Abstract
Precise genome editing using engineered nucleases can significantly facilitate biological studies and disease treatment. In particular, clustered regularly interspaced short palindromic repeats (CRISPR) with CRISPR-associated (Cas) proteins are a potentially powerful tool for modifying a genome by targeted cleavage of DNA sequences complementary to designed guide strand RNAs. Although CRISPR/Cas systems can have on-target cleavage rates close to the transfection rates, they may also have relatively high off-target cleavage at similar genomic sites that contain one or more base pair mismatches, and insertions or deletions relative to the guide strand. We have developed a bioinformatics-based tool, COSMID (CRISPR Off-target Sites with Mismatches, Insertions, and Deletions) that searches genomes for potential off-target sites (http://crispr.bme.gatech.edu). Based on the user-supplied guide strand and input parameters, COSMID identifies potential off-target sites with the specified number of mismatched bases and insertions or deletions when compared with the guide strand. For each site, amplification primers optimal for the chosen application are also given as output. This ranked-list of potential off-target sites assists the choice and evaluation of intended target sites, thus helping the design of CRISPR/Cas systems with minimal off-target effects, as well as the identification and quantification of CRISPR/Cas induced off-target cleavage in cells.
Keywords: bioinformatics, bulges, off-target, program, RISPR, specificity
Introduction
Genome editing has successfully created cell lines and animal models for biological and disease studies, and promises to enjoy a wide range of therapeutic applications.1 In particular, engineered nucleases creating DNA double-strand breaks or single-strand breaks (“nicks”) at specific genomic sequences greatly enhance the rate of genomic manipulation. Double-strand breaks repaired by the cellular non-homologous end joining pathway often induce insertions, deletions, and mutations, which are effective for gene disruptions and knockouts. Alternatively, when a donor DNA is supplied, double-strand breaks and DNA nicks can be repaired through homologous recombination, which incorporates the donor DNA and results in precise modification of the genomic sequence. Regardless of the DNA repair pathway, it is important to minimize off-target cleavage in order to reduce the detrimental effects of mutations and chromosomal rearrangements. Although zinc finger nucleases and TAL effector nucleases have the potential to enjoy a wide range of applications, they were found to cleave at off-target sites at detectible rates.2,3,4,5,6
Clustered regularly interspaced short palindromic repeats (CRISPR), the bacterial defense system using RNA-guided DNA cleaving enzymes,7,8,9,10,11,12,13 is an exciting alternative to zinc finger nucleases and TAL effector nucleases due to the ease of directing the CRISPR-associated (Cas) proteins (such as Cas9) to multiple gene targets by providing guide RNA sequences complementary to the target sites.14,15 Target sites for CRISPR/Cas9 systems can be found near most genomic loci; the only requirement is that the target sequence, matching the guide strand RNA, is followed by a protospacer adjacent motif (PAM) sequence.16,17,18 For Streptococcus pyogenes (Sp) Cas9, this is any nucleotide followed by a pair of guanines (marked as NGG). Studies on CRISPR/Cas9 systems suggested the possibility of high off-target activity due to nonspecific hybridization of the guide strand to DNA sequences with base pair mismatches at positions distal from the PAM region.15,19,20,21 For CRISPR/Cas9 systems, recent studies have confirmed levels of off-target cleavage comparable with the on-target rates,22,23,24,25 even with multiple mismatches to the guide strand in the region close to the PAM. It has been revealed that RNA guide strands containing insertions or deletions in addition to base mismatches can result in cleavage and mutagenesis at genomic target site with levels similar to that of the original guide strand.26 To our knowledge, these studies provided the first experimental evidence that genomic sites could be cleaved when the DNA sequences contain insertions or deletions compared with the CRISPR guide strand. These results have clearly demonstrated the need to identify potential off-target sites when choosing guide strand designs and examine off-target effects experimentally when using CRISPR/Cas systems in cells and/or animals.
Herein, the term “insertion” is used when the endogenous DNA sequence has one or more extra bases compared with the sequence of the guide strand (a DNA bulge). Similarly, the term “deletion” is used when the DNA sequence has one or more missing bases compared with the guide strand (a RNA bulge). The term “indels” indicates either insertions or deletions. Although insertions and deletions may be viewed as mismatches, we use the term “mismatch” exclusively for base-pair mismatch when the guide strand and the potential off-target sequence have the same length, but differ in base composition.
A number of CRISPR tools have been developed, including Cas Online Designer,23 ZiFit,27 CRISPR Tools,23 and Cas OFFinder,28 for different functions.23,28,29,30,31,32,33 However, none of these bioinformatics search tools has considered the off-target sites due to insertions or deletions between target DNA and guide RNA sequences, nor provide application-specific primers. As revealed by our recent experimental study, off-target cleavage could be detected in cells with 15 different insertions and deletions between the guide strand and genomic sequence, sometimes at rates higher than that of the perfectly matched guide strand.26 Therefore, it is important to have a bioinformatics tool to identify potential off-target sites that have insertions and/or deletions between the RNA guide strand and genomic sequences, in addition to base-pair mismatches. To address this unmet need, we have developed a bioinformatics-based tool, COSMID (CRISPR Off-target Sites with Mismatches, Insertions, and Deletions), to search genomes for potential CRISPR off-target sites (http://crispr.bme.gatech.edu). COSMID ranks the potential off-target sites based on the number and location of mismatches, allowing the selection of better target sites and/or experimental confirmation of off-target sites using the primers provided. Therefore, the COSMID off-target search tool may significantly aid the design and optimization of CRISPR guide strands by selecting the optimal target sites with minimum Cas-induced off-target cleavage and by facilitating the experimental confirmation of off-target activity.
