Abstract
Objectives
The aim of this study was to investigate whether 6 candidate serum miRNAs and their interactions with serum folate level were associated with the risk for pancreatic cancer (PC).
Method
A hospital-based case-control study including 74 incident PC cases and 74 controls was conducted. Serum folate and miRNAs were determined by radioimmunoassay and real-time quantitative polymerase chain reaction, respectively. Cell lines AsPC-1 and PANC-1 were used for in vitro study.
Results
MiR-16 was elevated (P = 0.030–0.043) and miR-103 was reduced (P = 0.018–0.020) in PC after adjustment for age, sex, and smoking; however, after additional adjustment for folate, only miR-103 was significantly different between cases and controls (P = 0.010). After converting the relative expression of miRNAs into binary variables and adjusting for age, sex, smoking, and folate, the subjects with low miR-103 or low miR-601 were observed to have a higher risk for PC, with odds ratios of 2.33 (95% confidence interval, 1.06–5.10) and 2.37 (95% confidence interval, 1.07–5.26), respectively. Multifactor dimensionality reduction analysis showed a significant interaction for miR-16, folate, and smoking (cross-validation consistency, 10/10; mean testing accuracy, 0.696; P = 0.013). Interaction between miR-16 and folate was also verified in the AsPC-1 cells.
Conclusion
Serum miR-103; miR-601; and interactions among serum miR-16, folate, and smoking are associated with PC.
Key Words: microRNA, folate, pancreatic cancer, risk
Pancreatic cancer (PC) ranks fourth among causes of cancer-related deaths in the United States.1 In China, the adjusted and age-standardized mortality from PC has increased from 1.75 and 2.18 per 100,000 in 1991 to 3.06 and 3.26 per 100,000 in 2000.2 Pancreatectomy provides the only potential for cure. However, most diagnoses of PC are made in advanced stages that are not suitable for resection, which leads to a 5-year survival rate of less than 5%.1 Therefore, investigating the etiology and identifying the risk factors for PC are essential for primary disease prevention.
As for most human cancers, environmental and genetic factors have been demonstrated to be involved in the carcinogenesis of PC. The progression of PC is paralleled by the successive accumulation of both genetic and epigenetic alterations, including alterations in the expression of microRNA (miRNA) molecules.3,4 The miRNAs represent a novel class of 18nt to 23nt endogenous small noncoding RNAs. Upon binding to their target mRNAs, they promote posttranscriptional gene silencing by either inhibiting the translation process or cleaving the target mRNAs.5 Consequently, abnormal expression of miRNAs could have an effect on crucial biological processes in cancer, such as cell cycle progression and apoptosis.6 In fact, alterations in miRNA expression have been detected in many types of human tumors, including PC. In 2008, intriguing studies revealed that stable miRNAs also could be detected in serum and/or plasma and therefore might serve as diagnostic biomarkers.7,8 Altered expression of serum miRNAs, including miR-155 and miR-221, has been detected in PC.9–12 Array analysis has also been used to profile the association of blood miRNA levels with different cancers, and miR-16, miR-320, miR-601, and miR-1228 were shown to be differentially expressed between PC cases and controls (P < 0.0001).13
The DNA methylation is believed to be one of the most important mechanisms underlying the regulation of miRNA.14,15 As a cofactor involving the synthesis of S-adenosylmethionine, folate plays an essential role in DNA methylation. Dietary folate deficiency induces hypomethylation, which has been linked to the increased development of cancers, including PC.16,17 Interestingly, the expression of certain miRNAs, including miR-103 and miR-107, was significantly changed under folate-deficient conditions in human lymphoblastoid cells.18 Furthermore, expression of many miRNAs in hepatomas induced by folate and methyl-deficient diets was significantly altered compared with that in the livers from age-matched rats on a normal diet.19 On the basis of these findings, we hypothesized that an association might exist between certain miRNAs and the levels of serum folate in the development of PC.
To examine this hypothesis, 6 miRNAs were selected as candidates for investigating the interaction between miRNAs and folate in a PC case-control study of 74 PC patients and 74 controls. Among them, miR-103 and miR-107 have been reported to be differentially regulated by folate deficiency,18 and miR-16,miR-320, miR-601, and miR-1228 were abnormally expressed in PC cases.13
MATERIALS AND METHODS
Study Subjects
This study consisted of 74 PC patients and 74 cancer-free controls. All subjects were unrelated Han Chinese. Patients were recruited between 2002 and 2004 at the Peking Union Medical College Hospital and Cancer Hospital, Chinese Academy of Medical Sciences. Only histopathologically diagnosed incident patients who underwent intent-to-cure surgery were included, and all blood samples were collected when the patients were admitted to the hospital before surgery. The standard of care of PC in China is postoperative chemoradiotherapy, so there is no biologic difference between resected disease and biopsy-only disease. Cancer-free controls were randomly recruited from the same hospitals during the same period as the patients enrolled and were frequency-matched to the cases on age (±5 years) and sex. Informed consent was obtained from each participant. Questionnaire surveys were conducted to obtain information on demographic data and smoking history.
Ethics Statement
This study was approved by the ethics committee of the Chinese Academy of Medical Sciences and Peking Union Medical College.
