Abstract
Purpose:
To investigate the feasibility of applying a new quantitative image analysis method to improve breast cancer diagnosis performance using dynamic contrast enhanced magnetic resonance imaging (DCE-MRI) by integrating background parenchymal enhancement (BPE) features into the decision making process.
Methods:
A dataset involving 115 DCE-MRI examinations was used in this study. Each examination depicts one identified suspicious breast tumor. Among them, 75 cases were verified as malignant and 40 were benign by the biopsy results. A computer-aided detection scheme was applied to segment breast regions and the suspicious tumor depicted on the sequentially scanned MR images of each case. We then computed 18 kinetic features in which 6 were computed from the segmented breast tumor and 12 were BPE features from the background parenchymal regions (excluding the tumor). Support vector machine (SVM) based statistical learning classifiers were trained and optimized using different combinations of features that were computed either from tumor only or from both tumor and BPE. Each SVM was tested using a leave-one-case-out validation method and assessed using an area under the receiver operating characteristic curve (AUC).
Results:
When using kinetic features computed from tumors only, the maximum AUC is 0.865 ± 0.035. After fusing with the BPE features, AUC increased to 0.919 ± 0.029. At 90% specificity, the tumor classification sensitivity increased by 13.2%.
Conclusions:
The proposed quantitative BPE features provide valuable supplementary information to the kinetic features of breast tumors in DCE-MRI. Their addition to computer-aided diagnosis methodologies could improve breast cancer diagnosis based on DCE-MRI examinations.
Keywords: breast cancer, dynamic contrast enhanced breast magnetic resonance imaging (DCE-MRI), computer-aided diagnosis (CAD), breast background parenchymal features
1. INTRODUCTION
Breast cancer is the most prevalent cancer in women worldwide.1 Although mammography is the most popular and cost-effective imaging modality in breast cancer screening, its detection sensitivity is not satisfactory in particular for women with dense breasts and carrying certain high risk genes. For example, mammographic sensitivity reduces from over 98% to a range of 30%–48% when BIRADS ratings of mammographic density increase from 1 to 4.2,3 The mammographic sensitivity is also lower (ranging from 16% to 40%) for women who carry the high risk genes of BRCA1 and/or BRCA2.4 In order to improve cancer detection and diagnostic performance, dynamic contrast enhanced magnetic resonance imaging (DCE-MRI) examination of breasts is currently used as an adjunct imaging modality to the mammography.5 In the current breast cancer screening programs, adoption of DCE-MRI in the high risk population has shown benefit over mammography and other imaging modalities. The superior performance of using DCE-MRI to increase the sensitivity of early invasive breast cancer detection was well demonstrated in a recent large multi-institutional study organized by the American College of Radiology Imaging Network.6 Using DCE-MRI enables to detect tumors that are not clearly defined or occulted on mammograms7 primarily due to the 3D imaging technology and dynamic biological and physiological properties revealed by the DCE-MRI modality. In the diagnostic breast imaging, DCE-MRI is often applied to map tumor size and estimate tumor pathological stage and grade, which is used as an initial guide for clinical treatment (e.g., lumpectomy) and/or follow-ups.8 However, despite the high sensitivity, the reported specificity of DCE-MRI in distinguishing between malignant and benign tumors varies greatly, ranging from 37% to 90%.9
The high false-positive recall or biopsy rate substantially reduces efficacy of breast cancer screening and early diagnosis, which generates negative consequences including long-term psychosocial consequences,10 cumulative radiation exposure,11 and waste of healthcare resources with associated high costs.12 Therefore, in order to improve efficacy of breast cancer screening and diagnosis, it is important to develop and apply more accurate and reliable image biomarkers to significantly increase discriminatory power in classification between malignant and benign breast tumors. In assisting interpretation of DCE-MRI examinations, both morphological (e.g., shape, margin, and texture) and kinetic (e.g., contrast enhancement curve) features have been analyzed and used. A number of studies have been conducted to explore and reveal the unique kinetic characteristics of breast tumors using the statistical pharmacokinetic models that were established and trained using the data acquired from the DCE-MRI examinations.13–15 However, due to the large heterogeneity of the kinetic contrast enhancement features of the breast tumors, visual classification of the tumors depicted on DCE-MRI images is a difficult task. As a result, great research effort has been made to develop computer-aided detection and diagnosis (CAD) schemes, which use pharmacokinetic and morphological features computed from an identified breast tumor to predict the likelihood of the tumor being malignant.16,17
Although the majority of previous CAD studies focused on computing features within the breast tumors detected and segmented from the DCE-MRI images, several recent studies have demonstrated that the background parenchymal enhancement (BPE) revealed from the breast DCE-MRI images might also contain valuable information to assist prediction of the breast cancer risk17 and affect detection and staging of breast cancers.18 In this study, our hypothesis is that BPE might also carry the valuable and supplementary information to the kinetic contrast enhancement features computed from the segmented breast tumors and thus help improve performance of using DCE-MRI examinations to detect breast cancers (or classify between malignant and benign MRI examinations). Unlike the previous studies that qualitatively assessed average BPE level of two breasts,18,19 we recently developed a fully automated CAD scheme to detect BPE and its difference (or asymmetry) between the left and right breasts.20 The purpose of this study is to test our hypothesis by integrating the quantitative kinetic image features computed from both breast tumor regions and BPE into a CAD scheme and then investigating feasibility of using this new quantitative image analysis approach to help improve performance of breast cancer diagnosis using CAD schemes of breast DCE-MRI images.
