Abstract
Efficient DNA double-strand break (DSB) repair is critical for the maintenance of genome stability. Unrepaired or misrepaired DSBs cause chromosomal rearrangements that can result in severe consequences, such as tumorigenesis. RAD6 is an E2 ubiquitin-conjugating enzyme that plays a pivotal role in repairing UV-induced DNA damage. Here, we present evidence that RAD6 is also required for DNA DSB repair via homologous recombination (HR) by specifically regulating the degradation of heterochromatin protein 1α (HP1α). Our study indicates that RAD6 physically interacts with HP1α and ubiquitinates HP1α at residue K154, thereby promoting HP1α degradation through the autophagy pathway and eventually leading to an open chromatin structure that facilitates efficient HR DSB repair. Furthermore, bioinformatics studies have indicated that the expression of RAD6 and HP1α exhibits an inverse relationship and correlates with the survival rate of patients.
INTRODUCTION
Double-strand breaks (DSBs) in DNA are considered the most deleterious types of DNA damage and pose a great threat to the integrity of the genome. Two pathways, homologous recombination (HR) and nonhomologous end joining (NHEJ), have evolved in mammals to repair the broken ends that characterize DSBs (1). The HR pathway is a precise repair pathway, wherein missing and damaged sequence information is copied from sister chromatids to catalyze the repair (2, 3). In contrast, the repair of DNA DSBs by NHEJ is more error prone and often leads to insertions, deletions, or other types of chromosomal rearrangements. The accumulation of DNA mutations, due to either unrepaired broken ends or improper repair, is thought to increase the incidence rate of cancer and other types of diseases (4, 5).
Mounting evidence indicates that the ubiquitination of DSB repair proteins plays an important role in regulating DSB repair in mammals (6–8). Ubiquitination is classified into two types, monoubiquitination and polyubiquitination, depending on the number of ubiquitin molecules that become posttranslationally attached to target proteins. Monoubiquitinated proteins have been shown to participate in nonproteolytic pathways such as receptor trafficking, signal transduction, gene transcription, and DNA repair, while the polyubiquitination of substrates often leads to protein degradation either through the 26S proteasome pathway or through the autophagy pathway (9–12).
Ubiquitination is catalyzed by a series of enzymes that includes the ubiquitin activation enzyme (E1), the ubiquitin-conjugating enzyme (E2), and the ubiquitin ligase (E3) (13). RAD6 is an E2 ubiquitin-conjugating enzyme with a well-described role in stimulating the repair of UV-induced DNA damage (7, 14). In budding yeast, RAD6 interacts with RAD18 to catalyze the monoubiquitination of proliferating cell nuclear antigen (PCNA) on lysine 164, thereby promoting the error-prone DNA damage repair pathway by recruiting low-fidelity polymerases. Interestingly, the interaction between the RAD6-RAD18 complex and the Ubc13-MMS2-Rad5 complex facilitates the polyubiquitination of PCNA on the same site, eventually activating the error-free repair pathway (15, 16). In addition, several reports indicate that RAD6 regulates protein degradation by cooperating with different E3 ligases (17–20). For instance, our previous studies have shown that the RAD6-MDM2 complex targets p53 for degradation both in Drosophila melanogaster and mammals (21, 22). Additionally, a previous report indicated that in response to ionizing radiation (IR)-induced DNA DSBs, mammalian RAD6 forms a complex with RNF168 that is rapidly recruited to DSBs (23). However, the exact mechanism by which RAD6 participates in the repair of DNA DSBs remains to be elucidated.
The regulation of chromatin structure is a highly dynamic process. The assembly and disassembly of chromatin frequently occur during DNA replication, gene transcription, DNA damage response, and DNA repair (24–26). Heterochromatin is characterized as a relatively condensed chromatin configuration, which often results in reduced transcriptional activities of euchromatic genes inserted into the region. The evolutionarily conserved heterochromatin protein 1 (HP1) family proteins are well known for their roles in heterochromatin formation and regulation in gene transcription in various species (27–32). Increasing evidence indicates that HP1 family proteins also participate in DNA damage response and repair (33–36). Recent work showed that the repair of double-strand breaks in heterochromatin requires moving outside HP1α domains to complete the recombination (37), and HP1α can inhibit Rad51 recruitment and strand invasion by cooperating with the Smc5-Smc6 complex in heterochromatin. In response to DNA DSBs, the local disassembly of HP1α at DSB sites is essential for the formation of Rad51 nucleoprotein filaments and thus the successful completion of HR repair (37).
Here, we show that RAD6 promotes HR-directed DNA DSB repair by regulating autophagy-mediated HP1α degradation and subsequent changes in chromatin structure. We observed an enhanced interaction between RAD6 and HP1α in response to X-ray irradiation. This interaction leads to the ubiquitination of HP1α at residue K154 by RAD6, which results in the autophagy-mediated degradation of HP1α and, subsequently, a loosened chromatin structure that is more permissive for the catalysis of HR. Additional bioinformatics analyses of the relationship between these two proteins indicate that RAD6 expression is negatively correlated with HP1α both in terms of expression level and survival rate in patients with lung cancer, supporting a role of RAD6 and HP1α in tumorigenesis.
MATERIALS AND METHODS
Cell culture and transfection.
The HL-7702 human normal liver cell line, the HeLa human cervical carcinoma cell line, and the HEK293 human embryonic kidney cell line were all cultured at 37°C in Dulbecco's modified Eagle's medium (DMEM; Gibco catalog no. 11960-044) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin (Gibco product no. 15070-063) in a 5% CO2 incubator. The transfection of constructs into cells was performed with Lipofectamine 2000 reagent (Invitrogen catalog no. 11668-019) according to the manufacturer's standard protocol.
The two reporter cell lines used for analyzing NHEJ and HR, HCA2-I9a and HCA2-H15C, were grown in DMEM (Gibco catalog no. 11960-044) supplemented with 10% fetal bovine serum, 1× nonessential amino acids (Gibco catalog no. 11140-050), and 1% penicillin and streptomycin (Gibco catalog no. 15070-063) in a Hera240i incubator with 5% CO2 and 3% O2 at 37°C. The cells were transfected with different plasmids or small interfering RNAs (siRNAs) with a Lonza 4D electroporator using the DT-130 program.
Plasmid constructs.
pCMV-Myc, pCMV-HA, and pEGFP-N1 (Clontech catalog no. 635689, 635690, and 6085-1) plasmids expressing RAD6A and RAD6B were constructed by cloning the RAD6A and RAD6B PCR products into the pCMV-Myc, pCMV-HA, and pEGFP-N1 vectors. A pDsRed2-N1 (Clontech catalog no. 632406) plasmid expressing HP1α was constructed by cloning HP1α cDNA into the pDsRed2-N1 vector. pET-42b(+) (Novagen catalog no. 70562-3) plasmids expressing RAD6A, RAD6B, and HP1α were constructed by cloning RAD6A, RAD6B, and HP1α cDNAs into the pET-42b(+) vector. The HP1α K154 mutant plasmids were generated using a site-directed mutagenesis kit (Agilent catalog no. 200519) according to the manufacturer's standard protocol.
RNA interference (RNAi) knockdown of RAD6A, RAD6B, and HP1α in cultured human cell lines.
siRNAs against RAD6A and HP1α were designed and synthesized by the GenePharm Company (Shanghai, China). RAD6B siRNA was purchased from Santa Cruz Biotechnology Company (catalog no. sc-106915). The introduction of siRNAs into cultured HL-7702, HeLa, or HEK293 cells was achieved using Lipofectamine 2000 reagent (Invitrogen catalog no. 11668-019) according to the manufacturer's protocol. For the HCA2 cell lines, 5 μl of 20 μM siRNA was electroporated into 1 × 106 cells twice with a 2-day interval. On day 2 after the second transfection, cells were harvested to analyze the knockdown efficiency or subjected to I-SceI transfection to analyze the repair efficiency.