Results
COSMID search algorithm
The COSMID algorithm is based on sequence homology; it searches a genome of interest for sites similar to CRISPR guide strands using the efficient FetchGWI search program that has powered search tools including TagScan34 and ZFN-site.35 FetchGWI operates on indexed genome sequences that are precompiled and stored (Supplementary Figure S1). It can identify genomic locations with sequences that match any of the series of search entries. FetchGWI saves run time by searching indexed files that represent the genome sequences, rather than the sequences themselves. There is one index entry for each nucleotide in the genome, which allows a rapid and exhaustive search. This is a key advantage of COSMID over BLAST and other programs that scan nonoverlapping words and may miss potential off-target sites.35 COSMID currently allows searching the human, mouse, Caenorhabditis elegans, and rhesus macaque genomes. Other genomes will be added upon request.
COSMID search web interface
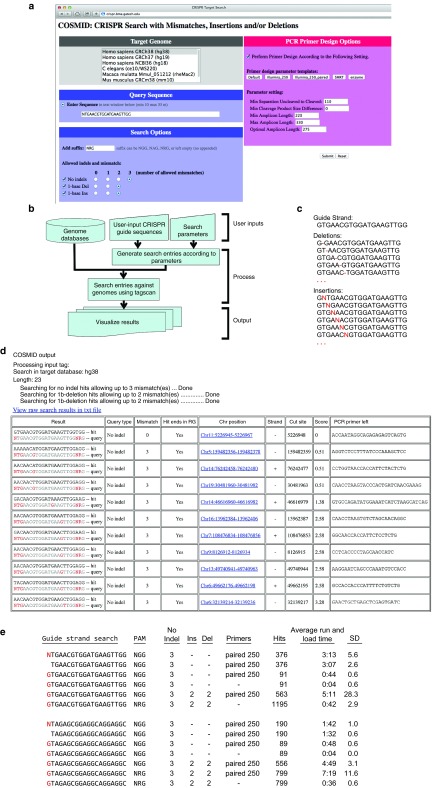
COSMID is an easy-to-use CRISPR off-target search tool with a web interface that allows directed and exhaustive genomic searches to identify potential off-target sites for guide strand choice or experimental validation. To perform a search, a user chooses the genome of interest from the list, and enters the guide strand and PAM sequences (Figure 1a). By clicking the appropriate selection buttons, a user can choose to include (i) ≤2 base mismatches with an insertion and/or deletion, or (ii) ≤3 base mismatches without any indels (Figure 1a). The user has the option to have primers as part of the output. Primers are designed by COSMID that are optimized to the specified criteria or to the defaults given for particular applications (Figure 1a). COSMID exhaustively scans the genome based on these input parameters (Figure 1b), allowing consideration of mismatches, insertions, and/or deletions (Figure 1c, Supplementary Figure S1). COSMID outputs a ranked list of perfectly matched (on-target site and possibly other sites) and partially matched (potential off-target) sites in the genome, their ranking score, along with reference sequences and primer designs that can be used for sequencing and/or mutation detection assays (Figure 1d). Each line of the output file describes one genomic locus matching the search criteria. A locus may appear on multiple lines if it can be modeled and found in multiple ways. Each hit is appropriately aligned to the query shown in the “Result” box (Figure 1d). DNA bases corresponding to mismatches and indels are shown in red in the query line. Similarly, ambiguity codes, such as N, are shown in red in the query line to identify the matching genomic bases. To the right of the “Result” box are boxes with the query type, number of mismatches, chromosomal position, score, primers, and other features. The web page showing COSMID output also includes links to test each primer pair and to reformat the output file as text or in a spreadsheet. The spreadsheet output allows thorough evaluation of the number and scores of the low-scoring sites that are predicted to be more likely off-target sites, which may provide important guidelines when evaluating and choosing guide strands and/or testing for true cleavage events using DNA samples from cells after CRISPR/Cas treatment.
Figure 1.
COSMID design, search steps and characteristics. (a) COSMID input consists of the guide sequence, type of PAM, allowed number of mismatches, insertions and deletions, genome of interest, and primer design parameters. (b) A flow chart showing the COSMID software design and the major steps in performing the search. (c) The search strings with insertions and deletions in the first six possible positions are shown. Alternate deletions of repeated bases are synonymous. (d) COSMID output in HMTL showing the genomic sites matching the user-supplied criteria in comparison to guide strand R-01 with chromosomal location. Scoring of the mismatches is provided for ranking, as are PCR primers and reference sequence. The right primers, in silico link, amplicon, and digest sizes are provided in the output, but not shown here. Links are provided to each location in the UCSC genome browser, and to the output file as a spreadsheet for further manipulation and primer ordering. (e) Run times were measured for COSMID using variations of guide strands R-01 and R-30, with and without a 5′G, using standard (NGG) or relaxed PAM (NRG). All runs included sites matching the guide strand with three or less mismatches without indels. More matching loci “hits” were identified by allowing single-base insertions or deletions together with ≤2 base mismatches. Allowing primer design increased the run times in proportion to the number of hits.
