Abstract
Epstein–Barr virus (EBV) causes several benign and malignant disorders of lymphoid and epithelial origin. EBV-related tumors display distinct patterns of viral latent gene expression, of which the BamHI-A rightward frame 1 (BARF1) is selectively expressed in carcinomas, regulated by cellular differentiation factors including ΔNp63α. BARF1 functions as a viral oncogene, immortalizing and transforming epithelial cells of different origin by acting as a mitogenic growth factor, inducing cyclin-D expression, and up-regulating antiapoptotic Bcl-2, stimulating host cell growth and survival. In addition, secreted hexameric BARF1 has immune evasive properties, functionally corrupting macrophage colony stimulating factor, as supported by recent functional and structural data. Therefore, BARF1, an intracellular and secreted protein, not only has multiple pathogenic functions but also can function as a target for immune responses. Deciphering the role of BARF1 in EBV biology will contribute to novel diagnostic and treatment options for EBV-driven carcinomas. Herein, we discuss recent insights on the regulation of BARF1 expression and aspects of structure-function relating to its oncogenic and immune suppressive properties. © 2013 The Authors. Reviews in Medical Virology published by John Wiley & Sons, Ltd.
INTRODUCTION
Epstein–Barr virus (EBV), a human gamma herpesvirus, infects over 90% of the world population and persists in its host for life, usually without complications. Primary infection frequently goes unnoticed early in life but may cause infectious mononucleosis if acquired during adolescence or adulthood. The virus initially infects submucosal B cells in the nasopharynx/oropharynx and transforms these into latently infected long lived memory cells, which are essential for virus persistence. EBV has dual tropism in vivo, infecting mainly B lymphocytes and epithelial cells 1–3. This dual tropism is reflected by its association with several lymphomas and carcinomas, where distinct viral gene products support viral genome maintenance and contribute to the oncogenic process 4–7. The immune system strongly responds to EBV-infected cells maintaining a lifelong well-balanced equilibrium, but the virus can escape into latency to evade elimination 7,8. This latent state, which is regulated by a small subset of viral genes, enables the virus to persist lifelong in the human host 4.
Since its discovery and proven oncogenic role in African Burkitt lymphoma 9,10, EBV has been found to play a diverse and complex role in multiple chronic and malignant diseases occurring worldwide 11,12. EBV causes most lymphoproliferative diseases that occur in immunocompromised individuals 13,14 and is causally linked to other lymphoid malignancies, i.c. 40–100% of classical Hodgkin's disease 14, 10–30% of non-Hodgkin lymphomas, and 100% of B-cell lymphomas in the elderly as well as extranodal T-cell/NK-cell lymphomas in immunocompetent persons 4,15. Moreover, EBV has a proven etiological role in certain epithelial malignancies, including 100% of undifferentiated nasopharyngeal carcinoma (NPC) 6,7,16 and approximately 10% of gastric cancer (GC) 4,17–19 worldwide. EBV-linked GC is considered a distinct disease entity compared with Helicobacter pylori-associated GC. EBV has been associated with breast cancer 20,21 and hepatocellular carcinoma 22,23, but supporting evidence is limited or contrary 24,25.
Besides two abundant small noncoding RNAs and about 44 distinct viral microRNAs that are widely expressed, the 172 000 basepair EBV genome contains >80 protein encoding reading frames that are largely silenced by epigenetic mechanisms 26. Only a few latent genes are expressed in EBV-related malignancies in distinct expression patterns, called latency types 4,26,27. Strikingly, EBV-positive epithelial malignancies show selective and abundant expression of a viral gene encoded in the BamHI-A rightward frame 1, encoding a polypeptide of Mr 29 000–33 000 called BARF1 28–31. In B cells and lymphomas, BARF1 expression is generally undetectable but can be induced during viral lytic replication 32,33.
In NPC, BARF1 may function as oncogene, parallel to the more widely studied latent membrane protein 1 (LMP1) 34. In EBV-positive GC, BARF1 is expressed in the absence of LMP1, possibly functioning as the main EBV oncogene in this disease 31. Since its first description, several distinct functions have been assigned to BARF1, varying from oncogenic to antiapoptotic and immune modulating. A schematic overview of BARF1 functions is given in Figure 1.
Figure 1.
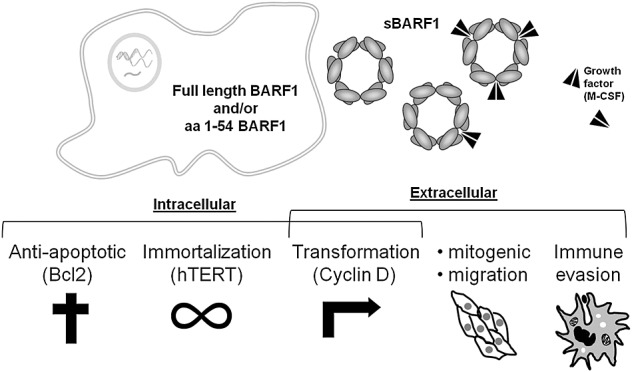
Putative functions of BamHI-A rightward frame 1. Schematic representation of functions ascribed to BamHI-A rightward frame 1
This review will discuss the role of BARF1 as a viral oncogene and immune modulator in EBV-driven carcinogenesis. It describes recent insights in BARF1 structure-function and its transcriptional regulation, as well as its potential use as a carcinoma-specific diagnostic factor.
BamHI-A rightward frame 1 protein structure
The first attempts to translate the BARF1 open reading frame (Figure 2A) in vitro revealed an intracellular monomeric polypeptide of Mr 26 000–33 00035,36, which was partially secreted in culture medium when linked to an immunoglobulin Fc-tail 37. Low level BARF1 protein expression was observed in Burkitt lymphoma cells induced into the lytic phase but not in uninduced cells. 38. Although initially a nuclear localization was reported 36, BARF1 protein was enriched in membrane fractions, and immunofluorescence analysis on fixed permeabilized cells showed a cytoplasmic Golgi localization 39–42. Later reports confirmed that the monomeric form of BARF1 is around Mr 29 000 with a lower band at Mr 25 000 40, and recent data show that BARF1 is distinctly glycosylated and rapidly and completely secreted as a soluble hexameric molecule BamH1-A rightward frame 1(sBARF1) when expressed in human epithelial cells 43–45. A recent study suggests cellular uptake of secreted sBARF1 with subsequent nuclear localization 46, which remains to be confirmed.
Figure 2.
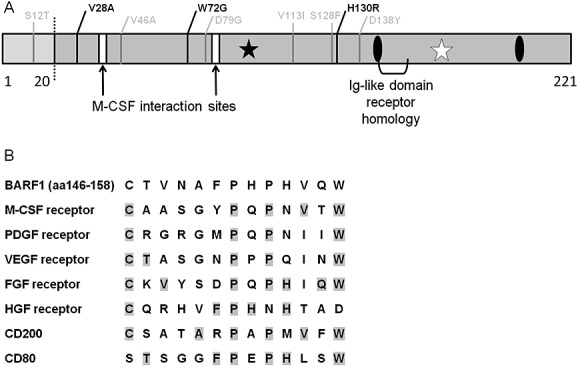
Mutations and homology domain of BamHI-A rightward frame 1(BARF1) protein. (A) Schematic representation of the BARF1 221 peptide. Left of the dotted line is the intracellular N-terminal part. Frequent amino acid mutations are depicted in black, rare mutations are depicted in gray. White bars; structural loops that interact with M-CSF, black asterisk; high mannose N-linked glycosylation at N95, white asterisk; predicted O-glycosylation site at T169, black ovals; C146 and C201 involved in folding and oligomerization. (B) BARF1 has sequence homology with a conserved domain found in several growth factor receptors. BARF1 aa146–158 is shown with homologous regions from the indicated receptors. Adapted from 37,48,53
The sBARF1 has high mannose N-linked glycosylation at Asn95, resulting in a sugar chain located at the inner side of the hexameric sBARF1 ring (Figure 3) 44,47–49. Asn95 N-glycosylation is essential for folding and secretion 47. An additional O-linked glycosylation site is present at Thr169, which carries a sialic end-group 44,48,49. Secreted BARF1 can be phosphorylated on both serine and threonine residues 49, but this remains unconfirmed 44. The cell-type expressing BARF1 may influence post-translational modifications, explaining small differences between publications. The human homolog of Drosophila tumorous imaginal disk 1 (hTid) was found to interact with BARF1, acting as a chaperone for proper folding and promoting its secretion as a monomeric protein 47. However, other studies on BARF1 expressing cell lines indicate rapid secretion of hexameric sBARF1, despite the presence of innate hTid 44,49. In human epithelial cells, most of the BARF1 translational product is rapidly and efficiently processed and cleaved from its putative aa1-20 leader sequence, yielding a secreted protein. Density gradient analysis revealed that sBARF1 is secreted as a hexameric complex of Mr 150 000–240 00044,48. In cells blocked for protein secretion by Golgi-modifiers monensin or brefeldin A, the BARF1 remains localized to perinuclear regions with reduced glycosylation, overlapping the endoplasmic reticulum. Upon release from blocking, BARF1 passes quickly through the Golgi system, paralleled by the addition of high mannose N-linked glycosylation, and can be detected in the cell membrane at later time points 44,47,49. The post-cleavage fate of the intracellular aa1-20 leader sequence remains undefined, but some important functions are assigned to this fragment 50.
Figure 3.
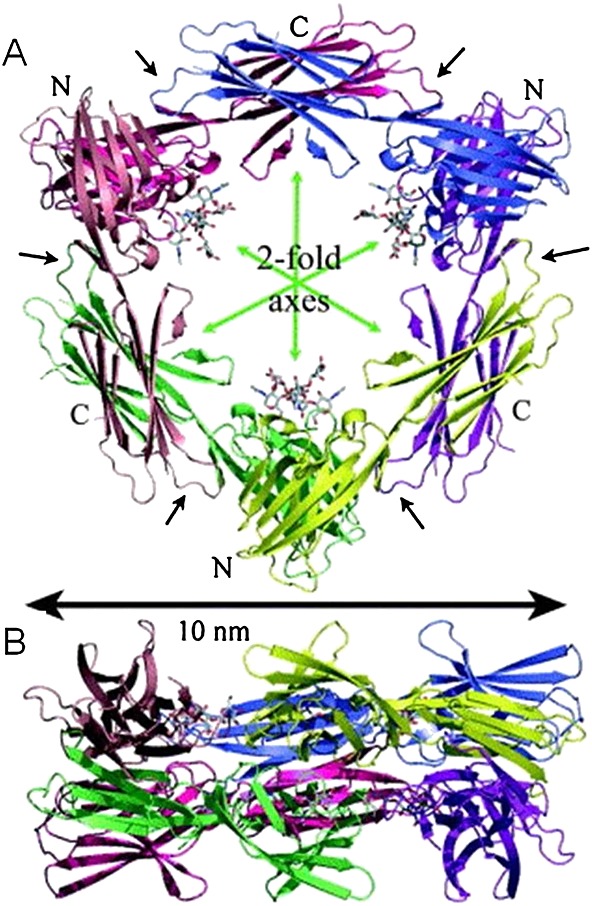
Cartoon of the soluble hexameric molecule BamH1-A rightward frame 1 (BARF1) hexameric structure. 48 (A) Top view of the BARF1 hexamer, the N-linked glycosylation is shown in a stick formation, (B) side view of the BARF1 hexamer
The BARF1 gene sequencing from NPC tumor samples revealed sequence variation, with 80.3% of samples having specific amino acid mutations compared with 33.3% in non-NPC isolates. The main substitutions found in Indonesian NPC samples are V29A, W72G, and H130R, but these conversions are not likely to change the protein tertiary structure or function 51. In northern Chinese samples, the V29A mutation is the most prominent conversion and is found more frequent in NPC than in GC and healthy controls 52. Four other amino acid changes (V46A, D79G, V113I, and D138Y) were detected in some samples (Figure 2A). None of the mutations is located in functional relevant domains of BARF1 (see in the succeeding text).
