Abstract
Background and Purpose
To demonstrate the accuracy across different acquisition and analysis methods, we evaluated the variability in hippocampal volumetric and surface displacement measurements resulting from two different MRI acquisition protocols.
Methods
Nine epilepsy patients underwent two independent T1-weighted magnetization prepared spoiled gradient sequences during a single 3T MRI session. Using high-dimension mapping-large deformation (HDM-LD) segmentation, we calculated volumetric estimates and generated a vector-based 3-dimensional surface model of each subject's hippocampi, and evaluated volume and surface changes, the latter using a cluster-based noise estimation model.
Results
Mean hippocampal volumes and standard deviations for the left hippocampi were 2750 (826) mm3 and 2782 (859) mm3 (p=0.13), and for the right hippocampi were 2558 (750) mm3 and 2547 (692) mm3 (p=0.76), respectively for the MPR1 and MPR2 sequences. Average Dice coefficient comparing overlap for segmentations was 86%. There was no significant effect of MRI sequence on volume estimates and no significant hippocampal surface change between sequences.
Conclusion
Statistical comparison of hippocampal volumes and statistically thresholded HDM-LD surfaces in TLE patients showed no differences between the segmentations obtained in the two MRI acquisition sequences. This validates the robustness across MRI sequences of the HDM-LD technique for estimating volume and surface changes in subjects with epilepsy.
Keywords: MRI, hippocampus, epilepsy
Introduction
Volumetric measurements based on magnetic resonance imaging (MRI) can detect hippocampal changes in different neuropsychiatric diseases due to overall hippocampal volume loss[1, 2]. Additionally, techniques analyzing hippocampal surface structure can detect sub-regional hippocampal pathophysiological changes, elucidating patterns of hippocampal structural change not evident with hippocampal volume measurements alone[3, 4]. MRI-based hippocampal imaging plays an important role in the diagnosis and treatment of epilepsy. In the clinical setting of an epileptic seizure history compatible with temporal lobe epilepsy (TLE), a significant hippocampal volume asymmetry is predictive of hippocampal atrophy (HA)[5, 6] and a favorable outcome after epilepsy surgery[7-9]. Recent studies have confirmed that automated methods of detection of hippocampal atrophy are highly predictive of the presence and laterality of hippocampal atrophy in TLE[10].
High-dimension mapping-large deformation (HDM-LD) is a computational technique which allows semi-automatic segmentation of the hippocampus. HDM-LD uses 107-108 parameters to transform grayscale image data and calculate vector-based three-dimensional surfaces of neuroanatomic structures, with resolution at the subvoxel level[11-13]. We have previously documented the validity of HDM-LD for hippocampal segmentation in patients with temporal lobe epilepsy, showing a 92.8% Dice coefficient for overlap in sequential hippocampal segmentations[14].
While the role of MRI-based hippocampal imaging is well-established in studies of epilepsy, validation of volumetric and surface measurement techniques, especially using different MRI platforms and/or sequences, remains understudied[15]. Reproducibility of volumetric and surface measurements is especially important in longitudinal or comparative studies, when repeated measurements are compared between different imaging sessions, or are acquired using different MRI scanners or imaging sequences. Past investigators have compared segmentation techniques for different scanner platforms[16] or different segmentation techniques on identical data sets[15, 17, 18]. In the current study, we evaluate the reproducibility of HDM-LD segmentation in subjects with epilepsy imaged during a single imaging session with two different volumetric MRI acquisition protocols, specifically testing the invariance of the technique across imaging sequences. In addition to comparison of volumetric measurements, we compare three-dimensional hippocampal surface structural changes using HDM-LD between the two imaging protocols.
Methods
Subject selection
Subjects were patients with a clinical diagnosis of temporal lobe epilepsy who underwent an MRI study for evaluation of their seizures. The cohort included 9 right-handed subjects (4 male, 5 female; 3 right TLE, 6 left TLE), with a mean age at scan of 46 years (range 21-66). Subjects were consecutively selected from our institutional series of patients who underwent two independent 3-dimensional T1-weighted spoiled gradient echo sequences during a single imaging session. Patients with space-occupying structural lesions, or who had undergone neurosurgical procedures involving the brain were excluded from the study. The study was approved by the local institutional review board at Washington University in St. Louis. A set of thirty-two healthy control image data sets (age range 18-65, 12 males) acquired under identical imaging conditions was used for the purpose of empiric surface noise estimation. All participants signed written consent to participate in the study.
Imaging protocol
Images were acquired with a Siemens MAGNETOM Trio 3T scanner (Erlangen, Germany). Two different T1-weighted magnetization-prepared spoiled gradient echo sequences were acquired for each patient for the purpose of hippocampal segmentation. These are indicated as MPR1 and MPR2 below and had the following characteristics: MPR1: voxel size 0.43 × 0.43 × 0.9 mm, TR (repeat time) 1570 ms, TE (echo time) 3.29 ms, TI (inversion time) 800 ms, flip angle 15 deg, FOV (field of view) 416 × 512 voxels (178.9 × 220.1 mm), coronal plane of acquisition; MPR2: 1 × 1 × 1 mm, TR 2400 ms, TE 3.16 ms, TI 1000 ms, flip angle 8 deg, FOV 256 ×256 voxels (256 × 256 mm), sagittal plane of acquisition. After reconstruction images were normalized to the same intensity range by matching high and low-percentile intensity values across the two sequences.