Coimmunoprecipitation.
Cells were transfected with hemagglutinin (HA)-RAD6A, RAD6B, or green fluorescent protein (GFP)-tagged LC3 using Lipofectamine 2000 reagent (Invitrogen catalog no. 11668-019). After 48 h, the cells were harvested, washed with ice-cold phosphate-buffered saline (PBS), resuspended in ATM lysis buffer (containing 100 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.2 mM EDTA, 20% glycerol, 0.4% NP-40, 2% Tween 20, and 0.2 mM phenylmethylsulfonyl fluoride [PMSF]) and sonicated on ice 10 times (3 s each), with a 20% efficiency. The cell lysates were incubated with a normal mouse IgG (Santa Cruz Biotechnology catalog no. sc-2025, as a negative control), anti-HA (Zhongshan Golden Bridge), or anti-GFP (Zhongshan Golden Bridge) antibody at 4°C overnight. Protein A/G-agarose beads (Santa Cruz Biotechnology catalog no. sc-2003) were subsequently added, and the solution was incubated for another 3 h, followed by centrifugation to harvest the agarose beads after they had been washed 5 times with lysis buffer. The precipitated proteins were released by boiling in loading buffer and resolved via SDS-PAGE (15%). Immunoblot analyses were performed with antibodies against HA, GFP, or HP1α.
Antibodies and Western blotting.
Antibodies against HA, Myc, Red, and GFP were purchased from Zhongshan Golden Bridge. An antibody against RAD6 was purchased from Santa Cruz Biotechnology (catalog no. sc-30078). Antibodies against HP1α, HP1β, and HP1γ were purchased from Cell Signaling Technology (catalog no. 2616, 2613, and 2619). An antiubiquitin antibody was purchased from R&D (catalog no. MAB701). All of the horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Zhongshan Golden Bridge.
Cells were lysed in ATM lysis buffer (containing 100 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.2 mM EDTA, 20% glycerol, 0.4% NP-40, 2% Tween 20, and 0.2 mM PMSF). The protein concentration in the supernatant was measured with a bicinchoninic acid (BCA) assay kit (Novagen catalog no. 71285-3). Then, the samples were loaded into a 15% gel to resolve the proteins. Different amounts of total protein were loaded in each experiment to facilitate the detection of different target proteins. After electrophoresis, the proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Amersham catalog no. 10600021) and hybridized with primary antibodies at a dilution of 1:2,000. The HRP-labeled secondary antibodies (Zhongshan Golden Bridge) were applied at a dilution of 1:4,000. An ECL detection system (Calbiochem catalog no. 345818) was used to detect the signals on the membranes.
RT-PCR assay.
Cells were lysed to isolate total RNA using TRIzol reagent (Invitrogen catalog no. 15596-026) according to the manufacturer's instructions. Reverse transcription (RT) was performed with a reverse transcription kit (TaKaRa catalog no. 2641A). Briefly, total RNA (5 μg) was reverse transcribed to synthesize cDNA in a volume of 20 μl using Moloney murine leukemia virus (MMLV) reverse transcriptase. In each 25-μl PCR mixture, 1 μl of cDNA was used, and the reaction was run for 20 to 25 cycles. The PCR products were loaded onto a 2% agarose gel, stained with ethidium bromide, and imaged.
GST pulldown assay.
Glutathione S-transferase (GST) fusion proteins (GST-RAD6A/B and GST-HP1α) were purified from Escherichia coli, quantified, and stored at −80°C. A 1-μg sample of each GST fusion protein was incubated overnight with HL-7702 whole-cell extracts. Sepharose 4B beads (GE Healthcare catalog no. 17-0756-01) were washed extensively with ATM lysis buffer and then added to the solutions, followed by incubation for another 3 h. The bound proteins were analyzed by Western blotting with the indicated antibodies.
In vivo ubiquitination assay.
HL-7702 cells were transfected with RAD6A/B plasmids or empty control plasmids. At 48 h posttransfection, an in vivo ubiquitination assay was performed under denaturing conditions. The cells were lysed with 100 μl of SDS lysis buffer containing 1% SDS, and the lysate was then boiled for 15 min. The resulting lysate was centrifuged for 15 min at 12,000 rpm at 4°C. The supernatant was diluted to 0.1% SDS with 900 μl of ATM lysis buffer. The lysate was subsequently incubated with a normal mouse IgG antibody (Santa Cruz Biotechnology catalog no. sc-2025) or anti-HP1α antibody (Cell Signaling Technology catalog no. 2616) at 4°C overnight. Protein A/G-agarose beads were then added to precipitate the bound proteins. The ubiquitination levels of HP1α were detected in Western blotting assays with antibodies against HP1α or ubiquitin (R&D catalog no. MAB701).
Autophagy analysis.
To induce cell starvation, cells were washed three times with PBS and incubated for 4 to 6 h in Hanks' balanced salt solution (HBSS; Invitrogen catalog no. 14025-076). To stimulate rapamycin-induced autophagy, cells were treated with complete medium containing 2 μM rapamycin at 37°C for 10 h. Autophagy was assessed based on LC3 cleavage. To inhibit starvation-mediated autophagy, cells were treated with HBSS containing 10 mM 3-methyladenine (3-MA; Sigma catalog no. M9281).
ChIP assay.
Chromatin immunoprecipitation (ChIP) was performed according to the published protocols from Upstate and our previous work (2, 3, 38, 39).
Bioinformatic analyses.
To assess the clinical relevance of RAD6 and HP1α, we performed two clinically based bioinformatic analyses by using the Oncomine (www.oncomine.org) and KM plotter online services to determine the correlations between expressional levels and patient survival (Kaplan-Meier plot) for cancer patients.
RESULTS
RAD6 interacts with heterochromatin protein HP1α.
Our previous yeast two-hybrid screen identified multiple RAD6-interacting proteins, including p53, and we showed that RAD6 regulates the degradation of p53 in both Drosophila and mammalian cells (21, 22). HP1α, an important regulator of heterochromatin formation and transcriptional gene silencing, was also found to be one of the interacting partners of RAD6 in our yeast two-hybrid screen (data not shown). Since both RAD6 and HP1α are known to play pivotal roles in DNA damage repair (33–35, 37), we wondered whether the interaction between RAD6 and HP1α is functionally linked in the DNA damage repair process.
To confirm the interaction between RAD6 and HP1α, we first studied the cytological colocalization of the two proteins in HL-7702 cells transfected with GFP-tagged RAD6 and DsRed-tagged HP1α plasmids. Consistent with our yeast two-hybrid screen result, we observed that RAD6 colocalized with HP1α, supporting the potential interaction between these two proteins (Fig. 1A). We then performed coimmunoprecipitation (Co-IP) experiments to validate this interaction. HL-7702 cells transfected with Myc-tagged RAD6 plasmids were lysed and subjected to Co-IP with an anti-Myc antibody. Western blotting revealed that RAD6 specifically interacts with HP1α but not with HP1β or HP1γ (Fig. 1B, top). In addition, a reverse Co-IP was performed using an anti-HP1α antibody in HL-7702 cells transfected with HA-tagged RAD6 plasmids. We observed that RAD6 was also precipitated by HP1α (Fig. 1B, bottom). We next performed endogenous Co-IP experiments using antibodies against either RAD6 or HP1α, and the interaction between these two factors was also confirmed (data not shown). Moreover, in vitro GST pulldown experiments using bacterially expressed GST-RAD6 and GST-HP1α also confirmed the interaction between the two proteins (data not shown).