Blood Samples and Folate Analyses
Whole blood samples were centrifuged at 3000 × g for 15 minutes at 4°C to completely remove the cellular fraction, and serum was collected. The serum supernatant was stored at −80°C until analysis. Serum folate levels were determined by radioimmunoassay using SimulTRAC-SNB Radioassay kit (ICN Pharmaceuticals, New York, NY) according to the manufacturer’s protocol, and all experiments were performed in duplicate. Folate levels of 3.0 ng/mL or greater were considered normal.
Cell Culture
The human pancreatic carcinoma AsPC-1 cell line was purchased from the Cell Resource Center of the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China); human pancreatic carcinoma PANC-1 cell line was purchased from American Type Culture Collection (ATCC, Rockville, MD). Cells were maintained in standard RPMI-1640 (Life Technologies, Carlsbad, CA) or folate-deficient RPMI 1640 (Life Technologies, Carlsbad, CA) at 37°C in a humidified incubator with 5% of CO2. Growth medium was supplemented with 10% of fetal bovine serum (Life Technologies, Carlsbad, CA) and 1% of penicillin-streptomycin. To eliminate intracellular folate sources, dialyzed fetal bovine serum (Life Technologies, Carlsbad, CA) was added to the folate-deficient medium. The cells and cell culture supernatant were harvested after 72 hours using TRIZOL reagent (Life Technologies, Carlsbad, CA) for total RNA isolation. To control for experimental variation in cell culture supernatants, synthetic Caenorhabditis elegans miR-39 (TaKaRa, Dalian, China) was spiked into each cell culture supernatant at a final concentration of 10−4 pmol/μL.
RNA Extraction and Reverse Transcription
The RNA was extracted from 200 μL of serum using TRIZOL reagent (Life Technologies, Carlsbad, CA) according to the manufacturer’s instruction. The concentration and purity of isolated RNA were tested using a Nanodrop ND-1000 (Thermo Scientific, Worcester, MA). An OD260/280 value close to 2.0 indicated high purity of RNA. Total RNA from each sample was diluted to a standard concentration of 20 ng/μL in RNAse-free distilled water, and 3 μl was reverse transcribed to cDNA in a final volume of 10 μl using RevertAid Reverse Transcriptase (Fermentas, Glen Burnie, MD), Recombinant Ribonuclease Inhibitor (TaKaRa, Dalian, China), dNTP mix (TaKaRa, Dalian, China), and stem-loop RT primers (Life Technologies, Carlsbad, CA).
Validation of Endogenous Reference for Serum miRNA Quantification
The miRNA expression levels from serum samples measured by real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR) are typically standardized using stable internal references. Because no universal endogenous references have been identified for serum miRNAs, miR-16, miR-191, and RNU6B were chosen as candidates based on previous studies.8,9,20 Eight serum samples were randomly selected from patients and controls. The expressions of the target miRNAs and reference miRNAs were detected by qRT-PCR, and the stability of the internal references was verified with NormFinder software (http://www.multid.se/genex/hs410.htm).21
Quantification of miRNAs
The miRNA expression was quantified by qRT-PCR with the StepOnePlus Real-Time PCR System (Life Technologies, Carlsbad, CA) using TaqMan miRNA probes (Life Technologies, Carlsbad, CA) according to the manual. The reactions were initiated in a 96-well optical plate at 95°C for 5 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The cycle threshold (Ct) was defined as the number of cycles required for the fluorescent signal to cross the threshold in qPCR. The relative expression levels of target miRNAs were expressed as ΔCt (ΔCt = Ctinternal reference − Cttarget miRNA).22 Reaction specificity was verified by including a no-template control in all experiments.
Statistical Analysis
Differences in the distribution of demographic characteristics were evaluated by a Student t test or χ2 test between cases and controls as appropriate. The Student t test was used to compare the differences in serum miRNA levels between PC patients and controls, the differences in miRNA levels between standard cell culture and folate-deficient cell culture, and the differential expression of miRNAs between standard and folate-deficient cell culture supernatant. Analysis of covariance was used to adjust age, sex, smoking, and serum folate status. P < 0.05 was considered significant.
Expression levels of each miRNA were classified into normal and abnormal expressions (dichotomous outcome) according to each 95% confidence interval (CI) of miRNA expression levels in control subjects. The upper limit of the CIs for the miRNAs was set as a cutoff point above which the miRNA was overexpressed. Conversely, the lower limit of the CI was set as a cutoff point for underexpression. The Pearson χ2 test was performed to determine whether there were significant differences in serum miRNA levels between patients and healthy controls. The associations between miRNA expression levels and PC risk were estimated by the odds ratios (ORs) and their 95% CIs using multivariate logistic regression analysis to adjust for age, sex, smoking, and serum folate status.