Hippocampal HDM-LD segmentation
We performed deformation segmentations as previously described[14]. The deformation segmentation procedure uses MRI-image volumetric segmentation information to transform a template segmentation of the left or right hippocampus (based on a single normal subject, not included in the study) into the corresponding hippocampus of an individual subject (control or patient). This generates a transform of the template hippocampus into the target individual hippocampus, yielding a volumetric estimate of the individual hippocampus as well as a vector-based three-dimensional surface of the individual hippocampus and its displacement from the template surface. This procedure was applied to each subject in the study, producing both volumetric and surface estimates for left and right hippocampi. Volume estimates for each hippocampus were computed directly from these volumetric representations. Dice coefficient was calculated as previously described[17]. Surface-based displacement from the template surface was computed for each individual subject. Average displacement surfaces were computed by averaging vertex-based displacement across subjects[19].
Statistical Approach
For volumetric measurements, analysis of the variance (ANOVA) and two-tailed paired t-test were used, with hippocampal volume as the dependent variable, MPR sequence and hippocampal side as within-subject factors, and individual subject as an across-subject factor. For surface deformation measurements, two-tailed two-sample t-test was computed for each vertex (or node) in the 3-dimensional hippocampal surface, after correction for multiple comparisons using a noise estimation model, yielding vertex-level statistical estimates of deformation from the template surface. Importantly, to control for multiple comparisons due to the large number of vertices in each hippocampal surface, estimation of vertex-level deformation noise, taking into account local coherence between neighboring vertices, was calculated empirically using an independent set of healthy control subjects (n = 32) that were scanned using the MPR2 sequence. Adapting iterative randomization methods routinely used for multiple comparison correction in volumetric imaging data[20], the probability of occurrence of suprathreshold clusters of neighboring vertices was modeled using the healthy data set, yielding maximum cluster-size cutoffs for pre-specified significance thresholds (e.g. α = 0.05 and 0.01). The 0.05 cutoff (666 contiguous vertices) was chosen for the purpose of maximizing the significance of potential variance across sequences and applied to the patient data, yielding a cluster-constrained correction for multiple comparisons across the hippocampal surface. Importantly, the control data set was used exclusively to derive an empiric estimation for the occurrence of suprathreshold clusters in the hippocampal surface in the normal population, i.e. it provided a conservative estimate of the maximum cluster of neighboring surface vertices that would show local shape changes by chance alone.
Results
Figure 1 demonstrates representative images from the MRP1 and MPR2 from subject 1, showing the coronal plane near the mid-section of the body of the hippocampus. The images serve to demonstrate differences in the acquisition protocols, including differences in resolution and gray-scale intensity contrasts.
Figure 1.
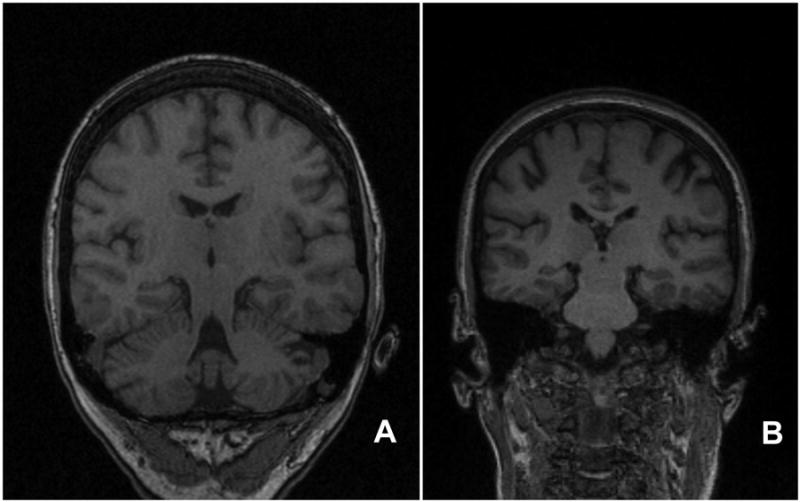
The figure shows representative images from the MRP1 and MPR2 sequences from subject 1. Figure A is the MPR1 image, and Figure B is the MPR2 image. Each image demonstrates the coronal plane near the mid-section of the body of the hippocampus. Because of differences in the angle of acquisition for each sequence, structures included in the representative images differ, such as inclusion of the cerebellum in panel A, but not in panel B. The images serve to demonstrate differences in the protocols, including different image resolution, gray-scale intensity contrast, and angle of acquisition of the images.
The hippocampal volumes as determined by the HDM-LD segmentation are reported in Table 1. A two-way within-subject ANOVA of these volumes revealed no significant differences or interactions between MPR protocols or hippocampal side (p > 0.05). In confirmatory fashion, a two-tailed paired t-test between MRI protocols produced non-significant p-values for each hippocampus (left: p=0.13, right: p=0.76; see Table 1). Hippocampal volumetric measurements, as shown in Table 1 and Figure 2, showed marked similarity of volumes in the MPR1 and MPR2 groups, in both the left and right hippocampi. The range of volume differences was 0.01-4.2% (mean 1.6%, SD 1.4%) for the left hippocampus and 1.5-11.3% (mean 3.6%, SD 3.1%) for the right hippocampus. Dice coefficient was similar for the right and left hippocampi, with 86% mean value (SD 1.9%) for the right hippocampus and 85% mean value (SD 4.1%) for the left hippocampus.
