Abstract
Iron is an essential, yet at elevated concentrations toxic trace element. To date, the mechanisms of iron sensing by eukaryotic iron-responsive transcription factors are poorly understood. The Saccharomyces cerevisiae transcription factor Yap5, a member of the Yap family of bZIP stress response regulators, administrates the adaptive response to high-iron conditions. Despite the central role of the iron-sensing process for cell viability, the molecule perceived by Yap5 and the underlying regulatory mechanisms are unknown. Here, we show that Yap5 senses high-iron conditions by two Fe/S clusters bound to its activator domain (Yap5-AD). The more stable iron-regulatory Fe/S cluster at the N-terminal cysteine-rich domain (n-CRD) of Yap5 is detected in vivo and in vitro. The second cluster coordinated by the C-terminal CRD can only be shown after chemical reconstitution, since it is bound in a labile fashion. Both clusters are of the [2Fe-2S] type as characterized by UV/visible (UV/Vis), circular dichroism, electron paramagnetic resonance (EPR), and Mössbauer spectroscopy. Fe/S cluster binding to Yap5-AD induces a conformational change that may activate transcription. The cluster-binding motif of the n-CRD domain is highly conserved in HapX-like transcription factors of pathogenic fungi and thus may represent a general sensor module common to many eukaryotic stress response regulators.
INTRODUCTION
Iron is a key trace element for virtually all organisms. It functions as an essential cofactor in electron transfer, metabolite biosynthesis, DNA metabolism, and protein translation. Although iron is highly abundant, its bioavailability is low due to its poor solubility under ambient conditions (1, 2). For microbial pathogens, iron acquisition from their host cells is frequently crucial for their virulence. Hence, mutations in microbial genes involved in iron acquisition or utilization are associated with the loss of pathogenicity (3, 4). On the other hand, high intracellular iron levels are both a source and an amplifier of reactive oxygen species and thus highly toxic.
Iron acquisition, transport and storage need to be tightly regulated, and disruption or deregulation of iron-related molecules can have significant health consequences. Hence, organisms have developed sophisticated and largely divergent strategies to acquire appropriate cellular iron levels and to avoid iron overloading (5–7). Except for vertebrates, which regulate cellular iron homeostasis mainly on the level of translation by the iron-regulatory proteins IRP1 and IRP2, transcriptional control of iron-responsive gene expression is widespread (7). In a large number of fungi, iron-responsive gene expression is conferred by the interplay of two conserved transcriptional repressors. Of these, the GATA-type transcription factor SreA regulates the expression of genes involved in iron uptake, while the basic leucine zipper (bZIP) transcription factor HapX regulates the iron-responsive expression of genes involved in iron-consuming pathways through interaction with the CCAAT-binding complex CBC (4, 8).
The model organism Saccharomyces cerevisiae utilizes three iron-responsive transcription factors. Aft1 and Aft2 play a central role in iron-dependent transcription activation of genes encoding components involved in cellular uptake and intracellular distribution of iron (9, 10). Yap5 is involved in the detoxification of excess iron by inducing the transcription of, e.g., CCC1 encoding the only known iron importer into the vacuole, the major metal storage organelle in fungi (11, 12). Yap5 belongs to a fungus-specific family of stress response regulators that contains eight members in S. cerevisiae (13–15). Yap proteins are related to AP-1 transcription factors that are widespread in higher eukaryotes (16, 17). These transcription factors share a conserved bZIP DNA-binding domain, which consists of a leucine zipper mediating dimerization, and an adjacent basic region that binds to specific short DNA sequences termed AP-1 sites or, in the case of Yap proteins, Yap response elements (YRE) (18). The S. cerevisiae Yap family members mediate the response to oxidative and nitrosative stress (Yap1), cadmium and arsenic stress (Yap1, Yap2, and Yap8), osmotic stress (Yap4 and Yap6), and iron overload (Yap1 and Yap5) (15).
The mechanisms of iron sensing by eukaryotic transcription factors are poorly understood, and the sensed molecules are largely unknown. Yap5 confers iron-responsive gene expression through a C-terminal activator domain that contains two short cysteine-rich sequence motifs (CRD) that are essential for transcriptional activation (11). Recent genetic evidence showed that S. cerevisiae cells with defects in the mitochondrial Fe/S cluster assembly (ISC) system are unable to induce genes controlled by Yap5 (19). The mitochondrial ISC and ISC export systems are essential for the maturation of all cellular Fe/S proteins (see Fig. S1 in the supplemental material) (20). Hence, this finding implies that either Yap5 harbors an iron-sulfur (Fe/S) cluster as an iron sensor or is activated through the interaction with a regulatory adapter Fe/S protein, similar to S. cerevisiae Aft1 or S. pombe Php4 and Fep1 that interact with the [2Fe-2S] glutaredoxin Grx4 (8, 21, 22) (see Fig. S1 in the supplemental material). In this work, we investigate the molecular mode of iron sensing by Yap5 and show that it accommodates two Fe/S cluster sensors that are essential for conferring iron-responsive gene expression. The Fe/S cluster binding motif of the N-terminal CRD (n-CRD) is highly conserved in fungal iron-responsive bZIP transcription factors, indicating a universal mode of iron sensing.
MATERIALS AND METHODS
Strains and growth conditions.
Yeast strains (background W303-1A) were cultivated in rich medium (yeast extract-peptone [YP]) or minimal medium containing all recommended supplements (SC), 2% (wt/vol) glucose (SD), or 2% (wt/vol) galactose (SGal) (23). Minimal medium lacking FeCl3 was obtained from ForMedium. For high-iron conditions, media were supplemented with 1 mM FeCl2 and 1 mM ascorbate. GAL1-10 promoter exchange strains (see Table S1 in the supplemental material) were depleted to critical protein levels by cultivation in SD medium for 40 h prior to analysis. Yap5 and Yap5-AD were expressed with a C-terminal Myc tag in S. cerevisiae from vector p426-TDH3 or p424-TDH3 (for information on plasmids, see Tables S2 and S3 in the supplemental material).
Purification and reconstitution of recombinant proteins.
Recombinant Yap5-AD (and mutants thereof) were expressed in Escherichia coli C41 (DE3) with an N-terminal His tag from vector pET15b (24) (see Tables S1 and S2 in the supplemental material). Proteins were purified by Ni-NTA chromatography from cell extracts of E. coli after cultivation in LB medium supplemented with 50 μM ferric ammonium citrate (FAC) and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 28°C for 16 h. For chemical Fe/S cluster reconstitution, purified proteins (200 μM final) were reduced with 5 mM dithiothreitol in 50 mM Tris–150 mM NaCl (pH 8.0) for 2 h under anaerobic conditions in a Coy type A anaerobic chamber. FAC and Li2S were added at 2- to 8-fold molar excesses, followed by incubation for 30 min, and reconstituted proteins were purified by gel filtration (PD10; GE Healthcare). Samples for Mössbauer spectroscopy were reconstituted with a 4-fold molar excess of Li2S and 57FeCl3. UV/visible (UV/Vis) spectra were recorded on a V550 spectrometer (Jasco), circular dichroism (CD) spectra were recorded on a J815 CD spectrometer (Jasco). Analytical gel filtration was carried out on a Waters HPLC system using a Shodex HW802.5 HPLC column at a flow rate of 0.8 ml/min in 50 mM Tris-HCl–250 mM NaCl (pH 8.0). Bovine serum albumin (67 kDa), ovalbumin (49 kDa), chymotrypsinogen (25 kDa), and a truncated version of Cia2 (17 kDa) were used as molecular mass markers.
Mössbauer spectroscopy.
Mössbauer spectra were recorded on oxidized 57Fe reconstituted Yap5-AD with alternating constant acceleration. The minimum experimental line width was 0.24 mm s−1 (full width at half-height). The sample temperature was controlled by an Oxford Instruments Variox cryostat. The γ-source (57Co/Rh, 1.8 GBq) was kept at room temperature. Isomer shifts are quoted relative to iron metal at 300 K. (For further information, see the supplemental material.)