Validation of COSMID searches for putative off-target cleavage sites
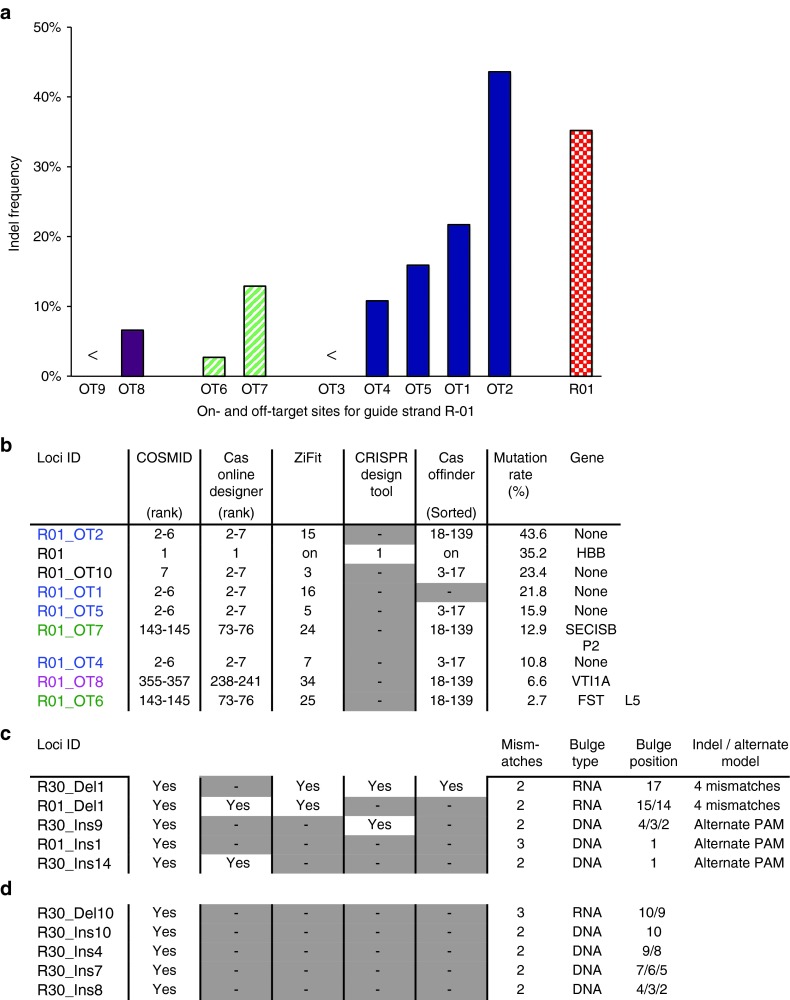
To validate COSMID predictions, mutation detection assays were performed to determine if off-target cleavage occurred at putative off-target sites identified by COSMID. A search for the guide strand R-01 (GTGAACGTGGATGAAGTTGG), which targets the human beta-globin gene24 gave 1,040 potential off-target sites in the human genome when allowing for up to three mismatches without any indels, and up to two mismatches with a one-base deletion or one-base insertion, adjacent to a NRG PAM (Figure 1a). Using primers as part of COSMID output, mutation detection assays were performed based on PCR amplification of the genomic loci36 after transfecting K-562 cells with a plasmid expressing Cas9 and guide strand R-01. A range of potential off-target sites without indels were studied in order to compare COSMID with other available bioinformatics tools. Of the 10 off-target sites tested, 8 sites, all with two mismatches, had off-target mutagenesis that could be detected by the T7EI mutation detection assay (Figure 2a, Supplementary Table S1), including an off-target site with higher activity than the on-target cleavage rate (44% versus 35%, Figure 2b). Similar to previous results obtained by us and others, the level of off-target activity was generally diminished at sites with mismatches closer to the PAM.19,20,21,22,23,24 Five different genomic sites with identical sequences, containing two mismatches respectively 14 and 19 bases from the PAM, had cleavage activities ranging from below the detection limit to 44%. The 10 sites chosen also contained two pairs of duplicated sites that had different mutation rates (13% versus 3%, and 7% versus below detection). The large variation in mutation rates at identical sequences, but different genomic regions may be due to the difference in gRNA/Cas9 accessibility and/or binding affinity at different genomic loci. This exemplifies the role genomic context can play in Cas9-induced cleavage and the difficulty in ranking off-target sites solely based on target sequences. Figure 2b lists these eight experimentally validated off-target sites in decreasing order of mutation rate (%), their ranking by COSMID, as well as that by other on-line CRISPR tools. In Figure 2b, a particular site is shown as a grey box if not found by a tool (e.g., R01_OT1 under “Cas OFFinder”). We further compared COSMID with other web tools for their ability to identify off-target sites that contain an extra bases (DNA bulge) or a missed base (RNA bulge) relative to the complementary genomic DNA sequence26 (Figure 2c). The off-target sites in Figure 2c can also be modeled as sites with four mismatches or noncanonical PAMs compared with the on-target site, though it is less likely that binding of Cas9 would occur without an NGG or NAG PAM. When an extra base is present in the genomic sequence, next to one or more of the same nucleotide, the DNA bulge may occur in multiple locations, such as in the off-target site R30_Ins9 where the additional G in the genomic sequence might be the first, second, or third of the three adjacent Gs, at locations 2, 3, or 4 nucleotides from the PAM (Supplementary Table S2). In addition to being modeled as having one insertion with two mismatches, this off-target site can be modeled as having three mismatches with a shift in the PAM from NGG to NAG. Further, the off-target site R01_Ins1 may be modeled as having a NAG PAM. Without a bulge, R30_Ins14 would need to have the unlikely GTA PAM, so it remains unclear how it was modeled by Cas Online Designer. Each site in Figure 2c,d is marked “yes” when found by COSMID (first column) or other search method; if any of the confirmed off-target site could not be identified by a search tool, it is shown as a grey box with a dash. Specifically, of the six off-target sites identified by COSMID (and previously sequence confirmed),26 Cas Online Designer, ZiFit, and CRISPR tools each only found two, and Cas OFFinder only found one. Figure 2d lists the sequence confirmed, off-target sites containing DNA or RNA bulges that could not be represented by other means. Each was identified by COSMID, but not by these search tools.