Cysteïnes 146 and 201 are involved in folding and oligomerization 47. Crystallographic analysis of sBARF1 (aa21–aa220) showed that sBARF1 forms hexameric rings in which three dimer molecules are interconnected head to tail, arranged in two layers (Figure 3). The C-terminal domains interact via hydrophobic contacts and hydrogen bonds, whereas the N-terminal domain also involves salt bridges representing a unique folding pattern 48. Early studies on BARF1 protein function were carried out with an Fc-tagged protein, which might have affected the 3D-hexameric conformation. The two N-terminal domains of the sBARF1 dimer bind each other similar to the dimerization domain of CD80, a co-stimulatory factor for T cells, but the residues forming the interaction have no similarities. The BARF1 protein has defined regions of homology to the tyrosine kinase receptor family (Figure 2B) and immunoglobulin super-family members CD80 and CD200, both having immunoregulatory and cell activating properties upon binding to their receptor 37,48,53. However, the relative orientations of the immunoglobulin-like domains differ between CD80 and BARF1 48. No study has yet addressed the latter homology at a functional level. The homology of BARF1 with the c-fms (the M-CSF receptor) will be discussed in detail later.
BamHI-A rightward frame 1 transcriptional regulation
The EBV gene expression is controlled by epigenetic modulation, and the majority of the EBV genome is methylated in latently infected cells 54. The BARF1 promoter region was found to be highly methylated, both in carcinoma and B-cell lines 55, indicating that BARF1 transcription activator(s) must be able to overcome methylation-induced repression. In EBV-infected B cells, BARF1 is considered an early lytic gene, whose expression is triggered by immediate early proteins BZLF1 (Z, Zta, ZEBRA, and EB1) and BRLF1 (R and Rta). Z and R are transcription factors that stimulate their own expression, reciprocally activate each other, and cooperatively induce expression of other early lytic viral proteins, allowing the virus to replicate 56,57. Z has enhanced ability to bind to methylated promoters 58. The Z-responsive elements in the BARF1 promoter are reversely oriented 59, and Z was found to only moderately transactivate the BARF1 promoter 55. R preferentially activates unmethylated lytic promoters; however, methylation does not inhibit DNA binding by R. Multiple potential R binding sites are present in the BARF1 promoter region 55, and Heilman et al. showed that R binds the bidirectional BALF2/BARF1 promoter 60. R was defined as the viral factor directly responsible for BARF1 transactivation upon lytic induction, and BARF1 mRNA can be detected within 6 h after transfection with an R expression vector 55, confirming its classification as early gene 61. A schematic view of BARF1 regulation is given in Figure 4.
Figure 4.
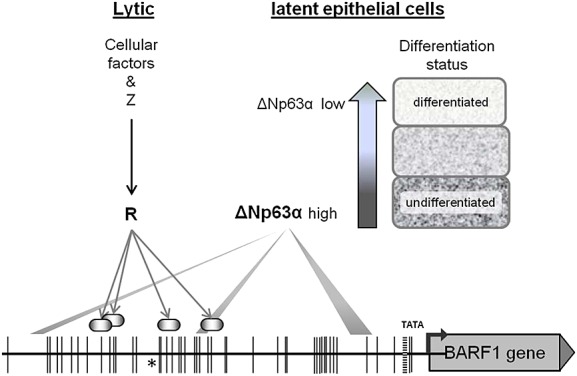
Schematic representation of BamHI-A rightward frame 1 (BARF1) transcriptional regulation. During lytic reactivation BARF1 is directly transactivated by the immediate early transcription factor R. Recent findings indicate that the differentiation marker ΔNp63α might be the cell type-specific transcription regulator enabling BARF1 transcription in latent in epithelial cells. The BARF1 promoter region is depicted up to −679 nucleotides relative to the BARF1 start-codon encoding methionine (ATG) start site and black vertical lines indicate methylation sites. Arrows point to the R binding sites in the BARF1 promoter 55, shaded bars indicate sites likely to be the most important for ΔNp63α BARF1 transactivation 62
Thus, during lytic replication, BARF1 expression is regulated by R, yet BARF1 is also expressed during latency. However, this occurs almost exclusively in undifferentiated epithelial tumors suggesting that BARF1 promoter activation is cell type-specific and may involve epithelial cell-specific transcription factors. Therefore, we recently explored potential candidates involved in epithelial-specific BARF1 expression. The BARF1 gene control region harbors multiple p53-p63-p73 family binding sites. Indeed, from this family of transcription factors, we identified the epithelial differentiation marker ΔNp63α as the major activator of BARF1 transcription in the epithelial background, functioning independently of virus lytic activation 62. ΔNp63α is a key regulator of epithelial cell development and differentiation 63, and overexpression of ΔNp63 is observed in nasopharyngeal carcinoma 64–66. Both LMP1 and LMP2A are involved in activation of ΔNp63α transcription 67–70. LMP1 down-regulates cellular microRNA 203, leading to higher levels of ΔNp63α 71. Whereas p63 is expressed in lymphoid cells, the dominant forms of p63 detected in B cells and lymphoma tissues are TA-p63 isoforms and not the ΔNp63α isoform 72. ΔNp63α is a stable factor during EBV latency in epithelial cells, and direct binding of ΔNp63α to the BARF1 control region enhances transcriptional activation in a reporter construct. However, overexpression of ΔNp63α was not sufficient to activate BARF1 expression in the context of the intact viral genome. Other cofactors might be required for full BARF1 upregulation by ΔNp63α in the epithelial cell background. The undifferentiated character of EBV-driven malignancies might be important to maintain expression of BARF1.
BamHI-A rightward frame 1 expression in Epstein–Barr virus-related malignancies
The BARF1 gene is consistently transcribed in EBV-linked carcinomas but virtually silent in EBV-driven lymphomas 29. The abundant transcriptional activity within the BamH1-A region of the EBV genome in NPC tumors was first described in 1989 73. Subsequent work dissected these transcripts into a family of BamH1-A rightward transcripts (BARTs), later identified as precursors of EBV miRNA's 74,75, differently spliced transcripts encoding putative proteins with speculative function, such as RK-BARF0, RPMS1, and A73 76–79, and a nonspliced transcript encoding the BARF1 protein.
The BARF1 transcription in NPC tissue was first demonstrated using reverse transcription polymerase chain reaction in a small cohort of North African NPC patients 80 and subsequently confirmed by nucleic acid sequence-based amplification (NASBA) (Figure 5), a technique allowing specific nonspliced RNA analysis in a homologous DNA background 28,29. BARF1 transcripts were also abundant in nasopharyngeal brushings of NPC patients, but not in healthy individuals, directly reflecting the presence of NPC cells in situ 30. More recently, BARF1 transcripts were detected in microdissected NPC tumor biopsies 81, establishing BARF1 expression deeper in the tumor.
Figure 5.
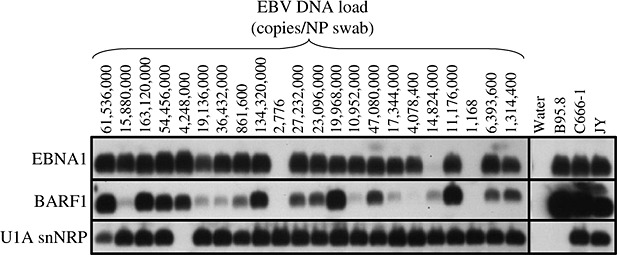
BamHI-A rightward frame 1 mRNA is abundantly detectable in nasopharyngeal (NP) brushings. 30 Epstein–Barr nuclear antigen 1 and BamHI-A rightward frame 1 RNA expression in NP brushings samples of NP carcinoma patients as determined by nucleic acid sequence-based amplification, in relation to Epstein–Barr virus DNA load. Shown is an autoradiogram of nucleic acid sequence-based amplification products hybridized with a radioactive-labeled internal oligonucleotide probe
Approximately 10% of all gastric adenocarcinomas comprise monoclonal EBV-positive cells 5,19. Zur Hausen et al. first demonstrated BARF1 expression in EBV-positive GC 31, later confirmed by Wang et al. 82. Seto et al. 17 showed that BARF1 in GC cells is expressed as a true latent gene. GC has a unique EBV latency pattern without expression of LMP1 leaving BARF1 as the major EBV oncogene in these tumors 31. In all carcinoma cells, BARF1 is expressed in absence of lytic cycle gene expression 17,83–85.
Nasal T-cell/NK-cell lymphoma is a rare and aggressive subtype of non-Hodgkin lymphoma (NHL), and particularly, the nasal type is closely associated with EBV 4,5,86–89. Zhang et al. found BARF1 mRNA in EBV-positive T-cell/NK-cell lines 89. Because these T-cell/NK-cell lines also expressed other early lytic genes, BARF1 expression may be induced by the immediate early gene R, which remains to be defined.
Several groups have investigated a role of EBV in breast cancer 20,21,90,91. In two studies, BARF1 mRNA was detected in roughly half of the tumors analyzed 20,91. However, EBV presence in breast cancer may be linked to sporadic lytic replication in ductal cells rather than associating with the proliferating tumor cells 24. A similar situation may hold for the link between EBV and hepatocellular carcinoma, where conflicting data have been published 23,25.
Although expression of BARF1 mRNA in NPC and GC biopsies is clearly described, detection of BARF1 protein proved to be more challenging. BARF1 protein was detected in whole tissue lysates in only two of seven NPC tissue samples by immunoblot analysis 17, and only one study managed to detect BARF1 protein by immunohistochemistry 83, showing uniform cytoplasmic and membrane staining of NPC biopsy tissue. However, this may be due to false-positive staining (unpublished data) 19. In our hands, despite having various monoclonal and polyclonal antibodies with excellent specificity to detect distinct linear and conformational epitopes of the BARF1 protein, suited for detection of recombinant BARF1 in formalin-fixed, paraffin-embedded insect cells, we were not able to detect BARF1 at the protein level in NPC or GC tissues, proven to have BARF1 mRNA expression 92. BARF1 protein is probably rapidly and completely secreted from tumor cells, as shown for sBARF1 transfected cells 40. In 2007, evidence for presence of sBARF1 in serum and saliva of NPC patients was provided 45, but this remains to be confirmed to date (see succeeding text).