2. MATERIALS AND METHOD
The DCE-MRI images used in this study were retrospectively collected from an existing clinical database at Zhejiang Cancer Hospital in the City of Hangzhou, Zhejiang Province, China. The dataset includes 115 DCE-MRI examinations acquired from 115 women who underwent breast cancer diagnosis at the hospital. All images selected in this dataset were acquired before any cancer treatment procedures (for diagnostic purpose). The dataset was divided into two case groups. Group one includes 75 cases depicted cancer (with biopsy and pathology-verified malignant tumors) and group two involves 40 cases that had highly suspicious tumors detected from a number of imaging examinations (i.e., mammography and DCE-MRI) but later proved by biopsy as benign. In addition, in this dataset, each of all malignant cases only depicts cancer in one breast and another breast is negative without any detectable tumors.
In this image dataset, the mean and standard deviations of the women’s age are 49.1 ± 8.4 and 41.6 ± 12.0 for the groups of cancer and benign cases, respectively, which indicate that the majority of women whose DCE-MRI examination images were selected in this dataset are relatively younger. Specifically, 57.3% (43/75) and 85% (34/40) of women were younger than 50 yr old in the cancer and benign case groups, respectively. Only one woman in cancer case group was over 70 yr old. In the cancer case group, the metastasis of cancer cells to the lymph nodes was detected in 38 women and not detected in the other 37 women.
Each breast DCE-MRI examination was originally performed using the following imaging examination protocol. The woman was scanned in prone position using a Siemens MRI machine (MAGNETOM Espree—Pink 1.5T) equipped with a dedicated eight-channel double-breast coil. Each examination included five series of sequential image scans. Two precontrast (or baseline) series of T1 and T2 weighted 3D image scans were first performed, followed by the injection of Gd-DTPA contrast agent intravenously with a dose of 0.2 mmol/kg body weight and a saline flush of 20 ml at the same injection flow rate of 2 ml/s. Then, after completion of the contrast agent injection, three post-contrast series of T1 weighted 3D MRI scans and data acquisitions were performed with a time interval of one minute. The first two postcontrast image scanning series, which were named as S-1 and S-2, used the same image scanning process and thus generated images with the same spatial resolution as the precontrast MRI scanning images (termed S-0) with a data matrix of 512 × 512 pixels. Specifically, all three (S-0–S-2) image series using T1 weighted 3D image scanning model included 88 image slides that were reconstructed using the same slice thickness representing the same in-depth resolution. The third postcontrast image scanning series (termed S-3) generated high in-depth resolution images, which involved 160 image slices. The detail image scanning protocol and the related parameters are summarized in Table I. In this study, only the images acquired from three DCE-MRI image scanning series (namely, S-0, S-1, and S-2) were selected and used in the quantitatively kinetic image feature computation and data analysis process.
TABLE I.