Figure 2.
Comparison of COSMID with other available tools in predicting off-target sites for guide strand R-01. (a) On- and off-target cleavage rates for guide strand R-01. Marked differences are seen in the cleavage rates at off-target sites with identical sequences, but different chromosomal locations, such as OT8 and OT9 (purple), OT6 and OT7 (green stripes), and OT1–OT5 (blue). The R-01 on-target indel rate is shown to the right (red pattern). (b–d) Comparison of COSMID with other available tools in predicting off-target sites for guide strand R-01. (b) Comparison of RO1 off-target sites that contain two mismatches. The cleavage rates at R-01 on-target site and off-target sites OT1–OT10 are listed by decreasing T7EI activity. OT3 and OT9 had activities below T7E1 detection limit. Sites with matching sequences (outside first base) have their names in bold with colors matching that shown in a. Annotated genes corresponding to the sites are listed. Off-target analysis was performed with different online search tools. If the genomic sites with measurable T7E1 activity (shown in a) were identified by a specific tool (such as Cas OFFinder), their rankings in its output (if sortable) are shown. Sites not in the output of that tool are indicated by a dash in a grey box. (c) Comparison of search results for off-target sites that contain deletions or insertions, in which sequence-verified off-target sites with insertions or deletions, which can also be modeled as loci with four mismatches or alternate PAM considered. (d) The sequence-verified off-target sites with insertions or deletions that cannot be modeled as four mismatches or alternate PAM can only be predicted by COSMID.
COSMID has better ability in identifying off-target sites with indels
Although a number of bioinformatics programs can be used for CRISPR designs, COSMID is unique in that it provides exhaustive genomic searches for off-target sites due to mismatches, deletions, and insertions, as well as providing primers for experimental validation of predicted off-target sites. The results shown in Figure 2b–d give examples of validated off-target sites identified by COSMID but not found by other search tools, including Cas Online Designer,23 ZiFit,27 CRISPR Tools,23 and Cas OFFinder,28 which have different functions, such as determining CRISPR guide sequences,30,31,32,37 scanning a genome for possible target sites, and comparing the potential off-target sites.23,33,38 In addition to providing optimized primer designs for sequencing and mutation detection for confirming putative off-target sites, COSMID also provides the reference sequence to facilitate sequencing. To illustrate, we compared the search results for two guide strands with validated activity and known off-target cleavage, including the guide strand R-01 that targets the human HBB gene, and the guide strand R-30 (GTAGAGCGGAGGCAGGAGC) that targets the human HIV co-receptor CCR5 gene.24,26 When the results of COSMID searches were compared with the output given by other existing search tools, we found that, when off-target sites contain insertions or deletions in addition to mismatches, only COSMID searches could identify all of the 10 sequence-validated off-target sites (Figure 2c,d and Supplementary Table S1). Note that the deletion contained in off-target sites R-01_Del1 or R-30_Del1 (Figure 2c) could be modeled as four mismatches, and the insertion in off-target sites R-01_Ins1, R-30_Ins9, or R-30_Ins14 (Figure 2c) could be modeled as having alternative PAMs. These alternative interpretations of the insertions and deletions for the sites shown in Figure 2c explain why some existing bioinformatics tools such as Cas Online Designer, ZiFit, CRISPR Tools, and Cas OFFinder could still identify some of the off-sites listed in Figure 2c, although these tools do not allow insertions or deletions to be considered in the searches. Since the insertions or deletions in off-target sites R-30_Del10, R-30_Ins4, R-30_Ins7, R-30_Ins8, R-30_Ins10 (Figure 2d) could not be modeled as either mismatches or having alternative PAM, they were not found by these tools.