ONCOGENIC PROPERTIES OF BamHI-A RIGHTWARD FRAME 1
The role of BamHI-A rightward frame 1 in host cell immortalization and transformation
Griffin et al. first suggested BARF1 to be involved in EBV-driven carcinogenesis 25 years ago 93. A BamHI-A fragment of the EBV genome, named P31 and later demonstrated to encode the BARF1 protein, was found to immortalize monkey kidney epithelial cells, surviving more than 200 passages and gaining the ability to grow in low-serum conditions. Subsequent studies showed that human epithelial cells could be immortalized by P31 as well 94,95. Moreover, mouse fibroblast lines NIH-3 T3 and Balb/c-3 T3 were immortalized upon transfection with a plasmid containing the putative BARF1 open reading frame, and importantly, BARF1 transfected cells induced tumor formation in newborn rodents 35. The related tumor tissues were BARF1 mRNA positive, but the BARF1 transgene was lost upon further in vitro passage. BARF1-immortalized monkey kidney epithelial cells were tumorigenic in severe combined immunodeficiency (SCID) mice but not in nude mice 96
To define which part of the BARF1 protein was essential for malignant transformation, Sheng et al. created six deletion mutants of the BARF1 gene 50. The first 54 amino acids at the N-terminus, including the membrane-spanning domain, were found to be responsible for the malignant transformation of BARF1 in rodent fibroblasts. In addition, this region was also responsible for the upregulation of the cellular antiapoptotic protein Bcl-2, mediating escape from senescence upon serum deprivation and mediating growth in soft agar. These results strongly suggest a role of BARF1 in immortalization and malignant transformation in vitro. Our unpublished data on the growth properties of stable BARF1-expressing human embryonic kidney epithelial (HEK-293) cells confirm the BARF1-driven increased growth in soft agar (Figure 6) and altered migration properties.
Figure 6.
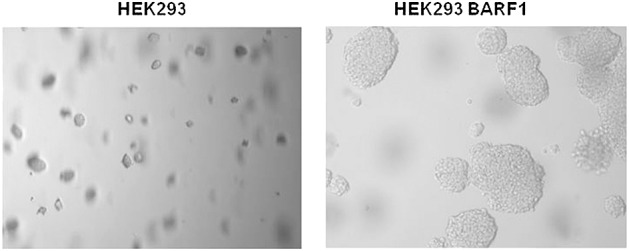
Anchorage-independent growth in soft agar. Unpublished results from our own group demonstrate contact independent growth of BamHI-A rightward frame 1 expressing 293HEK cells
Primate epithelial cells infected with NPC-derived EBV virions became immortalized, expressing both Epstein–Barr nuclear antigen 1 and BARF1, in contrast to their non-infected counterparts 97. Recently, Seto et al. 98 investigated the malignant phenotype of EBV-negative NPC cells, infected with a genetically modified EBV expressing BARF1 at increased levels from a SV40 promoter. In these cells, BARF1 expression was found to contribute to the tumorigenicity of EBV-infected NPC-cell lines in vivo. LMP1, the other known EBV oncogene, was not expressed in both epithelial models.
Although BARF1 expression can induce malignant transformation in several cell types, not all BARF1 immortalized primary epithelial cells were found to be tumorigenic 42,97,99. Recent studies showed that h-RAS coexpressing epithelial cell lines (PATAS and human epithelial cell line N69) become tumorigenic upon introduction of BARF1 suggesting that h-RAS expression synergizes with BARF1. After immortalization and transformation of PATAS cells by BARF1, activation of telomerase was observed, as well as activation of other signal pathways, including c-myc 100. These results suggest that the transforming abilities of BARF1 are partly mediated by activating pro-oncogenic cellular pathways 101. In BARF1-transfected human cervical cancer cells (Hela), activation of telomerase was observed, but not in macaque fibroblast-like cells, suggesting species differences in BARF1-induced telomerase activation 33,102. Intracellular pathways mediating BARF1-driven epithelial cell transformation and carcinogenesis should be further clarified to define the oncogenic role of BARF1.
Upregulation of the transcription factor and proto-oncogen c-myc was also described in the BARF1-immortalized EBV-negative human B-lymphocyte line Louckes 41. Inoculation of BARF1 transformed Louckes cells induced tumors with diffuse lymphoid phenotype in newborn rats and immune compromised mice, though not as aggressive as observed with transformed rodent fibroblasts. This process proved to be BARF1-specific, since cells loosing BARF1 expression were no longer tumorgenic. Also EBV-negative subclones of Burkitts lymphoma and EBV-negative Akata cell lines underwent malignant transformation upon transfection with BARF1 and were tumorgenic in SCID mice, like their EBV-positive Akata counterpart 103. Using a BARF1 knock-out virus, two studies described that BARF1 has a negligible role in the transformation or establishment of EBV latency in B cells. Cohen et al. 38 showed that addition of secreted BARF1 protein did not influence EBV transformation of B cells. Furthermore, an EBV virus with a BARF1-deleted genome was not impaired for B-cell transformation, and its related B-cell lines induced lymphoid tumors in SCID mice at the same rate as cells infected with wild-type (WT) EBV. In the closely related rhesus lymphocryptovirus (rhLCV) model, a minor role for BARF1 was found in establishing viral latency. rhLCV with truncated BARF1 was less efficient than WT virus for B-cell immortalization but only at low multiplicity of infection. Remarkably, this truncated BARF1 still contained the 54 N-terminal amino acids reported to be essential for malignant transformation of rodent fibroblasts 104. These findings underline the different mechanisms of EBV-induced lymphomagenesis and carcinogenesis, the former preferentially involving LMP1 as oncogene and the latter involving BARF1, either with or without LMP1.
BamHI-A rightward frame 1 as mitogenic growth factor
A fundamental characteristic of tumors is their enhanced growth rate compared with normal tissues. BARF1-transfected mouse fibroblasts grow twofold to fourfold faster than control cells 35. Similar results were found in human B cells 41 and primary monkey and human epithelial cells 50,94,95,100. In some studies, the N-terminus seems to be essential for this growth-stimulating effect and considered responsible for the transforming properties. Cells transfected with BARF1 lacking the amino acids 1-54 stopped proliferating after 4 days and at low-serum growth conditions 50. In other studies, autocrine cell growth was induced by secreted BARF1, which could be blocked by anti-BARF1 antibodies 105. Moreover, sera of mice-carrying BARF1-positive human NPC tumor cells and semi-purified sBARF1 from NPC patient sera were described to have mitogenic activities, which could be specifically blocked by BARF1 antibodies 45. Recently, uptake and cell cycle activation by sBARF1 was demonstrated in human keratinocytes 46.
These results indicate that BARF1 may exert its growth-stimulating effect both in cis and in trans. However, we and others could not confirm direct mitogenic effects of purified hexameric sBARF1 on either lymphoid or epithelial cells 106.
Transfection with BARF1 has been related to formation of double-minute (DM) extrachromosomal bodies, containing viral DNA 99, driving positive selection for DM-carrying cells. Furthermore, two recent microarray studies showed that BARF1 transfection into GC cell lines leads to deregulation of genes involved in cell proliferation, mitosis, and cell cycle regulation 107,108. However, in one study, cell proliferation of non-transfected GC cells was not affected by BARF1 107, whereas the other study found the BARF1-induced overexpression of Cyclin D1, a positive regulator of the cell cycle. In vivo, EBV-positive GC cells show upregulated Cyclin D1 protein expression 108. These findings indicate a possible role for BARF1 in cell cycle deregulation.
The antiapoptotic role of BamHI-A rightward frame 1 in Epstein–Barr virus-related malignancies
The oncogenic role of BARF1 may involve inducing escape from apoptosis. Cells transfected with full length BARF1 escape from apoptosis induced by serum starvation, unlike cells transfected with a N-terminal 1-54 BARF1 deletion mutant, suggesting BARF1 to modulate the intrinsic apoptosis pathway. Gene expression profiling of BARF1-positive GC cells revealed down-regulation of defined caspases and upregulation of the antiapoptotic Bcl-2 protein 50,107. By activating Bcl-2, the release of mitochondrial cytochrome-c is inhibited, which is necessary to recruit and activate effector caspases, the executioners of apoptosis 109. Upregulation of Bcl-2 prevents cells from death induced by cytotoxic drugs such as Taxol, an activator of the intrinsic apoptotic pathway. BARF1-expressing GC cells were found to be resistant to Taxol-induced apoptosis 107. An additional function of Taxol is to phosphorylate and inactivate Bcl-2 110,111, which can be overcome by BARF1-expressing cells. In vivo, upregulation of Bcl-2 expression was observed in EBV-associated GC and NPC 108,112,113. Because LMP1, also known to induce Bcl-2 expression, is not present in GC, this upregulation might directly relate to BARF1 expression 31,114.
Immune-modulating role of BamHI-A rightward frame 1
An immune-modulating role for BARF1 was first suggested by Strockbine et al. using basic local alignment search tool comparison of the BARF1 sequence to predict homologies to human proteins 37. A small region at aa146-158 within BARF1 showed homology to the extracellular domain of the c-fms proto-oncogene, a member of the tyrosine kinase receptor family, which includes platelet-derived growth factor receptor (PDGFR), fibroblast (FGFR), hepatocyte (HGFR), vascular endothelial (VEGFR), and macrophage colony stimulating factor receptor (M-CSFR) (Figure 2B). BARF1 was shown to bind a ligand on the cell membrane of activated peripheral blood T cells, identified as the membrane-associated splice variant of human M-CSF.
Macrophage colony stimulating factor is a pleiotropic cytokine important not only for regulating viability, proliferation, and differentiation of mononuclear phagocytes including monocytes and macrophages but also for the differentiation of Langerhans and other dendritic cell (DC) subsets 115–118. Macrophages are scavenger cells with pro-inflammatory and anti-inflammatory functions, which make them both ally and foe of tumor cells and a dangerous opponent for viruses.
Recombinant Fc-tagged monomeric BARF1 coprecipitated the α, β, and γ splice variants of M-CSF 37. These results were recently confirmed with native hexameric sBARF1 53. Recombinant BARF1 inhibits proliferation of mouse bone marrow cells 37, and sBARF1 inhibits proliferation of the M-CSF-dependent human myeloid leukemia cell line MUTZ-3 in a dose dependent fashion, whereas granulocyte M-CSF-induced proliferation remains unaffected 53. Tyrosine kinase analysis confirmed that sBARF1 inhibits M-CSFR downstream signaling 53. In the rhLCV model, similar M-CSF-inhibiting and cell-immortalizing abilities of rhBARF1 were demonstrated 106, suggesting an evolutionary conserved function of sBARF1 as selective M-CSF scavenger, interfering with innate and adaptive immune processes. The sBARF1/M-CSF ratio needed to completely block M-CSF-dependent MUTZ-3 growth revealed that the complex should consist of one hexamer sBARF1 binding three M-CSF dimers 53. This is in agreement with crystallography data of the sBARF1/M-CSF complex 48 as recently revealed by Shim et al. and Elegheert et al. 119,120.