EPR spectroscopy.
Yap5-AD and mutants thereof were reduced under anaerobic conditions by incubation with 2 mM dithionite for 3 min to achieve reduction of the Fe/S cluster(s) and immersed in liquid nitrogen. electron paramagnetic resonance (EPR) spectra were recorded using a Bruker EMX-6/1 spectrometer with cooling by an Oxford Instruments helium flow cryostat. Fe/S cluster concentrations were quantified by double integration of spectra acquired under nonsaturating conditions using a 1 mM Cu2+-EDTA standard. The EPR conditions were as follows: microwave frequency, 9.458 GHz; microwave power, 0.013 mW (Fig. 3D) and 0.0008, 0.013, 0.2, 0.2, and 0.2 mW (from the top to the bottom in Fig. 3E); modulation frequency, 100 kHz; modulation amplitude, 1.25 mT; temperature (T), 10 K (unless indicated otherwise).
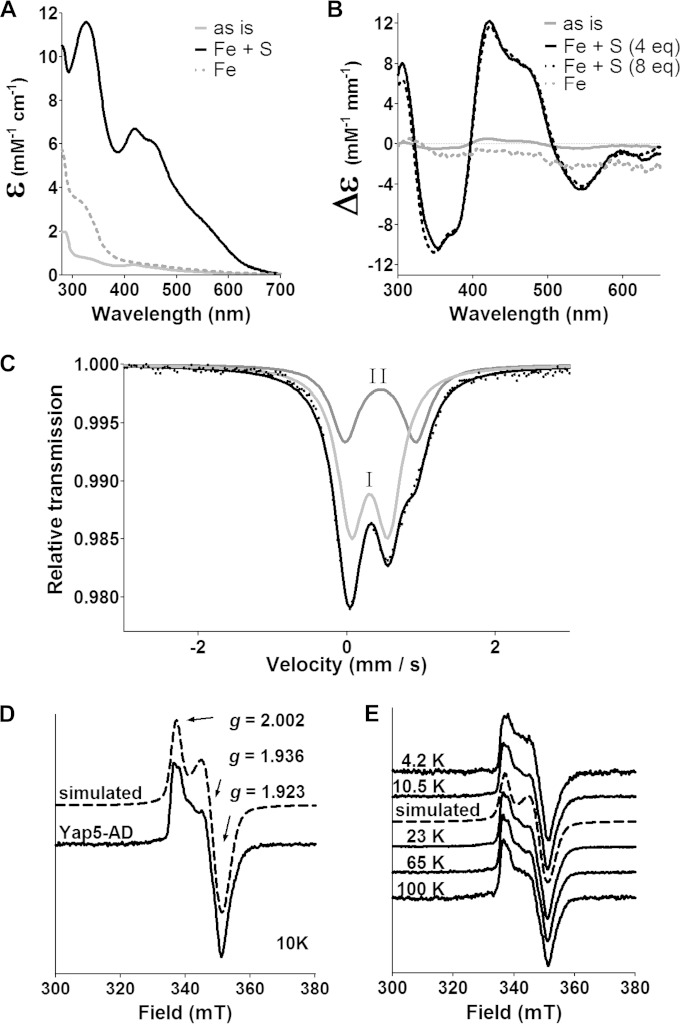
FIG 3.
Yap5-AD binds [2Fe-2S] clusters in vitro. (A, B) UV/Vis (A) and CD spectra (B) of Yap5-AD after isolation from E. coli (as is) and after chemical reconstitution with either Fe2+ and S2− ions (Fe + S) (4 and 8-fold molar excess, as indicated) or Fe2+ at 4-fold excess (Fe). (C) Zero-field Mössbauer spectrum of 57Fe reconstituted Yap5-AD recorded at 80 K. The black and gray lines represent a fit with two Lorentzian subspectra that can be assigned to [2Fe-2S]2+ (I) and [4Fe-4S]2+ (II) clusters (see the supplemental material). (D and E) Experimental EPR spectra and simulation of dithionite-reduced reconstituted Yap5-AD at different temperatures. Simulation parameters: microwave frequency, 9.458 GHz; g values of 2.002, 1.936, and 1.923 with line widths of 2.9, 5.0, and 5.5 mT.
Miscellaneous methods.
The following published methods were used: manipulation of DNA and PCR (25), transformation of yeast cells (26), immunostaining (27), in vivo labeling of yeast cells with 55FeCl3 (ICN) and measurement of 55Fe incorporation into Fe/S proteins by immunoprecipitation and scintillation counting (28), determination of promoter strength using luciferase (28), and quantification of nonheme iron and acid-labile sulfur (29). Antibodies were raised in rabbits against recombinant proteins. Antibodies against c-Myc were obtained from Santa Cruz.
RESULTS
Yap5 binds Fe/S clusters in vivo.
In order to analyze iron binding to Yap5 in its native environment in vivo, we utilized a truncated version of Yap5 that consisted of the activator domain and a C-terminal Myc-tag (Yap5-AD) (Fig. 1A). Yeast cells were radiolabeled with 55Fe, Yap5-AD was isolated by immunoprecipitation with anti-Myc antibodies, and 55Fe binding to Yap5-AD was quantified by scintillation counting. Significant amounts of 55Fe copurified with anti-Myc antibodies in cells overproducing Yap5-AD but not in cells lacking this protein (Fig. 1B). In order to distinguish whether Yap5-AD binds iron or an Fe/S cofactor, we tested whether iron binding to Yap5-AD depends on components of the mitochondrial Fe/S cluster assembly (ISC) and ISC export systems and the cytosolic iron-sulfur protein assembly (CIA) system (20, 30). Since the maturation of cellular Fe/S proteins, but not that of iron-only proteins, depends on the function of these three assembly systems (see Fig. S1 in the supplemental material), the dependence of iron binding on components of these systems is a strong indication that Yap5 may bind Fe/S clusters. 55Fe radiolabeling experiments were performed with GAL1-10 promoter exchange strains that deplete the regulated protein during cultivation in glucose-containing medium (Fig. 1B). Upon depletion of the mitochondrial ISC proteins Nfs1, Ssq1, and the export component Atm1 (see Fig. S1 in the supplemental material), iron binding to Yap5-AD was reduced in the respective Gal strains to 25 to 40% of wild-type levels. Since protein levels were, except for Gal-NFS1 cells, similar to those found for the wild type, this reduced iron binding capacity indicates that Yap5-AD binds Fe/S clusters (see Fig. S2 in the supplemental material). In contrast, 55Fe binding to Yap5-AD remained unaffected upon depletion of the CIA proteins Cfd1, Nbp35, Nar1, and Met18 (Fig. 1B). Since 55Fe binding to the cytosolic Fe/S protein Leu1 was strongly reduced under these conditions (see Fig. S2 in the supplemental material), these data show that iron incorporation into Yap5 does not involve the CIA system. A similar, ISC-dependent and CIA-independent maturation was observed previously for the cytosolic protein Grx4 which harbors a glutathione-coordinated bridging [2Fe-2S] cluster (21, 31).
