Abstract
The epigenetic writer Sir2 maintains the heterochromatin state of chromosome in three chromosomal regions, namely, the silent mating type loci, telomeres, and the ribosomal DNA (rDNA). In this study, we demonstrated the mechanism by which Sir2 is regulated under heat stress. Our study reveals that a transient heat shock causes a drastic reduction in the SIR2 transcript which results in sustained failure to initiate silencing for as long as 90 generations. Hsp82 overexpression, which is the usual outcome of heat shock treatment, leads to a similar downregulation of SIR2 transcription. Using a series of genetic experiments, we have established that heat shock or Hsp82 overexpression causes upregulation of CUP9 that, in turn, represses SIR2 transcription by binding to its upstream activator sequence. We have mapped the cis regulatory element of SIR2. Our study shows that the deletion of cup9 causes reversal of the Hsp82 overexpression phenotype and upregulation of SIR2 expression in heat-induced Hsp82-overexpressing cells. On the other hand, we found that Cup9 overexpression represses SIR2 transcription and leads to a failure in the establishment of heterochromatin. The results of our study highlight the mechanism by which environmental factors amend the epigenetic configuration of chromatin.
INTRODUCTION
Increasing amounts of evidence suggest that environmental factors lead to stable alteration in gene expression by modifying chromatin structure. The genome of the lower eukaryote Saccharomyces cerevisiae uses histone acetylation-deacetylation as one of the epigenetic mechanisms to control gene expression. Histone deacetylases (HDAC) are the transcriptional repressors which cause deacetylation of histones, thereby creating localized regions of repressed chromatin. They are categorized into three groups based on their homology to yeast proteins: RPD3 (class I), HDA1 (class II), and Sir2 (class III) (1, 2). Sir2 deacetylates histone H3 (at K9, K14, and K56) and H4 (particularly K16) to regulate telomeric heterochromatin structure in yeast (3, 4). Our previous studies have demonstrated that heat stress and concomitant overexpression of Hsp90 result in euchromatinization of silent subtelomeric chromatin by reducing the steady-state level of Sir2 (5).
Sir2 protein mediates silencing at the silent mating-type loci HML and HMR, telomeres, and ribosomal DNA locus through a series of protein-protein interactions. During telomere silencing, Rap1 and a Ku70/80 heterodimer, which are telomere binding proteins, recruit the Sir2/Sir4 complex (6). Sir2p deacetylates neighboring nucleosomes and facilitates the binding of Sir4 and Sir3 to hypoacetylated H3 and H4 (7, 8). Sir3 and Sir4 recruit additional Sir2, and thus, the renewal of this cycle causes the spread of the Sir complex along the chromosome (9). The propagation of silencing complex along the chromosome requires the NAD+-dependent histone deacetylase activity of Sir2. Mutation at the NAD+ binding pocket of Sir2 makes it severely defective in telomere silencing (10). Nicotinamide (NAM), which is generated as the by-product of the enzymatic reaction, acts as a noncompetitive inhibitor of Sir2 (11). It has been demonstrated that PNC1, which codes for nicotinamidase, acts as a positive regulator of Sir2 activity by causing deamidation of NAM and thus increasing the replicative life span of yeast (12). Surprisingly, how SIR2 gene is regulated at the transcription level under normal conditions or in response to different environmental cues is not understood at all.
Cup9 was originally identified as the gene that permits the cell to tolerate very high doses of copper, which are otherwise toxic. However, the mechanism of such an effect was not known (13). Published microarray experiments revealed that its transcription increases severalfold when cells are exposed to hypoxia and osmotic stress and when grown in the presence of alternate carbon sources (14, 15). Cup9 is a homeodomain transcriptional repressor having a high degree of identity with human PBX proteins (pre-B cell leukemia transcription factor) that are crucial for embryonic development (16). It also shows identity with the S. cerevisiae MATα2 locus (17). About 36 targets of Cup9 have been documented so far, among which the best characterized is the master peptide transporter (dipeptide and tripeptide) PTR2. It is known that Cup9, along with the corepressors Tup1 and Ssn6 (18), reduces peptide import in cells by repressing PTR2 transcription (19, 20).
Hsp90 is an evolutionarily conserved molecular chaperone found in organisms ranging from Escherichia coli (HtpG) to yeast (Hsc82 and Hsp82) to humans (Hsp90α and Hsp90β). It regulates diverse cellular functions by providing maturation to a specific group of proteins known as clients. A high-throughput evaluation by LUMIER assay (21) revealed that 60% of the Hsp90 clientele belongs to the kinase family, 30% belongs to the ubiquitin ligases, and about 7% are the transcription factors (22). Steroid hormone receptors are the most extensively characterized transcription factors that are chaperoned by Hsp90 (23). Unlike the other chaperones, such as Hsp70 and Hsp40, which act early in the folding process, the Hsp90 family interacts with the substrates at the later stages of protein folding (24, 25). It forms a multichaperone complex with Hop, Hip, Aha1, p23, CyP40, or FKBP and binds to the target proteins (26). Heat shock (HS) and other proteotoxic stresses trigger overexpression of Hsp90 due to the activation of the transcription factor Hsf1,which homotrimerizes and translocates to the nucleus from the cytoplasm and causes the transcriptional activation of Hsp90 (27, 28).
Previous experiments in our laboratory showed that Hsp82 (the yeast ortholog of Hsp90) homeostasis controls the abundance as well as activity of Sir2. The Hsp82 null condition not only leads to the reduced abundance of Sir2 but also results in the inactivation of Sir2 proteins. On the other hand, Hsp82 overexpression leads to the reduction of the steady-state level of Sir2 protein, though it does not affect the mating type silencing activity of Sir2 (5). In this report, we provide mechanistic insights into Hsp82 overexpression phenotype which occurs while cells are exposed to heat shock. Our work demonstrates that heat stress (or overexpression of Hsp82) leads to transcriptional downregulation of SIR2, which is inherited through successive generations before it returns to the normal level. Our work identified the transcriptional repressor Cup9, which negatively regulates SIR2 gene expression. We demonstrate that under heat shock as well as Hsp82 overexpression, Cup9 is upregulated and is recruited at the SIR2UAS region. Thus, our work explains the mechanism behind the alteration of the epigenetic state of chromatin in response to environmental cues such as heat stress.
MATERIALS AND METHODS
Plasmids.
The sequences of all the primers used in this study are given in Table 1. The reporter plasmid pCZ is a high-copy-number yeast expression vector having the LACZ reporter gene under the control of the CYC1 promoter. Using SLY20 genomic DNA as a template, we amplified 429 bp, 370 bp, 307 bp, and 200 bp of SIR2 upstream activator sequence (SIR2UAS) and cloned them individually in a LACZ reporter plasmid, replacing its original CYC1 promoter. The cloned vectors are referred to as 429UAS, 370UAS, 307UAS, and 200UAS, respectively, in this paper. We also made a vector by removing a built-in CYC1 promoter to obtain a promoterless control; it is referred to as pCZdelcyc1.
TABLE 1.