The potential miRNA-folate interactions were tested by a multiplicative model, an additive model, and the multifactor dimensionality reduction (MDR) method. A multiple logistic regression model was used to detect the potential multiplicative interactions, and the log likelihood ratio test was used to assess whether the model was significantly improved by adding an additional interaction (product) term.23 To test for additive interactions, 3 indexes were calculated using an R-program24 (Vienna, Austria): relative excess risk due to interaction, the attributable proportion due to interaction, and the synergy index (SI). The MDR, a nonparametric, model-free approach by MDR software (version 2.0, beta 8), and MDR permutation testing software (version 1.0, beta 2) were applied to identify possible high-order interactions associated with PC risk.25 The best candidate interaction model was selected according to testing accuracy and cross-validation consistency (CVC). Validation was derived empirically from permutation tests, which were considered statistically significant at the 0.05 level. All the variables identified in the best model by the MDR software were combined, and their ORs and 95% CIs in relation to PC risk were calculated using multivariate logistic regression analysis.
Statistical analyses were performed with Statistical Analysis System software (version 9.2; SAS Institute, Cary, NC), except for additive interaction analysis and MDR analysis.
RESULTS
Demographic Characteristics of the Study Population
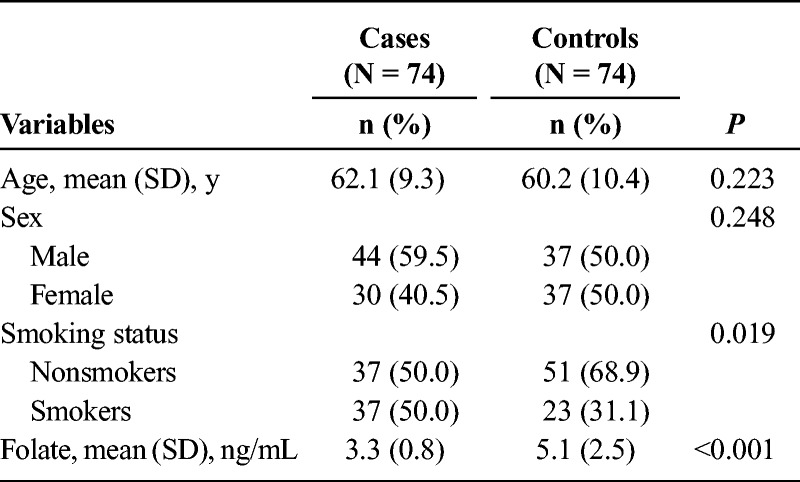
To determine whether an association exists between miRNA expression and PC, as well as a potential interaction between miRNAs and serum folate in PC, we assembled 74 PC patients and 74 sex-/age-matched control subjects. Demographic characteristics of study subjects were summarized in Table 1. There were no significant differences in the distribution of age and sex between the cases and controls. Among the study subjects, 59.5% of cases and 50% of controls were men, with a mean (SD) age of 62.1 (9.3) and 60.2 (10.4), respectively. More cases reported a smoking history than the controls (50% vs 31%, P = 0.019). Furthermore, examination of the serum folate demonstrated that levels for the cases were significantly lower than the controls (5.1 [2.5] vs 3.3 [0.8], P < 0.0001). These findings supported previous studies demonstrating that PC was associated with both an increased incidence of smoking and reduced folate levels.26
TABLE 1.
Distribution of Select Characteristics Among PC Patients and Controls
Validation of miR-191 as an Endogenous Reference for Quantification
To select an endogenous reference for the quantification of serum miRNAs, we examined 3 candidates (RNU6B, miR-16, and miR-191) using qRT-PCR of 16 randomly selected serum samples (8 healthy individuals and 8 PC patients). Data analysis with NormFinder yielded values of 0.352, 0.220, and 0.165 for RNU6B, miR-16, and miR-191, respectively. The arbitrary cutoff within NormFinder was 0.4, which suggested that all of these expression values were stable; however, the most stably expressed among the 3 candidates was miR-191, and thus, it was selected as an optimal reference for qRT-PCR.
Differential Expression of Serum miRNAs Among PC Patients and Controls
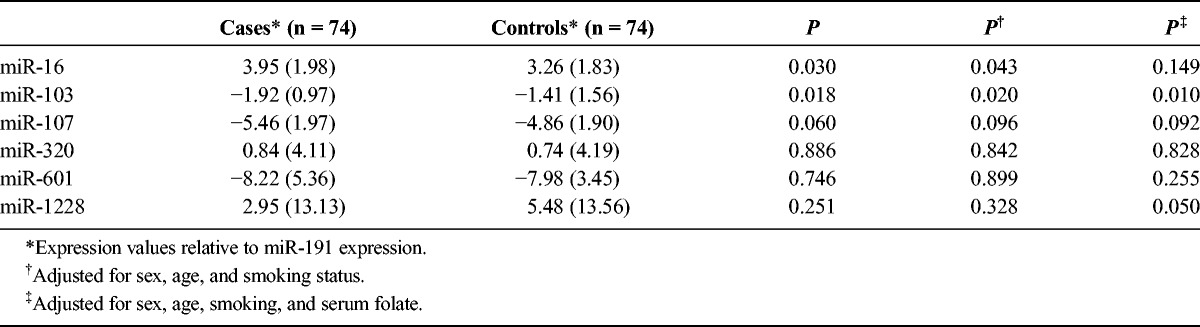
The 6 miRNAs were measured in the 148 serum samples (74 cases and 74 controls) relative to miR-191 expression. The mean (SD) expression of miR-16 among the cases was 3.95 (1.98), which was significantly higher than that in controls (3.26 [1.83]) (P = 0.030). Conversely, the expression of miR-103 in the cases was −1.92 (0.97), which was significantly lower than that in controls (−1.41 [1.56]) (P = 0.018). No significant differences between the 2 groups were observed for the other miRNAs (Table 2, first 3 columns). After adjusting for age, sex, and smoking status, consistent results were observed (Table 2, column P†). However, after adjusting for these 3 variables and serum folate status, a significance difference was observed only in miR-103 (P = 0.010) (Table 2, column P‡).
