Abstract
Purpose
To determine if neurochemical concentrations obtained at two MRI sites using clinical 3 T scanners can be pooled when a highly optimized, non-vendor short-echo, single voxel proton MRS pulse sequence is utilized in conjunction with identical calibration and quantification procedures.
Methods
A modified semi-LASER sequence (TE = 28 ms) was used to acquire spectra from two brain regions (cerebellar vermis and pons) on two Siemens 3 T scanners using the same B0 and B1 calibration protocols from two different cohorts of healthy volunteers (N=24–33 per site) matched for age and BMI. Spectra were quantified with LCModel using water scaling.
Results
The spectral quality was very consistent between the two sites and allowed reliable quantification of at least 13 metabolites in the vermis and pons compared to 3 – 5 metabolites in prior multi-site MRS trials using vendor-provided sequences. The neurochemical profiles were nearly identical at the two sites and showed the feasibility to detect inter-individual differences in the healthy brain.
Conclusion
Highly reproducible neurochemical profiles can be obtained on different clinical 3 T scanners at different sites, provided that the same, optimized acquisition and analysis techniques are utilized. This will allow pooling of multi-site data in clinical studies, which is particularly critical for rare neurological diseases.
Keywords: 3 Tesla, multi-site, reproducibility, spectroscopy
INTRODUCTION
Proton MR spectroscopy (1H MRS) is a non-invasive tool that allows the measurement of a wide range of biochemical compounds in the brain in both health and disease. The method has been demonstrated to be valuable in the evaluation of several common disorders of the central nervous system, including tumors, neonatal hypoxia-ischemia, inherited metabolic diseases, demyelinating disorders and infectious brain lesions (1). Because metabolic alterations are thought to precede structural changes, MRS is likely to provide dynamic biomarkers of neuronal dysfunction at an earlier stage of disease progression than structural MRI. This is of even greater importance for therapeutic approaches for which a neuroprotective effect is expected. However, unlike structural MRI, MRS has not gained widespread acceptance as a routine clinical tool for diagnostic and prognostic purposes (2,3). This is partially due to the relatively large variation in metabolite concentrations or ratios reported from different sites (4–9).
As higher magnetic fields are becoming routinely available, the increased sensitivity and resolution provided by them can highly benefit metabolite quantification (10–12) and facilitate robust clinical applications of the technique. However, with this potential, the need to standardize robust MRS acquisition and analysis methods is critical, as also emphasized by a recent MRS Consensus effort (1). Such standardized MRS methodology would allow pooling of data from multiple sites, which is particularly important for clinical research and clinical trials in rare diseases. Prior multi-site MRS trials primarily have utilized vendor-provided MRS sequences (PRESS and STEAM) and were able to quantify 3 to 5 metabolites, such as N-acetylaspartate (NAA), total creatine (tCr), total choline (tCho) and myo-inositol (Ins) at both 1.5T and 3T (6–9).
Recently, optimized short-echo sequences such as SPECIAL (12) and semi-LASER (13) were implemented on clinical platforms thereby allowing an extended neurochemical profile consisting of both singlet and J-coupled metabolites to be measured. Therefore there is a need to determine the between-site reproducibility of such profiles.
The aim of the present study was to examine the reproducibility of metabolite concentrations measured in two brain regions using a short-echo, single-shot, full-intensity sequence with identical experimental protocols at two different sites on clinical 3T scanners. A previously described semi-LASER sequence (14) was utilized to achieve lower apparent T2 relaxation, minimal J-coupling evolution and smaller chemical shift displacement errors relative to the standard PRESS sequence. Two relatively challenging brain regions were chosen for this two-site comparison: cerebellar vermis and pons (15).
METHODS
Two 3T whole-body Siemens Tim Trio (Siemens Medical Solutions, Erlangen, Germany) scanners were used in this study; one located at the Center for Magnetic Resonance Research (CMRR) in Minnesota and the other one at the Institut du Cerveau et de la Moelle (ICM) in Paris. Age- and BMI-matched healthy subjects (Table 1) were enrolled after giving informed consent according to procedures approved by the Institutional Review Board at CMRR and by the local ethics committee at ICM. The standard body RF coil was used for radiofrequency transmission and the 32-channel phased-array Siemens head coil was used for signal reception. Soft pads were used to hold each subject’s head in place to minimize head movement in the MR system. T1-weighted MPRAGE images (repetition time (TR) = 2530ms, echo time (TE) = 3.65 ms, flip angle = 7°, slice thickness = 1 mm, 224 slices, field-of-view = 256×176 mm2, matrix size = 256×256) were acquired to position the volume-of-interest (VOI) for MRS measurements. B0 shimming was achieved using an adiabatic version of FAST(EST)MAP (16), which is available as a work-in-progress (WIP) package on the Siemens system.
Table 1.
Demographics and spectroscopic parameters measured in two brain regions (values are given as mean ± SD).
| CMRR | ICM | P-values* | |
|---|---|---|---|
| Subjects scanned (N) | 24 | 33 | |
| Gender (male/female) | 13/11 | 15/18 | 0.52† |
| Age (years) | 53 ± 15 | 48 ± 13 | 0.19 |
| BMI (Kg/m2) | 26 ± 6 | 25 ± 4 | 0.47 |
| Vermis | N= 24 | N= 33 | |
| Water linewidth (Hz) | 8 ± 1 | 7 ± 1 | 0.06 |
| T2 tissue water (ms) | 80 ± 8 | 80 ± 7 | 0.93 |
| CSF fraction (%) | 11 ± 5 | 9 ± 4 | 0.04 |
| SNR of NAA‡ | 56 ± 5 | 59 ± 9 | 0.21 |
| Pons | N= 16 | N= 23 | |
| Water linewidth (Hz) | 8 ± 1 | 8 ± 1 | 0.57 |
| T2 tissue water (ms) | 67± 4 | 68 ±2 | 0.34 |
| CSF fraction (%) | 1 ±1 | 2 ±2 | 0.16 |
| SNR of NAA‡ | 27 ± 5 | 28 ± 6 | 0.49 |
SNR was measured in the frequency domain and no apodization functions were applied to the data.
unpaired, two-tailed student’s t-test except when noted otherwise.