The data imply that native sBARF1 interferes with human monocyte-macrophage differentiation 53. Specific surface markers of normal macrophages were down-regulated, and cell viability was reduced when cultured in the presence of sBARF1 (Figure 7(A,B)). A feature of macrophages is the release of interferon alpha upon virus infection to stimulate different immune cells 121–124. Recombinant BARF1 inhibited interferon alpha release from human mononuclear cells upon stimulation of toll-like receptor 3 (TLR3) with poly-IC, mimicking viral infection 38. Down-regulation of interferon alpha release may counteract immune responses to EBV-positive cells. Because the amount of tumor infiltrating macrophages is positively correlated with NPC prognosis 125,126, blocking of M-CSF by sBARF1 might be a possible mechanism for evasion from macrophage-mediated innate antitumor responses.
Figure 7.
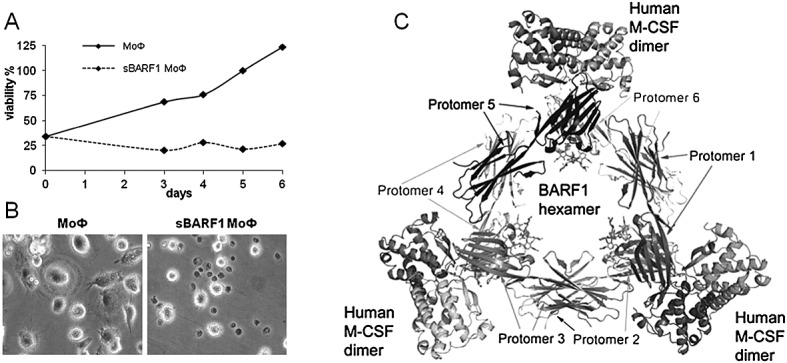
Soluble hexameric molecule BamH1-A rightward frame 1(sBARF1) is a macrophage colony stimulating factor (M-CSF) decoy receptor. (A) MTT proliferation test was used as readout for both increased cell numbers and level of differentiation. The inhibitory effect of sBARF1 on the monocyte to macrophage differentiation can be seen from day 3. The untreated cells showed increased viability, whereas the viability of sBARF1 treated macrophages remained the same in time. Adapted from 53. (B) sBARF1 negatively influences monocyte to macrophage differentiation in response to M-CSF, here shown are the differences in morphology and viability between normal monocyte derived macrophages (MoФ) and MoФ differentiated in the presence of sBARF1 (sBARF1 MoФ). Adapted from Hoebe et al. 53. (C) Structure of the BARF1/M-CSF complex as by Shim et al. 120. One sBARF1 hexamer can bind three human M-CSF dimers
During viral lytic replication, sBARF1 inhibition of M-CSF might affect monocyte/macrophage particle opsonization and degradation, resulting in virus transmission rather than virus inactivation 53,127,128. However, loss of BARF1 in the rhLCV model did not influence penetration of oral mucosa 104. The same study indicated that the immune-modulating role of BARF1 is important for acute and persistent EBV infection 104.
Recent studies have characterized the BARF1/M-CSF interaction site, unexpectedly revealing that this site is opposite to the M-CSF receptor interaction site (Figure 7C). Two BARF1 N-terminal loops form structural “tweezers” that grab M-CSF at the dimer interface 119,120. Mutations of the interface loops eliminated M-CSF-dependent MUTZ-3 growth 53. sBARF1 binds M-CSF at the beta sheets and not at the alpha helices like the M-CSF receptor, locking the M-CSF dimer in an unfavorable global orientation for receptor binding. Conflicting results exist on whether BARF1-complexed M-CSF is still able to bind (but not signal) the M-CSF receptor, possibly because BARF1 from different host backgrounds was used 119,120. A role for carbohydrate side-chains is suggested, indicating the need for using native glycosylated BARF1 molecules produced in a human cell background.
Next to the interaction with M-CSF and influence on the myeloid cell lineage, sBARF1 may affect other immune cells. BARF1 has 18% homology with CD80, also known as B7-1, suggesting that the BARF1 gene could have originated from CD80 during host-virus coevolution 48. B7-1 is a costimulator for T-cell activation or inhibition depending upon interaction with CD28 or CD152 (CTLA-4), respectively. Importantly, blockade of CD80 costimulation may provide yet another way for BARF1-positive cells to evade cellular immune responses. The possible T cell-silencing effect of sBARF1 needs to be further investigated.
Furthermore, a homology with CD200 was noted (M. Ressing and C. Kesmir, personal communication). Upon ligand-receptor binding, the CD200 receptor transmits inhibitory signals to the myeloid lineage 129,130, but CD200R is expressed on some T-cell subtypes as well 129. CD200 deficient mice are more readily immunologically activated compared with WT control mice 130, and lack of CD200 signaling is associated with an enhanced immune response to virus infection 131,132. Virus-encoded CD200 orthologues can be found in the genomes of multiple viruses, including herpes viruses, suggesting the existence of yet another viral means to modulate immune reactivity 133. Blockage of CD200 may also enhance tumor growth 134, but more research is needed to establish BARF1 effects on the CD200 receptor.
We conclude that sBARF1 may be important for carcinoma cells in mediating evasion from innate and adaptive immune responses. Next to its function as a M-CSF decoy receptor, BARF1 might have alternative immune-modulating functions that require further analysis. Interestingly, EBV-driven carcinomas are usually characterized by a significant infiltrate of activated T cells 135. However, in NPC, the lymphocytes from the tumor environment are functionally impaired 136, and the amount of lymphocyte infiltrate does not affect prognosis 137. In fact, the amount of tumor infiltrating activated granzyme-B expressing CD8+ (cytotoxic) T cells was shown to associate with worse prognosis, an effect linked to increased apoptosis-resistance of the tumor cells, possibly imposed by EBV gene products including either or both LMP1 and BARF1 138,139. The specificity of these T cells remains undetermined, but their presence illustrates the importance of immune evasion mechanisms in EBV-associated tumors.
BamHI-A rightward frame 1 and the human immune system
Although BARF1 has immune-modulating properties, its expression as nonself protein may trigger an immune response. Indirect immunofluorescence techniques have revealed antibody reactivity to BARF1-expressing cells in serum samples from NPC patients 39,41,42,140. With defined immunodominant BARF1 epitopes, only weak IgG responses could be measured in 21% of the NPC patients compared with 5% in the regional healthy controls, relative to the immunodominant viral capsid antigen and Epstein–Barr nuclear antigen 1 responses 140. IgG reactivity to BARF1 was higher than IgA responses 44,140, and the C-terminal extracellular domain is most immunogenic 44. Some healthy EBV carriers do have weak IgG responses to BARF1 possibly reflecting transcription of BARF1 during the lytic cycle during normal EBV persistence. Because of the low negative predictive value of the anti-BARF1 humoral response, this parameter has limited value for NPC diagnosis. It does however provide indirect proof that sBARF1 protein expression is elevated in NPC.
Orlova et al. 141 examined the immunogenic response to native hexameric sBARF1 and found that healthy EBV carriers and NPC patients have similar (possibly low level) IgG reactivity against sBARF1, in 66% and 70%, respectively. Unfortunately, BARF1 antibody responses were not correlated to those against viral capsid antigen and early antigen.
The BARF1 protein apparently has low immunogenicity in vivo, thus resembling LMP1 and LMP2 142. A prior study observed antibody-dependent cellular cytotoxicity against BARF1-transfected Raji cells using serum from NPC patients, but this study is poorly controlled for nonspecific interactions 143.
Martorelli et al. found that CD4+ and CD8+ T cells could be stimulated with BARF1 peptides, in particular when derived from the N-terminal 1-56 region and presented by professional antigen-presenting cells, like DCs 144. NPC patients reacted more strongly than healthy EBV-positive individuals, whereas EBV-negative individuals showed no reaction. This implies expression and (cross-) presentation of BARF1 in vivo triggering T-cell immunity, which needs to be further unraveled. Moreover, cells expressing BARF1 peptides were efficiently lysed by in vitro-pulsed cytotoxic CD8+ T cells (CTL), showing that BARF1 is a natural immunogen for T cells and that tumor cells expressing BARF1 may be susceptible to CTL killing. These studies were performed in the context of human leukocyte antigen (HLA)-A2, the most frequent HLA-type in Caucasians. Importantly, this HLA allele is significantly underrepresented in Italian NPC patients 145, suggesting protection conveyed through this HLA allele by means of cytotoxic T-cell control in vivo.
BamHI-A rightward frame 1 as target for diagnosis
Because BARF1 RNA is present in a high percentage of NPC and GC cells, it is considered a good diagnostic marker for detection of EBV-positive carcinoma cells in vivo. Despite having elevated levels of circulating EBV DNA, NPC patients have no detectable BARF1 mRNA in the circulation, excluding the presence of circulating viable NPC tumor cells 146. This is in line with the observation that circulating EBV DNA in NPC patients is highly fragmented and probably derives from apoptotic tumor cell fragments 147. The presence of BARF1 mRNA in situ in a high percentage of NPC patients is valuable for early-stage diagnosis and post-therapy monitoring of NPC. Stevens et al. demonstrated the diagnostic value of detecting BARF1 transcripts in noninvasive nasopharyngeal brushings reflecting the in situ presence of EBV-positive carcinoma cells 30. EBV DNA quantification in NP brushings combined with quantitative BARF1 mRNA detection may have promising diagnostic and prognostic utility and reduce the number of invasive NP biopsies.
The BARF1 protein may escape detection in situ because it is probably rapidly secreted from the tumor cells. Houali et al. developed a sandwich Enzyme ImmunoAssay (EIA) to measure BARF1 protein levels in sera of NPC patients. In young NPC patients, the BARF1 concentrations reached as much as 5000 ng/ml and in sera from adult patients at 500 ng/ml (n = 30) were detected 45. In addition, BARF1 was detected in saliva of NPC patients. However, the values reported are extremely high for a secreted factor. Serum levels of M-CSF in normal adults range from 1.7 to 8.4 ng/ml. The consequence of microgram levels of circulating sBARF1 would be major disruption of patient monocyte-macrophage pool. There are no indications that NPC patients have disturbed blood monocyte values. Importantly, no report has yet conformed the results of Houali et al., and our own efforts to detect sBARF1 using a variety of mouse and rabbit anti-BARF1 antibody combinations directed to conformational or linear epitopes on the BARF1 hexameric molecule have yielded mostly negative results in NPC sera, whereas the detection limit of the assay was around 5–10 ng/ml sBARF1 spiked in human serum (unpublished findings).
By mass spectrometry (MS)/MS, we have defined a peptide that could reliably be detected in sBARF1-spiked human serum, yet no such peptide could be detected in NPC sera (unpublished findings). Our results showed that by using trypsin digestion, a BARF1 peptide LGPGEEQVLIGR (aa51–62) is the main MS-identifier, and a quantitative targeted proteomics approach should be feasible to identify BARF1 values below 100 ng/ml in serum. No detailed data are available for BARF1 protein detection in GC blood samples, but it is unlikely to be much different from NPC.
Further evaluation and quantification of sBARF1 in serum as a carcinoma-specific diagnostic marker is needed. The addition of BARF1 mRNA or protein detection to current diagnostic screening efforts using EBV-IgA serology 148 or EBV DNA load testing may provide a new option for early tumor diagnosis, which is highly needed in both NPC and GC.