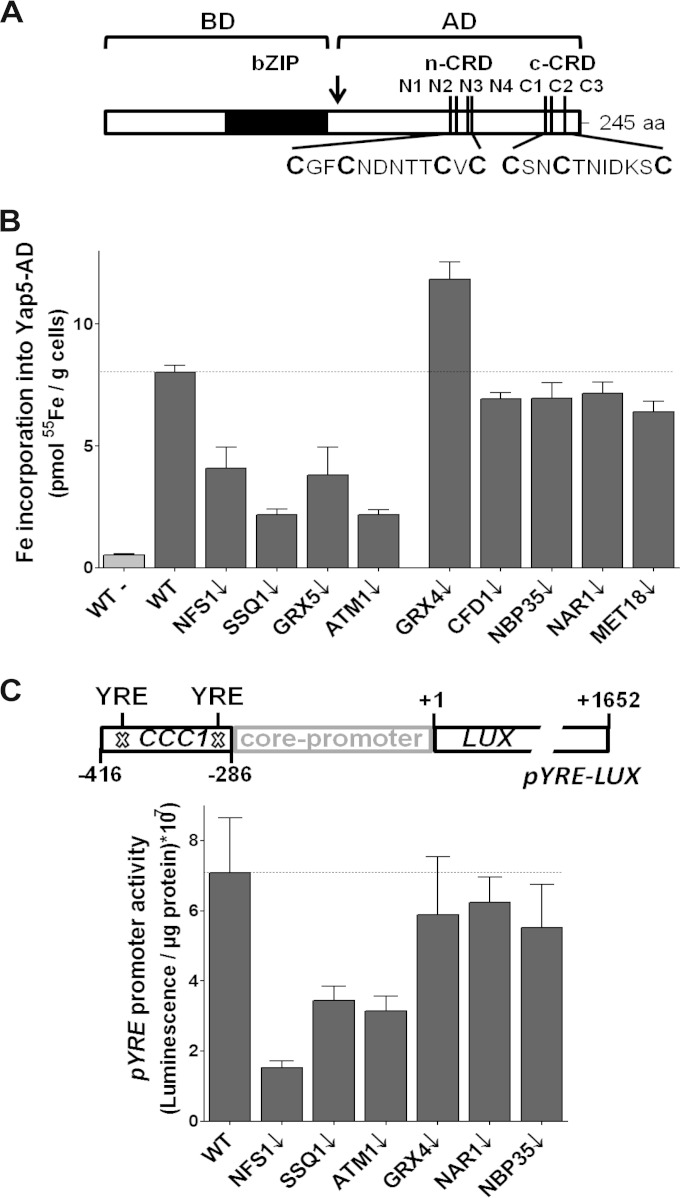
FIG 1.
Yap5 binds Fe/S clusters in vivo. (A) Schematic illustration of Yap5 which consists of a N-terminal bZIP DNA-binding domain (BD) and a C-terminal activator domain (AD). The arrow indicates the start of the activator domain that contains 7 conserved cysteines (N1 to N4 and C1 to C3) in two cysteine-rich domains (CRD). The primary sequence of the CRDs is depicted at the bottom. (B) Wild-type (WT) and depleted GAL1-10 promoter exchange strains of the indicated members of the ISC (left side) or the CIA machineries (right side) expressing Myc-tagged Yap5-AD were radiolabeled with 55Fe. 55Fe incorporation into Yap5-AD was determined by immunoprecipitation with α-Myc antibodies, followed by scintillation counting. Wild-type cells harboring the empty vector (WT−) served as controls. (C) WT and the indicated depleted GAL1-10 strains harboring the luciferase-based reporter construct pYRE-LUX were cultivated in SD medium supplemented with 1 mM FeCl2 for 12 h, and the luciferase activities were determined. The pYRE hybrid promoter consists of the two YREs of the CCC1 promoter and the LEU1 core promoter (panel C, top; see also Table S2 in the supplemental material). GAL1-10 strains were depleted to critical protein levels by cultivation in SD medium for 40 h prior to analysis. Error bars indicate the standard errors of the mean (SEM; n > 4).
Next, we explored whether the Yap5-dependent transcriptional activation of CCC1 depends on the cellular Fe/S protein biogenesis systems in a similar fashion as its Fe/S cluster binding. Since there is evidence that CCC1 is not controlled by Yap5 alone, we constructed a hybrid promoter that contains the two Yap response elements (YRE) of the CCC1 promoter fused to the core promoter of the LEU1 gene (Fig. 1C) (11, 32). This hybrid pYRE promoter was inserted in front of the luciferase reporter gene, and the resulting pYRE-LUX plasmid was transformed into various Gal-ISC and Gal-CIA cells. For quantification of promoter activities, ISC and CIA protein-depleted cells were cultivated for 12 h in minimal SD medium supplemented with excess FeCl2. pYRE promoter activities were 5-fold reduced in Gal-NFS1 and 2-fold lower in Gal-SSQ1 and Gal-ATM1 cells in comparison to the wild type (Fig. 1C). In contrast, Gal-CIA cells showed wild-type promoter activities. A similar regulation pattern was previously seen for the full-length CCC1 promoter (19) (see Fig. S2 in the supplemental material). This close correlation between Fe/S cluster binding and the promoter strength of a Yap5-dependent gene indicates that the Fe/S cluster is relevant for transcriptional activation by Yap5.
The yeast cytosolic glutaredoxins Grx3 and Grx4 play a critical role in the regulation of the iron-responsive transcription factor Aft1 (9) (21). In contrast, it was concluded previously that deletion of GRX3 and GRX4 does not directly affect Yap5-dependent transcription (19). Consistent with this, pYRE promoter activities remained at wild-type levels in Gal-GRX4/grx3Δ cells (Fig. 1C), while activities of the full CCC1 promoter were 5-fold higher than in the wild type (see Fig. S2 in the supplemental material). Since Gal-GRX4/grx3Δ cells accumulate large amounts of cytosolic iron, the strong induction of CCC1 indicates that the heavy metal detoxification response remains intact in cells with low levels of Grx3 and Grx4. Consistent with this conclusion, the higher intracellular iron concentration likely leads to an increased Fe/S cluster binding to Yap5, explaining why in vivo iron binding to Yap5-AD was ∼1.5-fold increased in Gal-GRX4/grx3Δ cells compared to wild-type cells (Fig. 1B). Taken together, these data show that the cytosolic glutaredoxins are not required for the maturation of Yap5's Fe/S clusters and therefore most likely not directly involved in the regulation of Yap5.
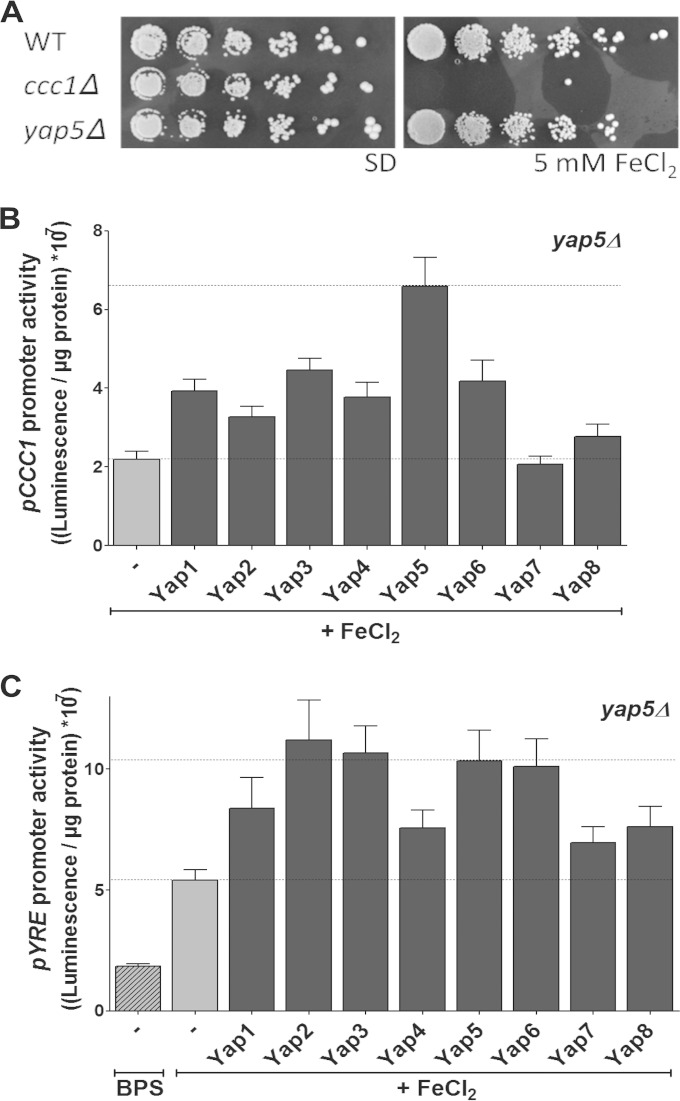
There are conflicting data in the literature whether Yap5 is essential or not for the regulation of CCC1 (11, 32). Likely, differences in laboratory strain backgrounds might (at least partially) explain these differences. Consistent with a nonessential role of Yap5, our yap5Δ deletion strain was able to induce CCC1 and was viable under high-iron conditions, unlike ccc1Δ cells (Fig. 2A and B). Since the various Yap proteins bind to similar promoter elements, it is likely that some of the other Yap members may also contribute to the overall regulation of CCC1. Indeed, Yap1-4 and Yap6 were able to partially induce the CCC1 promoter, when overproduced in yap5Δ cells (Fig. 2B). Their overproduction also induced the pYRE hybrid promoter, indicating that they function at least in part by binding to the YREs of the CCC1 promoter (Fig. 2C). Nevertheless, the induction of the full-length CCC1 promoter construct, which most likely more accurately reflects the situation in vivo, was highest upon overproduction of Yap5. Taken together, these data demonstrate that Yap5 is the dominant transcription factor for the induction of CCC1 and that the binding of an Fe/S cluster to Yap5 plays a central role in this process. Whether any of the other Yap proteins also utilize Fe/S clusters for regulation remains to be determined.