Chi-squared test.
Proton spectra were acquired using a modified semi-LASER sequence (TE = 28 ms, TR = 5 s, 64 averages) (14) from two VOIs: cerebellar vermis (10×25×25 mm3) and pons (16×16×16 mm3). Voxel placement was based on anatomical landmarks. The fourth ventricle, cervical spinal cord and the brainstem were used to separate the cerebellum. The surfaces, lobes, lobules and fissures of the cerebellum were then used as landmarks in positioning the voxel in the vermis. For pons VOI placement, the midbrain, fourth ventricle and the medulla were used as landmarks.
The semi-LASER sequence (14) used in this study is a more compact version of the originally published semi-LASER sequence (17). Briefly, the sequence consisted of a 2 ms asymmetric slice-selective 90° pulse (18) followed by two pairs of slice selective adiabatic full passage (AFP) pulses (4 ms duration, HS4 modulation, R25) (19), which were interleaved, rather than applied sequentially, to improve suppression of unwanted coherences with shorter spoiler gradient pulses. Water suppression was achieved with VAPOR, which was interleaved with outer volume suppression (OVS) to suppress unwanted coherences (18). A substantially lower chemical shift displacement error is obtained with the semi-LASER sequence (3.6% /ppm for the slice-selective 90° pulse and 2% /ppm for the AFP pulses) compared to the standard PRESS sequence provided on the Siemens platform (12–13% /ppm).
B1 levels required for localization pulses and for water suppression were adjusted for each voxel. Specifically, the RF power magnitude for the 90° asymmetric pulse was calibrated by monitoring the signal intensity whilst increasing the RF power and choosing the RF power setting that produced the maximum signal. The power for the AFP pulses was automatically set relative to the 90° pulse. A similar procedure was carried out for the water suppression calibration.
On the scanner, signals from individual coil elements were combined after correcting for phase shifts between elements and weighting them based on the coil sensitivities (20) to generate a free induction decay (FID). Each FID was then individually saved for shot-to-shot frequency and phase correction before averaging. Two non-suppressed water spectra were acquired: one for eddy current correction (the RF pulses of the VAPOR scheme were turned off) and one for use as reference for metabolite quantification (VAPOR and OVS schemes turned off in order to eliminate magnetization transfer effects). To evaluate the cerebrospinal fluid (CSF) contribution to each VOI, fully relaxed unsuppressed water signals were acquired at different TE’s ranging from 28–4000 ms (TR = 15 s) with the entire VAPOR and OVS scheme turned off (21).
All spectral processing was performed in Matlab by the same person prior to LCModel fitting. Eddy current correction was carried out first to correct for distorted line shapes and zero-order phase. Individual shots affected by subject motion (based on water suppression efficiency) were removed. Single-shot frequency correction was performed using a cross-correlation algorithm and phase correction was performed using a least-square fit algorithm. All steps were completely automated except for the removal of FIDs affected by motion. Finally the summed spectrum was referenced based on NAA resonance at 2.01 ppm.
Spectra were then analyzed with LCModel (22) with the water scaling option (version 6.3-0G). The model basis set was generated based on density matrix formalism as described before (23). The basis set also included macromolecule spectra, which were acquired using inversion-recovery technique in 4 healthy subjects (total averages = 928, TR = 2.5 s, inversion time, TI = 0.75 s, VOI = 15.6 mL, 5 ms duration HS5 inversion pulse, occipital cortex). Due to the shorter T1 relaxation time of the methylene protons of tCr at 3.93 ppm relative to other metabolite protons (24), this resonance was present in the metabolite-nulled macromolecule spectra and was removed using a Hankel singular value decomposition (HSVD) algorithm in Matlab. A 12.5 Hz Gaussian line broadening was also applied to the macromolecule spectra after incorporating a reference peak at 0 ppm (see supplementary material). No baseline correction, zero-filling or apodization functions were applied to the in vivo data prior to the analysis. LCModel fitting (supplementary material) was performed over the spectral range from 0.5–4.2 ppm.
Metabolite concentrations were determined after correcting for tissue water content and CSF contributions in the selected VOI using the water-scaling option in LCModel. The transverse relaxation times (T2) of tissue water and % CSF contribution to the VOI were obtained by fitting the integrals of the unsuppressed water spectra acquired in each VOI at different TE values with a bi-exponential fit (21), with the T2 of CSF fixed at 740 ms based on measurement of T2 of water in a small voxel located in ventricles with the same semi-LASER sequence (4 healthy subjects, TR = 15 s, VOI = 0.125–0.360 mL, twelve TE values ranging from 28–4000 ms), and three free parameters: T2 of tissue water, amplitude of tissue water, and amplitude of CSF water.
In order to obtain accurate metabolite concentrations, corrections must be made for T2 relaxation of both water and metabolites. In the case of semi-LASER, T2 relaxation is slowed due to the Carr-Purcell (CP) conditions, and T2 values under CP conditions must be used for quantification. For water, these values can be estimated by correcting the free precession T2 value measured for the tissue water signal at different echo-times by a fixed factor to account for CP effects. A previous study compared water T2 values measured with LASER and CP-LASER sequences at 4T and 7T (25). Extrapolating from that study, we assumed that the T2 of water under CP conditions is 1.5× longer than the measured free precession T2 at 3T. Signal loss due to T2 relaxation of metabolites was neglected since the apparent T2 is sequence-dependent. This assumption is justified by the fact that metabolites have longer T2 such that correction factors would be small at TE = 28 ms. Nonetheless this choice will result in somewhat underestimated metabolite concentrations relative to the true concentrations in tissue. A water content of 82% and 72% was used for vermis and pons, respectively (26,27).