The parameters of the DCE-MRI scanning protocol used in this study.
| Scanning parameter | Specification data |
|---|---|
| TR/TE | 4.4/1.6 ms |
| Flip angle | 12° |
| Slice thickness | 1.2 mm in S-0, S-1, and S-2 |
| 0.8 mm in S-3 | |
| Spatial resolution | 0.625 × 0.625 × 1.2 mm3 in S-0, S-1, and S-2 |
| 0.625 × 0.625 × 1.2 mm3 in S-3 |
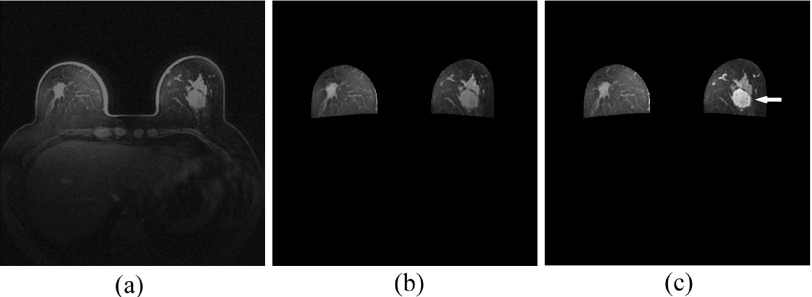
The center location of the suspicious breast tumor in each case was marked by the radiologists in this dataset. Using the marked tumor center location as an initial segmentation “seed,” the volumetric breast tumor boundary contour was automatically segmented by a computerized algorithm.21 The segmentation result was visually examined by our investigators and manually corrected (if needed). In this study, the manual correction of tumor boundary segmentation occurred in less than 25% cases. We also applied another computerized scheme to automatically segment the breast region depicted on all breast MR image slices, register sequential MRI scans (S-0–S-2) as previously reported.22 Figure 1 shows an example of a breast DCE-MRI slice and its CAD-segmented breast regions depicted on the image slice before and after contrast agent injection (S-0 and S-1). In this example, the left and right breast regions were automatically segmented and a breast tumor was identified. This image shows a tumor with higher contrast enhancement along with the BPE in both left and right breasts.
FIG. 1.
An example of a breast DCE-MRI slice (a) and its CAD-segmented breast regions depicted on the image slice before (S-0) and after (S-1) contrast agent injection [(b) and (c)] in which the nonbreast regions behind the chest wall are removed and a tumor is pointed by a white arrow on the S-1 image slice.
From the registered MRI sequential scans, we applied the computerized scheme to generate two image subtraction maps (namely, image maps of S-1 – S-0 and S-2 – S-0). Then, from these two sets of image maps, the computerized scheme initially computed 18 kinetic features (as summarized in Table II). Among them, six were tumor-related features computed from the voxels inside the segmented breast tumors. The other 12 were BPE features computed from the background parenchymal regions. From this BPE image map, the computerized scheme sorted and ranked voxels based on the computed BPE values. We then conducted experiment and data analysis to determine an optimal threshold (P%) to select percentage of voxels in the top BPE value ranking list in order to maximize the diagnostic value of using each BPE feature.22 Among the 12 BPE features, 6 were computed from one abnormal breast depicting the suspicious breast tumor, while other 6 were computed from the asymmetry (or difference) of the features between the two bilateral (left and right) breasts. These features represent (1) average contrast enhancement (P% = 0.17%) and the bilateral asymmetry (P% = 3.40%), (2) standard deviation of the contrast enhancement (P% = 6.54%) and the bilateral asymmetry (P% = 3.00%), and (3) the maximum contrast enhancement inside the tumor or background parenchymal regions and their bilateral asymmetry. Each feature was computed twice at two postcontrast enhancement points, namely, the subtraction maps generated by S-1 – S-0 and S-2 – S-0.
TABLE II.
Summary of 18 kinetic features computed from breast DCE-MRI images.
| Feature number | Feature definition | Computing method |
|---|---|---|
| Inside a tumor | ||
| 1 | Average contrast enhancement value | S-1 – S-0 |
| 2 | Average contrast enhancement value | S-2 – S-0 |
| 3 | Standard deviation of enhancement value | S-1 – S-0 |
| 4 | Standard deviation of enhancement value | S-2 – S-0 |
| 5 | The maximum enhancement value | S-1 – S-0 |
| 6 | The maximum enhancement value | S-2 – S-0 |
| From BP region | ||
| 7 | Average BPE in breast depicting tumor | S-1 – S-0 |
| 8 | Average BPE in breast depicting tumor | S-2 – S-0 |
| 9 | STD of BPE in breast depicting tumor | S-1 – S-0 |
| 10 | STD of BPE in breast depicting tumor | S-2 – S-0 |
| 11 | Asymmetry of average BPE in two breasts | S-1 – S-0 |
| 12 | Asymmetry of average BPE in two breasts | S-2 – S-0 |
| 13 | Asymmetry of STD of BPE in two breasts | S-1 – S-0 |
| 14 | Asymmetry of STD of BPE in two breasts | S-2 – S-0 |
| 15 | The maximum BPE in breast depicting tumor | S-1 – S-0 |
| 16 | The maximum BPE in breast depicting tumor | S-2 – S-0 |
| 17 | Asymmetry of maximum BPE in two breasts | S-1 – S-0 |
| 18 | Asymmetry of maximum BPE in two breasts | S-2 – S-0 |
Note: S-0, Images scanned before injection of contrast agent; S-1, The first image series scanned after contrast agent injection; S-2, The second image series scanned after contrast agent injection; STD, Standard deviation; BPE, Background parenchymal enhancement.