FIG 2.
Yap5 regulates the expression of CCC1, but not solely. (A) Serial dilutions of wild-type, ccc1Δ, and yap5Δ cells were spotted onto agar plates containing SD medium supplemented with or without 5 mM FeCl2 and incubated for 3 days at 30°C. (B and C) Luciferase activities were determined in yap5Δ cells harboring plasmids overproducing the indicated Yap proteins from a 2μ vector (22) and either pCCC1-LUX (B) or pYRE-LUX (C). Cells were grown for 12 h in SD medium supplemented with either 50 μM bathophenanthroline disulfonic acid (BPS) or 1 mM FeCl2. The luciferase activities were determined and normalized to protein concentration. Cells harboring the empty vector (−) served as a control. Error bars indicate the SEM (n > 4).
Purified Yap5 binds [2Fe-2S] clusters.
In order to further verify our conclusion that Yap5 binds Fe/S clusters, a His-tagged Yap5-AD protein was isolated from E. coli by affinity chromatography and analyzed by several spectroscopic techniques (see Fig. S3 in the supplemental material). Yap5-AD displayed a UV/Vis spectrum characteristic for Fe/S proteins that became much more intense upon chemical reconstitution with iron and S2− ions (Fig. 3A). The CD spectrum displayed a maximum at 422 nm and a plateau between 450 to 475 nm that strongly increased upon reconstitution and saturated upon reconstitution with a 4-fold molar excess of iron and S2− ions (Fig. 3B and see Fig. S4 in the supplemental material). The shapes of the CD spectra of reconstituted Yap5-AD were similar to those of the protein after isolation from E. coli (see Fig. S4 in the supplemental material). When Yap5-AD was incubated with Fe2+ in the absence of sulfide, the UV/Vis spectrum of the resulting protein lacked a discrete peak in the 300–350 nm region that indicates iron binding at a specific site (Fig. 3A). Furthermore, no CD signal was observed in the visible region (Fig. 3B). These data show that Yap5-AD binds Fe/S clusters, and yet most likely is not a mono- or dinuclear iron protein.
The zero-field Mössbauer spectrum of 57Fe-reconstituted Yap5-AD (4-fold excess of 57Fe and S2−) showed a superposition of two subspectra, I and II, with typical parameters characteristic for valence-localized Fe3+ in oxidized [2Fe-2S]2+ clusters and for mixed-valence Fe2.5+ found in oxidized [4Fe-4S]2+ clusters, respectively (33–35) (Fig. 3C). The experimental data could be fitted with simulated spectra for [2Fe-2S] and [4Fe-4S] cofactors at a molar ratio of 4:1 (see the supplemental material for further details). In agreement with the detection of diamagnetic Fe/S clusters by Mössbauer spectroscopy, reconstituted Yap5-AD was EPR silent. Upon dithionite reduction, however, a slightly rhombic EPR spectrum with simulated g values of 2.002, 1.936, and 1.923 appeared which integrated to maximally 0.2 cluster/Yap5-AD monomer (Fig. 3D). The low cluster occupancy is due to the lability of the clusters in the presence of dithionite. The EPR spectrum is typical for a reduced [2Fe-2S]1+ cluster. This is further supported by the fact that both shape and intensity at nonsaturating microwave power remained constant between 4.2 and 100 K (Fig. 3E). Taken together, Yap5-AD predominantly binds [2Fe-2S] clusters and yet may also incorporate small amounts of [4Fe-4S] clusters.
Both CRDs of Yap5 exhibit gene regulatory function, and yet only n-CRD stably binds a Fe/S cluster in vivo.
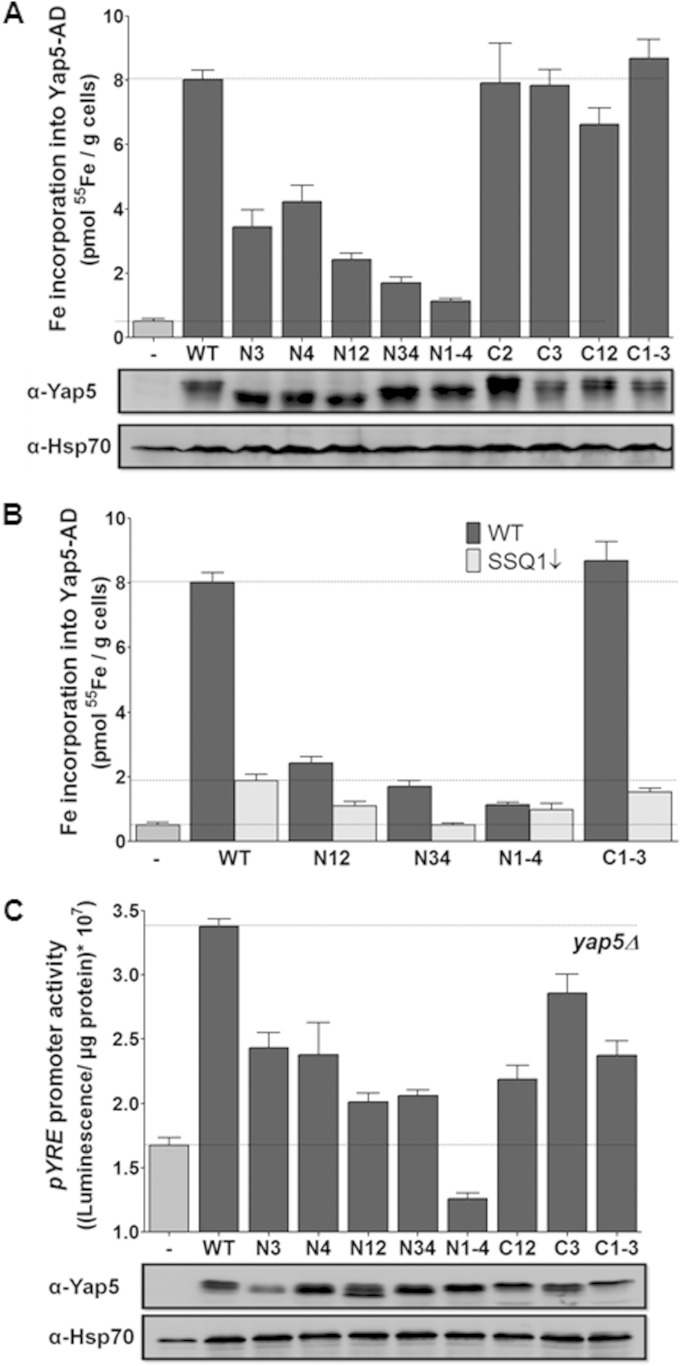
In order to identify the number of Fe/S clusters and their coordinating residues, a series of site-directed Yap5-AD mutant proteins were generated in which individual or combinations of up to four cysteine residues were substituted by alanine (see Fig. 1A). Upon expression of these mutated proteins in S. cerevisiae, their levels were similar to wild-type Yap5-AD (Fig. 4A). Replacement of single or double cysteine residues N1 to N4 of the n-CRD significantly reduced the in vivo 55Fe binding to Yap5-AD (Fig. 4A). Iron binding declined further to almost background levels when all four cysteine residues of the n-CRD were mutated. This was also observed when in vivo 55Fe binding experiments with this Yap5-AD mutant were carried out under anaerobic conditions. In contrast, replacement of the cysteine residues C1 to C3 of the c-CRD did not significantly affect iron binding to Yap5-AD, indicating that the 55Fe detected by our in vivo radiolabeling assay is bound to the n-CRD of Yap5-AD. The bound iron is part of an Fe/S cluster, since radiolabeling of the Yap5-AD/C1-3 variant in vivo declined strongly in the ISC assembly mutant Gal-SSQ1 (Fig. 4B). A similar Ssq1 dependence was seen for the weak 55Fe binding to the Yap5-AD variants N12 and N34, suggesting that Yap5-AD still may bind Fe/S clusters when the n-CRD is modified (Fig. 4B). This result suggests that also the c-CRD may bind Fe/S clusters, yet in comparison much more weakly than the n-CRD. Likely, most of the Fe/S clusters are lost during the immunoprecipitation procedure.