Extensive searches for HBB-targeted (R-01) and CCR5-targeted (R-30) guide strands
In addition to off-target sites of the same length as the guide strand but with mismatches, many similar sites exist in a genome with insertions (DNA bulges) and deletions (RNA bulges). As revealed recently, Cas9 can tolerate DNA and RNA bulges and induce cleavage at genomic loci with high rates, sometimes even higher than the target site.26 To further demonstrate the capabilities of COSMID, the guide strands R-01 and R-30 (refs. 24,26) were extensively analyzed using COSMID to search the human genome for sites similar to the R-01 or R-30 guide strands, having (i) up to three mismatches with no indels, (ii) up to two mismatches with a single-base insertion, and (iii) up to two mismatches with a single-base deletion. Since matching a guide strand's initial G is not essential, it was omitted in these searches. The off-target sites with a mismatched A at this position (OT1 and OT2) happened to have higher mutation rates than the three sites with a matching G (OT3–5) (Figure 2a). The outputs provided many possible off-target sites, including those with insertions or deletions (Supplementary Files S1–S4). The number of putative genomic off-target sites output by COSMID increased drastically when indels were allowed in the search. For example, allowing one-base insertions together with two mismatches increased the number of genomic sites adjacent to a NAG or NGG PAM ~3 and ~7 times for R-01 and R-30 respectively compared with those without indels and two mismatches (166 versus 49 for R-01 and 224 versus 34 for R-30, Table 1). When one-base deletions are allowed together with two mismatches, the number of genomic sites identified is even higher, ~18 and ~26 times higher for R-01 and R-30 respectively compared with those without indels (883 sites for R-01 and 883 sites for R-30) (Table 1). With one-base insertion or one-base deletion in addition to base mismatches, the number of unique loci found was greatly increased compared with the corresponding number without indels. For example, when a one-base deletion was allowed in addition to ≤2 mismatches, the unique off-target loci found by COSMID is 333 for R-01 and 761 for R-30 (Table 2).
Table 1. Comparison of search results for guide strands R-01 and R-30 with deletion or insertion permitted.
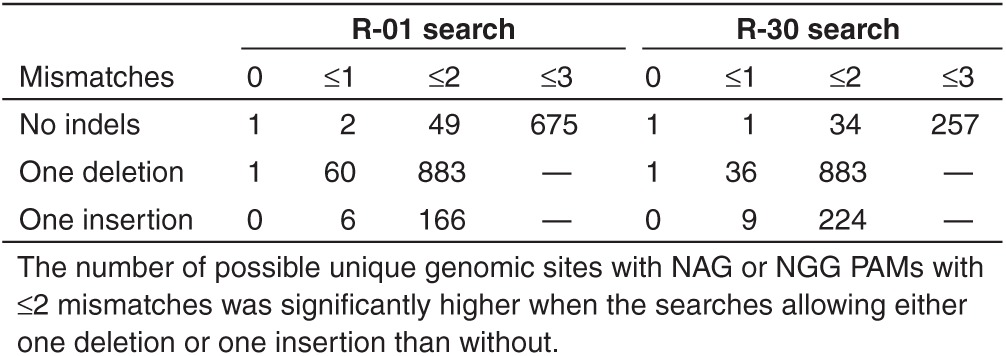
Table 2. The number of additional unique off-target loci identified for guide strands R-01 and R-30 when an insertion or deletion was allowed with ≤2 mismatches compared with the results when only searching with base mismatches.
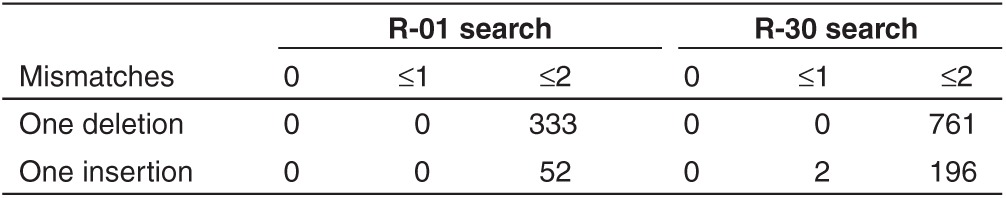
When allowing (i) up to three mismatches with no indels, or (ii) up to two mismatches with a one-base insertion, or (iii) up to two mismatches with a one-base deletion, COSMID searches of off-target sites for guide strands R-01 and R-30 with NRG PAM located 1,040 unique putative off-target sites for R-01 and 1,218 for R-30 (see the sorted tab in Supplementary Files S2 and S4). There were many identical sites located by multiple query types (examples shown in Supplementary Figure S2). The results varied between the two guide strands R-01 and R-30 (each targets a coding sequence), as can be expected in a nonrandom genome (Supplementary Figure S3). Specifically, we found that R-01 had a markedly larger number of matching sites with no indels. Of note was a particular 3-mismatch hit in 69 sites (Supplementary Files S1 and S2).
Run times
COSMID uses the TagScan algorithm to minimize run times while still performing exhaustive genome searches.34 With the primer design option off, the run times averaged 4 seconds for the guide strands without indels (Figure 1e). Run times were determined by inputting the guide strands to search the human genome, choosing either NGG or NRG for the PAM, and if insertions or deletions were allowed. Clearly, allowing insertions or deletions in addition to mismatches increases run time. For example, when searching with a 19-nt guide strand and an NRG PAM, and including two mismatches with either an insertion or an deletion resulted in run times averaging 42 seconds for R-01 and 36 seconds for R-30. The run times for the search with three mismatches without insertions or deletions were similar. We found that including primer design increased the run times proportional to the number of primer sets and reference sequences returned.