Primers used in this study
| Primer name | Sequence | Purpose |
|---|---|---|
| OSB 125 | 5′ ATC CTC GAG CTG CAA CTC CTC AAT GTG TC 3′ | Forward primer used to amplify 429-bp SIR2UAS |
| OSB 193 | 5′ ATC CTC GAG GTA TAT GCT TAT ATG CAT GCG 3′ | Forward primer used to amplify 370-bp SIR2UAS |
| OSB 194 | 5′ATC CTC GAG CCA AGC TAC ATC TAG CAC TC 3′ | Forward primer used to amplify 307-bp SIR2UAS |
| OSB 126 | 5′ ATC CTC GAG CTT TGG CCG CCA GTT GCG 3′ | Forward primer used to amplify 200-bp SIR2UAS |
| OSB 87 | 5′ ATC GGA TCC GGT CAT CCA GCT TTA ATG TGC CG 3′ | Common reverse primer used to amplify all above deletion constructs of SIR2UAS |
| OSB 203 | 5′ GAC GGA TCC ATG AAT TAT AAC TGC GAA ATA C 3′ | Forward primer used to amplify CUP9 for cloning in pESC-MYC-tagged vector |
| OSB 204 | 5′ CGA GTC GAC ATT CAT ATC AGG GTT GGA TAG 3′ | Reverse primer used to amplify CUP9 for cloning in pESC-MYC-tagged vector |
| OSB 164 | 5′ CTT TTA TGC TAA CAA CCT TCG AGA ATA GTT ACA TTC GAA GCG GAT CCC CGG GTT AAT TAA 3′ | Forward primer used for CUP9 knockout |
| OSB 165 | 5′ TAT AAT TAT ATG AAT ATT TAA GTA ATG CAT TGA TAA GTG AGA ATT CGA GCT CGT TTA AAC 3′ | Reverse primer used for CUP9 knockout |
| OSB 170 | 5′ AAG TTT CAT ACA TAA TTA ACA AAA TTC GTT TGT TGC GGG GCG GAT CCC CGG GTT AAT TAA 3′ | Forward primer used for SUM1 knockout |
| OSB 171 | 5′ TTT TAT CTA TTC TCG AAA CTG CCC CAA CGT ACG GAC CAG CGA ATT CGA GCT CGT TTA AAC 3′ | Reverse primer used for SUM1 knockout |
| OSB 173 | 5′ ACT GAA AAC GGT AAA GTA GGT TTG TTT AAA TTG ACT TAA GCG GAT CCC CGG GTT AAT TAA 3′ | Forward primer used for RIM101 knockout |
| OSB 174 | 5′ GCA AAG AAA CAA CTA AGA ATA AAA TAT CCG ACA ATC CAT AGA ATT CGA GCT CGT TTA AAC 3′ | Reverse primer used for RIM101 knockout |
| OSB 161 | 5′ CAA AAT CAT CCT TAT ATA ACC CTG GTA AGG TCC TTT TGT CCG GAT CCC CGG GTT AAT TAA 3′ | Forward primer used for SOK2 knockout |
| OSB 162 | 5′ GAT TAA AGT AAC ATA ATT ATC CAA GGA ATT CAT AGT TGT TGA ATT CGA GCT CGT TTA AAC 3′ | Reverse primer used for SOK2 knockout |
| OSB 196 | 5′ GCT GGA AGA ATT GAA AAA GCT ATC CAA CCC TGA TAT GAA TCG GAT CCC CGG GTT AAT TAA 3′ | Forward primer used to generate MYC tag at the portion of CUP9 corresponding to the C terminus at the chromosomal locus |
| OSB 197 | 5′ TAT AAT TAT ATG AAT ATT TAA GTA ATG CAT TGA TAA GTG AGA ATT CGA GCT CGT TTA AAC 3′ | Reverse primer used to generate MYC tag at the portion of CUP9 corresponding to the C terminus at the chromosomal locus |
| OSB 16 | 5′ TGA CCA AAC TAC TTA CAA CTC C 3′ | Forward primer used to amplify ACT1 for real-time RT-PCR |
| OSB 14 | 5′ TTA GAA ACA CTT GTG GTG AAC G 3′ | Reverse primer used to amplify ACT1 for real-time RT-PCR |
| OSB 131 | 5′ CTG ATT AAT CGT GAT CCC GTC 3′ | Forward primer used to amplify SIR2 for real-time RT-PCR |
| OSB 132 | 5′ CTT AGA GGG TTT TGG GAT GTT C 3′ | Reverse primer used to amplify SIR2 for real-time RT-PCR |
| OSB 121 | 5′ CAA CTG ATG GAA ACC AGC C 3′ | Forward primer used to amplify LACZ for real-time RT-PCR |
| OSB 122 | 5′ TTA CGC GAA ATA CGG GCA G 3′ | Reverse primer used to amplify LACZ for real-time RT-PCR |
| OSB 189 | 5′ CTA ATG ACA ACG CGA ATA ATA C 3′ | Forward primer used to amplify CUP9 for real-time RT-PCR |
| OSB 190 | 5′ CAA TTC ATA TCA GGG TTG GAT AG 3′ | Reverse primer used to amplify CUP9 for real-time RT-PCR |
| OSB 19 | 5′ ATC ACG AGT AAG GAT CAA AG 3′ | Forward primer used to amplify YFR057w for real-time RT-PCR |
| OSB 20 | 5′ TTA TGG CTT TGT TAC GCT TG 3′ | Reverse primer used to amplify YFR057w for real-time RT-PCR |
| OSB 62 | 5′ AAT CGG CGG ATG GGT TGG 3′ | Forward primer used to amplify HMLα for real-time RT-PCR |
| OSB 63 | 5′ TCA TTC TTT CTT CTT TGC CAG 3′ | Reverse primer used to amplify HMLα for real-time RT-PCR |
Using SLY20 genomic DNA as a template, we amplified CUP9 and cloned it in a 2μ C-terminally Myc-tagged vector, pESC-HIS (Agilent Technologies). This generated pESC-CUP9-MYC, which overexpresses CUP9 under the control of the GAL promoter.
Yeast strains.
Strains used in this study are listed in Table 2. 429UAS, 370UAS, 307UAS, and 200UAS deletion constructs along with the reporter plasmid without the CYC1 promoter were transformed into strain SLY20 to generate isogenic strains SLY57, SLY84, SLY83, SLY56, and SLY64, respectively. The HSP82 overexpression plasmid pRS313/HSP82 (5) was transformed into strains SLY20, SLY57, SLY84, SLY83, and SLY56, and colonies were selected on SC-his medium to generate isogenic strains SLY13, SLY61, SLY86, SLY85, and SLY60, respectively. The empty vector pHCA was also transformed into SLY20, SLY57, SLY84, SLY83, and SLY56, and colonies were selected on SC-his medium to generate isogenic strains SLY13C, SLY61C, SLY86C, SLY85C, and SLY60C, respectively.
TABLE 2.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| SLY20 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 | 5 |
| SLY12 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 sir2::KANr | 5 |
| SLY13C | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pHCA | This study |
| SLY13 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pHCA/HSP82 | 5 |
| SLY56 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pCZ/200UAS | This study |
| SLY57 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pCZ/429UAS | This study |
| SLY60C | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pCZ/200UAS, pHCA | This study |
| SLY60 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pCZ/200UAS, pHCA/HSP82 | This study |
| SLY61C | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pCZ/429UAS, pHCA | This study |
| SLY61 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pCZ/429UAS, pHCA/HSP82 | This study |
| SLY64 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pCZdelcyc1 | This study |
| SLY71 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 cup9::TRP1 | This study |
| SLY74 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 sok2::TRP1 | This study |
| SLY73 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 rim101::TRP1 | This study |
| SLY75 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 sum1::TRP1 | This study |
| SLY77C | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 cup9::TRP1 pHCA | This study |
| SLY77 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 cup9::TRP1 pHCA/HSP82 | This study |
| SLY80C | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 sok2::TRP1 pHCA | This study |
| SLY80 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 sok2::TRP1 pHCA/HSP82 | This study |
| SLY79C | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 rim101::TRP1 pRS313 | This study |
| SLY79 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 rim101::TRP1 pRS313/HSP82 | This study |
| SLY81C | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 sum1::TRP1 pRS313 | This study |
| SLY81 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 sum1::TRP1 pRS313/HSP82 | This study |
| SLY83 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pCZ/307UAS | This study |
| SLY84 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pCZ/370UAS | This study |
| SLY85C | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pCZ/307UAS, pHCA | This study |
| SLY85 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pCZ/307UAS, pHCA/HSP82 | This study |
| SLY86C | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pCZ/370UAS, pHCA | This study |
| SLY86 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pCZ/370UAS, pHCA/HSP82 | This study |
| SLY87 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 CUP9-13MYC-KANMX6 | This study |
| SLY88 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 CUP9-13MYC-KANMX6 pHCA/HSP82 | This study |
| SLY90 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pESC/CUP9MYC | This study |
| SLY91 | MATa leu2-3,112 trp1 ura3-1 ade2-1 his3-11,15 VIIL::ADE2 pESC(MYC) | This study |
Using plasmid pFA6a-TRP1 as a template (29), we amplified the TRP1 cassette with CUP9 flanking regions. The product was then integrated into SLY20 and selected on a medium lacking tryptophan to generate a Δcup9 strain, which is referred to as SLY71 in this paper. HSP82 overexpression plasmid pRS313/HSP82 and the empty vector were transformed into SLY71 to generate SLY77 and SLY77C, respectively. Using the same strategy, we created Δsum1, Δrim101, and Δsok2 strains, which are referred to as SLY75, SLY73, and SLY74, respectively, in this paper. We transformed HSP82 overexpression plasmid and the empty vector into SLY75, SLY73, and SLY74 to generate SLY81 and SLY81C, SLY79 and SLY79C, and SLY80 and SLY80C, respectively.