TABLE 2.
Serum Expression Levels of 6 miRNAs
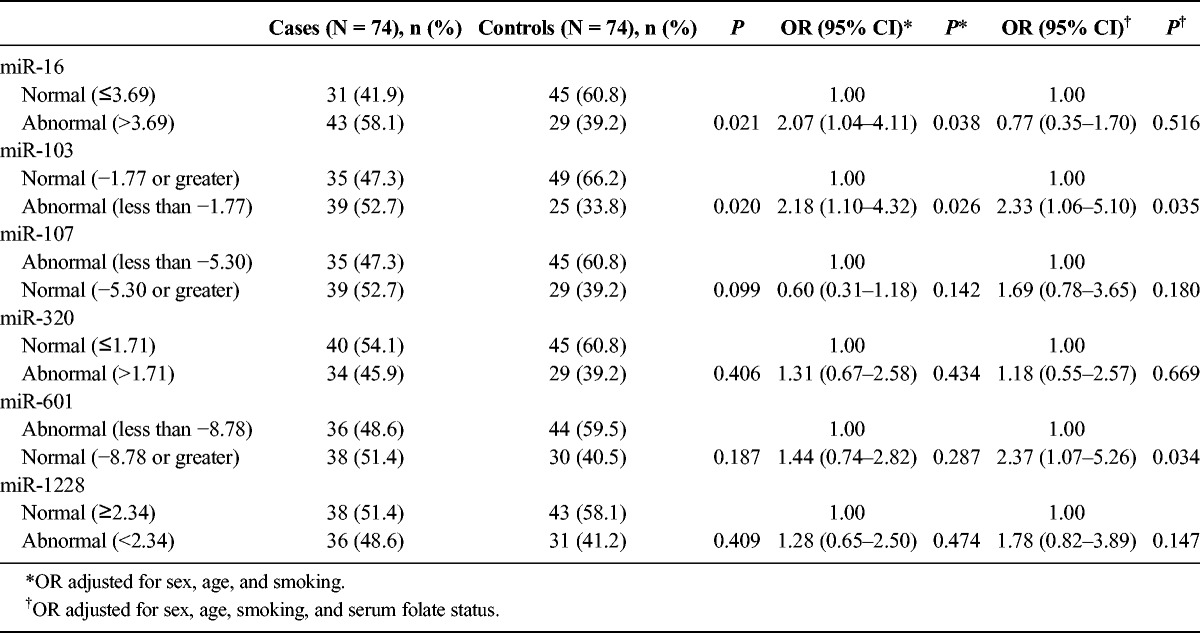
To further investigate the association between differential miRNA expression and PC, miRNAs were classified into normal or abnormal expression according to 95% CIs of miRNA expression levels of control subjects. The results confirmed that high miR-16 expression and low miR-103 expression were associated with significantly increased risk for PC after adjustment for age, sex and smoking status, with ORs of 2.07 (95% CI, 1.04–4.11) and 2.18 (95% CI, 1.10–4.32), respectively. However, after additionally adjusting for serum folate status, low expressions of miR-103 and miR-601 (but not high miR-16 expression) were associated with significantly increased risk for PC, with ORs of 2.33 (95% CI, 1.06–5.10) and 2.37 (95% CI, 1.07–5.26), respectively (Table 3). These results indicated that elevated miR-16 concomitant with decreased folate levels might increase the risk for PC, that reduced miR-103 expression was predictive of PC regardless of the folate status, and that miR-601was a risk factor for PC when considered in relation to folate status.
TABLE 3.