Metabolites that were quantified with Cramér-Rao lower bounds (CRLB) ≤ 50% from at least half of the spectra from a particular brain region were included in the neurochemical profile. In addition, if the correlation between two metabolites was very high (i.e. correlation coefficient r more negative than −0.7) in the majority of the spectra from a region, then only their sum was reported, e.g. tCr (creatine + phosphocreatine) and tCho (glycerophosphorylcholine + phosphorylcholine). If there was indication for pairwise correlation with r from −0.5 to −0.7, then the concentration sum of the pair was reported in addition to the individual metabolites’ concentrations, e.g. NAA, NAAG and total NAA (tNAA, NAA + NAAG), as recommended by the LCModel manual, Jan 2013 (22). Moreover, spectra with the associated water reference linewidth greater than 10 Hz were excluded due to trends observed in overestimating aspartate and ascorbate and underestimating glutamate in these spectra. Water linewidths > 10 Hz only occurred for spectra acquired from the pons region.
RESULTS
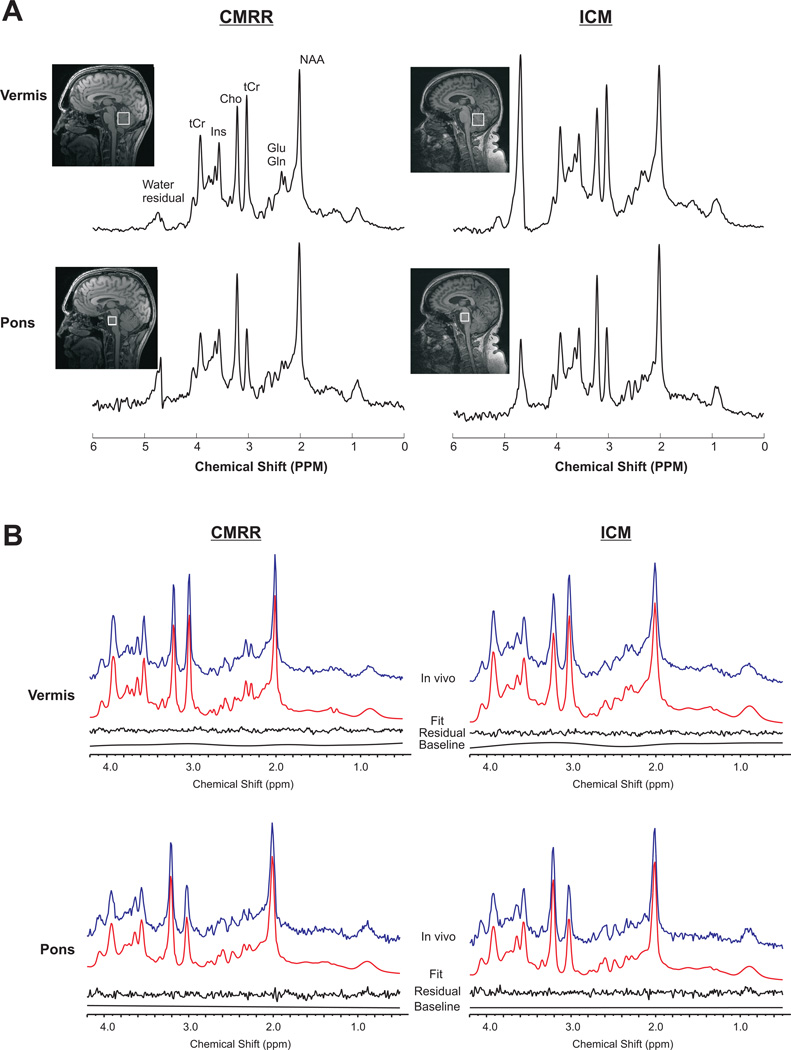
Using the modified semi-LASER sequence with identical parameters and identical B0 and B1 calibration protocols on two 3T scanners, consistently high quality 1H spectra with comparable peak signal-to-noise ratio (SNR) were obtained at both sites (Figure 1A, Table 1). No noticeable baseline distortions due to insufficient water suppression or contamination by signals from outside the voxel, such as out-of-phase lipids, were observed. The peak SNR in the pons was lower than in the vermis due to the smaller voxel size and lower sensitivity of the receive coil in this particularly deep brain region. Examples of LCModel fits obtained in both regions are illustrated in Figure 1B. No obvious residual was observed around 0.9–2ppm region suggesting that the macromolecule spectrum acquired from the occipital cortex is appropriate when fitting spectra from vermis and pons. This finding is consistent with a recent study (28) which showed that the differences in macromolecule signal between gray and white matter regions are relatively small and concluded that a general macromolecule baseline provides sufficiently accurate neurochemical profiles.
Figure 1.
A) Typical proton spectra obtained from the cerebellar vermis and pons in two different subjects at the two sites using semi-LASER (TE = 28 ms, TR = 5 s, 64 averages) at 3 T. The locations of the VOI are shown on the T1-weighted images. Spectra were processed with a 1 Hz exponential decay and 5 Hz Gaussian functions. Comparable spectral quality and pattern are apparent for each region at both sites. B) LCModel fits of the spectra shown in A without any apodization functions. From top to bottom: the in vivo spectrum, the fit, the residual after subtracting the fit from the in vivo spectrum and the baseline.
At CMRR, pons data were not collected from 3 subjects due to poor B0 shimming in two cases and subject movement in one case and 5 spectra were rejected due to the broad linewidth criteria. Similarly at ICM, pons data from 8 subjects were not acquired due to poor shimming and 2 spectra were rejected due to broad water linewidth. This was consistent with the know challenges with shimming in the brainstem (29) due to the presence of the sphenoid sinus. All spectra from the vermis were used in the final results (Table 1). Motion effects were minimal in this cohort of healthy volunteers and only one single-shot in the vermis (out of 64) was excluded from the sum in one subject due to motion.
No statistical differences were observed in the spectral quality metrics, i.e. water peak linewidth and SNR, as well as tissue water T2 values between the two sites in both brain regions (Table 1). CSF fraction was found to be slightly higher in the vermis VOI from CMRR relative to ICM (P = 0.04). As expected, the T2 of tissue water in pons was shorter compared to that in vermis consistent with the fact that pons consists mainly of white matter (30).