The CUP9 overexpression plasmid pESC-CUP9-MYC and the empty vector pESC-MYC were transformed into SLY20 to create SLY90 and SLY91, respectively.
In order to Myc tag the C-terminal end of the Cup9 protein expressed from the chromosomal locus, a 13MYC-KanMX6 cassette (29) was amplified with regions flanking this portion of CUP9. It was then integrated into SLY20 to generate CUP9 MYC-tagged strain SLY87. HSP82 overexpression plasmid pHCA/HSP82 was transformed into SLY87 to generate SLY88.
TPE color assay.
The SLY20 strain used for the TPE color assay is isogenic to W303a, having ADE2 marked at telomere VIIL. SLY13C, SLY12, SLY13, SLY77, SLY77C, SLY81, SLY79, SLY80, SLY90, and SLY91 cells were grown on appropriate medium, and the telomere position effect (TPE) assay was performed according to the protocol described previously (5).
Antibodies.
The anti-Hsp90 antibody (Calbiochem) and antiactin antibody (Abcam) were used at a 1:5,000 dilution. The anti-Sir2 antibody (Santa Cruz Biotechnology Inc., CA) and anti-Myc antibody (Abcam) were used at 1:200 and 1:8,000 dilutions, respectively. Horseradish peroxidase (HRP)-conjugated rabbit IgG (Santa Cruz Biotechnology Inc.) was used as a secondary antibody for Sir2 and Myc at a 1:10,000 dilution, and HRP-conjugated mouse IgG (Promega) was used as a secondary antibody for Hsp82 and actin at a 1:10,000 dilution. A chemiluminescence detection system (Pierce) was used to develop Western blots.
RNA isolation and real-time RT-PCR.
Total RNA was isolated by the acid-phenol method as described in our earlier paper (5). For real-time PCR, cDNA was diluted (1:50) and used for PCR using a reverse transcription-PCR (RT-PCR) kit (Roche). The real-time analysis was done using the Applied Biosystems 7500 fast real-time PCR system. Primers used for the amplification of 200- to 300-bp stretches at the portions of ACT1, SIR2, LACZ, CUP9, YFR067w, and HMLα corresponding to the C terminus are listed in Table 1. The threshold cycle (CT) value of the ACT1 transcript of each sample was used to normalize the corresponding CT values of SIR2, LACZ, CUP9, YFR057w, and HMLα transcripts. The normalized CT values of SIR2, LACZ, CUP9, YFR057w, and HMLα from different samples were compared to obtain ΔCT values. The relative levels of mRNA were estimated as 2−ΔΔCT. The mean values (±standard deviations [SD]) from three independent experiments were plotted using GraphPad Prism 6 software.
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed as described previously (30), with some modifications. A 50-ml quantity of cells was grown to an optical density at 600 nm (OD600) of 1.2 and cross-linked with 1% formaldehyde at 30°C for 15 min. Glycine at 2.5 M was added, and the cells were shaken for 5 min before being spun down and washed with PBS buffer (10 mM KH2PO4, 40 mM K2HPO4, 150 mM NaCl) containing dithiothreitol (DTT). The cells were then suspended in 2 ml of spheroplast buffer (18.2% sorbitol, 1% glucose, 0.2% yeast nitrogen base, 0.2% Casamino Acids, 25 mM HEPES [pH 7.4], 50 mM Tris, 1 mM dithiothreitol) along with 0.8 mg of lyticase and incubated at 30°C for 30 min to generate spheroplasts. The spheroplasts were first washed in 500 μl of ice-cold PBS buffer containing phenylmethylsulfonyl fluoride (PMSF). Then they were resuspended in HEPES–Triton X-100 buffer (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES [pH 6.5]) containing 0.5 mM PMSF and protease inhibitor cocktail (Roche) and spun down at 7,000 rpm for 7 min. Ultimately, the spheroplasts were resuspended in HEPES–NaCl buffer (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES [pH 6.5]) containing 0.5 mM PMSF and protease inhibitor cocktail and again centrifuged at 7,000 rpm for 7 min. Finally, the spheroplasts were resuspended in 100 μl of SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8.1]) containing 0.5 mM PMSF and protease cocktail inhibitor and sonicated (Elma; model-S-60H) to generate an average DNA fragment size of 0.5 to 1 kb. After centrifugation, approximately 1.1 ml of supernatant was added to 1 ml of IP dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris [pH 8.1], 167 mM NaCl, 0.5 mM PMSF, and protease cocktail inhibitor) and left on ice for 15 min to form the chromatin fraction. Immunoprecipitation was performed with 1 μg of anti-Myc antibody to precipitate Cup9. SIR2UAS was amplified using the primers OSB 125 and OSB 87 in a reaction volume of 50 μl using 1/75 of immunoprecipitates and 1/50 of input DNA. Samples were subjected to electrophoresis on 1.5% agarose. Cup9 binding was also measured at the ACT1 locus using the primers OSB 16 and OSB 14. The control antibody for ChIP was rabbit IgG.
Southern hybridization.
Telomere Southern blotting was carried out according to the protocol published earlier (31). Briefly, yeast cells were grown in yeast extract-peptone-dextrose (YPD) medium to a density of 1.5 × 107 cells per milliliter. Heat shock was carried out by exposing the cells to 39°C for 40 min and subsequently returning them to 30°C and then growing them for 7 days. The genomic DNA was isolated from the control as well as from the post-HS samples collected after 2 h and those collected on the 4th and 6th days. Equal amounts of genomic DNA from each sample were subjected to XhoI digestion. Subsequently, all the digested samples were electrophoresed on a 0.8% agarose gel and transferred to a nitrocellulose membrane. For probe preparation, a 625-bp poly(G·T)/poly(C·A) fragment (kindly provided by Arthur Lustig) was labeled with [α-32P]dCTP using a Deca label DNA labeling kit (Fermentas). The Southern blot was finally exposed to X-ray film and developed.
RESULTS
HS or Hsp82 overexpression induces transcriptional downregulation of SIR2.
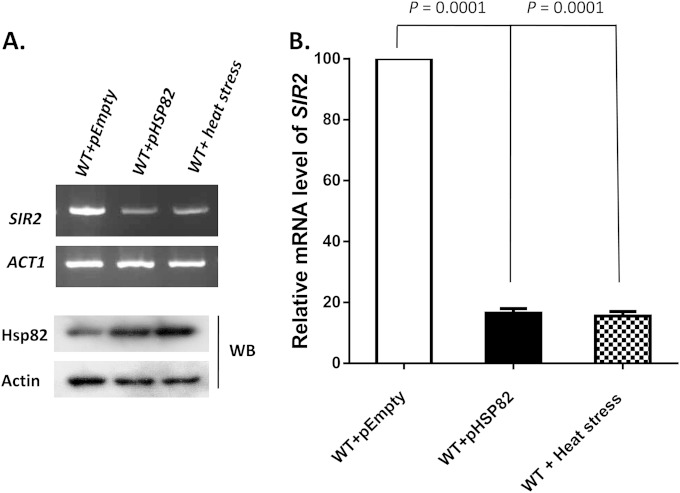
In an earlier study (5), we had established that when cells are exposed to heat shock (HS) (at 39°C for 40 min), the total cellular pool of Sir2 was reduced considerably. Similarly, Hsp82 overexpression, a general phenomenon associated with heat shock response, also caused drastic reduction in Sir2p in a dose-dependent manner. To investigate whether the reduction of Sir2p is merely at the protein level or extends to the transcript level as well, we analyzed SIR2 mRNA under heat stress. To this end, we exposed SLY20 cells to 39°C for 40 min, a condition that yields overexpression of Hsp82, and compared the level of SIR2 mRNA with that of the wild-type (WT) cells. We also transformed the cells with an Hsp82 overexpression plasmid (centromeric expression vector) (SLY13) and compared the level of SIR2 mRNA with that of the cells containing the empty expression vector (SLY13C). Under both conditions, we measured the levels of Hsp82 protein, which were higher than that of the control cells (Fig. 1A, bottom). The semiquantitative RT-PCR showed that Hsp82 overexpression (artificially or under heat stress) downregulated the level of SIR2 (Fig. 1A). Quantitative analysis by real-time RT-PCR revealed about a 5-fold reduction in the SIR2 transcript under the heat shock/Hsp82 overexpression condition compared to that in the control cells (Fig. 1B).