Concluding remarks
The BARF1 is a viral oncoprotein with pleiotropic functions, contributing to cell growth and survival as well as to immune modulation, allowing virus-producing cells and latently infected carcinoma cells to escape elimination. BARF1 mRNA is detected frequently in latently infected carcinomas and rarely observed in lymphomas except for a few cells induced into the viral lytic cycle 28,29,80. BARF1 stimulates cell growth and survival when transfected in epithelial cells 94,95, up-regulating the transcription factor and proto-oncogen c-myc 100, the chromosome stabilizer hTERT 100, the cell cycle regulator cyclin-D 108, and the antiapoptotic Bcl-2 protein 50,107, as well as other genes 108, indicating a transforming and stimulating role of BARF1 in the cell cycle. In combination with the putative role of BARF1 in preventing apoptosis and immune evasion, BARF1 is a potent multifunctional player in the carcinogenesis of NPC and GC. To further define sBARF1 as suggested, mitogenic growth factor 105 would require identification of a putative BARF1 receptor as well as a signaling pathway underlying this mitogenic effect. It also remains undefined to which extend structural, conformational, and post-translational modifications in BARF1, such as glycosylation and phosphorylation play a role in BARF1 function(s) 44,47–49. Insight in the exact mechanisms of BARF1-supported tumor cell growth can open the way for therapeutic intervention with specific antibodies or small molecule inhibitors.
Epstein–Barr virus-positive cells are in danger of being eliminated by the immune system. sBARF1 can function as a decoy receptor to functionally inhibit M-CSF, thereby acting as an immune modulator, interfering with antigen processing and innate danger-signaling functions of myeloid cells 37,38,53. Because BARF1 has homology with several secreted growth factors, further characterization of BARF1 binding partners may clarify the role of BARF1 as an immune modulator. Recent in vivo studies using a BARF1-truncated EBV-homologue rhLCV demonstrate that BARF1 is important for acute and persistent viral infection 104,106, which could be a suitable model to further explore BARF1 functions in vivo.
During lytic replication, BARF1 expression is directly regulated by the immediate early gene R and is expressed as an early gene 55,60. Without BARF1, B-cell immortalization is less effective, and viral loads during the acute phase of infection are lower 104,106. Interestingly, the first 54 amino acids described to be sufficient for transformation and not affecting the apoptotic pathway were not deleted in this truncated rhLCV virus, suggesting that BARF1 immune-modulating function is important for the initial EBV infection. Blocking formation of BARF1/M-CSF complexes may reduce immune evasion of EBV-positive cells, and the recent structural and functional definition of the BARF1/M-CSF interaction domain 53,119,120 might be the first step toward a novel therapeutic approach, either to minimize EBV viral amplification during acute infection or to treat patients with BARF1-expressing carcinomas.
The clear influence of tissue type on the expression of BARF1 in EBV-associated malignancies may reflect differences in BARF1 promoter regulation by host factors. The epithelial differentiation marker ΔNp63α may be responsible for BARF1 transactivation in latent epithelial cell types 62, but more research is needed to confirm these findings. The BARF1 promoter is highly methylated in B-cell and epithelial cell lines, which could influence transcription of the BARF1 gene 55. Thorough analysis of additional epigenetic events and/or transcription factors modifying BARF1 expression could identify the specific regulators in epithelial cells and carcinomas driving BARF1 expression.
Detection of BARF1 mRNA and protein in nasopharyngeal brushings and/or plasma may provide new options for early and specific diagnosis of NPC 30,45. Both diagnostic options need to be further explored for gastric carcinoma as well. It is clear that understanding the full range of activities of BARF1 in EBV persistence, immune modulation and oncogenesis is important for developing new treatment strategies for EBV-driven carcinomas.
CONFLICT OF INTEREST
The authors have no competing interest.
Glossary
- BARF-1
BamH1-A rightward frame 1
- BARTs
BamH1-A rightward transcripts
- CTL
CD8+ T cells
- DC
dendritic cell
- DM
double-minute
- FGFR
fibroblast growth factor receptor
- GC
gastric cancer
- HGFR
hepatocyte growth factor receptor
- hTid
homolog of Drosophila tumorous imaginal disk 1
- LMP1
latent membrane protein 1
- M-CSF
macrophage colony stimulating factor
- M-CSFR
macrophage colony stimulating factor receptor
- NASBA
nucleic acid sequence-based amplification
- NHL
non-Hodgkin lymphoma
- NPC
nasopharyngeal carcinoma
- PDGFR
platelet-derived growth factor receptor
- rhLCV
rhesus lymphocryptovirus
- sBARF1
soluble hexameric molecule BamH1-A rightward frame 1
- TLR3
toll-like receptor 3
- VEGFR
vascular endothelial growth factor receptor
REFERENCES
- 1.Rickinson AB. Epstein–Barr virus. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Wolters Kluwer Health; 2007. pp. 2655–2700. [Google Scholar]
- 2.Sixbey JW, Nedrud JG, Raab-Traub N, Hanes RA, Pagano JS. Epstein–Barr virus replication in oropharyngeal epithelial cells. New England Journal of Medicine. 1984;310:1225–1230. doi: 10.1056/NEJM198405103101905. [DOI] [PubMed] [Google Scholar]
- 3.Souza TA, Stollar BD, Sullivan JL, Luzuriaga K, Thorley-Lawson DA. Peripheral B cells latently infected with Epstein–Barr virus display molecular hallmarks of classical antigen-selected memory B cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18093–18098. doi: 10.1073/pnas.0509311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Middeldorp JM, Brink AA, van den Brule AJ, Meijer CJ. Pathogenic roles for Epstein–Barr virus (EBV) gene products in EBV-associated proliferative disorders. Critical Reviews in Oncology/Hematology. 2003;45:1–36. doi: 10.1016/s1040-8428(02)00078-1. [DOI] [PubMed] [Google Scholar]
- 5.Young LS, Rickinson AB. Epstein–Barr virus: 40 years on. Nature Reviews Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 6.Klein G, Giovanella BC, Lindahl T, Fialkow PJ, Singh S, Stehlin JS. Direct evidence for the presence of Epstein–Barr virus DNA and nuclear antigen in malignant epithelial cells from patients with poorly differentiated carcinoma of the nasopharynx. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:4737–4741. doi: 10.1073/pnas.71.12.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raab-Traub N. Epstein–Barr virus in the pathogenesis of NPC. Seminars in Cancer Biology. 2002;12:431–441. doi: 10.1016/s1044579x0200086x. [DOI] [PubMed] [Google Scholar]
- 8.Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: lessons from Epstein–Barr virus. Annual Review of Immunology. 2007;25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- 9.Paschos K, Smith P, Anderton E, Middeldorp JM, White RE, Allday MJ. Epstein–Barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathogens. 2009;5:e1000492. doi: 10.1371/journal.ppat.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorley-Lawson DA, Allday MJ. The curious case of the tumour virus: 50 years of Burkitt's lymphoma. Nature Reviews Microbiology. 2008;6:913–924. doi: 10.1038/nrmicro2015. [DOI] [PubMed] [Google Scholar]
- 11.Kutok JL, Wang F. Spectrum of Epstein–Barr virus-associated diseases. Annual Review of Pathology. 2006;1:375–404. doi: 10.1146/annurev.pathol.1.110304.100209. [DOI] [PubMed] [Google Scholar]
- 12.Thompson MP, Kurzrock R. Epstein–Barr virus and cancer. Clinical Cancer Research. 2004;10:803–821. doi: 10.1158/1078-0432.ccr-0670-3. [DOI] [PubMed] [Google Scholar]
- 13.Rezk SA, Weiss LM. Epstein–Barr virus-associated lymphoproliferative disorders. Human Pathology. 2007;38:1293–1304. doi: 10.1016/j.humpath.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 14.Saha A, Robertson ES. Epstein–Barr virus-associated B-cell lymphomas: pathogenesis and clinical outcomes. Clinical Cancer Research. 2011;17:3056–3063. doi: 10.1158/1078-0432.CCR-10-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapatai G, Murray P. Contribution of the Epstein–Barr virus to the molecular pathogenesis of Hodgkin lymphoma. Journal of Clinical Pathology. 2007;60:1342–1349. doi: 10.1136/jcp.2007.050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiology, Biomarkers & Prevention. 2006;15:1765–1777. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- 17.Seto E, Yang L, Middeldorp J, et al. Epstein–Barr virus (EBV)-encoded BARF1 gene is expressed in nasopharyngeal carcinoma and EBV-associated gastric carcinoma tissues in the absence of lytic gene expression. Journal of Medical Virology. 2005;76:82–88. doi: 10.1002/jmv.20327. [DOI] [PubMed] [Google Scholar]
- 18.Takada K. Epstein–Barr virus and gastric carcinoma. Molecular Pathology. 2000;53:255–261. doi: 10.1136/mp.53.5.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Beek J, zur Hausen A, Klein KE, et al. EBV-positive gastric adenocarcinomas: a distinct clinicopathologic entity with a low frequency of lymph node involvement. Journal of Clinical Oncology. 2004;22:664–670. doi: 10.1200/JCO.2004.08.061. [DOI] [PubMed] [Google Scholar]
- 20.Arbach H, Viglasky V, Lefeu F, et al. Epstein–Barr virus (EBV) genome and expression in breast cancer tissue: effect of EBV infection of breast cancer cells on resistance to paclitaxel (Taxol) Journal of Virology. 2006;80:845–853. doi: 10.1128/JVI.80.2.845-853.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fina F, Romain S, Ouafik L, et al. Frequency and genome load of Epstein–Barr virus in 509 breast cancers from different geographical areas. British Journal of Cancer. 2001;84:783–790. doi: 10.1054/bjoc.2000.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Wu BA, Zeng YM, et al. Epstein–Barr virus in hepatocellular carcinogenesis. World Journal of Gastroenterology. 2004;10:3409–3413. doi: 10.3748/wjg.v10.i23.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugawara Y, Mizugaki Y, Uchida T, et al. Detection of Epstein–Barr virus (EBV) in hepatocellular carcinoma tissue: a novel EBV latency characterized by the absence of EBV-encoded small RNA expression. Virology. 1999;256:196–202. doi: 10.1006/viro.1999.9619. [DOI] [PubMed] [Google Scholar]