Table 1. Hippocampal volumes as determined by HDM-LD segmentation.
| Hippocampal Volume (mm3) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Left Hippocampus | Right Hippocampus | |||||||
| Subject | MPR1 | MPR2 | % Diff | Dice (%) | MPR1 | MPR2 | % Diff | Dice (%) |
| 1 | 2598 | 2561 | 1.4 | 86 | 1504 | 1674 | 11.3 | 78 |
| 2 | 1964 | 2018 | 2.7 | 84 | 1317 | 1354 | 2.8 | 79 |
| 3 | 1424 | 1401 | 1.7 | 84 | 2318 | 2272 | 2.0 | 89 |
| 4 | 3062 | 3072 | 0.3 | 88 | 3128 | 3031 | 3.1 | 88 |
| 5 | 2789 | 2832 | 1.5 | 89 | 2911 | 2830 | 2.8 | 89 |
| 6 | 2871 | 2991 | 4.2 | 85 | 2493 | 2564 | 2.8 | 87 |
| 7 | 3279 | 3283 | 0.1 | 89 | 3450 | 3501 | 1.5 | 86 |
| 8 | 2411 | 2410 | 0.01 | 86 | 2606 | 2567 | 1.5 | 86 |
| 9 | 4349 | 4470 | 2.8 | 87 | 3295 | 3132 | 4.9 | 85 |
| mean: | 2750 | 2782 | 1.6 | 86 | 2558 | 2547 | 3.6 | 85 |
| SD: | 826 | 859 | 1.4 | 1.9 | 750 | 692 | 3.1 | 4.1 |
| p-value: | 0.13 | 0.76 | ||||||
Figure 2.
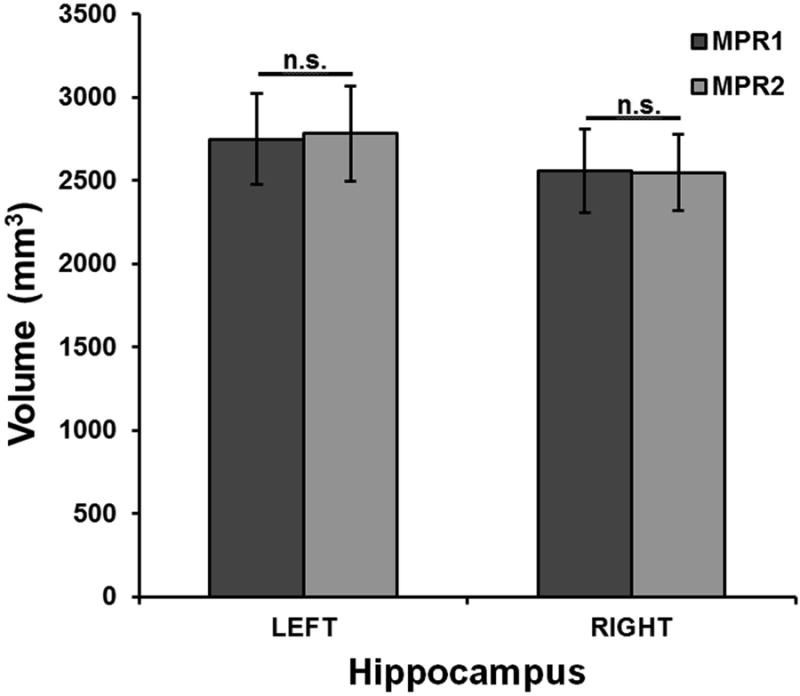
Volumetric differences between MPR1 and MPR2 sequences. Mean hippocampal volumes comparing the MPR1 and MPR2 sequences, with error bars showing 1 SD ranges in each group. Highly consistent volumetric measurements were obtained across MRI sequences.
Figure 2 highlights the similarity in volumetric measurements between the MPR1 and MPR2 groups. Figure 3 shows the variability in hippocampal volume estimates across sequences for individual subjects, demonstrating that the deviation in hippocampal volume estimates between the MRI protocols is minimal even at the individual level.
Figure 3.
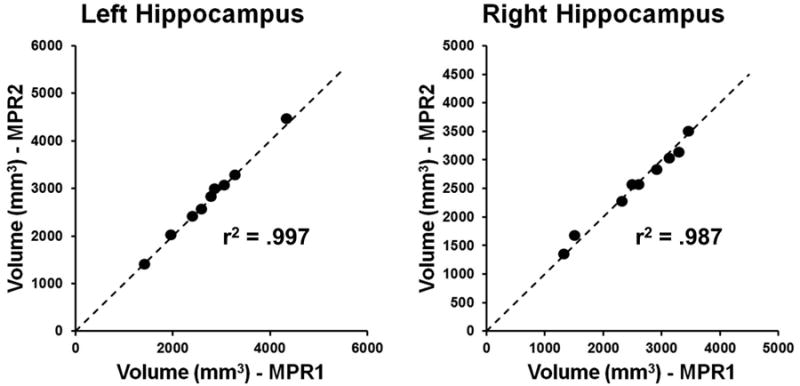
Variability between volumetric measurements for individual subjects. The x-axis for each graph represents the hippocampal volume estimate derived from the MPR1 sequence, while the y-axis for each graph represents the corresponding hippocampal volume estimate derived from the MPR2 sequence. Note that the dashed line represents a line of slope 1, i.e. the ideal correspondence between volume estimates across sequences. Also shown is the r2 value for the correlation between volume estimates for the two sequences.