After feature extraction and computation, we trained and applied a support vector machine (SVM) based statistical machine learning tool to fuse the computed kinetic features and test SVM performance in classifying between the malignant and benign DCE-MRI examinations. A publically available SVM software package (LIBSVM; http://www.csie.ntu.edu.tw/~cjin/libsvm) was used in this study to train and optimize the SVM classifiers. The SVM was trained and tested using a leave-one-case-out (LOCO) validation method.23 The performance assessment indices used in this study included the area under a receiver operating characteristic (ROC) curve (AUC) computed by rockit program (http://www.radiology.uchicago.edu/krl/) and the classification sensitivity at 90% specificity. We systematically compared the change of performance levels of the SVMs that were trained using the kinetic features computed from the segmented tumors only and the fused features computed from both inside the tumor and background parenchymal regions. A complete permutation searching method was applied in this study to identify a SVM with the highest AUC value using a set of quantitatively computed optimal image features.
3. RESULTS
After sorting the classification performance levels (the computed AUC values) of the SVMs, Table III summarizes two sets of the top ten SVMs and their classification performance levels. In one set, SVMs were trained using the kinetic image features computed from inside the tumors only, and in another set, SVMs were trained using the combination of both tumor and BPE features. The same testing image dataset and LOCO validation method were applied in the performance comparison. The results demonstrated that adding the BPE features of the DCE-MRI images into a CAD scheme (e.g., a SVM in this study) helped improve CAD classification performance. Using the kinetic features computed from inside tumors only, the top ten AUC values ranged from 0.828 to 0.865. After adding the BPE features, the top ten AUC values significantly increased to a range from 0.907 to 0.919 (p < 0.05). The comparison of these 20 SVMs listed in Table II indicated that adding BPE features into a CAD type scheme enabled to help improve CAD performance of classifying between the malignant and benign DCE-MRI examinations.
TABLE III.
Comparison of the ten top SVMs trained using the kinetic image features computed from either tumors only or the combination of both tumors and BPE features.
| Tumor features only | Fused tumor and BPE features | |||
|---|---|---|---|---|
| Performance order | AUC | Feature list | AUC | Feature list |
| 1 | 0.865 | 1, 2, 4 | 0.919 | 1, 2, 3, 4, 7, 10, 11, 12, 16, 17 |
| 2 | 0.857 | 1, 2, 4, 6 | 0.916 | 1, 2, 4, 7, 10, 15, 16, 17, 18 |
| 3 | 0.857 | 1, 2, 4, 5, 6 | 0.915 | 1, 2, 4, 7, 12, 16 |