- 24.Huang J, Chen H, Hutt-Fletcher L, Ambinder RF, Hayward SD. Lytic viral replication as a contributor to the detection of Epstein–Barr virus in breast cancer. Journal of Virology. 2003;77:13267–13274. doi: 10.1128/JVI.77.24.13267-13274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.zur Hausen A, van Beek J, Bloemena E, ten Kate FJ, Meijer CJ, van den Brule AJ. No role for Epstein–Barr virus in Dutch hepatocellular carcinoma: a study at the DNA, RNA and protein levels. J Gen Virol. 2003;84:1863–1869. doi: 10.1099/vir.0.19217-0. [DOI] [PubMed] [Google Scholar]
- 26.Brooks L, Yao QY, Rickinson AB, Young LS. Epstein–Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. Journal of Virology. 1992;66:2689–2697. doi: 10.1128/jvi.66.5.2689-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fahraeus R, Fu HL, Ernberg I, et al. Expression of Epstein–Barr virus-encoded proteins in nasopharyngeal carcinoma. International Journal of Cancer. 1988;42:329–338. doi: 10.1002/ijc.2910420305. [DOI] [PubMed] [Google Scholar]
- 28.Brink AA, Vervoort MB, Middeldorp JM, Meijer JM, van den Brule AJ. Nucleic acid sequence-based amplification, a new method for analysis of spliced and unspliced Epstein–Barr virus latent transcripts, and its comparison with reverse transcriptase PCR. Journal of Clinical Microbiology. 1998;36:3164–3169. doi: 10.1128/jcm.36.11.3164-3169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes DP, Brink AA, Vervoort MB, Middeldorp JM, Meijer CJ, van den Brule AJ. Expression of Epstein–Barr virus (EBV) transcripts encoding homologues to important human proteins in diverse EBV associated diseases. Molecular Pathology. 1999;52:97–103. doi: 10.1136/mp.52.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens SJ, Verkuijlen SA, Hariwiyanto B, et al. Noninvasive diagnosis of nasopharyngeal carcinoma: nasopharyngeal brushings reveal high Epstein–Barr virus DNA load and carcinoma-specific viral BARF1 mRNA. International Journal of Cancer. 2006;119:608–614. doi: 10.1002/ijc.21914. [DOI] [PubMed] [Google Scholar]
- 31.zur Hausen A, Brink AA, Craanen ME, Middeldorp JM, Meijer CJ, van den Brule AJ. Unique transcription pattern of Epstein–Barr virus (EBV) in EBV-carrying gastric adenocarcinomas: expression of the transforming BARF1 gene. Cancer Res. 2000;60:2745–2748. [PubMed] [Google Scholar]
- 32.Hatfull G, Bankier AT, Barrell BG, Farrell PJ. Sequence analysis of Raji Epstein–Barr virus DNA. Virology. 1988;164:334–340. doi: 10.1016/0042-6822(88)90546-6. [DOI] [PubMed] [Google Scholar]
- 33.Ooka T. Biological role of the BARF1 gene encoded by Epstein–Barr virus. In: Robertson ES, editor. Epstein–Barr Virus. Norwich, UK: Caister Academic Press; 2005. [Google Scholar]
- 34.Zheng H, Li LL, Hu DS, Deng XY, Cao Y. Role of Epstein–Barr virus encoded latent membrane protein 1 in the carcinogenesis of nasopharyngeal carcinoma. Cellular and molecular immunology. 2007;4:185–196. [PubMed] [Google Scholar]
- 35.Wei MX, Ooka T. A transforming function of the BARF1 gene encoded by Epstein–Barr virus. EMBO Journal. 1989;8:2897–2903. doi: 10.1002/j.1460-2075.1989.tb08438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang CX, Decaussin G, Daillie J, Ooka T. Altered expression of two Epstein–Barr virus early genes localized in BamHI-A in nonproducer Raji cells. Journal of Virology. 1988;62:1862–1869. doi: 10.1128/jvi.62.6.1862-1869.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strockbine LD, Cohen JI, Farrah T, et al. The Epstein–Barr virus BARF1 gene encodes a novel, soluble colony-stimulating factor-1 receptor. Journal of Virology. 1998;72:4015–4021. doi: 10.1128/jvi.72.5.4015-4021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen JI, Lekstrom K. Epstein–Barr virus BARF1 protein is dispensable for B-cell transformation and inhibits alpha interferon secretion from mononuclear cells. Journal of Virology. 1999;73:7627–7632. doi: 10.1128/jvi.73.9.7627-7632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Turenne-Tessier M, Jolicoeur P, Ooka T. Expression of the protein encoded by Epstein–Barr virus (EBV) BARF1 open reading frame from a recombinant adenovirus system. Virus Research. 1997;52:73–85. doi: 10.1016/s0168-1702(97)00101-9. [DOI] [PubMed] [Google Scholar]
- 40.de Turenne-Tessier M, Jolicoeur P, Middeldorp JM, Ooka T. Expression and analysis of the Epstein–Barr virus BARF1-encoded protein from a tetracycline-regulatable adenovirus system. Virus Research. 2005;109:9–18. doi: 10.1016/j.virusres.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Wei MX, Moulin JC, Decaussin G, Berger F, Ooka T. Expression and tumorigenicity of the Epstein–Barr virus BARF1 gene in human Louckes B-lymphocyte cell line. Cancer Research. 1994;54:1843–1848. [PubMed] [Google Scholar]
- 42.Wei MX, de Turenne-Tessier M, Decaussin G, Benet G, Ooka T. Establishment of a monkey kidney epithelial cell line with the BARF1 open reading frame from Epstein–Barr virus. Oncogene. 1997;14:3073–3081. doi: 10.1038/sj.onc.1201128. [DOI] [PubMed] [Google Scholar]
- 43.Fiorini S, Ooka T. Secretion of Epstein–Barr virus-encoded BARF1 oncoprotein from latently infected B cells. Virology Journal. 2008;5:70. doi: 10.1186/1743-422X-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoebe EK, Hutajulu SH, van Beek J, et al. Purified hexameric Epstein–Barr virus-encoded BARF1 protein for measuring anti-BARF1 antibody responses in nasopharyngeal carcinoma patients. Clinical and Vaccine Immunology. 2011;18:298–304. doi: 10.1128/CVI.00193-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houali K, Wang X, Shimizu Y, et al. A new diagnostic marker for secreted Epstein–Barr virus encoded LMP1 and BARF1 oncoproteins in the serum and saliva of patients with nasopharyngeal carcinoma. Clinical Cancer Research. 2007;13:4993–5000. doi: 10.1158/1078-0432.CCR-06-2945. [DOI] [PubMed] [Google Scholar]
- 46.Sakka E, Hausen AZ, Houali K, Liu H, Fiorini S, Ooka T. Cellular localization of BARF1 oncoprotein and its cell stimulating activity in human epithelial cell. Virus Res. 2013;174(1-2):8–17. doi: 10.1016/j.virusres.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Tam JP, Liu DX. Biochemical and functional characterization of Epstein–Barr virus-encoded BARF1 protein: interaction with human hTid1 protein facilitates its maturation and secretion. Oncogene. 2006;25:4320–4331. doi: 10.1038/sj.onc.1209458. [DOI] [PubMed] [Google Scholar]
- 48.Tarbouriech N, Ruggiero F, de Turenne-Tessier M, Ooka T, Burmeister WP. Structure of the Epstein–Barr virus oncogene BARF1. Journal of Molecular Biology. 2006;359:667–678. doi: 10.1016/j.jmb.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 49.de Turenne-Tessier M, Ooka T. Post-translational modifications of Epstein–Barr virus BARF1 oncogene-encoded polypeptide. Journal of General Virology. 2007;88:2656–2661. doi: 10.1099/vir.0.83058-0. [DOI] [PubMed] [Google Scholar]
- 50.Sheng W, Decaussin G, Sumner S, Ooka T. N-terminal domain of BARF1 gene encoded by Epstein–Barr virus is essential for malignant transformation of rodent fibroblasts and activation of BCL-2. Oncogene. 2001;20:1176–1185. doi: 10.1038/sj.onc.1204217. [DOI] [PubMed] [Google Scholar]
- 51.Hutajulu SH, Hoebe EK, Verkuijlen SA, et al. Conserved mutation of Epstein–Barr virus-encoded BamHI-A Rightward Frame-1 (BARF1) gene in Indonesian nasopharyngeal carcinoma. Infect Agent Cancer. 2010;5:16. doi: 10.1186/1750-9378-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Wang XF, Sun ZF, Luo B. Unique variations of Epstein–Barr virus-encoded BARF1 gene in nasopharyngeal carcinoma biopsies. Virus Research. 2012;166:23–30. doi: 10.1016/j.virusres.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 53.Hoebe EK, Le Large TY, Tarbouriech N, et al. Epstein–Barr virus-encoded BARF1 protein is a decoy receptor for macrophage colony stimulating factor and interferes with macrophage differentiation and activation. Viral Immunol. 2012;25(6):461–470. doi: 10.1089/vim.2012.0034. [DOI] [PubMed] [Google Scholar]
- 54.Ambinder RF, Robertson KD, Tao Q. DNA methylation and the Epstein–Barr virus. Seminars in Cancer Biology. 1999;9:369–375. doi: 10.1006/scbi.1999.0137. [DOI] [PubMed] [Google Scholar]
- 55.Hoebe EK, Wille C, Hopmans ES, et al. Epstein–Barr virus transcription activator R upregulates BARF1 expression by direct binding to its promoter, independent of methylation. J Virol. 2012;86(20):11322–11332. doi: 10.1128/JVI.01161-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Israel BF, Kenney SC. EBV lytic infection. In: Robertson ES, editor. Epstein–Barr Virus. Philadelphia: Caister Academic Press; 2011. pp. 571–611. [Google Scholar]
- 57.Kieff E. Epstein–Barr virus and its replication. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Wolters Kluwer Health; 2007. pp. 2603–2654. [Google Scholar]
- 58.Dickerson SJ, Xing Y, Robinson AR, Seaman WT, Gruffat H, Kenney SC. Methylation-dependent binding of the Epstein–Barr virus BZLF1 protein to viral promoters. PLoS Pathogens. 2009;5:e1000356. doi: 10.1371/journal.ppat.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergbauer M, Kalla M, Schmeinck A, et al. CpG-methylation regulates a class of Epstein–Barr virus promoters. PLoS Pathogens. 2010;6:e1001114. doi: 10.1371/journal.ppat.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heilmann AM, Calderwood MA, Portal D, Lu Y, Johannsen E. Genome-wide analysis of Epstein–Barr virus RTA DNA binding. Journal of Virology. 2012;86:5151–5164. doi: 10.1128/JVI.06760-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu CC, Jeng YY, Tsai CH, et al. Genome-wide transcription program and expression of the RTA responsive gene of Epstein–Barr virus. Virology. 2006;345:358–372. doi: 10.1016/j.virol.2005.09.064. [DOI] [PubMed] [Google Scholar]