FIG 1.
Interactions between RAD6 and HP1α. (A) HL-7702 cells transfected with RAD6A/B-GFP plasmids and the HP1α-Red plasmid were stained with 4′,6-diamidino-2-phenylindole (DAPI) for fluorescence microscopy analysis. (B) RAD6 interacts with HP1α. HL-7702 cells transfected with Myc-RAD6A/B plasmids were lysed to perform a Co-IP assay with an anti-Myc antibody. Western blotting against the indicated proteins was performed. A reverse Co-IP was conducted in HA-RAD6A/B-overexpressing HL-7702 cells, followed by Western blotting. (C) HEK293 cells were irradiated at 80 kV for 5 min in an X-ray irradiator, and the cells were then harvested at the indicated times postirradiation and lysed for Co-IP with an anti-RAD6 antibody, followed by Western blotting. (D and E) Diagram of the KillerRed (KR) reporter system. A hydrozoan-derived fluorescent protein, KillerRed, was used to generate reactive oxygen species (ROS)-induced DNA damage, including base damage and DNA single-strand and double-strand breaks, in defined genomic locations within living cells. KR was fused to the tetracycline repressor (tetR) or transcription activator (TA) and expressed in the U2OS TRE cell line, in which a plasmid containing the tetracycline response element (TRE) array was integrated into a specific genomic site in approximately 200 copies, to induce ROS DNA damage in heterochromatin or euchromatin. Through a protein-DNA interaction involving TRE and tetR, KR is bound at the TRE-integrated site in the genome. mCherry serves as a control to mark the integration site without DNA damage. The activation of KR in bulk cells was achieved by exposing cells to a 15-W Sylvania cool white fluorescent bulb for 10 min in a stage UVP (Uvland, CA). In a 10-min light exposure, 9,000 J was delivered to the whole dish; the final power delivered to the KR (approximately 1 μm2) spot upon light exposure was approximately 9 mJ/μm2. Cells were placed under a water bottle (height to light is 15 cm) to prevent a temperature increase.
The interaction between RAD6 and HP1α is enhanced in irradiated cells.
Because both RAD6 and HP1α are well known for their roles in DNA damage repair (7, 14–16, 23, 33–35, 37), we next wondered whether the interaction between these two proteins was modulated in response to DNA DSB damage signals induced by X-ray irradiation. RAD6 was therefore immunoprecipitated from the protein extracts of irradiated HEK293 cells with subsequent Western blotting to analyze the interactions between RAD6 and HP1α. Indeed, we observed an enhanced interaction between these two proteins at 2 h and 8 h after irradiation compared with that at 0 h after the induction of DNA DSBs (Fig. 1C), indicating that the interplay between RAD6 and HP1α likely participates in the regulation of DNA damage repair.
We also tested the dynamics of RAD6 using a microirradiation method. In agreement with previous reports (23), we found that RAD6 localized to DNA DSB sites after the induction of DNA damage (data not shown). To further determine whether the recruitment of RAD6 at DNA damage sites occurs in euchromatin and/or heterochromatin, a well-defined KillerRed reporter system (40) (Fig. 1D), which allowed us to introduce DNA damage specifically at euchromatin/euchromatin-like or heterochromatin/heterochromatin-like sites, was employed. As shown in Fig. 1E, we found that RAD6 was recruited to the DNA damage sites in both euchromatin and heterochromatin regions, and HP1α localized to the same DNA damage sites (Fig. 1E), supporting the conclusion that both RAD6 and HP1α associate with DNA damage repair in both types of chromatin. Collectively, these results further indicate a functional link between RAD6 and HP1α in DNA damage repair.
RAD6 and HP1α regulate HR-directed DNA DSB repair in an opposite fashion.
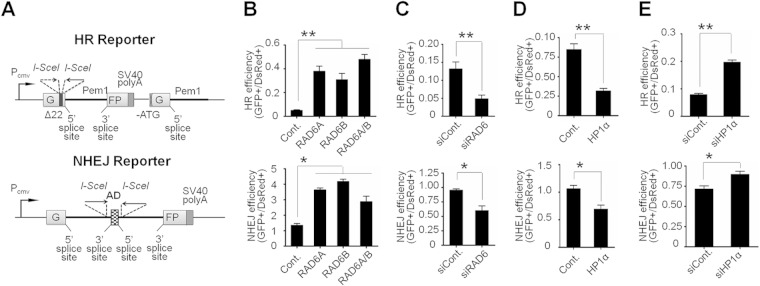
To examine the function of RAD6 in DNA DSB repair, we next employed two well-defined reporter cell lines to separately analyze the repair efficiency of HR and NHEJ (2) (Fig. 2A). Briefly, the HR reporter contains two mutated GFP-Pem1 copies. The first exon of GFP-Pem1 contains a 22-bp deletion combined with the insertion of two I-SceI recognition sites in inverted orientation for the induction of DSBs. The deletion ensures that GFP cannot be reconstituted by NHEJ. The two I-SceI sites result in incompatible ends after I-SceI transfection. The second copy of the GFP-Pem1 lacks a promoter, ATG, and the second exon of GFP. The intact construct cannot express GFP. However, upon induction of DSB by I-SceI transfection, the functional GFP gene will be reconstituted by gene conversion between the two mutated copies of the first GFP-Pem1 exon. The second copy of the GFP gene lacks the first ATG and the second exon. Therefore, crossover or single-strand annealing will not restore GFP activity. This design allows the exclusive detection of gene conversion, which is the predominant HR pathway in mammalian cells. The reporter cassette for detecting NHEJ contains the GFP gene with an artificially engineered 3-kb intron from the Pem1 gene. The Pem1 intron contains an adenoviral exon flanked by I-SceI endonuclease recognition sequences in inverted orientation. Digestion with I-SceI generates DSBs with incompatible DNA ends. An unrearranged NHEJ cassette is GFP negative, because the adenoviral exon disrupts the GFP open reading frame (ORF). Upon the induction of DSBs by the expression of I-SceI, the adenoviral exon is removed, NHEJ reconstitutes the functional GFP gene, and green cells can be quantified by flow cytometry (2).
FIG 2.
RAD6 and HP1α regulate HR in an opposing manner. (A) Diagrams of the reporters used for detecting the repair efficiencies of HR and NHEJ. The mechanisms of the reporters are described as previously reported (2). The reporter cassette for HR examination consists of two mutated copies of GFP-Pem1. In the first copy of GFP-Pem1, the first GFP exon contains an insertion of two I-SceI recognition sites in inverted orientation and a deletion of 22 nucleotides (nt) (Δ22), which ensures that GFP cannot be reconstituted by an NHEJ event. The second copy of GFP-Pem1 lacks a promoter, the first ATG, and the second exon of GFP. Upon induction of DSBs by I-SceI transfection, gene conversion events reconstitute an active GFP gene. The reporter cassette for NHEJ examination consists of a GFP gene under a cytomegalovirus (CMV) promoter with an engineered intron from the rat Pem1 gene. This intron is interrupted by an adenoviral exon (AD). The adenoviral exon is flanked by I-SceI recognition sites in inverted orientation for the induction of DSBs. In this construct, the GFP gene is inactive. However, the induction of a DSB and successful NHEJ will trigger the construct to be GFP+. (B and C) Roles of RAD6 in DNA DSB repair. The DNA DSB repair efficiency was calculated as described elsewhere (2). (D and E) Roles of HP1α in DNA DSB repair. *, P < 0.05; **, P < 0.01 (two-tailed unpaired t test; n > 3).