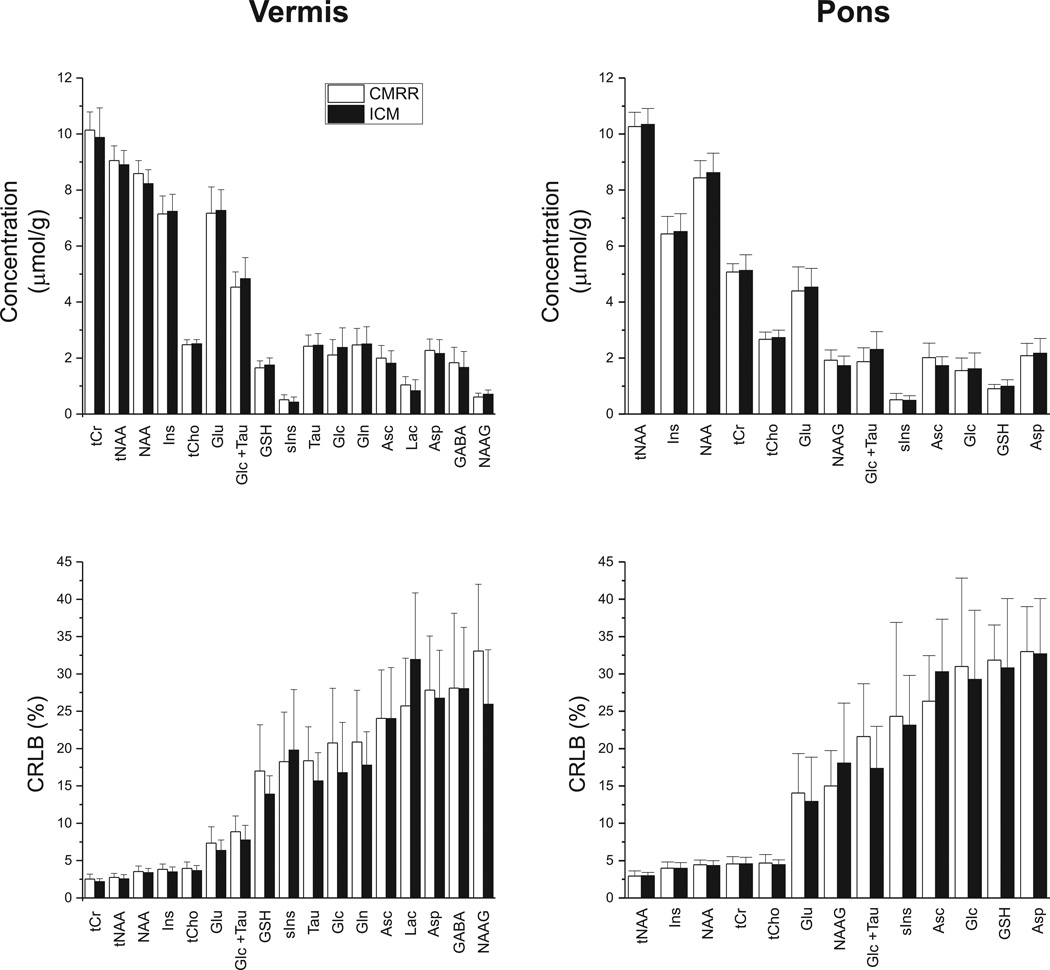
Using the unsuppressed water signal as an internal concentration reference and after correcting for T2 relaxation of water, tissue water and CSF contributions, the concentrations of 17 metabolites were determined in the vermis using the reliability criteria described in the Methods. Due to lower SNR and higher cross-correlation between metabolites in the pons compared to the vermis (supplementary material), 13 concentrations passed the same criteria in the pons (Figure 2).
Figure 2.
Mean metabolite concentrations (in µmol/g) and CRLB (in %) measured in the cerebellar vermis (N = 24 at CMRR, N = 33 at ICM) and pons (N =16 at CMRR, N = 23 at ICM) at the two sites. Error bars represent inter-subject SD. tNAA: total N-acetylaspartate, tCr: total creatine, tCho: total choline, Ins: myo-inositol, Glu: glutamate, Glc: glucose, Tau: taurine, GSH: glutathione, sIns: scyllo-inositol, Asc: ascorbate, Asp: aspartate, GABA: γ-aminobutyric acid, Lac: lactate, Gln: glutamine, NAAG: N-acetylaspartylglutamate.
The neurochemical profiles of the two brain regions were nearly identical between the two sites (Figure 2, Table 2). A comparison of the metabolite concentrations from ICM and CMRR in each brain region (i.e. vermis and pons) revealed no statistically significant difference between the two sites using the analysis of variance (ANOVA). Similarly, no differences were observed in the metabolite quantification precision (as determined by CRLB) between the sites (Figure 2), as expected based on the similar spectral quality (Table 1). In the vermis, the mean CRLB for all singlets (NAA, tNAA, tCr and tCho), Ins, Glu and Glc + Tau was less than 10% with the other metabolites having mean CRLBs less than 35%. In the pons, NAA, tNAA, tCr, tCho and Ins were quantified with mean CRLB less than 5% with other metabolites having mean CRLBs smaller than 35%.
Table 2.
Mean ± SD metabolite concentrations (in µmol/g) measured in the cerebellar vermis and pons at the two sites.