- 62.Hoebe EK, Wille C, Hagemeier SR, Middeldorp JM, Kenney SC, Greijer AE. Epstein–Barr virus gene BARF1 expression is regulated by the epithelial differentiation factor deltaNp63alpha in undifferentiated nasopharyngeal carcinoma. 2013. Submitted at JVI,. Ref Type: Unpublished Work. [DOI] [PMC free article] [PubMed]
- 63.Su X, Chakravarti D, Flores ER. p63 steps into the limelight: crucial roles in the suppression of tumorigenesis and metastasis. Nature Reviews Cancer. 2013;13:136–143. doi: 10.1038/nrc3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crook T, Nicholls JM, Brooks L, O'Nions J, Allday MJ. High level expression of deltaN-p63: a mechanism for the inactivation of p53 in undifferentiated nasopharyngeal carcinoma (NPC)? Oncogene. 2000;19:3439–3444. doi: 10.1038/sj.onc.1203656. [DOI] [PubMed] [Google Scholar]
- 65.Guo C, Pan ZG, Li DJ, et al. The expression of p63 is associated with the differential stage in nasopharyngeal carcinoma and EBV infection. Journal of Translational Medicine. 2006;4:23. doi: 10.1186/1479-5876-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yip YL, Tsao SW. Regulation of p63 expression in primary and immortalized nasopharyngeal epithelial cells. International Journal of Oncology. 2008;33:713–724. doi: 10.3892/ijo_00000057. [DOI] [PubMed] [Google Scholar]
- 67.Fotheringham JA, Mazzucca S, Raab-Traub N. Epstein–Barr virus latent membrane protein-2A-induced deltaNp63alpha expression is associated with impaired epithelial-cell differentiation. Oncogene. 2010;29:4287–4296. doi: 10.1038/onc.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morrison JA, Klingelhutz AJ, Raab-Traub N. Epstein–Barr virus latent membrane protein 2A activates beta-catenin signaling in epithelial cells. Journal of Virology. 2003;77:12276–12284. doi: 10.1128/JVI.77.22.12276-12284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.You S, Zhang F, Meng F, et al. EBV-encoded LMP1 increases nuclear beta-catenin accumulation and its transcriptional activity in nasopharyngeal carcinoma. Tumour Biology. 2011;32:623–630. doi: 10.1007/s13277-011-0161-x. [DOI] [PubMed] [Google Scholar]
- 70.Ruptier C, De GA, Ansieau S, et al. TP63 P2 promoter functional analysis identifies beta-catenin as a key regulator of DeltaNp63 expression. Oncogene. 2011;30:4656–4665. doi: 10.1038/onc.2011.171. [DOI] [PubMed] [Google Scholar]
- 71.Yu H, Lu J, Zuo L, et al. Epstein–Barr virus downregulates microRNA 203 through the oncoprotein latent membrane protein 1: a contribution to increased tumor incidence in epithelial cells. Journal of Virology. 2012;86:3088–3099. doi: 10.1128/JVI.05901-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fukushima N, Satoh T, Sueoka N, et al. Clinico-pathological characteristics of p63 expression in B-cell lymphoma. Cancer Science. 2006;97:1050–1055. doi: 10.1111/j.1349-7006.2006.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hitt MM, Allday MJ, Hara T, et al. EBV gene expression in an NPC-related tumour. EMBO Journal. 1989;8:2639–2651. doi: 10.1002/j.1460-2075.1989.tb08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cosmopoulos K, Pegtel M, Hawkins J, et al. Comprehensive profiling of Epstein–Barr virus microRNAs in nasopharyngeal carcinoma. Journal of Virology. 2009;83:2357–2367. doi: 10.1128/JVI.02104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cai X, Schafer A, Lu S, et al. Epstein–Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathogens. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen H, Smith P, Ambinder RF, Hayward SD. Expression of Epstein–Barr virus BamHI-A rightward transcripts in latently infected B cells from peripheral blood. Blood. 1999;93:3026–3032. [PubMed] [Google Scholar]
- 77.Smith PR, de Jesus O, Turner D, et al. Structure and coding content of CST (BART) family RNAs of Epstein–Barr virus. Journal of Virology. 2000;74:3082–3092. doi: 10.1128/jvi.74.7.3082-3092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Beek J, Brink AA, Vervoort MB, et al. In vivo transcription of the Epstein–Barr virus (EBV) BamHI-A region without associated in vivo BARF0 protein expression in multiple EBV-associated disorders. Journal of General Virology. 2003;84:2647–2659. doi: 10.1099/vir.0.19196-0. [DOI] [PubMed] [Google Scholar]
- 79.Thornburg NJ, Kusano S, Raab-Traub N. Identification of Epstein–Barr virus RK-BARF0-interacting proteins and characterization of expression pattern. Journal of Virology. 2004;78:12848–12856. doi: 10.1128/JVI.78.23.12848-12856.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sbih-Lammali F, Djennaoui D, Belaoui H, Bouguermouh A, Decaussin G, Ooka T. Transcriptional expression of Epstein–Barr virus genes and proto-oncogenes in north African nasopharyngeal carcinoma. Journal of Medical Virology. 1996;49:7–14. doi: 10.1002/(SICI)1096-9071(199605)49:1<7::AID-JMV2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 81.Hu C, Wei W, Chen X, et al. A global view of the oncogenic landscape in nasopharyngeal carcinoma: an integrated analysis at the genetic and expression levels. PLoS One. 2012;7:e41055. doi: 10.1371/journal.pone.0041055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y, Luo B, Yan LP, Huang BH, Zhao P. Relationship between Epstein–Barr virus-encoded proteins with cell proliferation, apoptosis, and apoptosis-related proteins in gastric carcinoma. World Journal of Gastroenterology. 2005;11:3234–3239. doi: 10.3748/wjg.v11.i21.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Decaussin G, Sbih-Lammali F, de Turenne-Tessier M, Bouguermouh A, Ooka T. Expression of BARF1 gene encoded by Epstein–Barr virus in nasopharyngeal carcinoma biopsies. Cancer Research. 2000;60:5584–5588. [PubMed] [Google Scholar]
- 84.Luo B, Wang Y, Wang XF, et al. Expression of Epstein–Barr virus genes in EBV-associated gastric carcinomas. World Journal of Gastroenterology. 2005;11:629–633. doi: 10.3748/wjg.v11.i5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luo B, Wang Y, Wang XF, Gao Y, Huang BH, Zhao P. Correlation of Epstein–Barr virus and its encoded proteins with Helicobacter pylori and expression of c-met and c-myc in gastric carcinoma. World Journal of Gastroenterology. 2006;12:1842–1848. doi: 10.3748/wjg.v12.i12.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kanavaros P, Lescs MC, Briere J, et al. Nasal T-cell lymphoma: a clinicopathologic entity associated with peculiar phenotype and with Epstein–Barr virus. Blood. 1993;81:2688–2695. [PubMed] [Google Scholar]
- 87.Kanegane H, Nomura K, Miyawaki T, Tosato G. Biological aspects of Epstein–Barr virus (EBV)-infected lymphocytes in chronic active EBV infection and associated malignancies. Critical Reviews in Oncology/Hematology. 2002;44:239–249. doi: 10.1016/s1040-8428(02)00115-4. [DOI] [PubMed] [Google Scholar]
- 88.Nava VE, Jaffe ES. The pathology of NK-cell lymphomas and leukemias. Advances in Anatomic Pathology. 2005;12:27–34. doi: 10.1097/01.pap.0000151318.34752.80. [DOI] [PubMed] [Google Scholar]
- 89.Zhang Y, Ohyashiki JH, Takaku T, Shimizu N, Ohyashiki K. Transcriptional profiling of Epstein–Barr virus (EBV) genes and host cellular genes in nasal NK/T-cell lymphoma and chronic active EBV infection. British Journal of Cancer. 2006;94:599–608. doi: 10.1038/sj.bjc.6602968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Labrecque LG, Barnes DM, Fentiman IS, Griffin BE. Epstein–Barr virus in epithelial cell tumors: a breast cancer study. Cancer Research. 1995;55:39–45. [PubMed] [Google Scholar]
- 91.Xue SA, Lampert IA, Haldane JS, Bridger JE, Griffin BE. Epstein–Barr virus gene expression in human breast cancer: protagonist or passenger? British Journal of Cancer. 2003;89:113–119. doi: 10.1038/sj.bjc.6601027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Beek J. EBV-bearing gastric carcinomas: expression patterns, pathogenesis and clinical features. 2005. Ref Type: Thesis/Dissertation.
- 93.Griffin BE, Karran L. Immortalization of monkey epithelial cells by specific fragments of Epstein–Barr virus DNA. Nature. 1984;309:78–82. doi: 10.1038/309078a0. [DOI] [PubMed] [Google Scholar]
- 94.Karran L, Teo CG, King D, et al. Establishment of immortalized primate epithelial cells with sub-genomic EBV DNA. International Journal of Cancer. 1990;45:763–772. doi: 10.1002/ijc.2910450432. [DOI] [PubMed] [Google Scholar]
- 95.Song LB, Zen MS, Zhang L, et al. Effect of EBV encoded BARF1 gene on malignant transformation of human epithelial cell line HBE. Ai Zheng. 2004;23:1361–1364. [PubMed] [Google Scholar]
- 96.Guo X, Sheng W, Zhang Y. Malignant transformation of monkey kidney epithelial cell induced by EBV BARF1 gene and TPA. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2001;15:321–323. [PubMed] [Google Scholar]
- 97.Danve C, Decaussin G, Busson P, Ooka T. Growth transformation of primary epithelial cells with a NPC-derived Epstein–Barr virus strain. Virology. 2001;288:223–235. doi: 10.1006/viro.2001.1072. [DOI] [PubMed] [Google Scholar]
- 98.Seto E, Ooka T, Middeldorp J, Takada K. Reconstitution of nasopharyngeal carcinoma-type EBV infection induces tumorigenicity. Cancer Research. 2008;68:1030–1036. doi: 10.1158/0008-5472.CAN-07-5252. [DOI] [PubMed] [Google Scholar]
- 99.Gao Y, Lu YJ, Xue SA, Chen H, Wedderburn N, Griffin BE. Hypothesis: a novel route for immortalization of epithelial cells by Epstein–Barr virus. Oncogene. 2002;21:825–835. doi: 10.1038/sj.onc.1205130. [DOI] [PubMed] [Google Scholar]
- 100.Jiang R, Cabras G, Sheng G, Zeng Y, Ooka T. Synergism of BARF1 with Ras induces malignant transformation in primary primate epithelial cells and human nasopharyngeal epithelial cells. Neoplasia. 2009;11:964–973. doi: 10.1593/neo.09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shay JW, Zou Y, Hiyama E, Wright WE. Telomerase and cancer. Human Molecular Genetics. 2001;10:677–685. doi: 10.1093/hmg/10.7.677. [DOI] [PubMed] [Google Scholar]
- 102.Shimizu Y, Ishida T, Ooka T. Inhibitory effects of Epstein–Barr virus BARF1 gene on the growth of macaque cells at preimmortalized stage. 2006. EBV Conference,. 9-6-2006. Ref Type: Conference Proceeding.