| 4 | 0.852 | 1, 2, 6 | 0.910 | 1, 2, 3, 4, 7, 9, 11, 16, 17 |
| 5 | 0.851 | 1, 2, 3, 4, 5, 6 | 0.909 | 1, 2, 3, 4, 6, 7, 9, 10, 15, 17 |
| 6 | 0.850 | 1, 2, 3, 4, 6 | 0.909 | 1, 2, 4, 5, 7, 9, 10, 15, 17 |
| 7 | 0.845 | 1, 2, 3 | 0.908 | 1, 2, 3, 4, 5, 7, 9, 10, 15, 17 |
| 8 | 0.844 | 1, 2 | 0.907 | 1, 2, 3, 4, 7, 9, 10, 11, 12, 16, 17 |
| 9 | 0.833 | 1, 2, 5, 6 | 0.907 | 1, 2, 4, 7, 11, 12, 16 |
| 10 | 0.828 | 3, 4 | 0.907 | 1, 2, 4, 7, 9, 10, 12, 16, 17 |
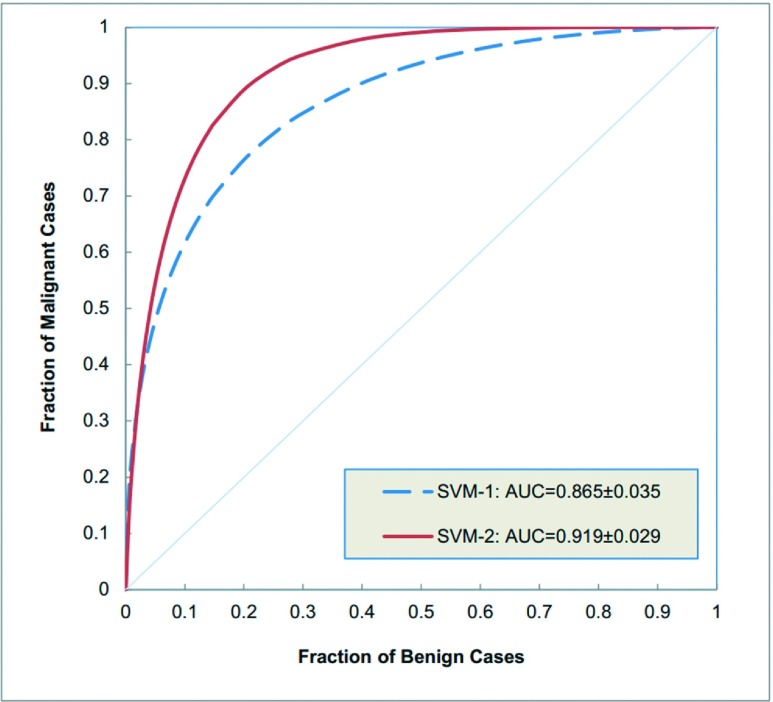
Figure 2 illustrates and compares two ROC curves generated by two SVMs listed in the top one position of two SVM sets of Table III, which were trained using either tumor based kinetic features only or fused features computed from tumor and BPE. When using the tumor-related kinetic features only, the top one SVM used three kinetic features and yielded the highest classification performance of AUC = 0.865. The standard error of the AUC is 0.035 and the 95% confidence interval was [0.785, 0.921]. When fusing the kinetic features computed from both the tumors and the parenchymal background, the top one SVM used ten kinetic features including four computed from the tumor and six computed from BPE. This SVM increased the classification performance to AUC = 0.919 with a standard error of 0.029 and a corresponding 95% confidence interval of [0.847, 0.962], which indicates more than 5% increase in AUC value as comparing to using the kinetic features computed from tumor only.
FIG. 2.
Comparison of two ROC-type performance curves generated by two SVM classifiers in which SVM-1 was built using three kinetic features computed only inside the segmented tumors and SVM-2 was built using ten kinetic features (four computed inside the tumor and six computed from the BPE features).
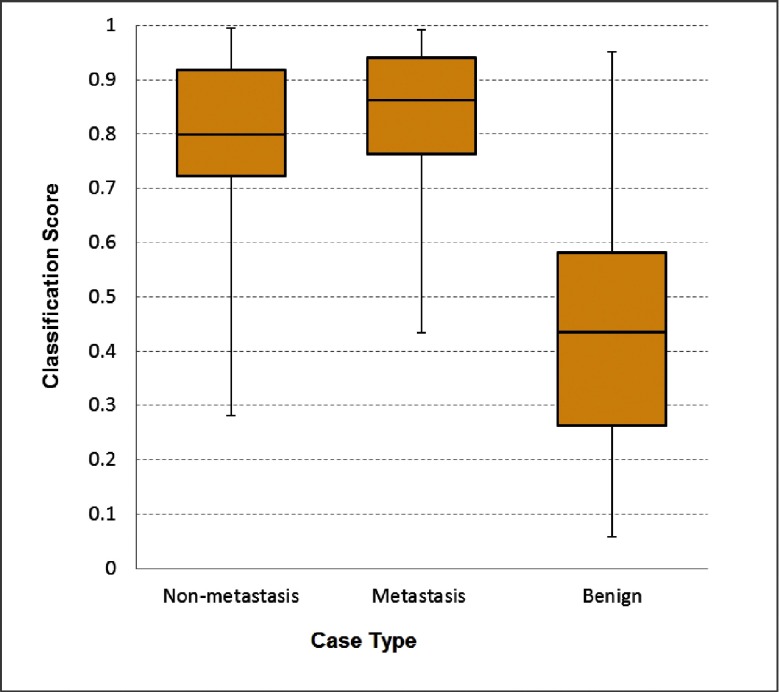
By separately sorting the classification scores generated by two top one SVMs listed in Table III, the classification sensitivities are 70.7% (53/75) and 80.0% (60/75), respectively, for the two SVM trained using the tumor-related kinetic features only and the fused tumor and BPE features at a 90% specificity level. The comparison indicated that adding BPE features increased the sensitivity by 13.2%. In addition, by dividing the cancer cases into two subgroups of 38 metastasis and 37 nonmetastasis cases, the results showed that classification performance of the SVM trained using the fused tumor and BPE features was higher in the metastasis case subgroup. As shown in Fig. 3, the median SVM classification scores are 0.862 and 0.799 for the metastasis and nonmetastasis cases subgroups, respectively. At 90% specificity, 84.2% (32/38) sensitivity was achieved in the metastasis case subgroup, while in the nonmetastasis case subgroup, the sensitivity was 75.7% (28/37). Both are higher than using only tumor-related features.