Effect of the 6 miRNAs and PC
Assessment of the Interaction Between Serum miRNAs and Folate Expression With PC by Multiplicative Interaction Analysis, Additive Interaction Analysis, and MDR Analysis
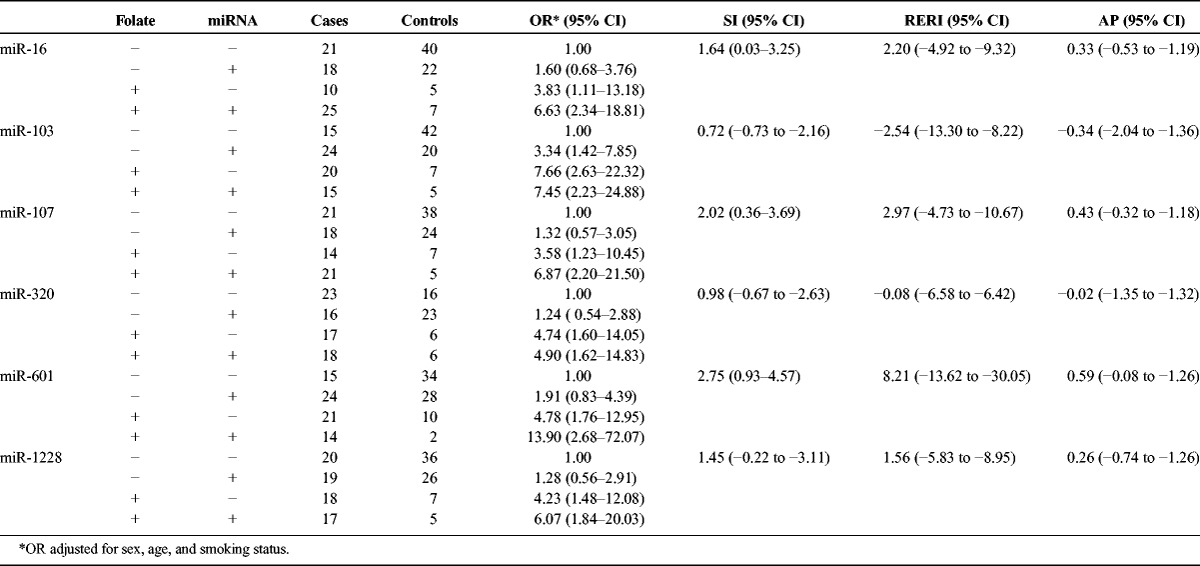
To further understand the relationship between miRNA expression, folate status, and PC, we assessed the miRNA and folate expression data by multiplicative interaction analysis, additive interaction analysis, and MDR analysis. For the multiplicative interaction analysis, a likelihood ratio test was used to assess whether the multiplicative interaction term for miRNA and folate could improve the logistic model. However, no multiplicative or additive interactions between miRNAs and folate were observed (Table 4).
TABLE 4.
Additive Interaction Analysis of Serum miRNA-Folate
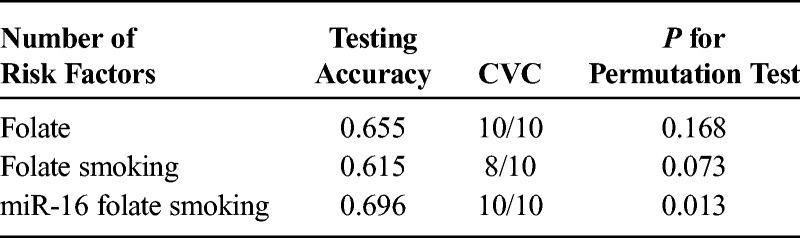
Next, MDR analysis was used to identify potential high-dimensional interactions between serum miRNAs and folate (Table 5). The best interaction model by MDR analysis was the 3-factor model consisting of miR-16, folate, and smoking (testing accuracy, 0.696; CVC, 10/10; permutation P = 0.013). The 3 factors identified in the model were combined according to the MDR software, and individuals carrying the combined risk stratum had a 4.06-fold increase for PC (95% CI, 1.24–13.26).
TABLE 5.
Interaction Models by MDR Analysis
Effects of Folate Deprivation on miRNA Expression in AsPC-1 Cells and Cell Culture Supernatant
Serum miRNAs that were differentially regulated in PC patients were likely to have originated from the pancreas, and therefore, we examined miRNA levels after folate deprivation in pancreatic carcinoma AsPC-1 and PANC-1 cells lines and their cell culture supernatant. The RNA was isolated from these 2 cell lines and cell culture supernatants after 72-hour culture in folate-deprived medium. The expression levels of miR-16, miR-103, and miR-601 were measured. The miR-16 was significantly up-regulated both in the folate-deprived AsPC-1 cell line (P < 0.001) and the culture supernatant of the folate-deprived AsPC-1 cell line (P = 0.004) (Fig. 1). Moreover, the high expression of miR-16 was also confirmed in folate-deprived PANC-1 cell line (P = 0.008), although no statistically significant difference of miR-16 was observed in the cell culture supernatant (Fig. 1). These findings verified that miR-16 could be regulated in human PC cells by folate deprivation.
FIGURE 1.
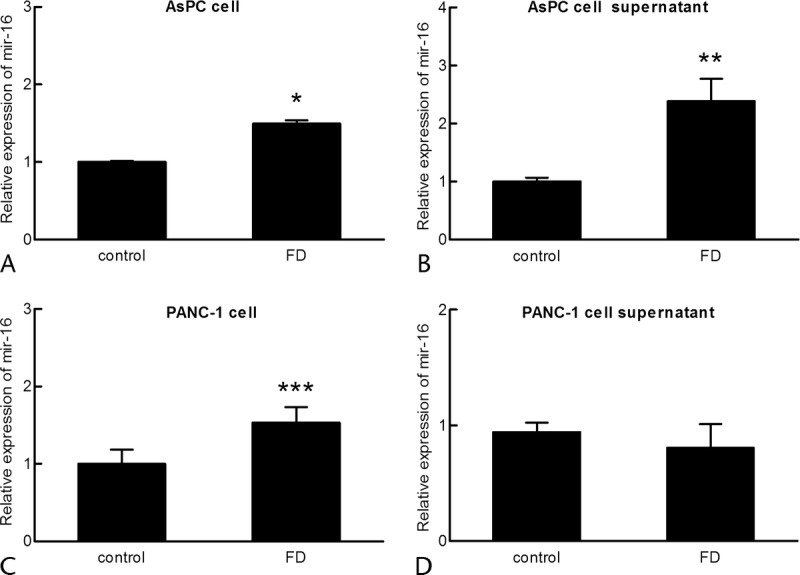
MiR-16 expression in PC cell lines and their culture supernatants. The expression level of miR-16 in PC cell lines and cell supernatants was assayed following 72-hour culture in normal medium (control) or folate-deficient medium (FD). Values were standardized to U6 or cel-miR-39 and normalized to 1 in the control cells. The mean (SD) from 3 biologically independent experiments is shown. *P < 0.001, **P = 0.004, ***P = 0.008. A, AsPC cell. B, AsPC cell supernatant. C, PANC-1 cell. D, PANC-1 cell supernatant.