The HDM-LD technique has been shown to yield reliable estimation of hippocampal surfaces in controls and patients with neurologic diseases. We applied this technique in conjunction with a noise-estimation model to assess the variability in hippocampal surface estimates across MRI sequences. Figure 4 shows the difference in average displacement in the left and right hippocampal surfaces between MRI sequences. Specifically, the color scale shows the absolute displacement (mean and S.D.), as projected on the MPR1 group surfaces, between the MPR1 and MPR2 hippocampal surfaces. Statistical comparison (unpaired two-tailed t-test, 8 d.f.) of differences between MPR1-derived and MPR2-derived surfaces, using the noise estimation model derived from the normal subject group discussed above, yielded no regions of significant displacement difference between MRI sequences (see Figure 4). Note that the majority of the surface shows less than 1-mm difference in the estimate between MRI sequences.
Figure 4.
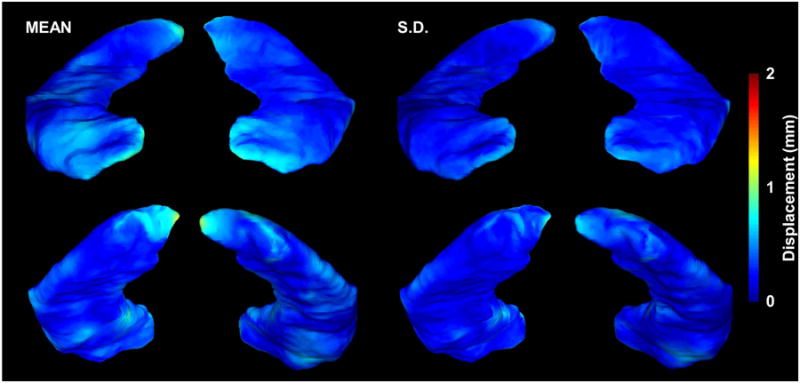
Mean and standard deviation of the displacement of the MPR2-derived hippocampal surface from the MPR1-derived surface. The color scale shows the mean (left) and standard deviation (right) of the displacement, as projected on the MPR1 group surfaces, between the MPR1 and MPR2 hippocampal surfaces. In the anterior view of the surfaces, the hippocampal head and uncinate gyrus of each hippocampal surface is visualized, showing that the margins of this hippocampal region show more relative displacement. In the posterior view, the hippocampal tail is well visualized, also showing a slightly greater displacement over the medial regions of the hippocampal tail.
In addition to local changes in hippocampal shape, overall hippocampal atrophy may itself affect the accuracy of the segmentation and volume estimation technique. To assess this for the HDM-LD technique in this study we computed the correlation between the mean volume estimate and the Dice coefficient for both the left and right hippocampi (see Figure 5). We noted a positive correlation across subjects between mean hippocampal size and Dice coefficient for both hippocampi, suggesting that our segmentation and estimation is more accurate and reliable for larger hippocampi.
Figure 5.
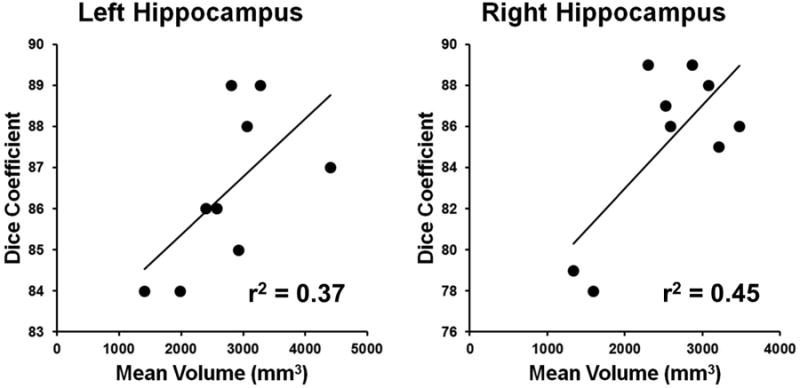
Correlation between mean hippocampal volume and Dice coefficient. The x axis shows the mean of the two hippocampal volume estimates for the MPR1 and MPR2 sequences for each individual, while the y axis shows the corresponding Dice coefficient. Also shown is the linear intercept of each correlation and the corresponding r2 value.