We first tested the effect of overexpressing RAD6 and found that the overexpression of RAD6A or RAD6B stimulated HR and NHEJ by approximately 6- to 7-fold and 2.5- to 3-fold, respectively (Fig. 2B). Furthermore, knocking down RAD6 reduced the efficiency of HR and NHEJ by approximately 63% and 37%, respectively (Fig. 2C), suggesting that RAD6 is required for HR, whereas it plays a minor role in NHEJ. Using the same strategy, we found that the overexpression or knockdown of HP1α influenced DNA DSB repair in an opposite manner to RAD6 (Fig. 2D and E). These results therefore support a functional interaction between Rad6 and HP1α in DSB repair.
To rule out the possibility that the depletion of HP1α increases the efficiency of I-SceI digestion, allowing the enzyme to induce more breaks and thereby leading to an artificially high repair efficiency, we performed real-time PCR to examine the cutting efficiency in the absence of HP1α. We found that knocking down HP1α resulted in a modest 20% increase in the I-SceI digestion of the reciprocal restriction site (data not shown), which could not account for the 2.5-fold increase in HR observed in the absence of HP1α. However, the increase in NHEJ induced by depleting HP1α was approximately 30%, suggesting that HP1α plays a major role in HR DNA repair.
RAD6 promotes HP1α degradation.
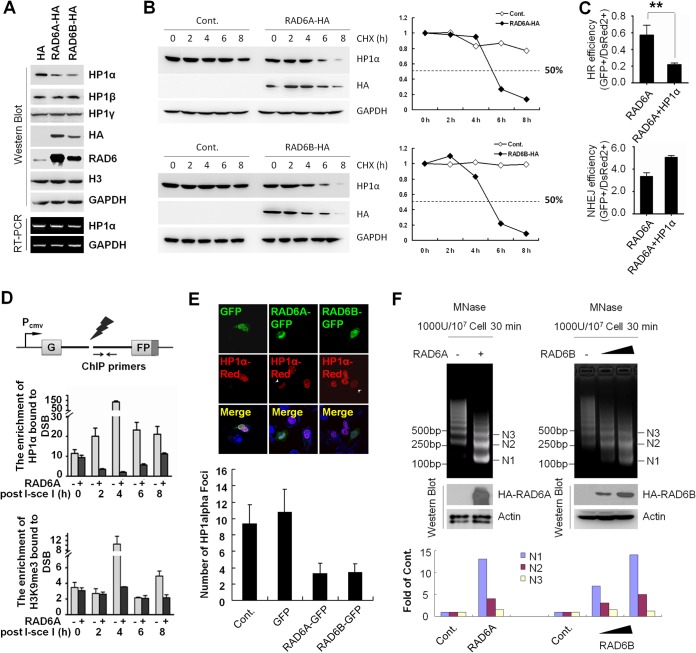
RAD6 is known as an E2 ubiquitin-conjugating enzyme capable of facilitating the degradation of various substrates (17–22). We therefore proposed that RAD6 potentially regulates HR by degrading HP1α through ubiquitination. To test this hypothesis, we first examined HP1α protein levels after overexpressing RAD6 in HL-7702 cells. In agreement with our hypothesis, RAD6 overexpression significantly downregulated HP1α protein levels, whereas no obvious changes in either HP1β or HP1γ protein levels were observed (Fig. 3A, top), supporting a specific role for RAD6 in HP1α degradation. In addition, RAD6 overexpression had no effect on HP1α mRNA levels (Fig. 3A, bottom, RT-PCR), implying that the suppressive effect on HP1α protein levels likely occurs through degradation at a posttranslational level.
FIG 3.
RAD6 stimulates HR by suppressing the recruitment of HP1α to damaged sites and promoting chromatin flexibility through HP1α degradation. (A and B) RAD6 promotes the degradation of HP1α at the posttranslational but not transcriptional level. (A) HA-RAD6A/B-transfected HL-7702 cells were lysed for Western blotting with the indicated antibodies. Total RNA was extracted to examine the mRNA levels of HP1α and gapdh via RT-PCR. (B) The HA-RAD6A/B plasmid-transfected HL-7702 cells were incubated with 50 μg/ml CHX and lysed. Western blotting was then conducted with the indicated antibodies. (C) Cooverexpressing HP1α and RAD6A significantly impaired the stimulatory effect of RAD6 on HR. **, P < 0.01 (two-tailed unpaired t test; n > 3). (D) Diagram of the site for which ChIP primers were designed (arrows). At different time points posttransfection, NHEJ reporter cells were harvested and lysed for ChIP assays with antibodies against HP1α and H3K9me3, followed by RT-PCR. (E) The abnormal HP1α distribution in RAD6-overexpressing cells. The number of HP1α foci was quantified in RAD6-transfected or untransfected cells. (F) RAD6A/B overexpression sensitizes HL-7702 cells to MNase digestion. After transfection, cells were digested with MNase, followed by the separation of different amounts of extracted genomic DNA (1×, 4×) on a 2% agarose gel. The results were quantified using Quantity One software. N1, N2, and N3 indicate the length of the DNA wrapped around approximately 1, 2, and 3 nucleosomes.
To determine the effect of RAD6 on the rate of HP1α degradation, we performed a chase assay. Empty control and HA-tagged RAD6 (RAD6A and RAD6B)-overexpressing HL-7702 cells were treated with 50 μg/ml cycloheximide (CHX), an inhibitor of eukaryotic translation, for the indicated times. We observed that the HP1α protein was more stable under control conditions, with a half-life of over 8 h, than in the presence of RAD6 overexpression, where its half-life dropped to below 6 h (Fig. 3B), confirming that RAD6 promotes the degradation of HP1α.
The degradation of HP1α by RAD6 is required for DNA DSB repair by HR.
To examine whether the degradation of HP1α by RAD6 affects DNA DSB repair, we cooverexpressed RAD6 and HP1α and then analyzed the efficiency of DNA DSB repair. Interestingly, this analysis revealed that cooverexpressing RAD6 and HP1α significantly inhibited the efficiency of HR in comparison to merely overexpressing RAD6 (Fig. 3C, top), whereas the cooverexpression of RAD6 and HP1α had no suppressive effect on the efficiency of NHEJ (Fig. 3C, bottom). Cumulatively, this suggests that the degradation of HP1α is a crucial step during the repair of DSBs by HR.
To provide direct evidence that the RAD6-induced removal of HP1α from chromatin is required for HR, we then analyzed the abundance of HP1α and H3K9me3, which is an HP1-associated heterochromatin marker (41), at DNA DSB sites in the presence or absence of RAD6 overexpression using a ChIP assay (3). When RAD6 was not overexpressed, HP1α and H3K9me3 rapidly accumulated at damage sites in response to DNA DSBs (Fig. 3D), possibly resulting in a more condensed chromatin structure to suppress HR (Fig. 2G and H), whereas much less HP1α and H3K9me3 accumulation appeared at broken ends upon RAD6 overexpression (Fig. 3D). Notably, the reduction in H3K9me3 seemed to be less dramatic than that of HP1α, indicating that the dynamics of HP1α and H3K9me3 on chromatin under DNA damage conditions are potentially varied. We also performed a ChIP assay with HP1α antibody in RAD6-knockdown cells, and an inverse relationship between RAD6 and HP1α was observed, compared with RAD6-overexpressing cells (data not shown). The results therefore support a direct role of RAD6 and HP1α in HR.