DISCUSSION
In this study, we explore whether an association exists between miRNAs and folate in PC. Because there were some debates over the optimal choice of an endogenous control for the detection of serum miRNAs, we first assessed 3 candidates to identify a suitable internal control in PC. The results revealed that miR-191 was the optimal endogenous control for the detection of serum miRNAs, which was consistent with a previous study.20 Within a cohort of 74 PC patients and 74 controls, we found that the subjects with high expression of miR-16 or low expression of miR-103 had a higher risk for PC after adjusting for age, sex, and smoking status with ORs of 2.07 (95% CI, 1.04–4.11) and 2.18 (95% CI, 1.10–4.32), respectively. However, the higher risk for PC could not be predicted for miR-16 after additional adjustment for folate status. Furthermore, subjects with low expression of miR-601 had a higher risk for PC with an OR of 2.37 (95% CI, 1.07–5.26), after adjusting age, sex, smoking, and serum folate status. The MDR analysis showed a significant interaction among miR-16, folate, and smoking status. We also demonstrated that, for cultured AsPC cells, miR-16 was significantly up-regulated in cell lysates and in cell culture supernatant after 72-hour growth in folate-deficient medium. In addition, a high level of miR-16 was observed in folate-deprived PANC-1 cell line.
Our identification of a role for miR-16 in PC is in accordance with other studies showing that high expression of miR-16 is associated with PC. Several studies found that miR-16 was overexpressed in PC tissues.12,27 The expression of miR-16 in PC patient serum was also higher than in controls.28 Functional studies showed that overexpression of miR-16 could reduce the survival of dendritic cells and thus diminish immune responses, which led to immune escape and limitless proliferation of cancer cells.29 The miR-16 targeted the silencing mediator for retinoid and thyroid hormone receptor and promoted nuclear factor-κB–regulated transactivation of the IL-8 gene, which played an important role in tumorigenesis and metastasis of various cancers including PC.30–32 After adjusting for age, sex, smoking, and serum folate status, the risk for high expression of miR-16 was not observed, which indicated an association between miR-16 and serum folate. This association was also supported by the MDR analysis and by the in vitro studies of folate deprivation in PC cell lines. In both PC cell lines, the expression of miR-16 was increased under a folate deprivation condition; it did indicate that low folate might play a role in the regulation of miR-16 expression. In cell culture supernatants, there were no consistent results from the 2 cell lines, and the disparity might be attributed to different mechanisms of RNA secretion in complicated cell-cell communications. The mechanism of miRNAs and folate is likely to involve effects on DNA methylation. Low expression of miR-16 in tumor tissue was associated with the methylation of DNA,33 and miR-16 increased when DNA methylation was inhibited.34 The most important biological function of folate is to provide a substrate for DNA methylation in the form of tetrahydrofolate. Thus, folate deficiency usually leads to whole genome dysregulation of methylation and abrogates the expression of some important tumor suppressors, including miRNAs, which ultimately may lead to tumorgenesis.35–38 Epidemiologic studies also indicated that folate deficiency was highly related to the risk for PC.16 In vitro and in vivo experiments showed that folate deficiency led to the dysregulation of miRNAs in both cancer cells and tumor tissues.18,19 In addition to folate deficiency, cigarette smoke could contribute to the alterations in miRNA expression.39 Nicotine had been shown to up-regulate miR-16 and miR-21.40 Furthermore, upon exposure to environmental cigarette smoke for 28 days, 26% of the miRNAs tested, most of which were involved in the proliferation and apoptosis of cancer cells, were up-regulated in the lungs of rats.41 Interestingly, cigarette smoke could result in a decrease of folate in serum.42 So, to identify the association between serum folate, miR-16, and PC without an effect of smoking status, we further analyzed the association between miR-16, folate, and PC among nonsmokers (data not shown) and found a potential additive interaction effect of folate deficiency and up-regulated miR-16 (SI [95% CI], 1.88 [−0.26 to 4.01]). In all, the abovementioned collective evidence suggested that miR-16 and folate deficiency might contribute to the progression of PC.