Conclusion
Our method of hippocampal segmentation uses the HDM-LD technique, which we have previously validated in a group of subjects with temporal lobe epilepsy with mesial temporal sclerosis[14]. HDM-LD uses a single template-based method, in which global landmarks and hippocampal landmarks are placed on each target MRI to provide an initial transformation from the reference template image to the target images. After the initial transformation step, the subsequent steps are fully automated and driven only by the volume data itself[14]. Several hippocampal segmentation techniques exist, including an HDM-LD technique using FreeSurfer segmentation to provide initial transformation parameters, obviating the use of landmarks[17]. Other approaches use multiple or probabilistic atlases, leveraging data from multiple subjects to derive probabilistic information for the location of the hippocampus in standard space, allowing for a fully automated technique[18].
Methods using either single template landmarked-based techniques (such as HDM-LD) or automated techniques have produced comparable results in normal subjects. One of the most common measures used to validate segmentation techniques is the Dice coefficient, which has been used to quantify overlap of segmentations[17]. When comparing Dice coefficients among coregistration techniques in normal hippocampi, landmark-based HDM-LD produced a Dice coefficient of 84%, while HDM-LD using FreeSurfer-initialized segmentation yielded a Dice coefficient of 75%[17]. With techniques using multiple or probabilistic atlases approaches, results are similar, with Dice coefficients ranging between 70-86%[16, 18, 21-23].
Validation studies in subjects with epilepsy generally show less accuracy with smaller, abnormal hippocampi. In hippocampi with mesial temporal sclerosis (MTS), HDM-LD yielded a Dice coefficient of 66%[14], while a probabilistic atlas-based approach showed a Dice coefficient of 77%[18]. Factors that lead to inaccurate segmentation of abnormal hippocampi likely include systematic errors of the techniques in the handling of smaller volumes[24, 25], operator-dependent variability in manual hippocampal segmentation of abnormal hippocampi[14, 26] (which is often the “gold standard” for validation of automated or semi-automated segmentation methods), and difficulties of the mapping/segmentation algorithm in defining hippocampal borders, especially in hippocampi with pathologic changes in size, shape and rotation[15, 27]. In the current study, the Dice coefficient was 86%. This finding is comparable to the highest Dice coefficient for normal subjects, and higher than those in previous studies of epilepsy subjects.
A recent study evaluated hippocampal segmentations of epilepsy subjects with matched healthy subject controls, comparing a region-growing algorithm constrained by anatomical priors (SACHA), a probabilistic-atlas based technique (FreeSurfer), and a multi-atlas technique (ANIMAL-multi)[15]. Investigators evaluated the performance of the algorithms when considering variables such as malrotation of the hippocampus (with respect to the collateral sulcus) or atrophy, comparing results to manual segmentations. The ANIMAL-multi technique demonstrated similar accuracy in epilepsy subjects as well as healthy controls, while both SACHA and FreeSurfer were less accurate in epilepsy subjects. When compared to manual volumetry, however, all three automated procedures underestimated the magnitude of atrophy in epilepsy subjects, highlighting the negative impact of developmental abnormalities (including malrotation) and atrophy on the segmentation of the hippocampus using automated methods. While progress continues in automated segmentation of the hippocampus, there remains a need to validate the techniques when used in subjects with pathologic changes, such as with epilepsy.
Aside from the limitations of semi-automated or automated segmentation techniques in yielding accurate hippocampal segmentations in a specific imaging environment, the issue of the generalizability or applicability of each technique, i.e. the reproducibility of the reported estimates across different acquisition parameters or imaging platforms, is also of significant concern[28]. Past studies have typically validated techniques in healthy subjects[29], typically with differences in hippocampal anatomic boundaries chosen or segmentation techniques[30]. Such differences have produced a wide range of “normal” hippocampal volumes across studies[31]. The ability to accurately quantify hippocampal volume and surface anatomy using different T1-weighted volumetric image acquisition protocols[14] or in different MRI scanners[3, 32, 33], is an issue of critical interest for patients with epilepsy. Our study design compared two T1-weighted sequences with different TR/TE/TI values, different planes of acquisition and different voxel size and shape (isometric vs. anisometric), allowing a qualitative assessment of the robustness of our volume-estimation technique across key factors such as differences in gray-white contrast, spatial resolution and voxel properties. Of note, acquiring both MRI sequences on the same MRI scanner during the same imaging session focused our inquiry on differences at the sequence level rather than at the scanner level. However, our study design does not address across-scanner factors such as scanner-specific intensity inhomogeneity or geometric distortion, which could negatively affect the segmentation accuracy. Also, while there were differences in the TR/TE/TI parameters of our two acquisition protocols, many T1-weighted volumetric acquisition protocols exist that use different parameter variations. Our results therefore do not formally address the effects of specific parameters on hippocampal volume estimation, especially when considering MRI studies acquiring data on multiple MRI scanners. Rather the main goal of our study was the direct comparison of two T1-weighted sequences that are typically used for imaging of patients and healthy control subjects and have significant differences in multiple imaging parameters. Additional studies will be needed to more formally and systematically address the effect of specific imaging parameters (e.g. voxel size) on hippocampal volume estimates.