| Metabolite | Vermis CMRR |
ICM | Pons CMRR |
ICM |
|---|---|---|---|---|
| tCr | 10.1 ± 0.7 | 9.9 ± 1.1 | 5.1 ± 0.3 | 5.1 ± 0.6 |
| tNAA | 9.1 ± 0.5 | 8.9 ± 0.5 | 10.3 ± 0.5 | 10.3 ± 0.6 |
| NAA | 8.6 ± 0.5 | 8.2 ± 0.5 | 8.4 ± 0.6 | 8.6 ± 0.7 |
| Ins | 7.1 ± 0.6 | 7.2 ± 0.6 | 6.4 ± 0.6 | 6.5 ± 0.6 |
| tCho | 2.5 ± 0.2 | 2.5 ± 0.1 | 2.7 ± 0.3 | 2.7 ± 0.3 |
| Glu | 7.2 ± 0.9 | 7.3 ± 0.7 | 4.4 ± 0.9 | 4.5 ± 0.7 |
| Glc+Tau | 4.5 ± 0.5 | 4.8 ± 0.8 | 1.9 ± 0.5 | 2.3 ± 0.6 |
| GSH | 1.6 ± 0.3 | 1.7 ± 0.3 | 0.9 ± 0.2 | 1.0 ± 0.2 |
| sIns | 0.5 ± 0.2 | 0.4 ± 0.2 | 0.5 ± 0.2 | 0.5 ± 0.2 |
| Tau | 2.4 ± 0.4 | 2.5 ± 0.4 | ||
| Glc | 2.1 ± 0.6 | 2.4 ± 0.7 | 1.5 ± 0.5 | 1.6 ± 0.6 |
| Gln | 2.5 ± 0.6 | 2.5 ± 0.6 | ||
| Asc | 2.0 ± 0.5 | 1.8 ± 0.4 | 2.0 ± 0.5 | 1.7 ± 0.3 |
| Lac | 1.0 ± 0.3 | 0.8 ± 0.4 | ||
| Asp | 2.3 ±0.4 | 2.2 ± 0.5 | 2.1 ± 0.4 | 2.2 ± 0.5 |
| GABA | 1.8 ± 0.5 | 1.7 ± 0.6 | ||
| NAAG | 0.6 ± 0.1 | 0.7 ± 0.2 | 1.9 ± 0.4 | 1.9 ± 0.4 |
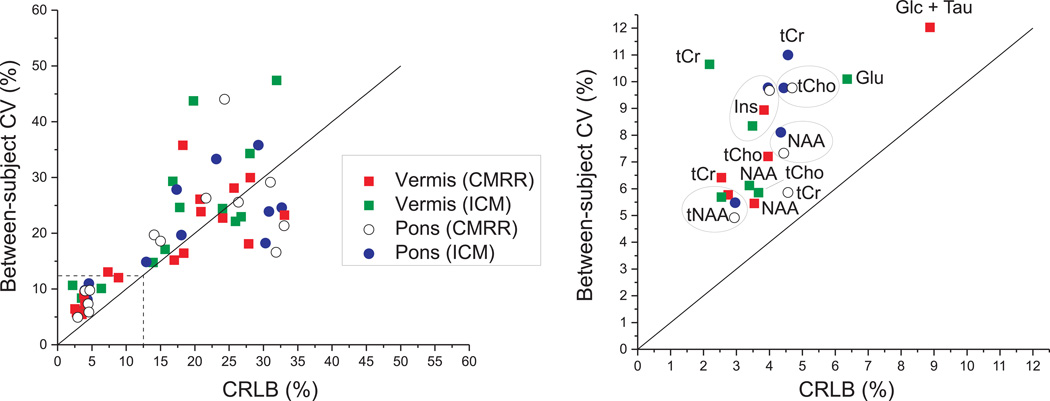
When the relationship between the mean CRLB and between-subject coefficients-of-variance (CV, SD/mean) was investigated for each brain region and each site (Figure 3), the CRLBs were consistently lower than between-subject CV for the most reliably quantified metabolites (CRLB <12%). For example, the between-subject CVs for tCr, tNAA, tCho, NAA and Ins were in the range of 6–12% in both vermis and pons, while their CRLBs were in the range of 2–5%, indicating that the method has precision to detect inter-individual differences in these metabolites in the healthy brain (Figure 3, right). For other metabolites, between-subject CVs and CRLBs were more comparable (metabolites that fall around the identity line in Figure 3), indicating the measurement errors were comparable to or higher than physiological, inter-individual differences.
Figure 3.
Relationship between mean CRLB and between-subject coefficients of variance (CV) for all metabolites reported in Figure 2: CV and CRLB ≤ 50% (left) and zoomed CV and CRLB between 0 and 12.5% (right). The solid line represents the identity line.
DISCUSSION
This study demonstrates that nearly identical neurochemical profiles consisting of 13–17 metabolites are obtained in two different brain regions in relatively large healthy cohorts by different operators at two MR sites. The acquisition of high quality MRS data from the cerebellum and brainstem is particularly challenging due to their caudal location in the head and broader intrinsic linewidths relative to other cerebral VOI (14,15). We were able to obtain high quality MRS data in a dual-site setting due the consistency of obtaining artifact-free short TE spectra using an in-house developed and highly optimized pulse sequence and identical B0 and B1 adjustment protocols.
The relative metabolite concentrations within and between the two VOIs were consistent with previous publications (14,31–34). Note that the concentrations reported are slightly underestimated relative to their true tissue values since the effects of T2 relaxation for metabolites were not taken into account. This approach was chosen since apparent T2s are pulse sequence- (35), brain region- (24) (also see water T2s in Table 1) and metabolite- dependent (36). While it is relatively straightforward to measure the water T2 in each VOI from all volunteers, acquisition of region-specific T2s for all metabolites in the reported profiles was both outside the scope of this project and is not feasible in routine clinical applications. Alternatively, a single correction factor could be identified based on literature values for a different pulse sequence, however, this would almost certainly be inaccurate for most metabolites reported. Note however that the systematic bias in metabolite concentrations resulting from omission of the metabolite T2 correction is inconsequential for multi-site investigations provided that the same assumptions are used in the analysis of all data.
Almost all multi-site trials on clinical scanners so far have only reported the concentrations or concentration ratios of tNAA, tCr, tCho and Ins (the latter only measured at short TE) using 1H spectra measured at short or long echo-times (6–9). Although the TE of semi-LASER used in this study is comparable to that of the vendor-provided PRESS sequence (shortest TE of 30 ms), the presence of the two pairs of 180° adiabatic pulses, which act as a Carr-Purcell pulse train, helps to preserve the J-modulation and signal intensity of metabolites (37). As such, the semi-LASER sequence enabled the quantification of at least 13 metabolites in the pons and vermis at each site, thereby showing the feasibility of consistently measuring J-coupled metabolites in addition to singlet metabolites between different sites.