FIG. 3.
Boxplots of the SVM-generated classification scores among the three subgroups of cases.
4. DISCUSSION
Breast DCE-MRI examinations are currently widely used in breast cancer detection and diagnosis. However, its performance (including sensitivity and/or specificity) heavily depends on the identification of effective and reproducible image features, which are often difficult to be reliably quantified subjectively. Thus, the computerized quantitative image feature analysis and CAD schemes can play an important role in assisting radiologists in interpreting breast DCE-MRI images. In this study, we developed and tested a new quantitative image feature detection and analysis scheme aiming to help improve performance of breast cancer diagnosis using DCE-MRI examinations. This study along with our new quantitative image analysis scheme has a number of unique characteristics. First, although a number of CAD schemes of DCE-MRI images including the commercialized CAD schemes have been previously developed and used in the clinical practice, BPE based kinetic features have never been tested and implemented in any existing CAD schemes. In this study, we demonstrated for the first time that BPE image features computed from the abnormal breast or the bilateral asymmetry of the left and right breasts provided the low-correlated and supplementary information to the kinetic features computed inside the detected breast tumors. For example, based on the top one SVM using ten fused kinetic image features (as shown in Table III), we retrained and tested a new SVM that only used six BPE features (deleting other four tumor-related features). The classification performance of this BPE-feature only SVM yielded AUC = 0.781 ± 0.045. Although this performance level is significantly lower than that generated by the SVM trained using the tumor-related kinetic features (also shown in Table III), the classification results using either the tumor-related or BPE features are not highly correlated (with correlation coefficient r < 0.5). Hence, fusion of the kinetic image features computed from both tumor and BPE enables to improve the overall classification performance of a CAD scheme of DCE-MRI examinations.
Second, unlike the previous studies that used a subjective and qualitative grading method (similar to BIRADS) to estimate the BPE values of the DCE-MRI examinations,18,19 which is likely to produce the large intra- and inter-reader variability, we developed and presented a fully automated method and scheme to quantitatively compute and measure the BPE image features. The quantitative image analysis approach provides us the flexibility to apply more effective image feature computation methods and extract optimal statistical kinetic features with higher discriminatory power. In this study, we detected BPE only from the abnormal breast that depicts the suspicious tumor and also the bilateral asymmetry of the BPE detected from the left and right breasts. Both computational methods enable to enhance the relevant BPE information surrounding the diseased breast by reducing the potential signal noise in the normal (negative) breast that does not depict suspicious tumors. In addition, unlike using only one subjectively rated BPE number, we computed and tested 12 BPE features in this study. Our study demonstrated that using an optimal set of multiple BPE features could help improve the performance of the CAD schemes (e.g., the SVM models used in this study).
From the experimental data analysis results, we also had a number of interesting observations. For example, based on the comparison of the frequency of the feature selection using a complete permutation searching method to train SVMs, we found that in the tumor-related features, the average contrast enhancement value had substantially higher discriminatory power than the maximum contrast enhancement value computed inside a tumor. The kinetic features computed from two postcontrast enhancement MRI scans (S-1 – S-0 and S-2 – S-0) are equally important and often selected together by the SVMs with higher performance (Table III). However, the majority of BPE features selected by the highly performed SVMs are typically computed only using the first postcontrast enhancement series (S-1 – S-0). This indicates that the BPE of the first postcontrast enhancement series plays a dominative role in assisting case or tumor classification. This simplifies the BPE feature computation process. In addition, although all SVMs were trained and tested using all cancer cases as one integrated group, we found that the SVM actually yielded higher classification performance in the subgroup of metastasis cases (Fig. 3), which may indicate that kinetic features computed from both inside the tumors and BPE can also provide the useful information to assist cancer staging and prognosis assessment.