In the current study we addressed the issue of accuracy of hippocampal segmentation and volume estimation using different MRI acquisition sequences during the same imaging session. Most past validation studies (as discussed above) have used different segmentation techniques on identical data sets to determine the accuracy of the involved algorithm[15, 17, 18]. Our study instead assessed the variability introduced by different T1-weighted MRI sequences on the same HDM-LD hippocampal segmentation technique. We evaluated both hippocampal volume and hippocampal surface estimates obtained from HDM-LD segmentations of the two types of T1-weighted MRI images. Mean volumes of both the left hippocampus and right hippocampus, as presented in Figure 2, showed no significant difference between the MPR1 and MPR2 imaging sequences. Interestingly, as highlighted in Figure 3, hippocampal size did not qualitatively appear to affect the variability in volume estimates between MRI sequences. The degree of volume differences between the groups was well within the volume differences reported in previous studies that used manual hippocampal segmentation[29]. For instance, mean percentage difference in hippocampal volumes using HDM-LD was 4.3% (SD 2.7%) in previous studies that compared sequential HDM-LD segmentations on the same MRI image[14]. In the current study, the range of volume differences were 0.01-4.2% (mean 1.6%, SD 1.4%) for the left hippocampus and 1.5-11.3% (mean 3.6%, SD 3.1%) for the right hippocampus. Also of notable interest, there was significant reproducibility of hippocampal volume estimates at the individual level, as exemplified in Figure 3, of potential interest for clinical settings where individualized assessment of hippocampal size is important.
In addition to comparing hippocampal volumes between MRI imaging sequences, we used HDM-LD segmentation to assess surface-level changes between MRI sequences. Statistical comparisons of vertex-based surface estimates are complicated by the large number of vertices that compose an estimated surface, especially for a complex structure such as the human hippocampus, making correction for multiple comparisons paramount when estimating differences between groups. Our empiric technique uses an assumption-free iterative randomization approach, inspired by similar work in volumetric noise estimation in functional magnetic resonance imaging[20], to produce a cluster-based noise estimation model using an independent set of hippocampal deformations from healthy control subjects. This estimates the probability of occurrence of suprathreshold clusters in the normal population yielding cluster-based cutoffs at different significance levels. Notably, no clusters of significant surface displacement between MRI sequences were found using this technique, suggesting that the changes in local surface estimates produced by different MRI sequences are likely small. Effectively, this comparison demonstrates that MRI-dependent surface changes are significantly less than changes observed in normal subjects, suggesting that hippocampal surface deformation differences produced from different MRI acquisition sequences would not be a significant factor when comparing groups of normal or diseased subjects. The validity of our findings is especially compelling given that our test subjects all had a history of epilepsy, which typically induces greater variability in hippocampal surface structure. As illustrated in Figure 4, relatively greater displacement is present in the region of the uncinate gyrus, as well as in the medial aspect of the hippocampal tail. Previous studies have shown that segmentations tend to show the most variability in these regions, likely because of local lack of gray-white differentiation and distinct anatomical hippocampal boundaries[14].
In addition to considerations regarding the effect of imaging parameters, actual hippocampal pathology itself may be responsible for local shape changes that may affect the accuracy of a segmentation and estimation technique. Kim and colleagues, for instance, showed that hippocampi of patients with malformations of cortical development differ from hippocampi of patients with TLE in terms of the extent and distribution of local shape changes [34]. Similarly, hippocampal malrotation specifically has been shown to have an effect on the success of automated segmentation and surface estimation algorithms [15]. While we did not formally estimate long-axis rotation or curvature in our data set, visual inspection did not reveal obvious examples of malrotation, even for the patients with significant atrophy. We cannot however exclude a potential effect of malrotation on our segmentation, volume and surface estimation technique when comparing patients with and without malrotation directly in a larger sample. Another source of potential variation is mere hippocampal atrophy, i.e. more atrophic hippocampi may be more difficult to segment via an automated or semi-automated method. In our study there was a positive correlation between mean hippocampal volume and Dice coefficient (Figure 5), suggesting that the HDM-LD technique is more accurate for less atrophic hippocampi (though it should be noted that the range of Dice coefficients was not very wide, suggesting that this effect is probably small). Of note, the reliability of volume estimates across imaging sequences remained quite good even for more atrophic hippocampi. Even though our sample size of subjects is relatively small, the reproducibility and reliability of findings between subjects is robust. The number of subjects included in this analysis is similar to those in previous validation studies of hippocampal segmentations [14, 18, 26].
At our institution, MPR1 is typically a sequence used for clinical studies, while MPR2 is typically a research sequence. Differences in sequences, including anisometric (MPR1) and isometric (MPR2) voxels, coronal (MPR1) and sagittal (MPR2) plane of acquisition, and different TR/TE/TI intervals, could all conceivably introduce variability in deformations. Other investigators have addressed differences in gray matter intensity characteristics[14, 18, 26], or have normalized intensities in their algorithm for hippocampal segmentation[27]. Prior to HDM-LD segmentation, our technique included a global normalization of each MRI image to the same intensity range by matching high and low-percentile intensity values across the two sequences, without specifically correcting for local signal intensity range differences within the hippocampus compared to extra-hippocampal structures. Overall, our findings indicate that factors of voxel size/shape, plane of acquisition, and contrast properties do not significantly affect the HDM-LD algorithm, in terms of both hippocampal volume and surface estimation, in subjects undergoing scans in the same MRI scanner.