Multi-site trials utilizing 1H MRS have been challenging due to large variations in reported metabolite concentrations between sites, even within each site. In such investigations the within-site CVs ranged between 2–30% and between-site CVs were between 2–35% for singlet metabolites (5,6,9). These reproducibility issues might be related to various factors such as the quality of the raw spectral data, number of subjects studied, reference used for quantification or the stability of the MR scanner. On the other hand, multi-site studies that have used metabolite ratios (e.g. tNAA/tCr or tNAA/tCho) have reported lower within-site CV of less than 10% (38,39). Although quantifying ratios does not require corrections for T2 and water content and therefore is easier, it does not provide a clear understanding on how individual metabolites change under different pathological conditions. For instance, tCr concentration, which is generally used as an internal reference, was reported to change in various neurological conditions (40,41).
In the present study, we took the more challenging approach of water scaling (also referred to as “absolute” quantification). The within-site CV for singlet metabolite concentrations was between 6–12% (Figure 3), which lies in the lower end of the CV range reported in prior multi-site investigations. Furthermore, the mean CRLBs, which indicate quantification precision, were substantially lower (3–5%) than CVs for these metabolites, indicating that the CV is dominated by between-subject differences rather than measurement errors. This demonstrates the feasibility of detecting inter-individual differences in the healthy brain at high field, consistent with our prior experience at 4T and 7T (11). Similarly, a recent two-site 3T MRS imaging (MRSI) study using semi-LASER reported mean CRLBs of 6% or less for tNAA, tCr, tCho and Ins in selected gray and white matter VOI in the cerebrum and also demonstrated lower within-subject variation in these metabolites than between-subject variation (13). No between-site CVs are reported in the present study since different subjects were scanned at each site. However since the % difference in metabolite concentrations between sites was very small, the between-site CV is also expected to be low.
The semi-LASER sequence was developed and optimized (i.e. spoiler gradients and OVS parameters) at CMRR prior to the transfer to the ICM site. The only requirements to successfully run the spectroscopy sequence was to adjust the first and second order shims using FAST(EST)MAP and to calibrate the RF power required for the 90° and water suppression pulses. This step was done in each study for each VOI location because the standard slice-based voltage adjustment done by the scanner once at the beginning of the scanning session often under/overestimates the RF power required in the selected VOI. The MRS data acquired at the two sites using widely available commercial hardware demonstrates that non-vendor, ready-to-use MRS sequences can be shared among sites, generating highly reproducible spectral quality. This is expected to facilitate robust, multi-site MRS trials where large numbers of datasets can be acquired in a relatively short time.
The main limitation of the current study was that the reproducibility of neurochemical profiles was tested at two sites utilizing an MR scanner and hardware from the same vendor. For more generalized conclusions, it is critical to investigate across-vendor reproducibility of neurochemical profiles in larger multi-site investigations. Such efforts were recently reported in abstract form (42). Another limitation was the need to manually initiate the voxel specific B1 calibrations; automating these steps as done in standard vendor-provided packages is feasible and will be important for seamless application in the clinical environment.
CONCLUSION
This dual-site study shows that a wide range of metabolites (singlet and J-coupled) can be quantified on clinical 3T scanners with highly reproducible neurochemical profiles using an in-house developed and highly optimized pulse sequence. These profiles can be pooled in multi-site investigations provided that the same acquisition and analysis techniques are utilized at all sites. Furthermore, within each site the between-subject coefficients of variance for singlet resonances and myo-inositol were substantially higher than their CRLBs, indicating precision to detect inter-individual differences in the healthy brain.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Diane Hutter, RN, for assistance with subject recruitment, Dr. Petr Bednařík for providing the measured T2 value of CSF, Dr. Ivan Tkáč for discussions about the LCModel control parameter choices, Dr. Lynn Eberly for the chi-square statistics and the staff of the Center for MR Research for maintaining and supporting the MR systems. This work was supported by the National Institute of Neurological Disorders and Stroke (NINDS) grant R01 NS070815, the Assistance Publique des Hôpitaux de Paris (APHP) and the program “Investissements d’avenir” ANR-10-IAIHU-06. The Center for MR Research is supported by National Center for Research Resources (NCRR) biotechnology research resource grant P41 RR008079, National Institute of Biomedical Imaging and Bioengineering (NIBIB) grant P41 EB015894 and the Institutional Center Cores for Advanced Neuroimaging award P30 NS076408.
