid immunoglobulins and nanobody technology. Vet Immunol Immunopathol. 2009;128:178–183. doi: 10.1016/j.vetimm.2008.10.299. [DOI] [PubMed] [Google Scholar]
- 16.Bird RE, et al. Single-chain antigen-binding proteins. Science. 1988;242:423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- 17.Skerra A, Pluckthun A. Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science. 1988;240:1038–1041. doi: 10.1126/science.3285470. [DOI] [PubMed] [Google Scholar]
- 18.Worn A, Pluckthun A. Stability engineering of antibody single-chain Fv fragments. Journal of molecular biology. 2001;305:989–1010. doi: 10.1006/jmbi.2000.4265. [DOI] [PubMed] [Google Scholar]
- 19.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shagin DA, et al. GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity. Mol Biol Evol. 2004;21:841–850. doi: 10.1093/molbev/msh079. [DOI] [PubMed] [Google Scholar]
- 21.Dorner T, Radbruch A. Antibodies and B cell memory in viral immunity. Immunity. 2007;27:384–392. doi: 10.1016/j.immuni.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Benner R, Hijmans W, Haaijman JJ. The bone marrow: the major source of serum immunoglobulins, but still a neglected site of antibody formation. Clin Exp Immunol. 1981;46:1–8. [PMC free article] [PubMed] [Google Scholar]
- 23.Becker RS, Knight KL. Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbits. Cell. 1990;63:987–997. doi: 10.1016/0092-8674(90)90502-6. [DOI] [PubMed] [Google Scholar]
- 24.Knight KL. Restricted VH gene usage and generation of antibody diversity in rabbit. Annu Rev Immunol. 1992;10:593–616. doi: 10.1146/annurev.iy.10.040192.003113. [DOI] [PubMed] [Google Scholar]
- 25.Conrath KE, et al. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob Agents Chemother. 2001;45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarez-Rueda N, et al. Generation of llama single-domain antibodies against methotrexate, a prototypical hapten. Mol Immunol. 2007;44:1680–1690. doi: 10.1016/j.molimm.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez-Martinez J, et al. Structure-function mapping of a heptameric module in the nuclear pore complex. The Journal of Cell Biology. 2012;196:419–434. doi: 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirchhofer A, et al. Modulation of protein properties in living cells using nanobodies. Nature structural & molecular biology. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 30.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 31.Ries J, Kaplan C, Platonova E, Eghlidi H, Ewers H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nature Methods. 2012;9:582–584. doi: 10.1038/nmeth.1991. [DOI] [PubMed] [Google Scholar]
- 32.Dolman NJ, Kilgore JA, Davidson MW. A review of reagents for fluorescence microscopy of cellular compartments and structures, part I: BacMam labeling and reagents for vesicular structures. Curr Protoc Cytom. 2013;Chapter 12(Unit 12):30. doi: 10.1002/0471142956.cy1230s65. [DOI] [PubMed] [Google Scholar]
- 33.DeGrasse JA, et al. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Molecular & cellular proteomics: MCP. 2009;8:2119–2130. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matz MV, et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nature biotechnology. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 35.Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 36.Xia NS, et al. Bioluminescence of Aequorea macrodactyla, a common jellyfish species in the East China Sea. Mar Biotechnol (NY) 2002;4:155–162. doi: 10.1007/s10126-001-0081-7. [DOI] [PubMed] [Google Scholar]
- 37.Goldflam M, Tarrago T, Gairi M, Giralt E. NMR studies of protein-ligand interactions. Methods in molecular biology. 2012;831:233–259. doi: 10.1007/978-1-61779-480-3_14. [DOI] [PubMed] [Google Scholar]
- 38.Georgescu J, Rehm T, Wiehler J, Steipe B, Holak TA. Backbone H(N), N, C(alpha) and C(beta) assignment of the GFPuv mutant. Journal of biomolecular NMR. 2003;25:161–162. doi: 10.1023/a:1022296413190. [DOI] [PubMed] [Google Scholar]
- 39.Khan F, Stott K, Jackson S. 1H, 15N and 13C backbone assignment of the green fluorescent protein (GFP) Journal of biomolecular NMR. 2003;26:281–282. doi: 10.1023/a:1023817001154. [DOI] [PubMed] [Google Scholar]
- 40.Zuiderweg ER. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 41.Battistutta R, Negro A, Zanotti G. Crystal structure and refolding properties of the mutant F99S/M153T/V163A of the green fluorescent protein. Proteins. 2000;41:429–437. doi: 10.1002/1097-0134(20001201)41:4<429::aid-prot10>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 42.Neri D, Momo M, Prospero T, Winter G. High-affinity antigen binding by chelating recombinant antibodies (CRAbs) Journal of molecular biology. 1995;246:367–373. doi: 10.1006/jmbi.1994.0091. [DOI] [PubMed] [Google Scholar]
- 43.Silverman J, et al. Multivalent avimer proteins evolved by exon shuffling of a family of human receptor domains. Nature biotechnology. 2005;23:1556–1561. doi: 10.1038/nbt1166. [DOI] [PubMed] [Google Scholar]
- 44.Vanlandschoot P, et al. Nanobodies(R): new ammunition to battle viruses. Antiviral Res. 2011;92:389–407. doi: 10.1016/j.antiviral.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Huang L, Muyldermans S, Saerens D. Nanobodies(R): proficient tools in diagnostics. Expert Rev Mol Diagn. 2010;10:777–785. doi: 10.1586/erm.10.62. [DOI] [PubMed] [Google Scholar]
- 46.Revets H, De Baetselier P, Muyldermans S. Nanobodies as novel agents for cancer therapy. Expert Opin Biol Ther. 2005;5:111–124. doi: 10.1517/14712598.5.1.111. [DOI] [PubMed] [Google Scholar]
- 47.Vincke C, et al. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem. 2009;284:3273–3284. doi: 10.1074/jbc.M806889200. [DOI] [PubMed] [Google Scholar]
- 48.Els Conrath K, Lauwereys M, Wyns L, Muyldermans S. Camel single-domain antibodies as modular building units in bispecific and bivalent antibody constructs. J Biol Chem. 2001;276:7346–7350. doi: 10.1074/jbc.M007734200. [DOI] [PubMed] [Google Scholar]
- 49.Jahnichen S, et al. CXCR4 nanobodies (VHH-based single variable domains) potently inhibit chemotaxis and HIV-1 replication and mobilize stem cells. Proc Natl Acad Sci U S A. 2010;107:20565–20570. doi: 10.1073/pnas.1012865107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roovers RC, et al. A biparatopic anti-EGFR nanobody efficiently inhibits solid tumour growth. Int J Cancer. 2011;129:2013–2024. doi: 10.1002/ijc.26145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulrichts H, et al. Antithrombotic drug candidate ALX-0081 shows superior preclinical efficacy and safety compared with currently marketed antiplatelet drugs. Blood. 2011;118:757–765. doi: 10.1182/blood-2010-11-317859. [DOI] [PubMed] [Google Scholar]
- 52.Ormo M, et al. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 53.Pletneva NV, et al. Yellow fluorescent protein phiYFPv (Phialidium): structure and structure-based mutagenesis. Acta Crystallogr D Biol Crystallogr. 2013;69:1005–1012. doi: 10.1107/S0907444913004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wall MA, Socolich M, Ranganathan R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat Struct Biol. 2000;7:1133–1138. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- 55.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 56.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 57.Alber F, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 58.Rout MP, et al. The yeast nuclear pore complex: composition, architecture, and transport mechanism. The Journal of cell biology. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dereeper A, Audic S, Claverie JM, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic acids research. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 62.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.