FIG 1.
Heat shock or Hsp82 overexpression induces transcriptional downregulation of SIR2. (A) (Top) Semiquantitative RT-PCR shows the SIR2 transcript in cells exposed to heat shock (39°C for 40 min) and in cells harboring an Hsp82 overexpression vector compared to that present in wild-type cells. (Bottom) Western blotting (WB) was done with anti-Hsp82 and antiactin antibodies. (B) The relative mRNA levels of SIR2 under the above-mentioned conditions (indicated on the x axis) were plotted after normalization with ACT1 mRNA. In each case, the mean value (±SD) from three independent experiments with three independent harvests of cells was calculated and was plotted using GraphPad Prism6 software. P values were calculated using the two-tailed Student t test.
Transient heat shock leads to transgenerational transmission of derepressed subtelomeric chromatin.
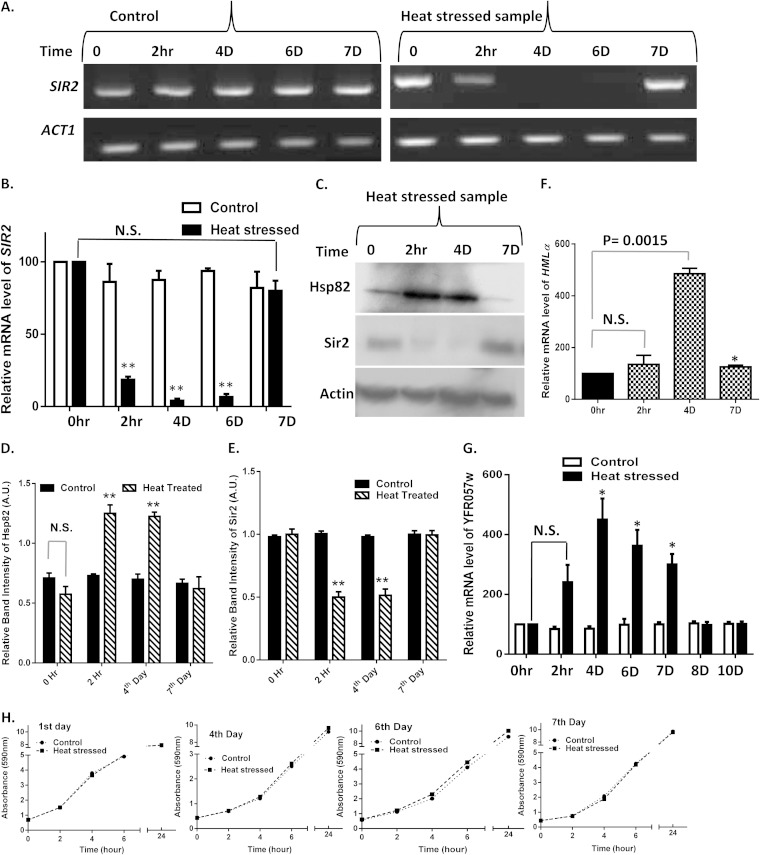
We focused on the effects of HS over multiple generations. To this end, we exposed cells of the SLY20 strain to heat stress at 39°C for 40 min and subsequently returned them to 30°C. These heat-stressed cells were maintained for several generations (up to 10 days), with regular medium changes every 24 h. In parallel, we maintained a wild-type culture which was not subjected to heat shock. We collected the total RNA before HS (0 h), 2 h post-HS, and thereafter at an interval of every 24 h. We repeated the semiquantitative RT-PCR three times with independent harvests of cells; the results of one of the representative experiments are presented in Fig. 2A. Our results showed that HS-mediated reduction in the SIR2 transcript continued through successive generations. The SIR2 transcript was barely observed in the 4th- and 6th-day HS samples. However, it started to return to a level comparable to that of the unstressed cells on the 7th day post-HS. The real-time RT-PCR result showed that the relative level of SIR2 mRNA had been reduced to nearly 25% and 15% in the 4th-day and 6th-day HS cultures, respectively. However, in the 7th-day HS culture, the SIR2 transcript level was comparable to that of the control (Fig. 2B). Our observation was supported by Western blotting with anti-Hsp82 and anti-Sir2 antibodies. Our results indicated that Hsp82 overexpression under heat stress led to a significant reduction in Sir2p in the 2 h and the 4th day post-HS. It is important to note that in the 7th-day HS sample, the Sir2p level went back to that of the unstressed cells, and there was a significant reduction of Hsp82 (Fig. 2C). The quantification of the relative band intensities from three independent experiments showed that the Hsp82p level increased 2.2 times in the 2 h post-HS, remained 2.1 times higher in the 4 days post-HS than the level of control, and returned to the level of the control after 7 days (Fig. 2D). Similarly, the relative band intensity of Sir2 was reduced by half in the 2 h post-HS and remained at that level up to 4 days and then returned to the level of the control cells on the 7th day (Fig. 2E). Our previous work showed that Hsp82 overexpression leads to derepression of telomere silencing, without any change in mating type silencing in yeast (5). We wanted to investigate whether a transient heat shock leads to any transgenerational mating type silencing defect in yeast. To that end, we monitored the HMLα transcript at various time intervals in the post-HS sample and compared it to that of the control cells, which were never exposed to heat shock. The real-time RT-PCR showed that the relative levels of HMLα in the 2-h post-HS sample were slightly higher than those in the control cells; however, they increased significantly (5 times) on the 4th day post-HS and ultimately returned to the normal level on the 7th day (Fig. 2F). To test the telomeric silencing activity of Sir2 in HS samples, we measured the transcription of subtelomeric gene YFR057w by real-time RT-PCR analysis. Under normal conditions, Sir2 represses the transcription of YFR057w by spreading through the subtelomeric ends of chromosome. Our results showed that there was no significant change in the relative mRNA level of YFR057w in the 2-h post-HS sample compared to that of the control. However, in the 4th-, 6th-, and 7th-day post-HS samples, the YFR057w transcript had increased 4.5-fold, 3.6-fold, and 3-fold, respectively, and it was repressed again from the 8th day onwards (Fig. 2G). We measured the growth of control and HS culture for 7 days, and the kinetics showed that they were dividing at the same rate (Fig. 2H).
FIG 2.