In summary, we evaluated the reproducibility of HDM-LD segmentation hippocampal volume and surface estimates in subjects with epilepsy, specifically testing the invariance of the technique across two T1-weighted volumetric imaging sequences. Statistical comparison of hippocampal volume estimates and of statistically-thresholded hippocampal surfaces showed no differences between MRI sequences. This suggests that the HDM-LD segmentation technique can provide robust estimates of both hippocampal volume and hippocampal surface in patients with hippocampal pathology, with reliability across imaging conditions.
Acknowledgments
This work was supported by the National Center for Advancing Translational Sciences [UL1TR000448, sub award KL2TR000450], and the Institute of Clinical and Translational Sciences at Washington University [UL1RR024992].
Reference List
- 1.Jack CR, Barkhof F, Bernstein MA, Cantillon M, Cole PE, DeCarli C, et al. Steps to standardization and validation of hippocampal volumetry as a biomarker in clinical trials and diagnostic criterion for Alzheimer's disease. Alzheimers & Dementia11. 2011;7:474–85. doi: 10.1016/j.jalz.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hogan RE, Wang L, Bertrand ME, Willmore LJ, Bucholz RD, Nassif AS, et al. MRI-based high-dimensional hippocampal mapping in mesial temporal lobe epilepsy. Brain. 2004;127:1731–40. doi: 10.1093/brain/awh197. [DOI] [PubMed] [Google Scholar]
- 3.Hogan RE, Carne RP, Kilpatrick CJ, Cook MJ, Patel A, King L, et al. Hippocampal deformation mapping in MRI negative PET positive temporal lobe epilepsy. J Neurol Neurosurg Psychiatry. 2008;79:636–40. doi: 10.1136/jnnp.2007.123406. [DOI] [PubMed] [Google Scholar]
- 4.Csernansky JG, Joshi S, Wang L, Haller JW, Gado M, Miller JP, et al. Hippocampal morphometry in schizophrenia by high dimensional brain mapping. Proc Natl Acad Sci USA. 1998;95:11406–11. doi: 10.1073/pnas.95.19.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lencz T, McCarthy G, Bronen RA, Scott TM, Inserni JA, Sass KJ, et al. Quantitative magnetic resonance imaging in temporal lobe epilepsy: relationship to neuropathology and neuropsychological function. Ann Neurol. 1992;31:629–37. doi: 10.1002/ana.410310610. [DOI] [PubMed] [Google Scholar]
- 6.Cascino GD. Clinical correlations with hippocampal atrophy. Magn Reson Imaging. 1995;13:1133–6. doi: 10.1016/0730-725x(95)02023-m. [DOI] [PubMed] [Google Scholar]
- 7.Jack CR, Jr, Sharbrough FW, Cascino GD, Hirschorn KA, O'Brien PC, Marsh WR. Magnetic resonance image-based hippocampal volumetry: correlation with outcome after temporal lobectomy. Ann Neurol. 1992;31:138–46. doi: 10.1002/ana.410310204. [DOI] [PubMed] [Google Scholar]
- 8.Arruda F, Cendes F, Andermann F, Dubeau F, Villemure JG, Jones-Gotman M, et al. Mesial atrophy and outcome after amygdalohippocampectomy or temporal lobe removal. Ann Neurol. 1996;40:446–50. doi: 10.1002/ana.410400314. [DOI] [PubMed] [Google Scholar]
- 9.Cascino GD. Clinical evaluation and noninvasive electroencephalography. Preoperative evaluation. Neuroimaging Clin N Am. 1995;5:547–58. [PubMed] [Google Scholar]
- 10.Farid N, Girard HM, Kemmotsu N, Smith ME, Magda SW, Lim WY, et al. Temporal Lobe Epilepsy: Quantitative MR Volumetry in Detection of Hippocampal Atrophy. Radiology. 2012;264:542–50. doi: 10.1148/radiol.12112638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen GE, Rabbitt RD, Miller MI. In: Prince J, editor. A deformable neuroanatomy textbook based on viscous fluid mechanics; Proceedings of the 27th Annual Conference on Information Sciences and Systems; Baltimore. The Johns Hopkins University; 1993. pp. 211–6. [Google Scholar]
- 12.Miller MI, Grenander U. Computational anatomy: an emerging discipline. Q Appl Math. 1998;5:617–94. [Google Scholar]
- 13.Miller MI, Christensen GE, Amit Y, Gernander U. Mathematical textbook for deformable neuroanatomies. Proc Natl Acad Sci USA. 1993;90:11944–8. doi: 10.1073/pnas.90.24.11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogan RE, Mark KE, Wang L, Joshi S, Miller MI, Bucholz RD. Mesial temporal sclerosis and temporal lobe epilepsy: MR imaging deformation-based segmentation of the hippocampus in five patients. Radiology. 2000;216:291–7. doi: 10.1148/radiology.216.1.r00jl41291. [DOI] [PubMed] [Google Scholar]
- 15.Kim H, Chupin M, Colliot O, Bernhardt BC, Bernasconi N, Bernasconi A. Automatic hippocampal segmentation in temporal lobe epilepsy: impact of developmental abnormalities. Neuroimage. 2012;59:3178–86. doi: 10.1016/j.neuroimage.2011.11.040. [DOI] [PubMed] [Google Scholar]