Discussion
Identifying off-target cleavage by CRISPR/Cas9 systems in a genome of interest is important, especially in treating human disease and creating model organisms, as CRISPR off-target cleavage22,23 can result in mutations, deletions, inversions, and translocations,24,39 inducing detrimental biological consequences and potentially causing disease. However, accurate and complete genome-wide analysis of off-target efforts is a daunting task, since unbiased sequencing of a full genome to determine off-target activity is very costly, and many nuclease-treated clones would have to be sequenced. Therefore, having a bioinformatics-based tool to predict and rank potential off-target cleavage sites can greatly aid the off-target analysis, and provide valuable guidance for guide strand designs. In particular, it is important to perform extensive bioinformatics searches for potential off-target sites that contain base mismatches, insertions, and deletions compared with the intended CRISPR target site.
As a novel CRISPR off-target search tool, COSMID can quickly and exhaustively search a genome for DNA sequences that partially match the target sequence of the guide strand, but contain insertions or deletions in addition to base mismatches. As shown in Table 2, a large number of potential off-target sites would be missed using search tools that only consider base mismatches, but not insertions or deletions. COSMID outputs potential off-target sites (“hits”) corresponding to allowed mismatches and indels, the PAM sequence and the chromosomal location of the hits. COSMID also outputs primer designs for experimental validation of the off-target sites. Further processing of the COSMID results from the output spreadsheets extends COSMIDs utility to different CRISPR/Cas platforms, including the use of Cas9 nickase pairs,40 Cas9/FokI fusion,41,42 and multiplexed targeting15 by searching for multiple (sometimes paired) sites within a user-input chromosomal proximity (instructions and example in Supplementary File S5). In addition to aiding the design of CRISPR/Cas systems for DNA cleavage, COSMID can be used to identify potential off-target sites of CRISPR activators, repressors, or other effector domains.43
The potential off-target sites given in the COSMID output can be tested experimentally using mutation detection assays36 or deep sequencing with genomic DNA harvested from cells treated by CRISPR/Cas. Mutation detection assays, including Surveyor and T7EI, are very commonly used to measure on- and off-target cleavage and mutagenesis.36 COSMID facilitates these assays by automatically designing primers to enable facile gel separation of the uncleaved and cleavage bands. The output also includes the genomic reference sequence for comparison to the sequencing results.
COSMID scores the potential off-target sites based on the number and location of base mismatches, allowing ranking of the more likely off-target sites. Due to limited experimental results, quantitatively considering the effect on off-target rates for sites with insertions or deletions, including their number, location, and combinations, is not possible at this point. Therefore, the scoring and ranking system in COSMID will continue to be refined as additional experimental results become available, which may suggest weighting for the effect of insertions or deletions in combination with base mismatches.
Bioinformatics based ranking of CRISPR/Cas off-target sites may be hindered by the effects of genomic context and DNA modifications. Identical genomic sites and duplicated sites may have dramatic differences in off-target activity (Figure 2a). The indel rate at off-target site R-01_OT2 was 44%, though other loci with the same complementary sequence have much less, or no activity, possibly due to nuclease blocking. The high level of variability of Cas9 cleavage at identical sites makes quantitative prediction and accurate ranking difficult, as the ranking may only indicate the possible effects with open context. Further experimental studies and modeling are therefore needed to incorporate the effects of chromatin condensation, DNA availability and other factors in the COSMID search algorithm.
Materials and Methods
COSMID search inputs. To perform a COSMID search, the genome of interest, guide strand, PAM sequence, and the number of base mismatches, insertions, and deletions allowed are specified (Figure 1a, Supplementary Figure S1). Three types of indel query are allowed: (i) the number of mismatches with no insertion or deletion (No indels); (ii) the number of mismatches in addition to a single-base deletion (Del); and (iii) the number of mismatches in addition to a single-base insertion (Ins). Up to three mismatches without indels, and up to two mismatches together with a one-base insertion or deletion could be chosen. If primers are desired, primer design parameter settings and parameter templates should also be entered (Figure 1a). PAM variants, such as NRG can be entered in the suffix box, as well as other PAM sequences.44 The spacer (Ns) and required nucleotides are entered into the suffix box, such as “NNNNGATT”,45 and include genomic sites with any nucleotide at the N positions in the output. Before performing the search, COSMID constructs a series of search entries according to the user-specified guide strand and search criteria (Figure 1b). The search entries include all insertions and deletions at each possible location (Figure 1c), and are subsequently used to perform rapid and accurate searches of the entire sequence of the interested genome, while allowing for the user-specified number of mismatches. These searches took ~4 seconds without primer design (Figure 1e).