Transient heat shock leads to transgenerational transmission of derepressed subtelomeric chromatin. (A) Wild-type cells were exposed to heat shock (39°C) for a period of 40 min and then returned to 30°C. They were allowed to grow for 10 days as described in Results. The SIR2 transcript profile was monitored after 2 h and on the 4th, 6th, and 7th days and compared with that of the control cells, which were not exposed to heat shock. The experiment was repeated three times; the results from one representative semiquantitative RT-PCR are presented. (B) Relative mRNA levels of SIR2 in normal and heat-stressed cells at different time points (as indicated on the x axis) were plotted. Error bars indicate SD (n = 3 experiments); asterisks indicate values significantly different from the control, as follows: **, P < 0.01, and *, P < 0.05. N.S., not significant. ACT1 was used as the normalization control. (C) A Western blot was developed with control and heat-stressed samples at different time intervals using anti-Sir2, anti-Hsp82, and antiactin antibodies. (D) Densitometric measurements of Hsp82 from three independent Western blots were plotted for control and heat-treated samples at the indicated time points. Error bars indicate SD. (E) Densitometric measurements of Sir2 from three independent Western blots were plotted with control (before heat shock) and heat-treated samples at the indicated time points. Error bars indicate SD. (F) Relative mRNA levels of HMLα in MATa haploids before and after heat shock at the time points given in the x axis are plotted. Error bars indicate SD (n = 3). *, P < 0.05. (G) Relative mRNA levels of YFR057w in wild-type cells and cells exposed to heat shock (39°C for 40 min) at different time points are shown. Error bars indicate SD (n = 3). *, P < 0.05. (H) Growth kinetics of wild-type and heat-stressed cells were monitored for 7 days. The graph represents a comparison between their growths on the 1st, 4th, 6th, and 7th days.
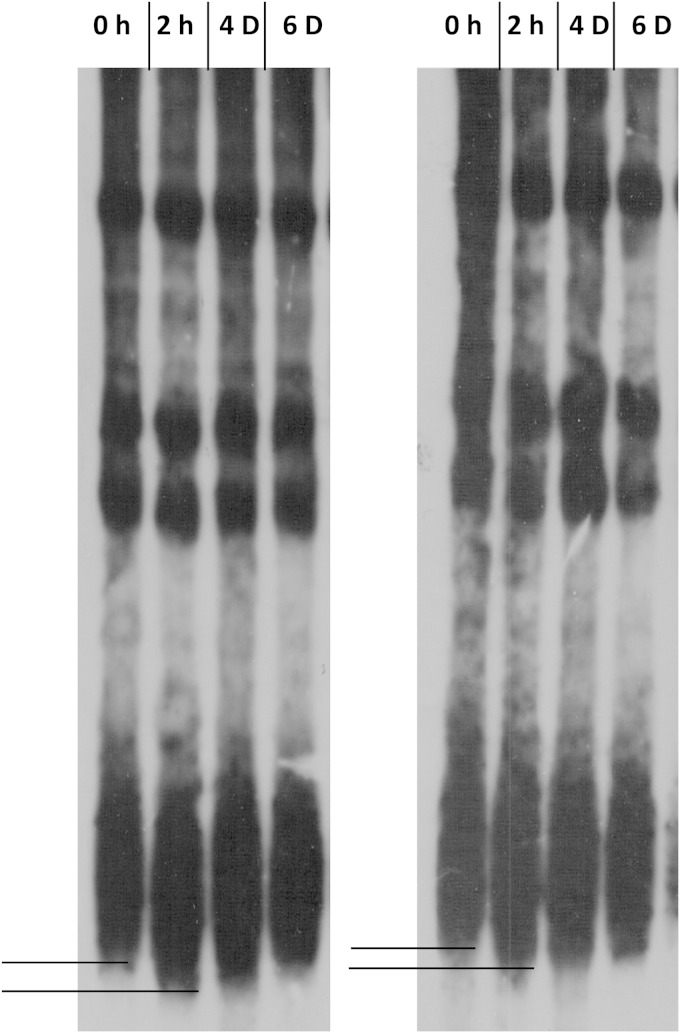
It was previously reported that prolonged heat shock (at 37°C) causes telomere shortening in yeast (32, 33). To understand whether telomere shortening is responsible for the derepression of subtelomeric chromatin, we wanted to find out whether transient heat shock also leads to such changes in the telomere structure. For that purpose, we monitored the length of the telomere for 7 days after a transient heat shock. We performed three independent experiments, and our results showed that transient heat shock leads to shortening of telomere length (Fig. 3). Telomere length remained short up to the 4th day and then returned to the wild-type length.
FIG 3.
Transient heat shock leads to telomere shortening in wild-type cells. Strain SLY20 was subjected to heat shock at 39°C for 40 min and then was grown at 30°C for 6 days. Genomic DNA was isolated at different time intervals (as marked at the top) and subjected to XhoI digestion, and telomere length was measured using Southern blot hybridization. The experiment was repeated with three independent colonies; two telomere blots are represented. The difference between the lengths of telomeres grown at 30°C and the 2-h post-HS sample is represented by solid lines.
Mapping the cis regulatory region of Sir2 affected by Hsp82 overexpression.
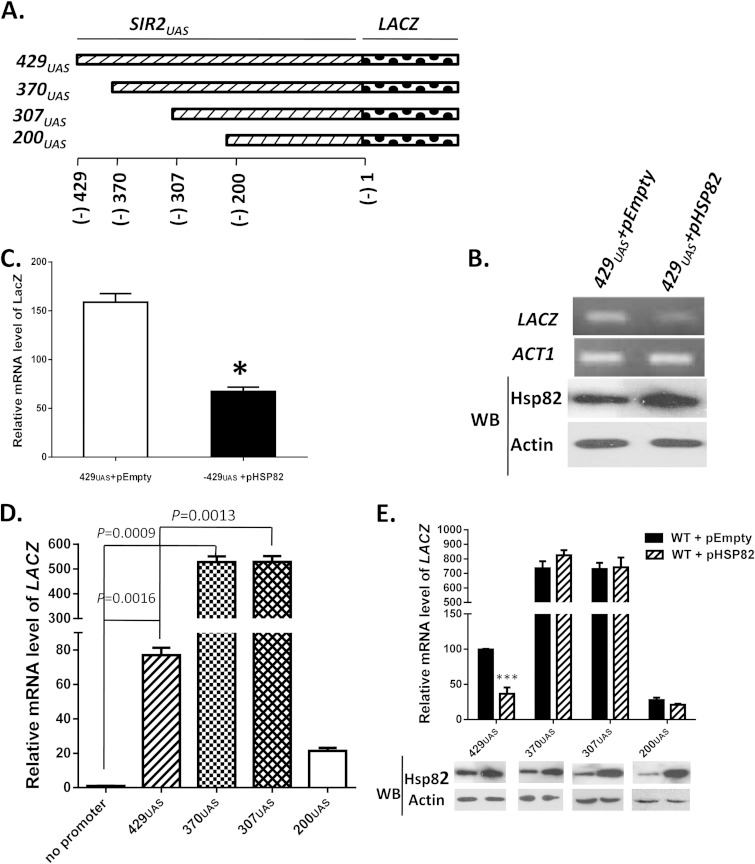
To further characterize the transcriptional repression of SIR2, we cloned the upstream regulatory element of SIR2 (−429 to −1) into a reporter plasmid carrying LACZ and named it 429UAS (Fig. 4A). Similarly, three more constructs (370UAS, 307UAS, and 200UAS) which spanned across the SIR2UAS (−370 to −1, −307 to −1, and −200 to −1) were generated, and they were individually cloned as the exclusive promoter region of the LACZ expression cassette. To provide a negative control, we generated a reporter plasmid without any promoter. Eventually all the constructs were transformed into SLY20 to generate isogenic strains SLY57 (429UAS), SLY84 (370UAS), SLY83 (307UAS), SLY56 (200UAS), and SLY64 (negative control), respectively. Expression of the LACZ gene from these constructs directly correlated with the transcriptional activity of different regions of the SIR2 upstream regulatory element.
FIG 4.
Mapping of the cis regulatory region of Sir2 that is affected by Hsp82 overexpression. (A) Upstream activator sequences of SIR2, 429, 370, 307, and 200 bp, were cloned in the upstream region of LACZ to generate four reporter plasmids, namely, 429UAS, 370UAS, 307UAS, and 200UAS. (B) (Top) 429UAS reporter plasmid was transformed into wild-type cells and cells harboring overexpressing Hsp82. Semiquantitative RT-PCR shows the relative levels of the LACZ transcript between wild-type cells and cell harboring Hsp82 overexpression plasmid. (Bottom) Western blotting was done with anti-Hsp82 and antiactin antibodies. (C) Real-time RT-PCR shows relative levels of LACZ mRNA between wild-type cells and cells bearing the Hsp82 overexpression plasmid. Error bars indicate SD (n = 3 experiments); the asterisk indicates a value significantly different from the control (P < 0.05). (D) A LACZ reporter plasmid without any promoter was transformed into wild-type cells and included in the experiment. Real-time RT-PCR was used to compare the relative mRNA levels of LACZ in cells harboring four different reporter plasmids along with cells having no promoter. P values were calculated using the two-tailed Student t test. (E) (Top) In cells with plasmids bearing each of the four reporter constructs, Hsp82 overexpression plasmid was transformed. Real-time RT-PCR shows how the relative abundance of LACZ was affected in the presence of the HSP82 overexpression plasmid. The relative abundances of mRNA from these constructs were plotted after normalization against ACT1 mRNA. Each bar represents mean mRNA level (±SD) from three independent experiments. ***, P < 0.001. (Bottom) Western blotting was done using anti-Hsp82 and antiactin antibodies.