Abbreviations
- CMRR
Center for Magnetic Resonance Research
- ICM
Institut du Cerveau et de la Moelle
REFERENCES
- 1.Öz G, Alger J, Barker P, Bartha R, Bizzi A, Boesch C, Bolan P, Brindle K, Cudalbu C, Dincer A, Dydak U, Emir U, Frahm J, Gonzalez R, Gruber S, Gruetter R, Gupta R, Heerschap A, Henning A, Hetherington H, Howe F, Huppi P, Hurd R, Kantarci K, Klomp D, Kreis R, Kruiskamp M, Leach M, Lin A, Luijten P, Marjanska M, Maudsley A, Meyerhoff D, Mountford C, Nelson S, Ozduman K, Necmettin P, Pan J, Peet A, Poptani H, Posse S, Pouwels P, Ratai E, Ross B, Scheenen T, Schuster C, Soher B, Tkac I, Vigneron D, Kauppinen R. Clinical Proton MR Spectroscopy in Central Nervous System Disorders: The MRS Consensus Group. Radiology. 2014;270(3):658–679. doi: 10.1148/radiol.13130531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wattjes MP. Structural MRI. International Psychogeriatrics. 2011;23(Supplement S2):S13–S24. doi: 10.1017/S1041610211000913. [DOI] [PubMed] [Google Scholar]
- 3.Hentschel F, Kreis M, Damian M, Krumm B, Frolich L. The clinical utility of structural neuroimaging with MRI for diagnosis and differential diagnosis of dementia: a memory clinic study. Int J Geriat Psychiatry. 2005;20(7):645–650. doi: 10.1002/gps.1333. [DOI] [PubMed] [Google Scholar]
- 4.Komoroski RA, Kotrla KJ, Lemen L, Lindquist D, Diaz P, Foundas A. Brain metabolite concentration ratios in vivo: multisite reproducibility by single-voxel 1H MR spectroscopy. Magn Reson Imaging. 2004;22(5):721–725. doi: 10.1016/j.mri.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Traber F, Block W, Freymann N, Gur O, Kucinski T, Hammen T, Ende G, Pilatus U, Hampel H, Schild H, Heun R, Jessen F. A multicenter reproducibility study of single-voxel 1H-MRS of the medial temporal lobe. European Radiology. 2006;16:1096–1103. doi: 10.1007/s00330-005-0108-y. [DOI] [PubMed] [Google Scholar]
- 6.Jessen F, Gur O, Block W, Ende G, Frolich L, Hammen T, Wiltfang J, Kucinski T, Jahn H, Heun R, Maier W, Kolsch H, Kornhuber J, Traber F. A multicenter 1H-MRS study of the medial temporal lobe in AD and MCI. Neurology. 2009;72(20):1735–1740. doi: 10.1212/WNL.0b013e3181a60a20. [DOI] [PubMed] [Google Scholar]
- 7.Chard DT, Parker GJM, Griffin CMB, Thompson AJ, Miller DH. The reproducibility and sensitivity of brain tissue volume measurements derived from an SPM-based segmentation methodology. J Magn Reson Imaging. 2002;15(3):259–267. doi: 10.1002/jmri.10064. [DOI] [PubMed] [Google Scholar]
- 8.Keevil SF, Barbiroli B, Brooks JCW, Cady EB, Canese R, Carlier P, Collins DJ, Gilligan P, Gobbi G, Hennig J, Kugel H, Leach MO, Metzler D, Mlynarik V, Moser E, Newbold MC, Payne GS, Ring P, Roberts JN, Rowland IJ, Thiel T, Tkac I, Topp S, Wittsack HJ, Wylezinska M, Zaniol P, Henriksen O, Podo F. Absolute metabolite quantification by in vivo NMR spectroscopy: II. a multicentre trial of protocols for in vivo localised proton studies of human brain. Magn Reson Imaging. 1998;16(9):1093–1106. doi: 10.1016/s0730-725x(98)00118-0. [DOI] [PubMed] [Google Scholar]
- 9.Vavasour I, Laule C, Meyers S, Mädler B, Harris T, Li D, Traboulsee A, MacKay A. Cross-Site Reproducibility of 1H-MRS. Proc Intl Soc Mag Reson Med. 2010;18:2008. [Google Scholar]
- 10.Deelchand DK, Iltis I, Henry P-G. Improved quantification precision of human brain short echo-time 1H magnetic resonance spectroscopy at high magnetic field: A simulation study. Magn Reson Med. 2013 doi: 10.1002/mrm.24892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tkac I, Oz G, Adriany G, Ugurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at high magnetic fields: metabolite quantification at 4T vs. 7T. Magn Reson Med. 2009;62(4):868–879. doi: 10.1002/mrm.22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mekle R, Mlynárik V, Gambarota G, Hergt M, Krueger G, Gruetter R. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magn Reson Med. 2009;61(6):1279–1285. doi: 10.1002/mrm.21961. [DOI] [PubMed] [Google Scholar]
- 13.Wijnen JP, van Asten JJA, Klomp DWJ, Sjobakk TE, Gribbestad IS, Scheenen TWJ, Heerschap A. Short echo time 1H MRSI of the human brain at 3T with adiabatic slice selective refocusing pulses; reproducibility and variance in a dual center setting. J Magn Reson Imaging. 2010;31(1):61–70. doi: 10.1002/jmri.21999. [DOI] [PubMed] [Google Scholar]
- 14.Öz G, Tkac I. Short-echo, single-shot, full-intensity proton magnetic resonance spectroscopy for neurochemical profiling at 4 T: Validation in the cerebellum and brainstem. Magn Reson Med. 2011;65(4):901–910. doi: 10.1002/mrm.22708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Öz G. Spectroscopy in health and disease. In: Manto M, Gruol D, Schmahmann J, Koibuchi N, Rossi F, editors. Handbook of the Cerebellum and Cerebellar Disorders. Springer; 2013. pp. 712–733. [Google Scholar]
- 16.Gruetter R, Tkac I. Field mapping without reference scan using asymmetric echo-planar techniques. Magn Reson Med. 2000;43(2):319–323. doi: 10.1002/(sici)1522-2594(200002)43:2<319::aid-mrm22>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 17.Scheenen TWJ, Klomp DWJ, Wijnen JP, Heerschap A. Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med. 2008;59(1):1–6. doi: 10.1002/mrm.21302. [DOI] [PubMed] [Google Scholar]
- 18.Tkac I, Starcuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med. 1999;41(4):649–656. doi: 10.1002/(sici)1522-2594(199904)41:4<649::aid-mrm2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 19.Tannus A, Garwood M. Improved Performance of Frequency-Swept Pulses Using Offset-Independent Adiabaticity. J Magn Reson, Series A. 1996;120:133–137. [Google Scholar]
- 20.Natt O, Bezkorovaynyy V, Michaelis T, Frahm J. Use of phased array coils for a determination of absolute metabolite concentrations. Magn Reson Med. 2005;53(1):3–8. doi: 10.1002/mrm.20337. [DOI] [PubMed] [Google Scholar]