We also recognized that this was a very preliminary study with a number of limitations. First, the size of the image dataset is small and relatively unbalanced with a case ratio of approximately 1–2 between the benign and malignant cases. Hence, the robustness of this study results needs to be further tested using much large and diverse image datasets in the future studies. Second, only two postcontrast MRI scan series were used and a limited number of kinetic features were computed from inside tumors and BPE. Third, no other tumor morphological or texture features were computed and involved in our automated tumor classification scheme. Despite these limitations, our preliminary experimental results were encouraging and we demonstrated a new concept of fusing BPE kinetic features into a CAD scheme or a quantitative image feature classification process to improve performance of classifying between malignant and benign DCE-MRI examinations. Although comparing with many previously developed and reported CAD schemes of DCE-MRI images, the approach and image features used in our scheme are relatively simple and easy to compute, our scheme does not directly compete with the existing CAD schemes. The new approach investigated in this study is not limited to our own CAD scheme (or SVM based classifier). It should also be relatively easily applicable to the other existing or future new CAD schemes of DCE-MRI images.
ACKNOWLEDGMENTS
This work is supported in part by the grants from National Natural Science Foundation of China (61271063), 973 Program (2013CB329502), National Distinguished Young Research Scientist Award (60788101), Science and Technology Program of Zhejiang Province (2013C33164), and Grant No. CA160205 from the National Cancer Institute, National Institutes of Health.
REFERENCES
- 1.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., and Forman D., “Global cancer statistics,” CA-Cancer J. Clin. 61, 69–90 (2001). 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 2.Mandelson M. T., Oestreicher N., Porter P. L., White D., Finder C. A., Taplin S. H., and White E., “Breast density as a predictor of mammographic detection: Comparison of interval-and screen-detected cancers,” J. Natl. Cancer Inst. 92, 1081–1087 (2000). 10.1093/jnci/92.13.1081 [DOI] [PubMed] [Google Scholar]
- 3.Berg W. A., Gutierrez L., Nessaiver M. S., Carter W. B., Bhargavan M., Lewis R. S., and Ioffe O. B., “Diagnostic accuracy of mammography clinical examination, US, and MR imaging in preoperative assessment of breast cancer,” Radiology 233, 830–849 (2004). 10.1148/radiol.2333031484 [DOI] [PubMed] [Google Scholar]
- 4.Warner E., Plewes D. B., Hill K. A., Causer P. A., Zubovits J. T., Jong R. A., Cutrara M. R., Deboer G., Yaffe M. J., Messner S. J., Meschino W. S., Piron C. A., and Narod S. A., “Surveillance of BRCA1 and BRCA2 mutation carriers with magnetic resonance imaging, ultrasound, mammography, and clinical breast examination,” JAMA, J. Am. Med. Assoc. 292, 1317–1325 (2004). 10.1001/jama.292.11.1317 [DOI] [PubMed] [Google Scholar]
- 5.Saslow D., Boetes C., Burke W., Harms S., Leach M. O., Lehman C. D., Morris E., Pisano E., Schnall M., Sener S., Smith R. A., Warner E., Yaffe M., Andrews K. S., and Russell C. A., “American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography,” CA-Cancer J. Clin. 57, 75–89 (2007). 10.3322/canjclin.57.2.75 [DOI] [PubMed] [Google Scholar]
- 6.Berg W., Zhang Z., Lehrer D., Jong R., Pisano E., Barr R., Bohm-Velez M., Mahoney M., Evans W., Larsen L., Morton M., Mendelson E., Farria D., Cormack J., Marques H., Adams A., Yeh N., and Gabrielli G., “Detection of breast cancer with addition of annual screening ultrasound or a single screening MRI to mammography in women with elevated breast cancer risk,” JAMA, J. Am. Med. Assoc. 307, 1394–1404 (2012). 10.1001/jama.2012.388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warner E., Messersmith H., Causer P., Eisen A., Shumak R., and Plewes D., “Systematic review: Using magnetic resonance imaging to screen women at high risk for breast cancer,” Ann. Intern. Med. 148, 671–679 (2008). 10.7326/0003-4819-148-9-200805060-00007 [DOI] [PubMed] [Google Scholar]