RAD6 overexpression leads to an altered chromatin structure.
Because HP1α is known to maintain compacted heterochromatin structures (42, 43), we therefore speculated that the degradation of HP1α by RAD6 leads to a more open chromatin organization. To test this hypothesis, we examined the subcellular distribution pattern of HP1α and the sensitivity of extracted chromatin to micrococcal nuclease (MNase). In HL-7702 cells overexpressing GFP-RAD6 and Red-HP1α, we observed a marked decrease in HP1α focus number, and the HP1α distribution in the nucleus was notably dispersed, relative to that of control cells (Fig. 3E). Additionally, due to the forced change in chromatin structure by RAD6 overexpression, an increased number of cells with abnormal nuclei was observed (data not shown). Furthermore, RAD6 overexpression resulted in more digested nucleosomes when treated by MNase, an enzyme preferentially digesting linker DNA between nucleosomes of chromatin, confirming that the presence of RAD6 leads to an open chromatin structure that is more accessible to MNase (Fig. 3F).
The regulation of HP1α degradation by RAD6 occurs via the autophagy-lysosome pathway.
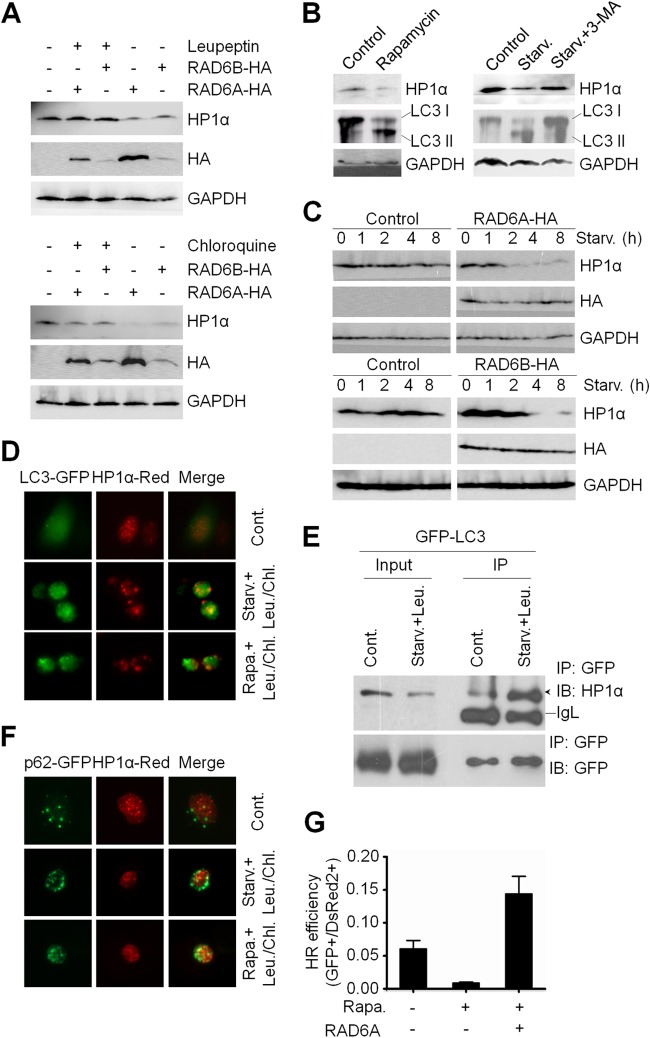
In eukaryotic cells, two major pathways—the ubiquitin-proteasome and the lysosomal proteolysis pathways—have been reported to be responsible for protein degradation. To determine which pathway mediates the degradation of HP1α by RAD6, we blocked the lysosome pathway using either leupeptin or chloroquine, which are lysosome inhibitors, and checked the protein levels of HP1α upon RAD6 overexpression. We found that inhibiting the lysosome rescued the degradation of HP1α (Fig. 4A). However, blocking the ubiquitin-proteasome pathway using MG132 did not restore HP1α protein levels in RAD6-overexpressing cells (data not shown). To rule out the possibility that the failure to observe HP1α restoration upon MG132 treatment was due to MG132 losing effectiveness, we utilized p53, which is known to be degraded through the proteasome pathway, as a control to confirm the efficacy of MG132 treatment. The elevated p53 level upon MG132 treatment indicates that MG132 functioned normally (data not shown), suggesting that the degradation of HP1α by RAD6 is mediated by lysosomal proteolysis.
FIG 4.
RAD6 regulates HP1α degradation through the autophagy-lysosome pathway. (A) Inhibiting lysosome activity rescued the RAD6-mediated decrease in HP1α protein levels. HA-RAD6A/B plasmid-transfected HL-7702 cells were treated with leupeptin or chloroquine, and proteins were then extracted for subsequent Western blotting. (B) Autophagy downregulates HP1α protein levels. HL-7702 cells were treated with rapamycin or cultured under starvation conditions for 8 h in the presence or absence of 3-MA. The cells were lysed for Western blotting. (C) RAD6 accelerates autophagy-mediated HP1α protein degradation. (D) Starvation and rapamycin promote the colocalization of HP1α and LC3. HL-7702 cells cotransfected with LC3-GFP and HP1α-Red were starved or treated with rapamycin. The cells were then stained with DAPI and analyzed via confocal microscopy. (E) Enhanced interaction between LC3 and HP1α in response to starvation. HL-7702 cells transfected with LC3-GFP were cultured under normal or starvation conditions and then lysed for Co-IP with anti-GFP, followed by Western blotting. (F) Starvation and rapamycin promote the colocalization of HP1α and p62. HL-7702 cells cotransfected with p62-GFP and HP1α-Red were starved or treated with rapamycin and stained with DAPI prior to analysis via confocal microscopy. (G) Effects of autophagy and RAD6 on HR repair. The HR repair efficiency was calculated as described elsewhere (2).
Because autophagy is involved in the lysosome-mediated protein degradation (44), we tested whether the observed HP1α degradation by RAD6 occurs through the autophagy pathway. HL-7702 cells were starved for 8 h or incubated with 2 μM rapamycin for 10 h to activate autophagy. Cells were then lysed and subjected to Western blotting. Indeed, activating autophagy downregulated HP1α protein levels. Inhibiting autophagy with 3-methyladenine (3-MA), a class III phosphoinositide 3-kinase inhibitor, rescued the decline of HP1α levels under starvation conditions (Fig. 4B). To further determine whether RAD6 affects autophagy-mediated HP1α degradation, we examined the half-life of HP1α under starvation conditions in the presence or absence of RAD6 overexpression. We observed that RAD6 overexpression indeed shortened the half-life of HP1α to less than 4 h under starvation conditions (Fig. 4C), suggesting that RAD6 promotes autophagy-mediated HP1α degradation.
LC3 is a marker of autophagosomes (10, 45), and we sought to determine whether HP1α and LC3 colocalized during starvation or rapamycin treatment. Immunofluorescence staining of cells transfected with LC3-GFP and HP1α-DsRed revealed that the induction of autophagy stimulated the colocalization of HP1α and LC3 (Fig. 4D). Co-IP experiments further confirmed that autophagy promoted the interaction between HP1α and LC3 (Fig. 4E), supporting the conclusion that the degradation of HP1α is executed through the autophagy pathway.