To test whether SIR2UAS is affected by Hsp82 overexpression, we compared LACZ transcription of 429UAS in the presence and the absence of Hsp82 overexpression. Semiquantitative RT-PCR revealed that overexpression of Hsp82 resulted in a significant reduction of the LACZ transcript in the strain carrying both 429UAS and pHSP82 (SLY61) compared to that in the strain carrying 429UAS and pEmpty (SLY61C) (Fig. 4B). Western blotting (Fig. 4B, bottom) confirmed overexpression of Hsp82 in SLY61 compared to SLY61C. Quantitative analysis by real-time RT-PCR revealed that the LACZ transcript was reduced 2.4-fold under the Hsp82 overexpression condition compared to the wild type (Fig. 4C). In order to find the repressor binding site in SIR2UAS, we quantified the relative levels of LACZ mRNA in cells harboring various deletion constructs. The transcription of LACZ in SLY64 was considered the baseline. We found about an 80-fold increase in LACZ in the cells carrying full-length (429UAS) upstream activator sequence of SIR2 compared to that of the negative control (no promoter). However, LACZ transcription further increased (7-fold) in 370UAS and in 307UAS (Fig. 4D). This provides evidence of a repressor binding site in the region spanning bp −429 to −369 of SIR2UAS. 200UAS, however, displayed about a 4-fold reduction in LACZ transcription compared to that of 429UAS. Next, we aimed to narrow down the cis regulatory element of SIR2 that is regulated via heat stress. For this, we mimicked the heat shock condition by transforming an Hsp82 overexpression plasmid in the cells carrying individual constructs. As a control, we transformed the empty plasmid in cells carrying different LACZ fusion constructs. Real-time RT-PCR data showed about a 2-fold reduction in LACZ transcription in cells carrying 429UAS along with Hsp82 overexpression (Fig. 4E). However, Hsp82 overexpression in 370UAS, 307UAS, and 200UAS did not affect the relative mRNA level of LACZ compared to that of cells having the empty plasmid. Western blotting in each of the four fusion constructs showed overexpression of Hsp82 (Fig. 4E, bottom). Together, these data suggest that the region spanning bp −429 to −369 of SIR2 is crucial for its regulation during Hsp82 overexpression.
Bioinformatics prediction of transcription factor binding to SIR2UAS.
We analyzed the 429 bp upstream of SIR2 regulatory region for transcription factor binding sites of Saccharomyces cerevisiae. The analysis for finding transcription factors was performed by the statistical method (34) employed in the widely used TRAP (transcription factor affinity prediction) web tool (35). TRAP was developed to predict transcription factor binding affinities to DNA. Based on the analysis with TRAP, eight transcription factors were found to have high binding affinity, namely, Cup9, Rim101, Sok2, Tod6, Phd1, Tec1, Dot6, and Sum1, out of which four are transcriptional repressors. The transcription factors from both the databases TRANSFAC (36) and JASPAR (37), with their ranks, are shown in Table 3. From the data, it is evident that Cup9 shows highest binding affinity (P value, 0.007). Its binding sequence belongs to the region from −411 to −402.
TABLE 3.
Transcription factors from both of the databases JASPAR and TRANSFAC
| Serial no. | Rank | Matrix no. | Name of position-specific matrix in databasesa | P valueb | Sequencec |
|---|---|---|---|---|---|
| 1 | 1 | M01549 | F$CUP9_01 | 0.00742 | TCCTCAATGTGTCAATTAAC |
| 2 | MA0288.1 | CUP9 | 0.00791 | AATGTGTCA | |
| 2 | 3 | M01030 | F$RIM101_01 | 0.0176 | CCAAGCTA |
| 3 | 4 | M01621 | F$SOK2_01 | 0.0278 | GCCTGCAACT |
| 5 | MA0385.1 | SOK2 | 0.0329 | TATATGCATGCG | |
| 4 | 6 | MA0350.1 | TOD6 | 0.0341 | ATTTTTCCCTCATCGGCACAT |
| 5 | 7 | M01523 | F$PHD1_01 | 0.0343 | ATGCTTATATGCATGCGCATA |
| 6 | 8 | M01534 | F$TEC1_01 | 0.0345 | TTGCCAAAATTCTTGCTTTC |
| 7 | 9 | M01537 | F$DOT6_01 | 0.0355 | ATTTTTCCCTCATCGGCACAT |
| 10 | MA0351.1 | DOT6 | 0.0363 | ATTTTTCCCTCATCGGCACAT | |
| 8 | 11 | MA0398.1 | SUM1 | 0.0424 | TTAATTTAT |
Names beginning with “F” are from the TRANSFAC database; all other names are from the JASPAR database.
Probability of observing a certain or higher affinity in a given sequence. An accurate P value computation for the TRAP tool scores allows determination of which factors are the most likely to regulate a given target gene. It is set to normalize an observed affinity for a random-sequence model and to give a statistical meaning to the statement that one factor binds stronger than another.
Sequence of the transcription factor binding site.
Reversal of the heat shock/Hsp82 overexpression phenotype in cup9 deletion strain.