- 21.Ernst T, Kreis R, Ross BD. Absolute Quantitation of Water and Metabolites in the Human Brain. I. Compartments and Water. J Magn Reson, Series B. 1993;102(1):1–8. [Google Scholar]
- 22.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6):672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 23.Deelchand DK, Henry P-G, Ugurbil K, Marjanska M. Measurement of transverse relaxation times of J-coupled metabolites in the human visual cortex at 4 T. Magn Reson Med. 2012;67(4):891–897. doi: 10.1002/mrm.23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traber F, Block W, Lamerichs R, Gieseke J, Schild HH. 1H metabolite relaxation times at 3.0 Tesla: Measurements of T1 and T2 values in normal brain and determination of regional differences in transverse relaxation. J Magn Reson Imaging. 2004;19(5):537–545. doi: 10.1002/jmri.20053. [DOI] [PubMed] [Google Scholar]
- 25.Bartha R, Michaeli S, Merkle H, Adriany G, Andersen P, Chen W, Ugurbil K, Garwood M. In vivo 1H2O T2+ measurement in the human occipital lobe at 4T and 7T by Carr-Purcell MRI: detection of microscopic susceptibility contrast. Magn Reson Med. 2002;47(4):742–750. doi: 10.1002/mrm.10112. [DOI] [PubMed] [Google Scholar]
- 26.Siegel GJ. Basic neurochemistry: molecular, cellular and medical aspects. Lippincott-Raven Publishers; 1999. [Google Scholar]
- 27.Randall L. Chemical topography of the brain. J Biol Chem. 1938;124(2):481–488. [Google Scholar]
- 28.Schaller Bt, Xin L, Gruetter R. Is the macromolecule signal tissue-specific in healthy human brain? A 1H MRS study at 7 tesla in the occipital lobe. Magn Reson Med. 2013 doi: 10.1002/mrm.24995. [DOI] [PubMed] [Google Scholar]
- 29.Pouwels PJW, Frahm J. Regional metabolite concentrations in human brain as determined by quantitative localized proton MRS. Magn Reson Med. 1998;39(1):53–60. doi: 10.1002/mrm.1910390110. [DOI] [PubMed] [Google Scholar]
- 30.Frahm J, Bruhn H, Gyngell ML, Merboldt KD, Hanicke W, Sauter R. Localized proton NMR spectroscopy in different regions of the human brain in vivo. Relaxation times and concentrations of cerebral metabolites. Magn Reson Med. 1989;11(1):47–63. doi: 10.1002/mrm.1910110105. [DOI] [PubMed] [Google Scholar]
- 31.Öz G, Hutter D, Tkac I, Clark HB, Gross MD, Jiang H, Eberly LE, Bushara KO, Gomez CM. Neurochemical alterations in spinocerebellar ataxia type 1 and their correlations with clinical status. Mov Disord. 2010;25(9):1253–1261. doi: 10.1002/mds.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker EH, Basso G, Barker PB, Smith MA, Bonekamp D, Horska A. Regional apparent metabolite concentrations in young adult brain measured by 1H MR spectroscopy at 3 Tesla. J Magn Reson Imaging. 2008;27(3):489–499. doi: 10.1002/jmri.21285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michaelis T, Merboldt KD, Bruhn H, Hanicke W, Frahm J. Absolute concentrations of metabolites in the adult human brain in vivo: quantification of localized proton MR spectra. Radiology. 1993;187(1):219–227. doi: 10.1148/radiology.187.1.8451417. [DOI] [PubMed] [Google Scholar]
- 34.Mascalchi M, Brugnoli R, Guerrini L, Belli G, Nistri M, Politi LS, Gavazzi C, Lolli F, Argenti G, Villari N. Single-voxel long TE 1H-MR spectroscopy of the normal brainstem and cerebellum. J Magn Reson Imaging. 2002;16(5):532–537. doi: 10.1002/jmri.10189. [DOI] [PubMed] [Google Scholar]
- 35.Michaeli S, Garwood M, Zhu XH, DelaBarre L, Andersen P, Adriany G, Merkle H, Ugurbil K, Chen W. Proton T2 relaxation study of water, N-acetylaspartate, and creatine in human brain using Hahn and Carr-Purcell spin echoes at 4T and 7T. Magn Reson Med. 2002;47(4):629–633. doi: 10.1002/mrm.10135. [DOI] [PubMed] [Google Scholar]
- 36.Xin L, Gambarota G, Mlynarik V, Gruetter R. Proton T2 relaxation time of J-coupled cerebral metabolites in rat brain at 9.4 T. NMR Biomed. 2008;21(4):396–401. doi: 10.1002/nbm.1205. [DOI] [PubMed] [Google Scholar]
- 37.Allerhand A. Analysis of Carr--Purcell Spin-Echo NMR Experiments on Multiple-Spin Systems. I. The Effect of Homonuclear Coupling. J Chem Phys. 1966;44(1):1–9. [Google Scholar]
- 38.Currie S, Hadjivassiliou M, Wilkinson I, Griffiths P, Hoggard N. Magnetic Resonance Spectroscopy of the Normal Cerebellum: What Degree of Variability Can Be Expected? The Cerebellum. 2013;12:205–211. doi: 10.1007/s12311-012-0415-1. [DOI] [PubMed] [Google Scholar]
- 39.Lee PL, Yiannoutsos CT, Ernst T, Chang L, Marra CM, Jarvik JG, Richards TL, Kwok EW, Kolson DL, Simpson D, Tang CY, Schifitto G, Ketonen LM, Meyerhoff DJ, Lenkinski RE, Gonzalez RG, Navia BA. A multi-center 1H MRS study of the AIDS dementia complex: Validation and preliminary analysis. J Magn Reson Imaging. 2003;17(6):625–633. doi: 10.1002/jmri.10295. [DOI] [PubMed] [Google Scholar]
- 40.Vrenken H, Barkhof F, Uitdehaag BMJ, Castelijns JA, Polman CH, Pouwels PJW. MR spectroscopic evidence for glial increase but not for neuro-axonal damage in MS normal-appearing white matter. Magn Reson Med. 2005;53(2):256–266. doi: 10.1002/mrm.20366. [DOI] [PubMed] [Google Scholar]
- 41.Öz G, Iltis I, Hutter D, Thomas W, Bushara KO, Gomez CM. Distinct Neurochemical Profiles of Spinocerebellar Ataxias 1, 2, 6, and Cerebellar Multiple System Atrophy. The Cerebellum. 2011;10:208–217. doi: 10.1007/s12311-010-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Bank BL, Emir UE, Boer VO, van Asten JJA, Wijnen JP, Kan HE, Öz G, Klomp DWJ, Scheenen TWJ. Multi-center Reproducibility of Short Echo Time Single Voxel 1H MRS of the Human Brain at 7T with Adiabatic Slice-Selective Refocusing Pulses. Proc Intl Soc Mag Reson Med. 2013:3982. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.