Our results also indicated that subjects with low expression of miR-103 had a higher risk for PC after adjusting for age, sex, smoking, and folate status. Recent studies also have supported an essential role for miR-103 in the development of various cancers as a tumor suppressor gene. For example, miR-103 could inhibit the proliferation and migration of nonsmall lung cancer cells by targeting the 3′ UTR of protein kinase C-ε.43 In addition, overexpression of miR-103 in neuroblastoma cells reduced proliferation and promoted differentiation by targeting the inhibitor of DNA binding-2, suggesting that its alteration might be involved in neural cancer development.44
After adjusting for age, sex, smoking, and folate status, the subjects with low expression of miR-601 were also observed to have a higher risk for PC. Although abnormal expression of miR-601 had not been reported in other PC studies, the expression of miR-601 had been shown to be reduced in the serum of colorectal cancer patients.45 The precise mechanisms needed further investigations, given that miR-601 can regulate multiple signaling pathways.46
The role of serum miRNAs as mediators in cell-cell communication is needed to be explored further. Studies have shown that serum miRNAs might be attributed to a complicated origin, including cancer cells, satellite cells, immune cells, endothelial cells, and so on.47 As one of the heterogeneous tumors, PC also consists of these types of cells,48 and it is essential to keep its intercellular communication. Serum miRNAs might play a role of cell-cell communication.49 Actually, miRNAs are stable against RNase in serum, although the mechanism still remains poorly elucidated. Several studies have identified that miRNAs could be packaged in microvesicles, exosomes, apoptotic bodies, Argonaute 2 (Ago2) complexes, and high-density lipoproteins.50–54 The miR-16 has been found partly bound to Ago2 in plasma and serum human samples, and the combination might facilitate a high stability of the miRNA-Ago2 complex.53 The mir-16 we detected in cell culture supernatant might be secreted by AsPC-1 cells as these forms. Furthermore, studies have suggested that serum miRNAs could be delivered to recipient cells and play a role in new cells.54,55 Therefore, further researches on the field may provide more valuable findings.
In conclusion, the interaction of abnormal serum miRNA and folate seems to positively correlate with increased risk for PC, suggesting a role for folic acid supplementation in PC. Expanded cohort studies and additional experimental evidences may elucidate the underlying biological mechanisms for this association.
Footnotes
Yao Tian and Yibo Xue contributed equally to this work.
This study was supported by the National Natural Science Foundation of China (NSFC-81041079).
The authors declare no conflict of interest.
REFERENCES
- 1. Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008; 58: 71– 96. [DOI] [PubMed] [Google Scholar]
- 2. Wang L, Yang GH, Lu XH, et al. Pancreatic cancer mortality in China (1991–2000). World J Gastroenterol. 2003; 9: 1819– 1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007; 297: 1901– 1908. [DOI] [PubMed] [Google Scholar]
- 4. Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007; 26: 4442– 4452. [DOI] [PubMed] [Google Scholar]
- 5. Valencia-Sanchez MA, Liu J, Hannon GJ, et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006; 20: 515– 524. [DOI] [PubMed] [Google Scholar]
- 6. Negrini M, Nicoloso MS, Calin GA. MicroRNAs and cancer—new paradigms in molecular oncology. Curr Opin Cell Biol. 2009; 21: 470– 479. [DOI] [PubMed] [Google Scholar]
- 7. Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008; 105: 10513– 10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008; 18: 997– 1006. [DOI] [PubMed] [Google Scholar]
- 9. Wang J, Chen J, Chang P, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila). 2009; 2: 807– 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kawaguchi T, Komatsu S, Ichikawa D, et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer. 2013; 108: 361– 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Papaconstantinou IG, Manta A, Gazouli M, et al. Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas. 2013; 42: 67– 71. [DOI] [PubMed] [Google Scholar]
- 12. Tang S, Bonaroti J, Unlu S, et al. Sweating the small stuff: microRNAs and genetic changes define pancreatic cancer. Pancreas. 2013; 42: 740– 759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Keller A, Leidinger P, Bauer A, et al. Toward the blood-borne miRNome of human diseases. Nat Methods. 2011; 8: 841– 843. [DOI] [PubMed] [Google Scholar]
- 14. Lodygin D, Tarasov V, Epanchintsev A, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008; 7: 2591– 2600. [DOI] [PubMed] [Google Scholar]
- 15. Weber B, Stresemann C, Brueckner B, et al. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle. 2007; 6: 1001– 1005. [DOI] [PubMed] [Google Scholar]
- 16. Oaks BM, Dodd KW, Meinhold CL, et al. Folate intake, post-folic acid grain fortification, and pancreatic cancer risk in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Am J Clin Nutr. 2010; 91: 449– 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Larsson SC, Hakansson N, Giovannucci E, et al. Folate intake and pancreatic cancer incidence: a prospective study of Swedish women and men. J Natl Cancer Inst. 2006; 98: 407– 413. [DOI] [PubMed] [Google Scholar]
- 18. Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006; 66: 10843– 10848. [DOI] [PubMed] [Google Scholar]
- 19. Kutay H, Bai S, Datta J, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006; 99: 671– 678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu Z, Dong J, Wang LE, et al. Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis. 2012; 33: 828– 834. [DOI] [PubMed] [Google Scholar]