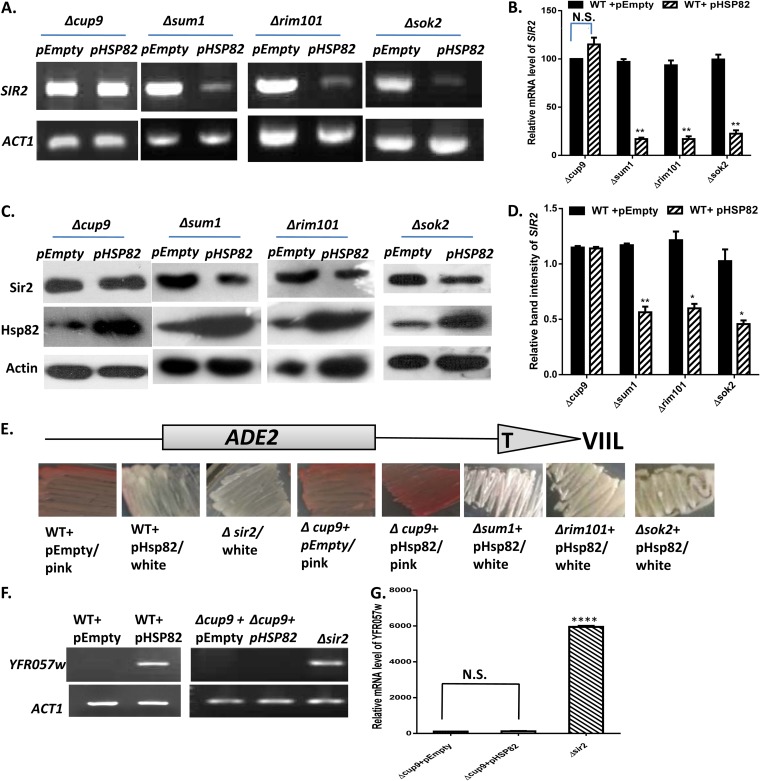
Based on the bioinformatics analysis, we characterized all four repressors, namely, Cup9, Sum1, Rim101, and Sok2, to identify the putative repressor of SIR2 transcription. We constructed four deletion strains, namely, Δcup9, Δrim101, Δsok2, and Δsum1 mutants, and screened each of them using various genetic experiments in an Hsp82 overexpression background. Semiquantitative RT-PCR showed no significant reduction in SIR2 transcript in the Δcup9 strain (SLY77C) carrying the empty vector or the Hsp82 overexpression plasmid (SLY77) (Fig. 5A). On the other hand, the Δsok2, Δsum1, and Δrim101 strains displayed significant reductions in SIR2 transcription upon Hsp82 overexpression. Real-time RT-PCR also displayed no significant reduction in relative mRNA levels of SIR2 in the Δcup9 strain with and without the HSP82 overexpression plasmid (Fig. 5B). However, other deletion strains exhibited considerable reductions in the SIR2 transcript with HSP82 overexpression similar to that in the wild type. By Western blotting, we observed the presence of a comparable amount of Sir2p in the cup9 deletion strain harboring the Hsp82 overexpression plasmid and that having an empty vector (Fig. 5C). However, Sir2p was considerably reduced under Hsp82 overexpression in Δsum1, Δrim101, and Δsok2 cells compared to those carrying the empty plasmid. Relative band intensity (with respect to actin) revealed about a 50% reduction of Sir2p in the Δsum1, Δrim101, and Δsok2 strains carrying the Hsp82 overexpression plasmid, but it remained unaltered in the Δcup9 strain with Hsp82 overexpression (Fig. 5D). These data are indicative of Cup9 being the mediator through which Hsp82 regulates SIR2 transcription. We wanted to monitor which of the deletion strains abrogate Hsp82 overexpression-mediated derepression of subtelomeric genes. With this in mind, we performed two independent functional assays of Sir2 using two different subtelomeric genes. First, we monitored the Sir2 function by a color assay scoring subtelomeric ADE2 expression. The Δcup9, Δsum1, Δrim101, and Δsok2 strains were generated in an isogenic background of SLY20 in which the telomere region of chromosome VIIL was marked with ADE2. The wild-type and Δcup9 strains both showed a pink color phenotype due to the silencing of the ADE2 gene, whereas the Δsir2 strain exhibited a white color phenotype correlating with derepression of ADE2. However, Hsp82 overexpression in the Δcup9 strain retained the pink color phenotype, as opposed to the white color phenotype observed during Hsp82 overexpression in the WT as well as in the Δsum1, Δrim101, and Δsok2 strains (Fig. 5E). In order to understand whether the maintenance of heterochromatinization is locus specific or not, we compared the levels of the YFR057w transcript, which is located near the chromosome VIR telomere. Our data showed that although Hsp82 overexpression caused derepression of YFR057w in wild-type cells, the Δcup9 strain did not display any silencing defect, as YFR057w remained silent even in the presence of Hsp82 overexpression (Fig. 5F). To score the silencing activity of Sir2p in a more quantitative manner, we performed real-time RT-PCR analysis, which showed no significant alteration in the YFR057w transcript in Δcup9 cells with and without Hsp82 overexpression (Fig. 5G). Our results imply that out of the four transcriptional repressors, deletion of only CUP9 restores wild-type-like Sir2 function under the Hsp82 overexpression condition. In other words, we observed increased expression of Sir2p specifically in the cup9 knockout strain during Hsp82 overexpression, which correlated well with the maintenance of Sir2 silencing function.
FIG 5.
Reversal of heat shock/Hsp82 overexpression phenotype in the cup9 deletion strain. (A) Δcup9, Δsum1, Δrim101, and Δsok2 strains were generated as described in Materials and Methods. An Hsp82 overexpression plasmid was transformed into each of the four deletion strains, and semiquantitative RT-PCR displays no alteration in the SIR2 transcript in the Δcup9 strain having an HSP82 overexpression background; ACT1 acted as the normalization control. (B) Real-time RT-PCR data for the relative quantity of SIR2 mRNA between the above-mentioned strains (presented on the x axis). Each bar represents the mean mRNA level (±SD) from three independent experiments. P values were calculated using the two-tailed Student t test. **, P < 0.01. (C) Western blot analysis was done with the protein extracted from the above-mentioned strains using antiactin, anti-Hsp82, and anti-Sir2 antibodies. (D) Densitometric measurements of Sir2 (after normalization with actin) from three independent experiments were plotted for the strains indicated on the x axis. Error bars indicate SD. **, P < 0.01; *, P < 0.05. (E) ADE2 reporter gene located at chromosome VIIL was used for the telomere silencing assay. Wild-type cells, wild-type cells carrying an Hsp82 overexpression plasmid, Δsir2 cells, Δcup9 cells, and Δcup9, Δsum1, Δrim101, and Δsok2 cells each carrying the Hsp82 overexpression plasmid were grown and plated as described in Materials and Methods. It should be noted that Δsum1, Δrim101, and Δsok2 strains carrying Hsp82 overexpression plasmid behave like wild-type cells carrying the Hsp82 plasmid. They are different from Δcup9 cells carrying the Hsp82 overexpression plasmid, which show intense pink coloration. (F) A telomere silencing assay was done with YFR057w localized adjacent to telomere VI-R. Semiquantitative RT-PCR was done to study the expression of YFR057w in wild-type cells with and without the Hsp82 overexpression plasmid and in the Δcup9 strain in the presence and absence of the Hsp82 overexpression plasmid. The Δsir2 strain was used as a control. The ACT1 transcript was measured as a loading control. (G) Real-time RT-PCR was done to quantify the relative abundance of YFR057w in the Δcup9 strain with or without Hsp82 overexpression plasmid and compare it with the same in the Δsir2 strain. ****, P < 0.0001.
Cup9 expression and its binding to SIR2UAS are enhanced by heat shock and Hsp82 overexpression.
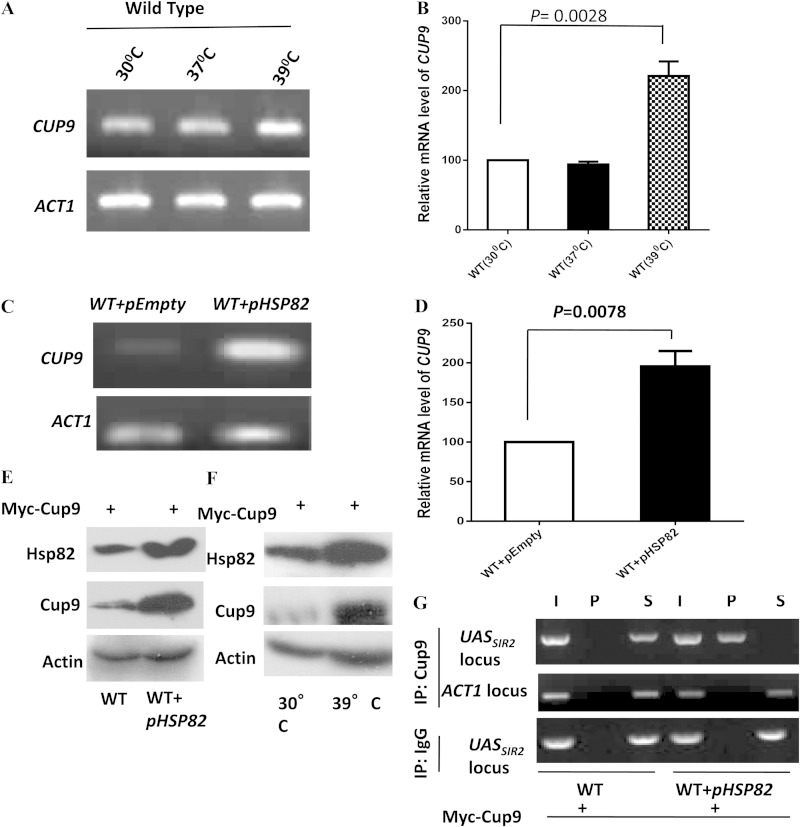
From the previous experiment, it was apparent that Hsp82-mediated transcriptional downregulation of Sir2 is dependent on Cup9. That led us to estimate the steady-state level of Cup9 under heat stress. We grew the cells at three different temperatures, 30°C, 37°C, and 39°C, and observed that the level of the CUP9 transcript was upregulated at 39°C (Fig. 6A). Real-time RT-PCR data showed that there was no change in the CUP9 transcript with an increase in temperature from 30°C to 37°C. However, at 39°C, CUP9 was upregulated 2.5-fold (Fig. 6B). We also monitored the level of the CUP9 transcript under the Hsp82 overexpression condition and observed that it had significantly increased compared to that of the wild type (Fig. 6C). Real-time RT-PCR analysis showed more than 2-fold upregulation of the CUP9 transcript under the Hsp82 overexpression condition (Fig. 6D). Our observation was further corroborated by the endogenous level of Cup9 protein under the Hsp82 overexpression condition. We tagged CUP9 with MYC at the chromosomal locus. Western blot analysis showed very low levels of Cup9-Myc in normal cells. However, an increased expression of Cup9-Myc was associated with overexpression of Hsp82 (Fig. 6E) as well as found under heat stress (Fig. 6F). Next, we used chromatin immunoprecipitation (ChIP) to analyze Cup9 recruitment at the upstream regulatory region of SIR2 in the presence of Hsp82 overexpression. We used Hsp82-overexpressing cells in which Cup9 (Cup9-Myc) was abundantly present and used anti-Myc antibody to immunoprecipitate chromatin-bound Cup9. Under the Hsp82 overexpression condition, we observed a bright signal of Cup9 specifically at the upstream regulatory element of SIR2 but not on the control ACT1 locus (Fig. 6G).