Although multi-base deletions (RNA bulges) and insertions (DNA bulges) could be tolerated,26 they are less common, and search for a wide range of insertions and deletions will likely result in a very large number of returned sites. Therefore, COSMID only allows searches for single-base insertions and deletions in the DNA sequence compared with the guide strand (Figure 1a). For the potential off-target sites, the search algorithm allows some ambiguities (such as N for any nucleotide). Ambiguities included in the search string are marked in red in the HTML results (as are mismatches and indels), but are not counted toward the user-specified mismatch limits. The use of ambiguities allows the inclusion of the matching genomic base with the output sequences. One possibility is to include an “N” in positions that can have substitutions, such as the first base in a guide strand that is often a G primarily to aid in transcription, but does not need to match the complementary target sequence.23,24,46 One can leave off this base when performing a search, or include a 5′ N in the search string, which allows COSMID to output and align to the “N,” the corresponding 5′ bases at each locus.
COSMID search outputs. COSMID outputs all genomic sequences that match the user-supplied search criteria in comparison with the entered guide strand. The first column of the HTML output shows the genomic sequence (“hit”) aligned to the query sequence with matches shown in black. Nucleotides that are not a direct match are colored red, including mismatches, insertions, and deletions (Figure 1d). Ambiguities in the query sequence, such as the N in the PAM sequence NGG, are also shown in red, though they do not count as mismatches. The second column lists the query type, including (i) no deletion or insertion (No indel), (ii) deletions (Del), or (iii) insertions (Ins). This column indicates if there are insertions or deletions, and specifies the indel positions as the number of nucleotides away from the PAM. The third column lists the number of mismatched bases between the query and target sequences. When two repeated bases appear in the guide strand, a deletion of either one of them in the target sequence gives the same query sequence, so the ambiguity is noted in the query column. The fourth column indicates if the PAM in the hit ends in RG, as NGG is the Cas9 PAM with the highest activity, followed by NAG.23 This column helps in ruling out genomic sites with unlikely PAMs. This function must be added to the excel spreadsheet for other PAMs. The fifth, sixth, and seventh columns contain respectively the chromosomal location of the matching sequence, its strand and the chromosomal location of the cleavage site. The predicted cleavage position is based on the fact that Cas9 primarily cleaves both DNA strands three nucleotides from the PAM.14 The HTML links included in the COSMID output are directed to the chromosomal sites in the UCSC genome browser. This allows determination of the gene that best matches the target sequence and if the target site is in an exon, intron, or other region. This information is helpful as mutations may be better tolerated in regions that are noncoding and nonfunctional.
The output is grouped by query types, including (i) genomic sites with base mismatches, but no insertions or deletions (No indels), (ii) sites with deletions (Del), and (iii) sites with insertions (Ins) between the query and potential off-target sites (Figure 1d). Within each category, sites with mismatches further from the PAM are listed first, which are more likely to result in off-target cleavage.22,23,24 The same genomic location may satisfy two or more search criteria, such as those sites that satisfy the mismatched base limit without and with an insertion or deletion. For example, mismatches at the base farthest from the PAM and deletions of this base will give the same set of genomic locations. This can also occur when the guide strand contains consecutively repeated bases. Since genomic locations can be specified through multiple criteria (examples shown in Supplementary Figure S2), they are listed in each of the corresponding groupings to aid further evaluation and scoring. Duplicate sites can be removed in the spreadsheet, as described below.
COSMID also outputs the potential off-target sites identified in a spreadsheet to allow for further processing, such as sorting by attributes or adding weight matrixes to rank the most likely off-target sites. The accumulation of additional experiments on CRISPR off-target activity will allow creation of a more predictive scoring system. It is thought that mutations in the PAM are least well tolerated followed by sites closest to the PAM; however, little is known about how the guide strand sequence influences these effects.20,22,23,24 The spreadsheet can also be used to indicate duplicate genomic sequences found using different search criteria, as mentioned above. The output list of off-target sites allows a user to compare the number and score of off-target sites for the input target sites.
COSMID's primer design function is used to assay for off-target cleavage after cells or animals are treated with CRISPR guide strands and nuclease. Primers are designed that fit the criteria needed for the particular assay or sequencing platform using an automated primer pair design process, not found in other CRISPR programs. The algorithm was developed for the zinc finger nucleases and TAL effector nucleases off-target search program PROGNOS and found to give a single specific band in ~93% of amplifications.47 The automated primer design alleviates the need for the iterative steps of primer design and verification of the resulting fragment sizes, that slow primer design, especially for mutation detection assays where the cleavage product sizes determine how easily the cleavage bands can be distinguished on gels. The recommended parameters for use in Surveyor assays resolved on 2% agarose gels are: Minimum Distance Between Cleavage Bands—100 bp, Minimum Separation Between Uncleaved and Cleaved Products—150 bp. Users can also input the number of bases the cleavage site must be from each amplicon's edge to ensure sequencing coverage depending on the different sequencing platforms. For single molecule, real-time (SMRT) sequencing, the recommended parameters are: Minimum Distance Between Cleavage Bands—0, Minimum Separation Between Uncleaved and Cleaved Products—125 bp. The output primers can be easily modified in the spreadsheet, such as to add flanking sequences for additional amplification and/or barcodes for sequencing.