Previous work has shown that p62 recognizes polyubiquitinated proteins and binds to Atg8 on the autophagosome membrane to target proteins to autophagosomes for degradation (46, 47). In agreement with these reports, autophagy induced by starvation and rapamycin treatment enhanced the colocalization between HP1α-DsRed and p62-GFP in HL-7702 cells (Fig. 4F), suggesting that p62 participates in targeting HP1α to autophagosomes for degradation. Intriguingly, in response to autophagy triggered by rapamycin, HR declined by approximately 86%; however, overexpressing RAD6A in the presence of rapamycin stimulated HR by 20-fold, implying an essential role for RAD6A in HR during autophagy (Fig. 4G).
RAD6 regulates HP1α ubiquitination at lysine 154.
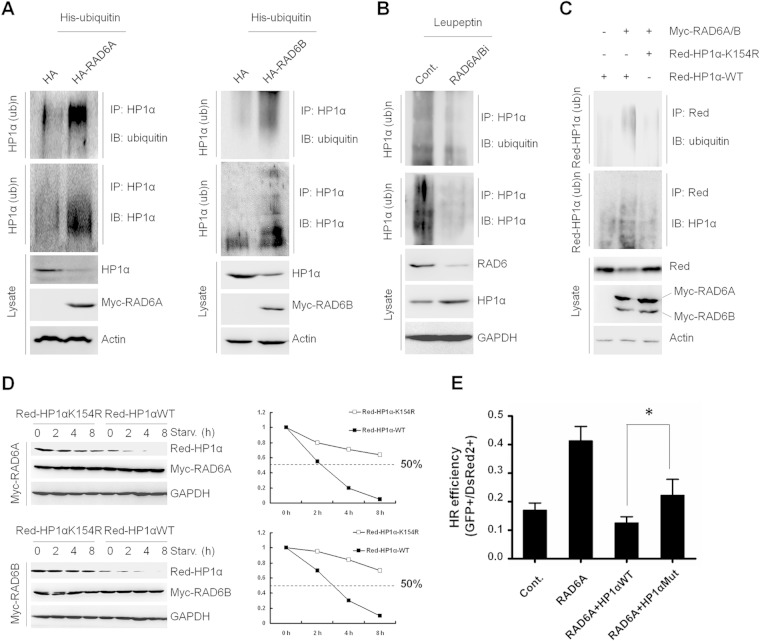
Several factors have been shown to be modified by RAD6, eventually leading to their degradation (18, 21, 22). Given that RAD6 promotes HP1α degradation, we sought to determine whether RAD6 promotes the ubiquitination of HP1α. We performed an in vivo ubiquitination assay and found that the overexpression of RAD6 promoted the polyubiquitination of HP1α (Fig. 5A), whereas knocking down endogenous RAD6 impaired the polyubiquitination of HP1α (Fig. 5B), suggesting that RAD6 indeed promotes the ubiquitination of HP1α.
FIG 5.
Ubiquitination of HP1α at K154 by RAD6 is essential for DNA repair via HR. (A) RAD6 overexpression promotes HP1α ubiquitination. HL-7702 cells transfected with an empty control plasmid or HA-tagged RAD6 (RAD6A and RAD6B) were lysed and subjected to an in vivo ubiquitination assay (see Materials and Methods). (B) Knockdown of RAD6 expression inhibits HP1α ubiquitination. Control or RAD6-specific siRNA-transfected HL-7702 cells were lysed and subjected to an in vivo ubiquitination assay. (C) The HP1α K154R mutant inhibits HP1α ubiquitination by RAD6. Samples that had undergone Co-IP with anti-Red were immunoblotted with antiubiquitin and anti-HP1α to detect the levels of ubiquitinated HP1α. (D) The K154 site of HP1α is essential for autophagy-mediated HP1α degradation upon RAD6 overexpression. WT or K154R mutant HP1α plasmid-transfected HL-7702 cells were starved for the indicated times. Cells were then harvested and subjected to Western blotting assays with the indicated antibodies. (E) Cooverexpressing RAD6A and HP1α K154R significantly suppressed HR. The HR repair efficiency was calculated as described elsewhere (2). *, P < 0.05 (two-tailed unpaired t test; n > 3).
PCNA is ubiquitinated at K164 by RAD6 (15). Based on a comparison of the protein sequences of HP1α and the PCNA sequence surrounding K164 in conjunction with PhosphoSitePlus software to predict potential ubiquitination sites, we hypothesized that HP1α K154 is a candidate site for ubiquitination by RAD6. Indeed, an HP1α K154R mutant could not be ubiquitinated by RAD6 in vivo (Fig. 5C, top). In addition, overexpressing RAD6 did not affect the HP1α K154R protein levels but greatly reduced the HP1α wild-type (WT) expression levels in HEK293 cells (Fig. 5C, bottom). We then compared the stability of HP1α K154R and HP1α WT upon starvation, and we found that, as expected, the mutant is more resistant to degradation (Fig. 5D).
The above-given results all indicated that RAD6 facilitates the degradation of HP1α by ubiquitination at K154 through the autophagy-lysosome pathway. We next examined whether the HP1α K154R mutant retains a suppressive role in HR in the presence of RAD6 overexpression. The results showed that the cooverexpression of the HP1α K154R mutant and RAD6 significantly suppressed HR in comparison to RAD6 overexpression alone (Fig. 5E).
RAD6 and HP1α are negatively correlated in cancer.
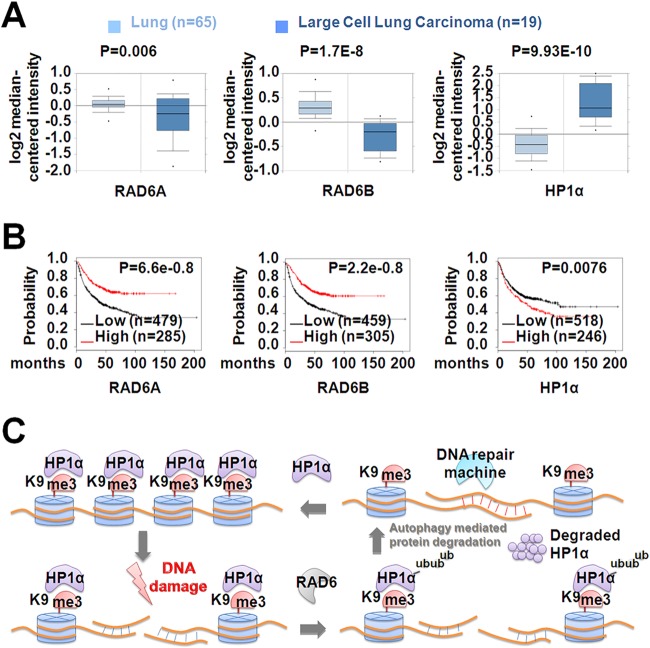
DNA DSB repair efficiency is essential for the maintenance of genome integrity. We next examined the clinical relevance of RAD6 and HP1α in tumorigenesis. Our analysis revealed that the expression levels of RAD6 and HP1α were negatively correlated between normal lung tissues and lung cancer tissues. That is, RAD6 was downregulated in lung cancer tissues compared to that in normal lung tissues, whereas HP1α was upregulated in lung cancer tissues (Fig. 6A). Moreover, RAD6 expression was negatively correlated with poor patient survival in lung cancer patients, whereas HP1α expression was positively correlated with poor survival (Fig. 6B).
FIG 6.