- 16.Han X, Fischl B. Atlas renormalization for improved brain MR image segmentation across scanner platforms. IEEE Trans Med Imaging. 2007;26:479–86. doi: 10.1109/TMI.2007.893282. [DOI] [PubMed] [Google Scholar]
- 17.Khan AR, Wang L, Beg MF. FreeSurfer-initiated fully-automated subcortical brain segmentation in MRI using Large Deformation Diffeomorphic Metric Mapping. Neuroimage. 2008;41:735–46. doi: 10.1016/j.neuroimage.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chupin M, Hammers A, Liu RS, Colliot O, Burdett J, Bardinet E, et al. Automatic segmentation of the hippocampus and the amygdala driven by hybrid constraints: method and validation. Neuroimage. 2009;46:749–61. doi: 10.1016/j.neuroimage.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hogan RE, Bucholz RD, Joshi S. Hippocampal deformation-based shape analysis in epilepsy and unilateral mesial temporal sclerosis. Epilepsia. 2003;44:800–6. doi: 10.1046/j.1528-1157.2003.45902.x. [DOI] [PubMed] [Google Scholar]
- 20.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage. 2006;33:115–26. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 22.Barnes J, Foster J, Boyes RG, Pepple T, Moore EK, Schott JM, et al. A comparison of methods for the automated calculation of volumes and atrophy rates in the hippocampus. Neuroimage. 2008;40:1655–71. doi: 10.1016/j.neuroimage.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Pohl KM, Bouix S, Nakamura M, Rohlfing T, McCarley RW, Kikinis R, et al. A hierarchical algorithm for MR brain image parcellation. IEEE Trans Med Imaging. 2007;26:1201–12. doi: 10.1109/TMI.2007.901433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartzokis G, Altshuler LL, Greider T, Curran J, Keen B, Dixon WJ. Reliability of medial temporal lobe volume measurements using reformatted 3D images. Psychiatry Research. 1998;82:11–24. doi: 10.1016/s0925-4927(98)00007-9. [DOI] [PubMed] [Google Scholar]
- 25.Goodkin DE, Ross JS, Medendorp SV, Konecsni J, Rudick RA. Magnetic resonance imaging lesion enlargement in multiple sclerosis. Disease-related activity, chance occurrence, or measurement artifact? Arch Neurol. 1992;49:261–3. doi: 10.1001/archneur.1992.00530270075021. [DOI] [PubMed] [Google Scholar]
- 26.Haller JW, Banerjee A, Christensen GE, Gado M, Joshi S, Miller MI, et al. Three-dimensional hippocampal MR morphometry with high-dimensional transformation of a neuroanatomic atlas. Radiology. 1997;202:504–10. doi: 10.1148/radiology.202.2.9015081. [DOI] [PubMed] [Google Scholar]
- 27.Kim H, Mansi T, Bernasconi N, Bernasconi A. Surface-based multi-template automated hippocampal segmentation: Application to temporal lobe epilepsy. Medical Image Analysis. 2012;16:1445–55. doi: 10.1016/j.media.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Hasboun D, Chantome M, Zouaoui A, Sahel M, Deladoeuille M, Sourour N, et al. MR determination of hippocampal volume: comparison of three methods. AJNR Am J Neuroradiol. 1996;17:1091–8. [PMC free article] [PubMed] [Google Scholar]
- 29.Jack CR, Jr, Bentley MD, Twomey CK, Zinsmeister AR. MR imaging-based volume measurements of the hippocampal formation and anterior temporal lobe: validation studies. Radiology. 1990;176:205–9. doi: 10.1148/radiology.176.1.2353093. [DOI] [PubMed] [Google Scholar]
- 30.Watson C, Jack CR, Jr, Cendes F. Volumetric magnetic resonance imaging. Clinical applications and contributions to the understanding of temporal lobe epilepsy. Arch Neurol. 1997;54:1521–31. doi: 10.1001/archneur.1997.00550240071015. [DOI] [PubMed] [Google Scholar]
- 31.Jack CR, Theodore WH, Cook M, McCarthy G. MRI-based hippocampal volumetrics: data acquision, normal ranges, and optimal protocol. Magn Reson Imaging. 1995;13:1057–64. doi: 10.1016/0730-725x(95)02013-j. [DOI] [PubMed] [Google Scholar]
- 32.Ogren JA, Wilson CL, Bragin A, Lin JJ, Salamon N, Dutton RA, et al. Three-dimensional surface maps link local atrophy and fast ripples in human epileptic hippocampus. Ann Neurol. 2009;66:783–91. doi: 10.1002/ana.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogren JA, Bragin A, Wilson CL, Hoftman GD, Lin JJ, Dutton RA, et al. Three-dimensional hippocampal atrophy maps distinguish two common temporal lobe seizure-onset patterns. Epilepsia. 2009;50:1361–70. doi: 10.1111/j.1528-1167.2008.01881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H, Mansi T, Bernasconi N. Disentangling hippocampal shape anomalies in epilepsy. Frontiers in Neurology. 2013;4 doi: 10.3389/fneur.2013.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]