- 103.Sheng W, Decaussin G, Ligout A, Takada K, Ooka T. Malignant transformation of Epstein–Barr virus-negative Akata cells by introduction of the BARF1 gene carried by Epstein–Barr virus. Journal of Virology. 2003;77:3859–3865. doi: 10.1128/JVI.77.6.3859-3865.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ohashi M, Fogg MH, Orlova N, Quink C, Wang F. An Epstein-Barr virus encoded inhibitor of colony stimulating factor-1 signaling is an important determinant for acute and persistent EBV infection. PLoS Pathogens. 2012;8:e1003095. doi: 10.1371/journal.ppat.1003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sall A, Caserta S, Jolicoeur P, Franqueville L, de Turenne-Tessier M, Ooka T. Mitogenic activity of Epstein–Barr virus-encoded BARF1 protein. Oncogene. 2004;23:4938–4944. doi: 10.1038/sj.onc.1207607. [DOI] [PubMed] [Google Scholar]
- 106.Ohashi M, Orlova N, Quink C, Wang F. Cloning of the Epstein–Barr virus-related rhesus lymphocryptovirus as a bacterial artificial chromosome: a loss-of-function mutation of the rhBARF1 immune evasion gene. Journal of Virology. 2011;85:1330–1339. doi: 10.1128/JVI.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Q, Tsao SW, Ooka T, et al. Anti-apoptotic role of BARF1 in gastric cancer cells. Cancer Letters. 2006;238:90–103. doi: 10.1016/j.canlet.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 108.Wiech T, Nikolopoulos E, Lassman S, et al. Cyclin D1 expression is induced by viral BARF1 and is overexpressed in EBV-associated gastric cancer. Virchows Archiv. 2008;452:621–627. doi: 10.1007/s00428-008-0594-9. [DOI] [PubMed] [Google Scholar]
- 109.Cotter TG. Apoptosis and cancer: the genesis of a research field. Nature Reviews Cancer. 2009;9:501–507. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- 110.Rodi DJ, Janes RW, Sanganee HJ, Holton RA, Wallace BA, Makowski L. Screening of a library of phage-displayed peptides identifies human Bcl-2 as a taxol-binding protein. Journal of Molecular Biology. 1999;285:197–203. doi: 10.1006/jmbi.1998.2303. [DOI] [PubMed] [Google Scholar]
- 111.Ganansia-Leymarie V, Bischoff P, Bergerat JP, Holl V. Signal transduction pathways of taxanes-induced apoptosis. Current Medicinal Chemistry. Anti-Cancer Agents. 2003;3:291–306. doi: 10.2174/1568011033482422. [DOI] [PubMed] [Google Scholar]
- 112.Chang MS, Lee HS, Jung EJ, Kim CW, Lee BL, Kim WH. Cell-cycle regulators, Bcl-2 and NF-kappaB in Epstein–Barr virus-positive gastric carcinomas. International Journal of Oncology. 2005;27:1265–1272. [PubMed] [Google Scholar]
- 113.Kume T, Oshima K, Shinohara T, et al. Low rate of apoptosis and overexpression of Bcl-2 in Epstein–Barr virus-associated gastric carcinoma. Histopathology. 1999;34:502–509. doi: 10.1111/j.1365-2559.1999.00686.x. [DOI] [PubMed] [Google Scholar]
- 114.Henderson S, Rowe M, Gregory C, et al. Induction of Bcl-2 expression by Epstein–Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell. 1991;65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- 115.Chitu V, Stanley ER. Colony-stimulating factor-1 in immunity and inflammation. Current Opinion in Immunology. 2006;18:39–48. doi: 10.1016/j.coi.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 116.Hume DA. Macrophages as APC and the dendritic cell myth. Journal of Immunology. 2008;181:5829–5835. doi: 10.4049/jimmunol.181.9.5829. [DOI] [PubMed] [Google Scholar]
- 117.MacDonald KP, Rowe V, Bofinger HM, et al. The colony-stimulating factor 1 receptor is expressed on dendritic cells during differentiation and regulates their expansion. Journal of Immunology. 2005;175:1399–1405. doi: 10.4049/jimmunol.175.3.1399. [DOI] [PubMed] [Google Scholar]
- 118.Pixley FJ, Stanley ER. CSF-1 regulation of the wandering macrophage: complexity in action. Trends in Cell Biology. 2004;14:628–638. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 119.Elegheert J, Bracke N, Pouliot P, et al. Allosteric competitive inactivation of hematopoietic CSF-1 signaling by the viral decoy receptor BARF1. Nat Struct Mol Biol. 2012;19(9):938–947. doi: 10.1038/nsmb.2367. [DOI] [PubMed] [Google Scholar]
- 120.Shim AH, Chang RA, Chen X, Longnecker R, He X. Multipronged attenuation of macrophage-colony stimulating factor signaling by Epstein–Barr virus BARF1. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12962–12967. doi: 10.1073/pnas.1205309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ganeway CA, Travers P, Walport M, Shlomchik MJ. 2001. Immunobiology, Garland Science,
- 122.Levy DE, Garcia-Sastre A. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine & Growth Factor Reviews. 2001;12:143–156. doi: 10.1016/s1359-6101(00)00027-7. [DOI] [PubMed] [Google Scholar]
- 123.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Weber F, Haller O. Viral suppression of the interferon system. Biochimie. 2007;89:836–842. doi: 10.1016/j.biochi.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gallo O, Bianchi S, Giannini A, Gallina E, Libonati GA, Fini-Storchi O. Correlations between histopathological and biological findings in nasopharyngeal carcinoma and its prognostic significance. Laryngoscope. 1991;101:487–493. doi: 10.1288/00005537-199105000-00008. [DOI] [PubMed] [Google Scholar]
- 126.Peng J, Ding T, Zheng LM, Shao JY. Influence of tumor-associated macrophages on progression and prognosis of nasopharyngeal carcinoma. Ai Zheng. 2006;25:1340–1345. [PubMed] [Google Scholar]
- 127.GuerreiroCacais AO, Li L, Donati D, et al. Capacity of Epstein–Barr virus to infect monocytes and inhibit their development into dendritic cells is affected by the cell type supporting virus replication. Journal of General Virology. 2004;85:2767–2778. doi: 10.1099/vir.0.80140-0. [DOI] [PubMed] [Google Scholar]
- 128.Tugizov S, Herrera R, Veluppillai P, Greenspan J, Greenspan D, Palefsky JM. Epstein–Barr virus (EBV)-infected monocytes facilitate dissemination of EBV within the oral mucosal epithelium. Journal of Virology. 2007;81:5484–5496. doi: 10.1128/JVI.00171-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Minas K, Liversidge J. Is the CD200/CD200 receptor interaction more than just a myeloid cell inhibitory signal? Critical Reviews in Immunology. 2006;26:213–230. doi: 10.1615/critrevimmunol.v26.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Barclay AN, Wright GJ, Brooke G, Brown MH. CD200 and membrane protein interactions in the control of myeloid cells. Trends in Immunology. 2002;23:285–290. doi: 10.1016/s1471-4906(02)02223-8. [DOI] [PubMed] [Google Scholar]
- 131.Rygiel TP, Rijkers ES, de Ruiter T, et al. Lack of CD200 enhances pathological T cell responses during influenza infection. Journal of Immunology. 2009;183:1990–1996. doi: 10.4049/jimmunol.0900252. [DOI] [PubMed] [Google Scholar]
- 132.Karnam G, Rygiel TP, Raaben M, et al. CD200 receptor controls sex-specific TLR7 responses to viral infection. PLoS Pathogens. 2012;8:e1002710. doi: 10.1371/journal.ppat.1002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stack G, Stacey MA, Humphreys IR. Herpesvirus exploitation of host immune inhibitory pathways. Viruses. 2012;4:1182–1201. doi: 10.3390/v4081182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rygiel TP, Meyaard L. CD200R signaling in tumor tolerance and inflammation: a tricky balance. Current Opinion in Immunology. 2012;24:233–238. doi: 10.1016/j.coi.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 135.van Beek J, zur Hausen A, Snel SN, et al. Morphological evidence of an activated cytotoxic T-cell infiltrate in EBV-positive gastric carcinoma preventing lymph node metastases. The American Journal of Surgical Pathology. 2006;30:59–65. doi: 10.1097/01.pas.0000176428.06629.1e. [DOI] [PubMed] [Google Scholar]
- 136.Li J, Zeng XH, Mo HY, et al. Functional inactivation of EBV-specific T-lymphocytes in nasopharyngeal carcinoma: implications for tumor immunotherapy. PLoS One. 2007;2:e1122. doi: 10.1371/journal.pone.0001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jayasurya A, Bay BH, Yap WM, Tan NG. Lymphocytic infiltration in undifferentiated nasopharyngeal cancer. Archives of Otolaryngology – Head & Neck Surgery. 2000;126:1329–1332. doi: 10.1001/archotol.126.11.1329. [DOI] [PubMed] [Google Scholar]
- 138.Oudejans JJ, Harijadi H, Kummer JA, et al. High numbers of granzyme B/CD8-positive tumour-infiltrating lymphocytes in nasopharyngeal carcinoma biopsies predict rapid fatal outcome in patients treated with curative intent. The Journal of Pathology. 2002;198:468–475. doi: 10.1002/path.1236. [DOI] [PubMed] [Google Scholar]
- 139.Oudejans JJ, Harijadi A, Cillessen SA, et al. Absence of caspase 3 activation in neoplastic cells of nasopharyngeal carcinoma biopsies predicts rapid fatal outcome. Modern Pathology. 2005;18:877–885. doi: 10.1038/modpathol.3800398. [DOI] [PubMed] [Google Scholar]
- 140.Paramita DK, Fatmawati C, Juwana H, et al. Humoral immune responses to Epstein–Barr virus encoded tumor associated proteins and their putative extracellular domains in nasopharyngeal carcinoma patients and regional controls. Journal of Medical Virology. 2011;83:665–678. doi: 10.1002/jmv.21960. [DOI] [PubMed] [Google Scholar]
- 141.Orlova N, Fogg MH, Carville A, Wang F. Antibodies to lytic infection proteins in lymphocryptovirus-infected rhesus macaques: a model for humoral immune responses to Epstein–Barr virus infection. Clinical and Vaccine Immunology. 2011;18:1427–1434. doi: 10.1128/CVI.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Meij P, Leen A, Rickinson AB, et al. Identification and prevalence of CD8(+) T-cell responses directed against Epstein–Barr virus-encoded latent membrane protein 1 and latent membrane protein 2. International Journal of Cancer. 2002;99:93–99. doi: 10.1002/ijc.10309. [DOI] [PubMed] [Google Scholar]
- 143.Tanner JE, Wei MX, Alfieri C, et al. Antibody and antibody-dependent cellular cytotoxicity responses against the BamHI A rightward open-reading frame-1 protein of Epstein–Barr virus (EBV) in EBV-associated disorders. Journal of Infectious Diseases. 1997;175:38–46. doi: 10.1093/infdis/175.1.38. [DOI] [PubMed] [Google Scholar]
- 144.Martorelli D, Houali K, Caggiari L, et al. Spontaneous T cell responses to Epstein–Barr virus-encoded BARF1 protein and derived peptides in patients with nasopharyngeal carcinoma: bases for improved immunotherapy. International Journal of Cancer. 2008;123:1100–1107. doi: 10.1002/ijc.23621. [DOI] [PubMed] [Google Scholar]
- 145.Pasini E, Caggiari L, Dal ML, et al. Undifferentiated nasopharyngeal carcinoma from a nonendemic area: protective role of HLA allele products presenting conserved EBV epitopes. International Journal of Cancer. 2009;125:1358–1364. doi: 10.1002/ijc.24515. [DOI] [PubMed] [Google Scholar]
- 146.Stevens SJ, Verkuijlen SA, Hariwiyanto B, et al. Diagnostic value of measuring Epstein–Barr virus (EBV) DNA load and carcinoma-specific viral mRNA in relation to anti-EBV immunoglobulin A (IgA) and IgG antibody levels in blood of nasopharyngeal carcinoma patients from Indonesia. Journal of Clinical Microbiology. 2005;43:3066–3073. doi: 10.1128/JCM.43.7.3066-3073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lo KW, Huang DP. Genetic and epigenetic changes in nasopharyngeal carcinoma. Seminars in Cancer Biology. 2002;12:451–462. doi: 10.1016/s1044579x02000883. [DOI] [PubMed] [Google Scholar]
- 148.Fachiroh J, Paramita DK, Hariwiyanto B, et al. Single-assay combination of Epstein–Barr virus (EBV) E. Journal of Clinical Microbiology. 2006;44:1459–1467. doi: 10.1128/JCM.44.4.1459-1467.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