FIG 4.
Yap5 binds a Fe/S cluster sensor at the n-CRD in vivo. (A) Yap5-AD variants in which the cysteine residues of the n-CRD (N1 to N4) or the c-CRD (C1 to C3) were substituted by alanine (or serine in N4) either alone or in combination were expressed in wild-type (WT) cells. Iron binding was determined by 55Fe radiolabeling and immunoprecipitation with anti-Myc-antibodies as described in Fig. 1. Protein levels of Yap5-AD were determined by immunostaining. Hsp70 served as a loading control. (B) Iron binding to the indicated Yap5-AD variants was determined in wild-type cells (WT) and depleted Gal-SSQ1 cells (SSQ1↓) by radiolabeling with 55Fe as described above. (C) Transcriptional activities of the indicated full-length Yap5 variants were determined in yap5Δ cells harboring reporter plasmid pYRE-LUX as described in Fig. 1. Protein levels of the full-length Yap5 fused to a Myc tag were determined by immunostaining. Hsp70 served as a loading control. Cells harboring the empty vectors (−) served as controls in panels A to C. Error bars indicate the SEM (n > 4).
To analyze whether both Fe/S cluster binding regions show gene regulatory activity, we measured the activation of the Yap5-inducible pYRE promoter by these mutant proteins (Fig. 4C). Promoter activities declined significantly upon substitution of single cysteine residues of the n-CRD, and even dropped below the levels for yap5Δ cells, when all four cysteine residues were simultaneously replaced. This parallel decline in both Fe/S cluster binding and Yap5 function as an iron-responsive transcriptional activator is a strong indication that Yap5 carries an iron-sensing Fe/S cluster at its n-CRD. Moreover, pYRE promoter activities also declined upon substitution of the cysteine residues of the c-CRD (Fig. 4C). Similar gene expression data were observed previously with the full-length CCC1 promoter (11). Since all Yap5 variants were expressed similarly to the wild-type protein, these findings show that also the c-CRD exerts gene regulatory activity. This regulatory activity may also be transmitted by an Fe/S cluster, even though this cluster is hardly detectable by our in vivo radiolabeling assay. The Fe/S cluster associated with c-CRD may therefore be rather labile.
Recombinant Yap5 binds Fe/S cluster at both CRDs.
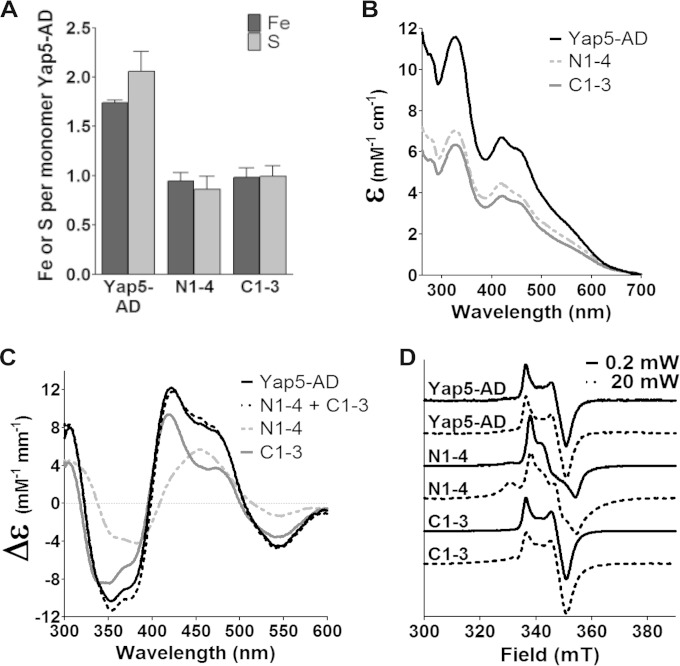
To investigate the number of Fe/S clusters to Yap5-AD in more detail, we purified recombinant versions of Yap5-AD/C1-3 and Yap5-AD/N1-4 and reconstituted the proteins chemically with iron and S2− ions. Both mutant proteins contained about 1 Fe and 1 S per monomer, i.e., about half of the wild-type Yap5-AD (Fig. 5A), suggesting that both mutants bind an Fe/S cluster. The proteins were then analyzed spectroscopically. Consistent with the iron und sulfide determination, reconstituted Yap5-AD/C1-3 displayed a UV/Vis spectrum characteristic for Fe/S proteins and yet with only half of the intensity of the spectrum of wild-type Yap5-AD (Fig. 5B). The CD spectrum of Yap5-AD/C1-3 displayed a maximum at 419 nm and a shoulder at 470 nm and its shape resembled that of the CD spectrum of wild-type Yap-AD (Fig. 5C). Furthermore, the EPR spectrum of dithionite-reduced Yap5-AD/C1-3 resembled closely that of wild-type Yap5-AD. The EPR spectra of both proteins remained unchanged upon variation of the microwave power, a further indication that the n-CRD part of Yap5-AD harbors a [2Fe-2S] cluster (Fig. 5D). Thus, the analyses of Yap5-AD/C1-3 in vivo and in vitro uniformly show that Yap5-AD binds a [2Fe-2S] cluster at the n-CRD.
FIG 5.
Yap5-AD binds [2Fe-2S] clusters at both CRDs. (A to C) Fe and acid-labile S2− contents (A) and UV/Vis (B) and CD (C) spectra of wild-type Yap5-AD, Yap5-AD/N1-4, and Yap5-AD/C1-3 after reconstitution with iron and S2− ions. The sum of the CD spectra of Yap5-AD/N1-4 and Yap5-AD/C1-3 is included in panel C. (D) EPR spectra of dithionite-reduced, reconstituted wild-type Yap5-AD, Yap5-AD/N1-4, and Yap5-AD/C1-3 at the indicated microwave powers (T = 10 K). Error bars indicate the SEM (n > 4).
Yap5-AD/N1-4 that lacks the Fe/S cluster ligands at the n-CRD also displayed an UV/Vis spectrum characteristic for Fe/S proteins after chemical reconstitution in vitro. The intensity of its UV/Vis spectrum was again only half of that of wild-type Yap5-AD (Fig. 5B). The CD spectrum of Yap5-AD/N1-4 displayed a broad peak with a maximum at 455 nm of low intensity (Fig. 5C). Although this spectrum differed from that of wild-type Yap5-AD, the sum of the CD spectra of Yap5-AD/C1-3 and Yap5-AD/N1-4 was similar in shape and intensity to that of wild-type Yap5-AD (Fig. 5C). Surprisingly, the EPR spectrum of dithionite-reduced Yap5-AD/N1-4 revealed two species which neither occurred in wild-type nor in Yap5-AD/C1-3 (Fig. 5D). The microwave power and temperature dependencies of the EPR spectrum of Yap5-AD/N1-4 suggest that the c-CRD accommodates a mixture of [2Fe-2S] and, possibly, small amounts of [4Fe-4S] centers. We therefore presume that the c-CRD accommodates a [2Fe-2S] cluster that, in the absence of the n-CRD cluster, has electronic properties different from those in wild-type Yap5-AD. Taken together, these observations suggest that Yap5-AD binds one [2Fe-2S] cluster at the n-CRD and one at the c-CRD.