RAD6 and HP1α are negatively correlated with each other during carcinogenesis. (A and B) Correlation between RAD6A/B and HP1α in terms of expression levels and survival rates in lung cancer patients. The clinical data were acquired online and subjected to bioinformatic analysis (see Materials and Methods). (A) RAD6A/B expression is significantly lower in lung tumor tissues than that in normal tissues, whereas the expression of HP1α in tumor tissues is significantly enhanced. (B) The survival rates of patients with lung tumors were positively correlated with the RAD6A/B expression level but negatively correlated with the HP1α expression level. (C) The model of HR regulation by RAD6 through the degradation of HP1α.
Taken together, our study suggests that RAD6-mediated HP1α degradation is critical for DNA DSB repair through HR. RAD6 interacts physically with HP1α, ubiquitinates HP1α at K154, and further degrades HP1α through the autophagy-lysosome pathway. In response to DNA DSBs, the interaction between these two factors is enhanced and the degradation of HP1α is accelerated, resulting in a less compacted chromatin structure that facilitates the recruitment of HR-associated factors and the repair of damaged DNA (Fig. 6C).
DISCUSSION
How cells choose between NHEJ and HR to repair DNA DSBs has been the subject of recent studies (1); however, the role of chromatin context in the choice between these two pathways remains to be understood. The tightly regulated dynamics of higher-order chromatin structure are thought to be essential to the DNA damage response and repair, as most DNA damage is repaired in the context of less compacted chromatin. Repair by NHEJ simply ligates the broken ends, which possibly does not require a relatively decondensed chromatin structure to occur (2, 48). However, the process of end resection, which produces single-stranded DNA to facilitate the subsequent HR steps, is required for HR-mediated repair (48). Understanding how higher-order chromatin structures are regulated in response to DNA DSB repair is essential to answering this question.
Although previous work has indicated that HP1α may rapidly arrive at broken ends, we found that the rapid recruitment of HP1α upon the induction of DNA DSBs plays an inhibitory role in HR. In normal fibroblasts with regular cell cycle checkpoints, NHEJ repairs three quarters of DNA DSBs, whereas HR is responsible for the remaining quarter (49). The biological significance of suppressing HR while tipping repair toward NHEJ by HP1α is most likely the avoidance of potential toxic recombination, because this may cause the large-scale loss of genetic information given the abundance of repetitive sequences in mammalian genomes. However, when NHEJ fails to repair these breaks due to various reasons, for instance a complex configuration formed at broken DNA ends, HR may take over the repair process. At this stage, the depletion of chromatin regulators, such as heterochromatin protein HP1α, thereby leading to an open chromatin structure, is likely a necessary step for ensuing repair by HR.
In this work, we found that RAD6, an E2 ubiquitin-conjugating enzyme that is required for histone H2B monoubiquitination, H3K4me3, and H3K79me3 (19, 50–52), specifically interacted with and caused the degradation of the HP1α protein. The interaction was enhanced in response to DNA DSBs at a late time point, when HR, rather than NHEJ, occurs (Fig. 1C). This result was also supported by our biochemical analysis, as well as a powerful system with a fluorescence-based tetracycline repressor-tagged KillerRed, a homolog of green fluorescent protein that functions as a photosensitizer and generates natural reactive oxygen species (ROS) upon light irradiation, to induce ROS DNA damage in heterochromatic or euchromatic structures. We showed that both RAD6 and HP1α accumulated to the DSB sites in response to the induction of DSBs (Fig. 1D and E). Our ChIP assays further show a degradative effect on HP1α in the presence of RAD6 on chromatin in response to DNA DSBs (Fig. 3D).
Notably, the RAD6-induced enrichment and reduction of HP1α on chromatin does not fully overlap H3K9me3, a well-defined binding partner of HP1α in heterochromatin (41, 53), at early times during DNA damage (2 h after I-SceI transfection). HP1α seemed to be more quickly recruited to DNA damage sites than H3K9me3 (Fig. 3D). When overexpressing RAD6, the reduction in HP1α was also more dramatic than that of H3K9me3, indicating that the dynamics of HP1α and H3K9me3 at the DNA damage site were likely differentially regulated. This rapid recruitment of HP1α to the damage sites at early time points postinduction of DNA DSBs is possibly mediated by DNA damage response complexes, such as HP1/ORC-associated protein (HOAP) and Mre11-Rad50-Nbs (MRN) proteins (54, 55). In fact, previous studies also suggested that the presence of HP1 on chromatin does not always depend on the presence of H3K9me3 in both mammals and Drosophila (56, 57). Further studies are required to elucidate the detailed mechanism.
Our finding that RAD6 specifically regulates the ubiquitination of HP1α, resulting in the degradation of HP1α through the autophagy-mediated pathway, is also supported by recent work showing that RAD6 is a critical regulator of parkin-mediated mitophagy through its role as an E2 ubiquitin-conjugating enzyme (58). In addition, our previous work also demonstrated that RAD6/dRad6 is required for the degradation of p53, a tumor suppressor, in both Drosophila and mammalian cells (21, 22), which all support that the dosage of RAD6 is critical in tumorigenesis. Why HP1α is specifically degraded in response to autophagy activation remains to be determined. We suspect that in response to autophagy, cells may have to degrade HP1α to maintain proper genomic integrity, for example, by facilitating DNA DSB repair by HR. Nonetheless, in addition to the crucial role of RAD6 in sensing DNA DSBs (59, 60), our observations provide a novel link between the RAD6-mediated HP1 ubiquitination and DNA DSB repair.
ACKNOWLEDGMENTS
This work was supported by the 973 program of the Ministry of Science and Technology of China (grant no. 2011CB965300, 2013CB967600, 2013CB967500, 2014CB964600, 2014CB964603, and 2014CB965001), the National Natural Science Foundation of China (grant no. 31330043, 31201051, 91419304, 31371396, and 91319306), and the Shanghai Municipal Natural Science Foundation (grant no. 12ZR1433100, and 12ZR1433000).
We thank Li Yu and Zhijie Chang for kindly providing the GFP-LC3 and GFP-p62 plasmids.