CRISPR transfection and mutation detection assays. The on- and off-target cleavage activity of Cas9 and guide strand R-01 was measured using the mutation rates resulting from the imperfect repair of double-stranded breaks by non-homologous end joining. An amaxa Nucleofector 4D was used to transfect 200,000 K-562 cells with 1 μg px330 expressing R-01 sgRNA, following manufacturer's instructions. The genomic DNA was harvested after 3 days using QuickExtract DNA extraction solution (Epicentre, Madison, WI), as described.36 On- and off-target loci were amplified using AccuPrime Taq DNA Polymerase High Fidelity (Life Technologies, Carlsbad, CA) following manufacturer's instructions for 40 cycles (94 °C, 30 seconds; 52–60 °C, 30 seconds; 68 °C, 60 seconds) in 50 µl reactions containing 1 µl of the cell lysate, and 1 µl of each 10 µmol/l amplification primer. The T7EI mutation detection assays were performed, as per manufacturers protocol,48 with the digestions separated on 2% agarose gels (Figure 2a) and quantified using ImageJ (Figure 2b).36 This guide strand was shown to have on-target cleavage at beta-globin and off-target cleavage at delta-globin,24 so a range of off-target sites were chosen, including two pairs of identical sites (OT6–OT7 and OT8–OT9) and five identical sites (OT1–OT5) to test for off-target mutations and evaluate the role of genomic context on cleavage and mutation rates. It is hoped that increased cellular data, such as provided in ENCODE for some cell lines, may prove useful in this regard.
SUPPLEMENTARY MATERIAL File S1. COSMID HTML output for R-01, HBB-directed guide strand searching the human genome allowing 3 mismatches with no indels, 2 mismatches with one insertion or deletion. File S2. COSMID spreadsheet output for R-01, HBB-directed guide strand searching the human genome allowing 3 mismatches with no indels, 2 mismatches with one insertion or deletion. File S3. COSMID HTML output for R-30, CCR5-directed guide strand searching the human genome allowing 3 mismatches with no indels, 2 mismatches with one insertion or deletion. File S4. COSMID spreadsheet output for R-30, CCR5-directed guide strand searching the human genome allowing 3 mismatches with no indels, 2 mismatches with one insertion or deletion. File S5. COSMID spreadsheet output for use with paired CRISPR systems, such as nickases and FokI fusions. Figure S1. Description of the search string processing by COSMID and examples showing the search strings, and portions of the web results and spreadsheet output for a search of the human genome using guide strand R-01. Figure S2. Two examples of genomic sites identified using different search queries for R-30. Figure S3. Chromosomal locations of COSMID search results for (A) R-01 and (B) R-30 with up to three mismatches (red) or with up to two mismatches and either an insertion or a deletion (blue). Table S1. Genomic sequences and chromosomal positions of the off-target sites tested using the mutation detection assay in Figure 2. Table S2. Sequence-verified off-target sites with mismatches and 1-base insertion (Ins) or deletion (Del).
Acknowledgments
This work was supported by the National Institutes of Health through a Nanomedicine Development Center Award (PN2EY018244 to G.B.).
Supplementary Material
References
- Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu TI, Cathomen T. Quantification of zinc finger nuclease-associated toxicity. Methods Mol Biol. 2010;649:237–245. doi: 10.1007/978-1-60761-753-2_14. [DOI] [PubMed] [Google Scholar]
- Ramirez CL, Certo MT, Mussolino C, Goodwin MJ, Cradick TJ, McCaffrey AP, et al. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 2012;40:5560–5568. doi: 10.1093/nar/gks179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson L, Usal C, Ménoret S, Leung E, Niles BJ, Remy S, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151 Pt. 8:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155 Pt. 3:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Shah SA, Erdmann S, Mojica FJ, Garrett RA. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38 suppl.:W462–468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Cheng Z, Kong L, Zhu Z, Lin S, Gao G, et al. CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics. 2014;30:1180–1182. doi: 10.1093/bioinformatics/btt764. [DOI] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau C, Gonnet M, Le Romancer M, Nicolas J. CRISPI: a CRISPR interactive database. Bioinformatics. 2009;25:3317–3318. doi: 10.1093/bioinformatics/btp586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseli C, Ambrosini G, Bucher P, Jongeneel CV. Indexing strategies for rapid searches of short words in genome sequences. PLoS One. 2007;2:e579. doi: 10.1371/journal.pone.0000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Ambrosini G, Iseli C, Bucher P, McCaffrey AP. ZFN-site searches genomes for zinc finger nuclease target sites and off-target sites. BMC Bioinformatics. 2011;12:152. doi: 10.1186/1471-2105-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronda C, Pedersen LE, Hansen HG, Kallehauge TB, Betenbaugh MJ, Nielsen AT, et al. Accelerating genome editing in CHO cells using CRISPR Cas9 and CRISPy, a web-based target finding tool. Biotechnol Bioeng. 2014;11:1604–1616. doi: 10.1002/bit.25233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Wang Z, Hu Y, Wu Y, Luo Z, Yang Z, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013;41:e141. doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S, Maier LK, Stoll B, Brendel J, Fischer E, Pfeiffer F, et al. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J Biol Chem. 2012;287:33351–33363. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine EJ, Cradick TJ, Zhao CL, Lin Y, Bao G. An online bioinformatics tool predicts zinc finger and TALE nuclease off-target cleavage. Nucleic Acids Res. 2013;42:e42. doi: 10.1093/nar/gkt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.