- 8.Ikeda D. M., Baker D. R., and Daniel B. L., “Magnetic resonance imaging of breast cancer: Clinical indications and breast MRI reporting system,” J. Magn. Res. Imaging 12, 975–983 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Rankin S. C., “MRI of the breast,” Br. J. Radiol. 73, 806–818 (2000). 10.1259/bjr.73.872.11026854 [DOI] [PubMed] [Google Scholar]
- 10.Brodersen J. and Siersma V. D., “Long-term psychosocial consequences of false-positive screening mammography,” Ann. Fam. Med. 11, 106–115 (2013). 10.1370/afm.1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linet M. S., Slovis T. L., Miller D. L., Kleinerman R., Lee C., Rajaraman P., and de Gonzalez A. B., “Cancer risks associated with external radiation from diagnostic imaging procedures,” CA-Cancer J. Clin. 62, 75–100 (2012). 10.3322/caac.21132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buist D. S. M., Anderson M. L., Haneuse S. J., Sickles E. A., Smith R. A., Carney P. A., Taplin S. H., Rosenberg R. D., Geller B. M., Onega T. L., Monsees B. S., Bassett L. W., Yankaskas B. C., Elmore J. G., Kerlikowske K., and Miglioretti D. L., “Influence of annual interpretive volume on screening mammography performance in the United States,” Radiology 259, 72–84 (2011). 10.1148/radiol.10101698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W., Giger M. L., Lan L., and Bick U., “Computerized interpretation of breast MRI: Investigation of enhancement-variance dynamics,” Med. Phys. 31, 1076–1082 (2004). 10.1118/1.1695652 [DOI] [PubMed] [Google Scholar]
- 14.Nie K., Chen J. H., Yu H. J., Chu Y., Nalcioqlu O., and Su M. Y., “Quantitative analysis of tumor morphology and texture features for diagnostic prediction in breast MRI,” Acad. Radiol. 15, 1513–1525 (2008). 10.1016/j.acra.2008.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karahaliou A., Vassiou K., Arikidis N. S., Skiadopoulos S., Kanavou T., and Costaridou L., “Assessing heterogeneity of tumor enhancement kinetics in dynamic contrast-enhanced MRI for breast cancer diagnosis,” Br. J. Radiol. 83, 296–306 (2010). 10.1259/bjr/50743919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W., Giger M. L., Newstead G. M., Bick U., Jansen S. A., Li H., and Lan L., “Computerized assessment of breast tumor malignancy using DCE-MRI,” Acad. Radiol. 17, 822–829 (2010). 10.1016/j.acra.2010.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vignati A., Giannini V., Luca M. D., Morra L., Persano D., Carbonaro L. A., Bertotto I., Martincich L., Regge D., Bert A., and Sardanelli F., “Performance of a fully automatic tumor detection system for breast DCE-MRI,” J. Magn. Reson. Imaging 34, 1341–1351 (2011). 10.1002/jmri.22680 [DOI] [PubMed] [Google Scholar]
- 18.King V., Brooks J. D., Bernstein J., Reiner A. S., Pike M. C., and Morris E. A., “Background parenchymal enhancement at breast MR imaging and breast cancer risk,” Radiology 260, 50–60 (2011). 10.1148/radiol.11102156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uematsu T., Kasami M., and Watanabe J., “Does the degree of background enhancement in breast MRI affect the detection and staging of breast cancer?,” Eur. J. Radiol. 21, 2261–2267 (2011). 10.1007/s00330-011-2175-6 [DOI] [PubMed] [Google Scholar]
- 20.Yang Q., Li L., Zhang J., Shao G., and Zheng B., “A computerized global MR image feature analysis scheme to assist diagnosis of breast cancer: A preliminary assessment,” Eur. J. Radiol. 83, 1086–1091 (2014). 10.1016/j.ejrad.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C. and Li L., “3D segmentation of masses in DCE-MRI images using FCM and adaptive MRF,” Proc. of SPIE, 9034, Medical Imaging 2014: Image Processing. 10.1117/12.2042834 [DOI] [Google Scholar]
- 22.Yang Q., Li L., Zhang J., Shao G., Zhang C., and Zheng B., “Computer-aided diagnosis of breast DCE-MRI images using bilateral asymmetry of contrast enhancement between two breasts,” J. Digital Imaging 27, 152–160 (2014). 10.1007/s10278-013-9617-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q. and Doi K., “Reduction of bias and variance for evaluation of computer-aided diagnostic schemes,” Med. Phys. 33, 868–875 (2006). 10.1118/1.2179750 [DOI] [PubMed] [Google Scholar]