REFERENCES
- 1.Haber JE. 2000. Partners and pathways repairing a double-strand break. Trends Genet 16:259–264. doi: 10.1016/S0168-9525(00)02022-9. [DOI] [PubMed] [Google Scholar]
- 2.Mao Z, Bozzella M, Seluanov A, Gorbunova V. 2008. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. 2011. SIRT6 promotes DNA repair under stress by activating PARP1. Science 332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieber MR. 2008. The mechanism of human nonhomologous DNA end joining. J Biol Chem 283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 5.Thompson LH, Schild D. 2001. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res 477:131–153. doi: 10.1016/S0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 6.Bennett EJ, Harper JW. 2008. DNA damage: ubiquitin marks the spot. Nat Struct Mol Biol 15:20–22. doi: 10.1038/nsmb0108-20. [DOI] [PubMed] [Google Scholar]
- 7.Bergink S, Jentsch S. 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 8.Panier S, Durocher D. 2009. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair 8:436–443. doi: 10.1016/j.dnarep.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 10.Kirkin V, McEwan DG, Novak I, Dikic I. 2009. A role for ubiquitin in selective autophagy. Mol Cell 34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 11.Kraft C, Peter M, Hofmann K. 2010. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol 12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 12.Ulrich HD, Walden H. 2010. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol 11:479–489. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 13.Weissman AM. 2001. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 14.Game JC, Chernikova SB. 2009. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair 8:470–482. doi: 10.1016/j.dnarep.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 16.Torres-Ramos CA, Yoder BL, Burgers PM, Prakash S, Prakash L. 1996. Requirement of proliferating cell nuclear antigen in RAD6-dependent postreplicational DNA repair. Proc Natl Acad Sci U S A 93:9676–9681. doi: 10.1073/pnas.93.18.9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dohmen RJ, Madura K, Bartel B, Varshavsky A. 1991. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc Natl Acad Sci U S A 88:7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sancar G, Sancar C, Brügger B, Ha N, Sachsenheimer T, Gin E, Wdowik S, Lohmann I, Wieland F, Höfer T, Diernfellner A, Brunner M. 2011. A global circadian repressor controls antiphasic expression of metabolic genes in Neurospora. Mol Cell 44:687–697. doi: 10.1016/j.molcel.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Sung P, Berleth E, Pickart C, Prakash S, Prakash L. 1991. Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J 10:2187–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watkins JF, Sung P, Prakash S, Prakash L. 1993. The extremely conserved amino terminus of RAD6 ubiquitin-conjugating enzyme is essential for amino-end rule-dependent protein degradation. Genes Dev 7:250–261. doi: 10.1101/gad.7.2.250. [DOI] [PubMed] [Google Scholar]
- 21.Chen S, Wang DL, Liu Y, Zhao L, Sun FL. 2012. RAD6 regulates the dosage of p53 by a combination of transcriptional and posttranscriptional mechanisms. Mol Cell Biol 32:576–587. doi: 10.1128/MCB.05966-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, Wei HM, Lv WW, Wang DL, Sun FL. 2011. E2 ligase dRad6 regulates DMP53 turnover in Drosophila. J Biol Chem 286:9020–9030. doi: 10.1074/jbc.M110.190314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C, Wang D, Wu J, Keller J, Ma T, Yu X. 2013. RNF168 forms a functional complex with RAD6 during the DNA damage response. J Cell Sci 126:2042–2051. doi: 10.1242/jcs.122945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akey CW, Luger K. 2003. Histone chaperones and nucleosome assembly. Curr Opin Struct Biol 13:6–14. doi: 10.1016/S0959-440X(03)00002-2. [DOI] [PubMed] [Google Scholar]
- 25.Eitoku M, Sato L, Senda T, Horikoshi M. 2008. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol Life Sci 65:414–444. doi: 10.1007/s00018-007-7305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palomera-Sanchez Z, Zurita M. 2011. Open, repair and close again: chromatin dynamics and the response to UV-induced DNA damage. DNA Repair 10:119–125. doi: 10.1016/j.dnarep.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 27.De Lucia F, Ni JQ, Vaillant C, Sun FL. 2005. HP1 modulates the transcription of cell-cycle regulators in Drosophila melanogaster. Nucleic Acids Res 33:2852–2858. doi: 10.1093/nar/gki584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eissenberg JC, Elgin SC. 2000. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev 10:204–210. doi: 10.1016/S0959-437X(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 29.Eissenberg JC, Elgin SC. 2014. HP1a: a structural chromosomal protein regulating transcription. Trends Genet 30:103–110. doi: 10.1016/j.tig.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lomberk G, Wallrath L, Urrutia R. 2006. The heterochromatin protein 1 family. Genome Biol 7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smallwood A, Estève PO, Pradhan S, Carey M. 2007. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev 21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng W, Ball AR Jr, Yokomori K. 2010. HP1: heterochromatin binding proteins working the genome. Epigenetics 5:287–292. doi: 10.4161/epi.5.4.11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. 2008. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature 453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 34.Ayoub N, Jeyasekharan AD, Venkitaraman AR. 2009. Mobilization and recruitment of HP1: a bimodal response to DNA breakage. Cell Cycle 8:2945–2950. doi: 10.4161/cc.8.18.9486. [DOI] [PubMed] [Google Scholar]
- 35.Dinant C, Luijsterburg MS. 2009. The emerging role of HP1 in the DNA damage response. Mol Cell Biol 29:6335–6340. doi: 10.1128/MCB.01048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, Aten JA, Fousteri MI, Jansen G, Dantuma NP, Vermeulen W, Mullenders LH, Houtsmuller AB, Verschure PJ, van Driel R. 2009. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol 185:577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. 2011. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu LP, Ni JQ, Shi YD, Oakeley EJ, Sun FL. 2005. Sex-specific role of Drosophila melanogaster HP1 in regulating chromatin structure and gene transcription. Nat Genet 37:1361–1366. doi: 10.1038/ng1662. [DOI] [PubMed] [Google Scholar]
- 39.Ni JQ, Liu LP, Hess D, Rietdorf J, Sun FL. 2006. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev 20:1959–1973. doi: 10.1101/gad.390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lan L, Nakajima S, Wei L, Sun L, Hsieh CL, Sobol RW, Bruchez M, Van Houten B, Yasui A, Levine AS. 2014. Novel method for site-specific induction of oxidative DNA damage reveals differences in recruitment of repair proteins to heterochromatin and euchromatin. Nucleic Acids Res 42:2330–2345. doi: 10.1093/nar/gkt1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng JC, Karpen GH. 2009. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 5:e1000435. doi: 10.1371/journal.pgen.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon SH, Workman JL. 2011. The changing faces of HP1: from heterochromatin formation and gene silencing to euchromatic gene expression: HP1 acts as a positive regulator of transcription. Bioessays 33:280–289. doi: 10.1002/bies.201000138. [DOI] [PubMed] [Google Scholar]
- 43.Wallrath LL, Elgin SC. 2012. Enforcing silencing: dynamic HP1 complexes in Neurospora. Nat Struct Mol Biol 19:465–467. doi: 10.1038/nsmb.2291. [DOI] [PubMed] [Google Scholar]
- 44.Mizushima N. 2007. Autophagy: process and function. Genes Dev 21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 45.Tanida I, Ueno T, Kominami E. 2004. LC3 conjugating system in mammalian autophagy. Int J Biochem Cell Biol 36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjørkøy G, Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24135. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 48.Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 49.Mao Z, Bozzella M, Seluanov A, Gorbunova V. 2008. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair 7:1765–1771. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. 2009. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. 2007. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 52.Robzyk K, Recht J, Osley MA. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 53.Lachner M, O'Sullivan RJ, Jenuwein T. 2003. An epigenetic road map for histone lysine methylation. J Cell Sci 116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 54.Ciapponi L, Cenci G, Ducau J, Flores C, Johnson-Schlitz D, Gorski MM, Engels WR, Gatti M. 2004. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr Biol 14:1360–1366. doi: 10.1016/j.cub.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 55.Gao G, Walser JC, Beaucher ML, Morciano P, Wesolowska N, Chen J, Rong YS. 2010. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J 29:819–829. doi: 10.1038/emboj.2009.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Figueiredo ML, Philip P, Stenberg P, Larsson J. 2012. HP1a recruitment to promoters is independent of H3K9 methylation in Drosophila melanogaster. PLoS Genet 8:e1003061. doi: 10.1371/journal.pgen.1003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horáková AH, Bártová E, Galiová G, Uhlírová R, Matula P, Kozubek S. 2010. SUV39h-independent association of HP1 beta with fibrillarin-positive nucleolar regions. Chromosoma 119:227–241. doi: 10.1007/s00412-009-0252-2. [DOI] [PubMed] [Google Scholar]
- 58.Haddad DM, Vilain S, Vos M, Esposito G, Matta S, Kalscheuer VM, Craessaerts K, Leyssen M, Nascimento RM, Vianna-Morgante AM, De Strooper B, Van Esch H, Morais VA, Verstreken P. 2013. Mutations in the intellectual disability gene Ube2a cause neuronal dysfunction and impair parkin-dependent mitophagy. Mol Cell 50:831–843. doi: 10.1016/j.molcel.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 59.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim H, Chen J, Yu X. 2007. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]