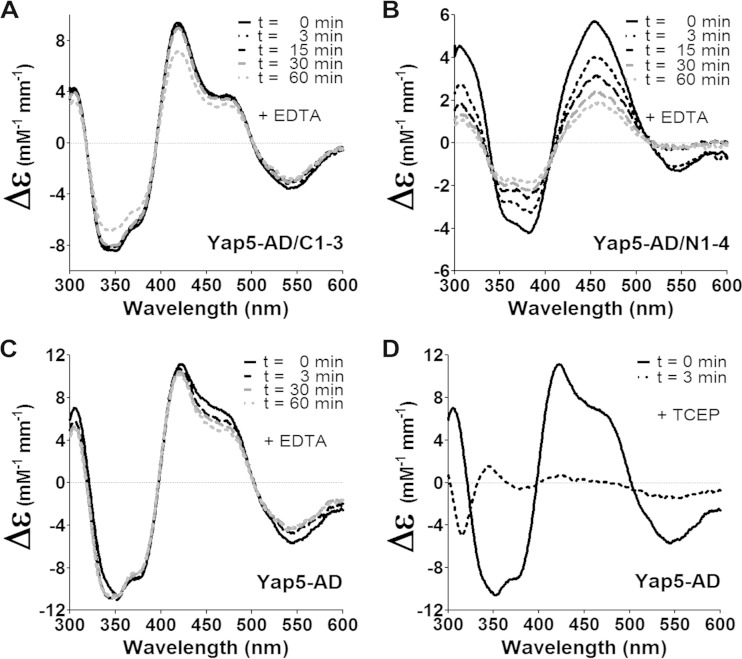
We next asked why the Fe/S cluster at the c-CRD is not detectable in vivo. To this end, we compared the stability of the two Fe/S clusters in the presence of metal chelators. Indeed, while the CD spectra of Yap5-AD/C1-3 remained unaffected for at least 30 min., the CD signal for the c-CRD cluster of Yap5-AD/N1-4 was already significantly diminished after 3 min. in the presence of EDTA (Fig. 6A and B). Consistently, the plateau between 450 and 475 nm of the CD spectrum of wild-type Yap5-AD that carries a large spectral contribution of the Fe/S cluster at the c-CRD was more affected by EDTA treatment than the maximum at 422 nm, which is mostly due to the Fe/S cluster at the n-CRD (Fig. 6C). The apparent instability of the Fe/S cluster at the c-CRD likely explains why we were unable to directly demonstrate its presence by in vivo 55Fe radiolabeling.
FIG 6.
The Fe/S cluster bound at the c-CRD is less stable than that bound at the n-CRD. CD spectra of reconstituted Yap5-AD/C1-3 (A), Yap5-AD/N1-4 (B), and wild-type Yap5-AD (C) were recorded after treatment with 20 mM citrate and 1 mM EDTA (pH 7.0) for the indicated times. (D) CD spectra of reconstituted Yap5-AD were recorded before and after 3 min incubation with 50 μM TCEP.
Yap5-AD undergoes a conformational change upon Fe/S cluster binding.
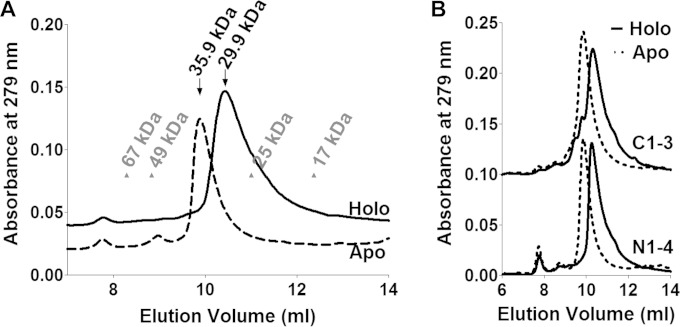
Reconstituted Yap5-AD incorporated about 2 Fe and 2 S per monomer, yet binds two Fe/S clusters (see Fig. 5A). To investigate whether wild-type Yap5-AD is a homodimer that shares the two [2Fe-2S] cofactors, we determined its oligomeric structure. Reconstituted wild-type Yap5-AD eluted at 30 kDa from a gel filtration column indicating a homodimer (Fig. 7A). Likewise, the reconstituted holoforms of Yap5-AD/N1-4 and Yap5-AD/C1-3 displayed apparent molecular masses of 30 kDa (Fig. 7B). In order to reveal whether the bound Fe/S clusters are responsible for the dimerization of Yap5-AD, we determined the molecular mass of the apoprotein. The Fe/S clusters on Yap5-AD were destroyed within minutes in the presence of the thiol-active reductant Tris(2-carboxyethyl)phosphine (TCEP) (Fig. 6D). The TCEP-treated apoprotein eluted at 36 kDa from a gel filtration column, which is again consistent with a dimeric structure (Fig. 7A). The increase in hydrodynamic radius indicates that apo-Yap5-AD adopts a more open conformation than holo-Yap5-AD. The same switch between an open apoform and a more compact holoform was observed for the two mutant proteins (Fig. 7B). Apparently, Fe/S cluster binding to only one of the CRDs is sufficient to induce a tighter physical packing of Yap5-AD. Likely, this Fe/S cluster-induced conformational change may trigger the conversion of Yap5 to a transcriptional activator.
FIG 7.
The Yap5-AD dimer undergoes a conformational change upon Fe/S cluster binding. (A and B) Gel filtration elution profiles of wild-type Yap5-AD (A), Yap5-AD/C1-3 (B, top; offset by an optical density of 0.1), and Yap5-AD/N1-4 (B, bottom) after chemical reconstitution (Holo) and after treatment with 50 μM TCEP (Apo). Elution positions of molecular mass markers are indicated by arrowheads in panel A. Arrows indicate the deduced molecular masses of Yap5-AD.
DISCUSSION
The molecular understanding of factors involved in iron-responsive transcriptional regulation in eukaryotes is still in its infancy (4, 7, 8). Both their underlying mechanisms and their sensed molecules are usually not well known. In some cases, genetic or in vitro analyses point to the putative identity of the sensed molecules, but reliable information is mostly missing (19, 22, 36, 37). Our study demonstrates that the activator domain of the bZIP stress response regulator Yap5 of S. cerevisiae binds two Fe/S clusters that sense toxic intracellular iron levels. Fe/S cluster association induces a conformational change in the dimeric activator domain that likely is of gene regulatory importance. The resulting transcriptional induction of, among other genes, CCC1, encoding a vacuolar divalent metal transporter eventually results in the sequestering of excess iron into the vacuole for detoxification (11, 12).
In bacteria, transcription factors with Fe/S clusters are widespread und used for the sensing of diverse environmental stimuli (38). Their sensitivity to oxygen or reactive oxygen species makes Fe/S clusters ideally suited to function as sensors. Despite this perfect aptitude, sensing Fe/S clusters are rare in eukaryotes. An exception is the comprehensively studied vertebrate iron-regulatory protein 1 (IRP1) that uses Fe/S cluster association-dissociation to regulate iron homeostasis by posttranscriptional binding to mRNAs of genes involved in cellular iron uptake and storage (6, 7). For eukaryotic transcription factors, virtually all evidence for physical binding of iron or Fe/S cofactors is based on in vitro analyses of recombinant proteins (22, 36, 37). The iron response regulators SRE from Neurospora crassa and Sre1 from Histoplasma capsulatum bind substoichiometric amounts of iron when purified from E. coli (36, 37). S. cerevisiae Aft2 coordinates [2Fe-2S] clusters upon chemical reconstitution in vitro (22). While being suggestive, the physiological relevance of these in vitro observations is usually difficult to test due to the low abundance of these transcription factors in vivo.