- 21. Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004; 64: 5245– 5250. [DOI] [PubMed] [Google Scholar]
- 22. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008; 3: 1101– 1108. [DOI] [PubMed] [Google Scholar]
- 23. Zhou XJ, Lu XL, Nath SK, et al. Gene-gene interaction of BLK, TNFSF4, TRAF1, TNFAIP3, and REL in systemic lupus erythematosus. Arthritis Rheum. 2012; 64: 222– 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kallberg H, Ahlbom A, Alfredsson L. Calculating measures of biological interaction using R. Eur J Epidemiol. 2006; 21: 571– 573. [DOI] [PubMed] [Google Scholar]
- 25. Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003; 19: 376– 382. [DOI] [PubMed] [Google Scholar]
- 26. Jarosz M, Sekula W, Rychlik E. Influence of diet and tobacco smoking on pancreatic cancer incidence in poland in 1960–2008. Gastroenterol Res Pract. 2012; 2012: 682156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y, Li M, Wang H, et al. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg. 2009; 33: 698– 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu J, Gao J, Du Y, et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer. 2012; 131: 683– 691. [DOI] [PubMed] [Google Scholar]
- 29. Min S, Liang X, Zhang M, et al. Multiple tumor-associated microRNAs modulate the survival and longevity of dendritic cells by targeting YWHAZ and Bcl2 signaling pathways. J Immunol. 2013; 190: 2437– 2446. [DOI] [PubMed] [Google Scholar]
- 30. Zhou R, Li X, Hu G, et al. miR-16 targets transcriptional corepressor SMRT and modulates NF-kappaB-regulated transactivation of interleukin-8 gene. PLoS One. 2012; 7: e30772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mian BM, Dinney CP, Bermejo CE, et al. Fully human anti-interleukin 8 antibody inhibits tumor growth in orthotopic bladder cancer xenografts via down-regulation of matrix metalloproteases and nuclear factor-kappaB. Clin Cancer Res. 2003; 9: 3167– 3175. [PubMed] [Google Scholar]
- 32. Finnerty JR, Wang WX, Hebert SS, et al. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010; 402: 491– 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009; 10: 704– 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hanoun N, Delpu Y, Suriawinata AA, et al. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin Chem. 2010; 56: 1107– 1118. [DOI] [PubMed] [Google Scholar]
- 35. Bandres E, Agirre X, Bitarte N, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009; 125: 2737– 2743. [DOI] [PubMed] [Google Scholar]
- 36. Hulf T, Sibbritt T, Wiklund ED, et al. Discovery pipeline for epigenetically deregulated miRNAs in cancer: integration of primary miRNA transcription. BMC Genomics. 2011; 12: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lujambio A, Ropero S, Ballestar E, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007; 67: 1424– 1429. [DOI] [PubMed] [Google Scholar]
- 38. Toyota M, Suzuki H, Sasaki Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008; 68: 4123– 4132. [DOI] [PubMed] [Google Scholar]
- 39. Russ R, Slack FJ. Cigarette-Smoke-Induced Dysregulation of MicroRNA Expression and Its Role in Lung Carcinogenesis. Pulm Med. 2012; 2012: 791234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shin VY, Jin H, Ng EK, et al. NF-kappaB targets miR-16 and miR-21 in gastric cancer: involvement of prostaglandin E receptors. Carcinogenesis. 2011; 32: 240– 245. [DOI] [PubMed] [Google Scholar]
- 41. Izzotti A, Calin GA, Arrigo P, et al. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009; 23: 806– 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Okumura K, Tsukamoto H. Folate in smokers. Clin Chim Acta. 2011; 412: 521– 526. [DOI] [PubMed] [Google Scholar]
- 43. Garofalo M, Romano G, Di Leva G, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2012; 18: 74– 82. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44. Annibali D, Gioia U, Savino M, et al. A new module in neural differentiation control: two microRNAs upregulated by retinoic acid, miR-9 and -103, target the differentiation inhibitor ID2. PLoS One. 2012; 7: e40269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Q, Huang Z, Ni S, et al. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS One. 2012; 7: e44398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ohdaira H, Nakagawa H, Yoshida K. Profiling of molecular pathways regulated by microRNA 601. Comput Biol Chem. 2009; 33: 429– 433. [DOI] [PubMed] [Google Scholar]
- 47. Allegra A, Alonci A, Campo S, et al. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review). Int J Oncol. 2012; 41: 1897– 1912. [DOI] [PubMed] [Google Scholar]
- 48. Lou E, Subramanian S, Steer CJ. Pancreatic cancer: modulation of KRAS, MicroRNAs, and intercellular communication in the setting of tumor heterogeneity. Pancreas. 2013; 42: 1218– 1226. [DOI] [PubMed] [Google Scholar]
- 49. Iguchi H, Kosaka N, Ochiya T. Secretory microRNAs as a versatile communication tool. Commun Integr Biol. 2010; 3: 478– 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008; 10: 1470– 1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007; 9: 654– 659. [DOI] [PubMed] [Google Scholar]
- 52. Zernecke A, Bidzhekov K, Noels H, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009; 2: ra81. [DOI] [PubMed] [Google Scholar]
- 53. Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011; 108: 5003– 5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011; 13: 423– 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grange C, Tapparo M, Collino F, et al. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011; 71: 5346– 5356. [DOI] [PubMed] [Google Scholar]