Our current work on the bZIP stress response regulator Yap5 of S. cerevisiae combined in vivo and in vitro approaches to study its high-iron sensing function. We document that both CRDs of the transcriptional activator domain bind a [2Fe-2S] cluster. The cluster bound to the n-CRD was visible by both in vivo and in vitro methods. Substitution of the cysteine residues of the n-CRD resulted in loss of iron binding to Yap5-AD in vivo and a parallel decrease in iron-responsive transcriptional activation by Yap5. This correlation demonstrates that the n-CRD accommodates an Fe/S cluster sensor that is essential for Yap5's function as a transcriptional activator. The maturation of this Fe/S cluster is unusual, because it depends on the mitochondrial ISC systems but not on the cytosolic CIA machinery (see Fig. S1 in the supplemental material). A similar CIA-independent maturation was observed for the cytosolic Grx4 which harbors a [2Fe-2S] cluster (21, 31). Furthermore, the maturation of the Fe/S cluster at the n-CRD was also independent of the cytosolic monothiol glutaredoxins Grx3 and Grx4 and cells depleted for these glutaredoxins displayed elevated levels of CCC1 expression. Apparently, unlike S. cerevisiae Aft1 and Aft2, the Fe/S clusters on Grx3 and Grx4 play no role for the regulation of Yap5 (8, 21, 22) (see Fig. S1 in the supplemental material).
The EPR spectra of purified wild-type Yap5-AD and Yap5-AD/C1-3, in which the cysteine residues of the c-CRD were substituted, were characteristic for [2Fe-2S] centers. Mössbauer spectroscopy indicated iron binding mainly in form of [2Fe-2S] clusters. These in vitro results uniformly show that Yap5 binds a [2Fe-2S] cluster sensor at its n-CRD. Yap5's n-CRD domain includes a CGFC motif that functions as a Fe/S-cluster coordination site in glutaredoxins (39). bZIP transcription factors bearing a CGFCX5CXC motif as found in Yap5's n-CRD are wide-spread in fungi (see Fig. S5 in the supplemental material). These proteins include members with homology to HapX, the iron-responsive transcriptional corepressor in ascomycetes (4) and the oxidative stress response regulator Yap1 from Ustilago maydis (40). These findings suggest that the n-CRD represents a novel structural domain that likely functions as a conserved binding site for Fe/S cluster sensors in fungal stress-responsive regulators. Consistent with this conclusion, a recent study demonstrates that the CGFCX5CXC motif in Aspergillus fumigatus HapX is essential for activation of the A. fumigatus CCC1 homolog (41).
Yap5-AD binds a second Fe/S cluster at its c-CRD which, due to its inherent instability, was visible by in vitro methods only. Purified recombinant Yap5-AD/N1-4, in which the four cysteine residues of the n-CRD were substituted by alanines and hence did not bind the N-terminal cluster, displayed spectral characteristics of [2Fe-2S] clusters. In addition, the CD spectrum of the sum of the individual CD spectra of Yap5-AD/N1-4 and Yap5-AD/C1-3 closely resembles that of wild-type Yap5-AD, and these mutant proteins each incorporated only half the amount of iron and sulfide compared to wild-type Yap5-AD. The shapes of the CD and EPR spectra of Yap5-AD/N1-4 were different from those of Yap5-AD/C1-3, suggesting that both Fe/S clusters are characteristically different. Furthermore, the C-terminal cluster may have a slightly altered electronic environment in the absence of the N-terminal cluster, as indicated by subtle differences in the EPR spectra and the absence of the characteristic peak values of the cluster at the c-CRD in the EPR spectrum of wild-type Yap5-AD. The fact that the EPR spectrum of the wild-type Yap5-AD is not exactly a composite of the individual spectra of the two mutant proteins suggests that both clusters are close to each other in wild-type Yap5-AD (9 to 15 Å) so that magnetic coupling can occur, which results in the partial cancellation of the EPR spectrum of the c-CRD cluster (42). We were unable to reliably detect the C-terminal Fe/S cluster directly by in vivo 55Fe radiolabeling under both aerobic and anaerobic conditions. However, Yap5-AD variants with substitution of the cysteine residues of the n-CRD displayed a weak remnant iron binding in vivo, a finding that may suggest that the c-CRD may also bind Fe/S clusters in its native host, albeit weakly. Fe/S cluster coordination at this region might be relevant in vivo because cysteine exchanges at the c-CRD impaired the iron-responsive transcriptional activation by Yap5, albeit moderately in comparison to mutations of the n-CRD (11; the present study). Hence, the involvement of c-CRD in transcriptional regulation may be triggered by [2Fe-2S] cluster association, even though an indirect, Fe/S cluster-independent influence of this sequence motif on Fe/S cluster binding to n-CRD cannot entirely be excluded. The labile character of the Fe/S cluster at the c-CRD in vitro nicely fits to a dynamic assembly-disassembly behavior which is ideal for a regulatory function. We therefore suggest that also the c-CRD of Yap5 fulfils a regulatory function and may use dynamic Fe/S cluster binding for iron sensing. Notably, the sequence motif of Yap5's c-CRD is rarely found in other fungal transcription factors, indicating that its function is more specialized in nature (see Fig. S5 in the supplemental material) and may be used for fine-tuning of the sensing process.
How do the Fe/S cluster sensors activate Yap5's function as a transcriptional activator? Yap5 binds to Yap response elements in an iron-independent manner (11). Although this constitutive DNA binding is not typical for Yap family members (15, 43), this observation renders the idea of an Fe/S cluster-induced DNA binding unlikely. bZIP and AP-1 transcription factors are bound to DNA as dimers (16). Since the activator domain of Yap5 is homodimeric in vitro, Yap5-AD may serve as a physiologically relevant model to study the mode of operation of this transcription factor. Our gel filtration data show that the apparent size of Yap5-AD is smaller in the Fe/S cluster-bound holoform than in its apoform, indicating that the binding of the Fe/S cluster sensor induces a conformational change that results in a tighter packing of the activator domain. This Fe/S cofactor-induced conformational change may convert Yap5 to a transcriptional activator, for instance, by exposing binding sites for RNA polymerases or transcription initiation factors. Since the conformational switch of Yap5 involves the compaction of its two interacting monomers, the Yap5-AD dimer may operate like a shell that opens and closes in response to changes in intracellular iron levels. This shell-like mode of operation can be best achieved by Fe/S clusters that bridge two halves of a homodimer and pull them closer together. The stoichiometry of two [2Fe-2S] clusters per dimer supports the idea that Yap5-AD binds bridging Fe/S clusters. The fact that Fe/S cluster binding to one of the CRDs is sufficient to induce a tighter packing of Yap5-AD fully fits to this mechanical model for transcriptional activation of Yap5.
Supplementary Material
ACKNOWLEDGMENTS
This study was generously supported by the Deutsche Forschungsgemeinschaft (SFB 593 and 987 and GRK 1216), von Behring-Röntgen Stiftung, the LOEWE program of the state Hessen, and Max-Planck Gesellschaft.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01033-14.
REFERENCES
- 1.Lyons TJ, Eide DJ. 2007. Transport and storage of metal ions in biology, p 57–77. In Bertini I, Gray HB, Stiefel EI, Valentine JS (ed), Biological inorganic chemistry. University Science Books, Sausalito, CA. [Google Scholar]
- 2.Gitlin JD, Lill R. 2012. Special issue: cell biology of metals. Biochim Biophys Acta 1823:1405. doi: 10.1016/j.bbamcr.2012.07.008. [DOI] [Google Scholar]
- 3.Weinberg ED. 2009. Iron availability and infection. Biochim Biophys Acta 1790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Schrettl M, Haas H. 2011. Iron homeostasis: Achilles' heel of Aspergillus fumigatus? Curr Opin Microbiol 14:400–405. doi: 10.1016/j.mib.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson GJ, Vulpe CD. 2009. Mammalian iron transport. Cell Mol Life Sci 66:3241–3261. doi: 10.1007/s00018-009-0051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. 2010. Two to tango: regulation of mammalian iron metabolism. Cell 142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 7.Dlouhy AC, Outten CE. 2013. The iron metallome in eukaryotic organisms. Metal Ions Life Sci 12:241–278. doi: 10.1007/978-94-007-5561-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Labbe S, Khan MG, Jacques JF. 2013. Iron uptake and regulation in Schizosaccharomyces pombe. Curr Opin Microbiol 16:669–676. doi: 10.1016/j.mib.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Outten CE, Albetel AN. 2013. Iron sensing and regulation in Saccharomyces cerevisiae: ironing out the mechanistic details. Curr Opin Microbiol 16:662–668. doi: 10.1016/j.mib.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Philpott CC, Leidgens S, Frey AG. 2012. Metabolic remodeling in iron-deficient fungi. Biochim Biophys Acta 1823:1509–1520. doi: 10.1016/j.bbamcr.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Bagley D, Ward DM, Kaplan J. 2008. Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol Cell Biol 28:1326–1337. doi: 10.1128/MCB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Jia X, Ward DM, Kaplan J. 2011. Yap5 protein-regulated transcription of the TYW1 gene protects yeast from high iron toxicity. J Biol Chem 286:38488–38497. doi: 10.1074/jbc.M111.286666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodrigues-Pousada CA, Nevitt T, Menezes R, Azevedo D, Pereira J, Amaral C. 2004. Yeast activator proteins and stress response: an overview. FEBS Lett 567:80–85. doi: 10.1016/j.febslet.2004.03.119. [DOI] [PubMed] [Google Scholar]
- 14.Herrero E, Ros J, Belli G, Cabiscol E. 2008. Redox control and oxidative stress in yeast cells. Biochim Biophys Acta 1780:1217–1235. doi: 10.1016/j.bbagen.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Rodrigues-Pousada C, Menezes RA, Pimentel C. 2010. The Yap family and its role in stress response. Yeast 27:245–258. doi: 10.1002/yea.1752. [DOI] [PubMed] [Google Scholar]
- 16.Hurst HC. 1995. Transcription factors 1: bZIP proteins. Protein Profile 2:101–168. [PubMed] [Google Scholar]
- 17.Reinke AW, Baek J, Ashenberg O, Keating AE. 2013. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science 340:730–734. doi: 10.1126/science.1233465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandes L, Rodrigues-Pousada C, Struhl K. 1997. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol 17:6982–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Miao R, Bertram S, Jia X, Ward DM, Kaplan J. 2012. A role for iron-sulfur clusters in the regulation of transcription factor Yap5-dependent high iron transcriptional responses in yeast. J Biol Chem 287:35709–35721. doi: 10.1074/jbc.M112.395533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, Uzarska MA, Webert H, Wilbrecht C, Muhlenhoff U. 2012. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta 1823:1491–1508. doi: 10.1016/j.bbamcr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Muhlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E, Lillig CH, Lill R. 2010. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab 12:373–385. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poor CB, Wegner SV, Li H, Dlouhy AC, Schuermann JP, Sanishvili R, Hinshaw JR, Riggs-Gelasco PJ, Outten CE, He C. 2014. Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2. Proc Natl Acad Sci U S A 111:4043–4048. doi: 10.1073/pnas.1318869111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherman F. 2002. Getting started with yeast. Methods Enzymol 350:3–41. doi: 10.1016/S0076-6879(02)50954-X. [DOI] [PubMed] [Google Scholar]
- 24.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol 185:60–89. doi: 10.1016/0076-6879(90)85008-C. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Russel DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 26.Gietz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96. doi: 10.1016/S0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 27.Harlow E, Lane D. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 28.Molik S, Lill R, Muhlenhoff U. 2007. Methods for studying iron metabolism in yeast mitochondria. Methods Cell Biol 80:261–280. doi: 10.1016/S0091-679X(06)80013-0. [DOI] [PubMed] [Google Scholar]
- 29.Balk J, Pierik AJ, Netz DJ, Muhlenhoff U, Lill R. 2004. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J 23:2105–2115. doi: 10.1038/sj.emboj.7600216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Netz DJ, Mascarenhas J, Stehling O, Pierik AJ, Lill R. 2014. Maturation of cytosolic and nuclear iron-sulfur proteins. Trends Cell Biol 24:303–312. doi: 10.1016/j.tcb.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Mapolelo DT, Randeniya S, Johnson MK, Outten CE. 2012. Human glutaredoxin 3 forms [2Fe-2S]-bridged complexes with human BolA2. Biochemistry 51:1687–1696. doi: 10.1021/bi2019089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pimentel C, Vicente C, Menezes RA, Caetano S, Carreto L, Rodrigues-Pousada C. 2012. The role of the Yap5 transcription factor in remodeling gene expression in response to Fe bioavailability. PLoS One 7:e37434. doi: 10.1371/journal.pone.0037434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunham WR, Bearden AJ, Salmeen IT, Palmer G, Sands RH, Orme-Johnson WH, Beinert H. 1971. The two-iron ferredoxins in spinach, parsley, pig adrenal cortex, Azotobacter vinelandii, and Clostridium pasteurianum: studies by magnetic field Mössbauer spectroscopy. Biochim Biophys Acta 253:134–152. doi: 10.1016/0005-2728(71)90240-4. [DOI] [PubMed] [Google Scholar]
- 34.Beinert H, Holm RH, Munck E. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 35.Schünemann V, Winkler H. 2000. Structure and dynamics of biomolecules studied by Mössbauer spectroscopy. Rep Prog Phys 63:263–353. doi: 10.1088/0034-4885/63/3/202. [DOI] [Google Scholar]
- 36.Harrison KA, Marzluf GA. 2002. Characterization of DNA binding and the cysteine rich region of SRE, a GATA factor in Neurospora crassa involved in siderophore synthesis. Biochemistry 41:15288–15295. doi: 10.1021/bi0204995. [DOI] [PubMed] [Google Scholar]
- 37.Chao LY, Marletta MA, Rine J. 2008. Sre1, an iron-modulated GATA DNA-binding protein of iron-uptake genes in the fungal pathogen Histoplasma capsulatum. Biochemistry 47:7274–7283. doi: 10.1021/bi800066s. [DOI] [PubMed] [Google Scholar]
- 38.Crack JC, Green J, Hutchings MI, Thomson AJ, Le Brun NE. 2012. Bacterial iron-sulfur regulatory proteins as biological sensor-switches. Antioxid Redox Signal 17:1215–1231. doi: 10.1089/ars.2012.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouhier N. 2010. Plant glutaredoxins: pivotal players in redox biology and iron-sulphur centre assembly. New Phytol 186:365–372. doi: 10.1111/j.1469-8137.2009.03146.x. [DOI] [PubMed] [Google Scholar]
- 40.Molina L, Kahmann R. 2007. An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 19:2293–2309. doi: 10.1105/tpc.107.052332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gsaller F, Hortschansky P, Beattie SR, Klammer V, Tuppatsch K, Lechner BE, Rietzschel N, Werner ER, Vogan AA, Chung D, Muhlenhoff U, Kato M, Cramer RA, Brakhage AA, Haas H. 2014. The Janus transcription factor HapX controls fungal adaptation to both iron starvation and iron excess. EMBO J pii:e201489468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathews R, Charlton S, Sands RH, Palmer G. 1974. On the nature of the spin coupling between the iron-sulfur clusters in the eight-iron ferredoxins. J Biol Chem 249:4326–4328. [PubMed] [Google Scholar]
- 43.Delaunay A, Isnard AD, Toledano MB. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J 19:5157–5166. doi: 10.1093/emboj/19.19.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.