FIG 6.
Heat shock and Hsp82 overexpression induce Cup9 expression and lead to its association with SIR2UAS. (A) Wild-type cells were divided into three groups: one grown at 30°C, another exposed to 37°C for 2 h, and the last exposed to 39°C for 40 min. Semiquantitative RT-PCR results for all groups show the CUP9 transcript profile at the different temperatures. ACT1 acted as the normalization control. (B) Real-time RT-PCR shows the quantitative abundance of the CUP9 transcript at 39°C compared to that at 30°C. Each bar represents mean mRNA level (±SD) from three independent experiments. P values were calculated using the two-tailed Student t test. (C) Wild-type cells and cells bearing the Hsp82 overexpression plasmid were used to assess the level of the CUP9 transcript by employing semiquantitative RT-PCR. The experiment was repeated three times; data from one representative experiment are presented here. (D) Real-time RT-PCR reveals the quantitative abundance of the CUP9 transcript in cells bearing HSP82 overexpression plasmid compared to that in the wild type. (E) In wild-type cells, CUP9 was MYC tagged at the chromosomal locus as described in Materials and Methods. Proteins isolated from wild-type cells and cells having the Hsp82 overexpression plasmid were subjected to Western blot analysis using anti-Myc (Cup9), anti-Hsp82, and antiactin antibodies. (F) The same cells were subjected to 39°C for 40 min, and heat-treated and untreated cells both were subjected to immunoblotting using antiactin, anti-Hsp82, and anti-Myc antibodies. (G) ChIP assays were performed using CUP9 MYC-tagged cells in the absence and presence of Hsp82 overexpression plasmid. Anti-Myc antibodies were used with control IgG (immunoglobulin G). Input (I), immunoprecipitated DNA (P), and supernatant (S) were amplified by semiquantitative RT-PCR with primers that covered SIR2UAS. The experiment was repeated twice; data from one representative experiment are presented. I, P, and S DNA were also amplified using primers that cover ACT1, which acted as a negative control.
Cup9 overproduction reduces the endogenous level as well as the function of Sir2.
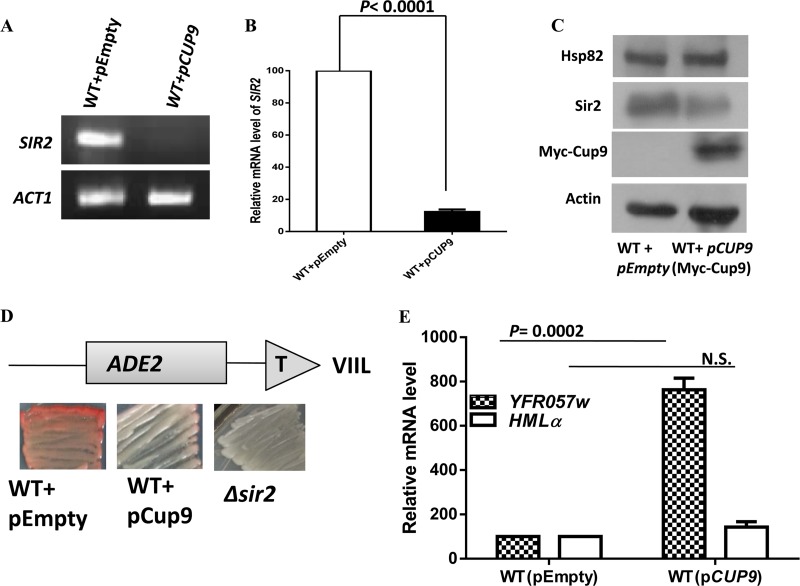
In order to explore whether the endogenous level and activity of Sir2 are directly regulated by Cup9, we analyzed them under the Cup9 overexpression condition. To that end, Cup9 overexpression plasmid pESC-CUP9 and the empty vector pESC were transformed into wild-type strain SLY20 to generate SLY90 and SLY91, respectively. This vector overexpresses Myc-tagged Cup9 under the control of a galactose-inducible promoter. The SIR2 transcript was quantified in those backgrounds by semiquantitative RT-PCR. The results showed that Cup9 overexpression entirely diminished the SIR2 transcript in SLY90 compared to that in the SLY91 strain (Fig. 7A). Real-time RT-PCR analysis showed a nearly 10-fold reduction of the SIR2 transcript in a Cup9 overexpression background (Fig. 7B). Our results were further confirmed after estimating Sir2p in a Cup9 overexpression background. Western blot analysis showed that Cup9 overexpression caused a modest reduction in the Sir2p level specifically, without any alteration to the Hsp82 or Act1 protein level (Fig. 7C). We subsequently investigated the silencing function of Sir2 under the Cup9 overexpression condition using three independent assays. We observed that SLY90 cells having a reduced level of Sir2 developed into white colonies, indicating derepression of the subtelomeric ADE2 gene, whereas SLY91 containing the empty vector developed as pink colonies due to the silencing of ADE2 (Fig. 7D). Also, quantification of the YFR057w transcript by real-time RT-PCR showed a nearly 8-fold increase in transcripts in cells harboring the Cup9 overexpression plasmid compared to those in the WT (Fig. 7E). We also compared the HMLα transcript by real-time RT-PCR in a Cup9 overexpression background but observed no significant change. These results indicate that Cup9 overexpression brings down the Sir2 level moderately, as a result of which the silencing activity of Sir2 at a hidden mating locus is maintained, though subtelomeric silencing activity is diminished.
FIG 7.
Cup9 overproduction reduces the endogenous level as well as the function of Sir2. (A) Cup9 was induced by adding galactose to a final concentration of 2%. The overnight culture grown in galactose medium was used as a secondary inoculum in galactose-containing medium and was grown for an additional 5 h (until the mid-log phase) before the isolation of RNA. The effect of Cup9 overexpression on SIR2 transcription was measured by semiquantitative RT-PCR. (B) Real-time RT-PCR quantification of SIR2 mRNA levels in Cup9-overexpressing cells relative to those of the wild-type cells is shown as the average of three experiments. Error bars indicate SD. P values were calculated using the two-tailed Student t test. ACT1 mRNA was used as the normalization control. (C) The overnight culture grown in galactose medium was used for Western blot analysis. The immunoblot shows the reduction of endogenous Sir2 protein upon Cup9 overexpression. The Cup9 overexpression vector harbors C-terminally Myc-tagged Cup9. (D) The ADE2 reporter gene at telomere VIIL was used for the telomere silencing assay. Wild-type, Cup9-overexpressing, and Δsir2 cells were grown and plated as described in Materials and Methods. Pink colonies indicate a silenced ADE2 gene, and white colonies indicate transcriptionally active ADE2. (E) Cup9 overexpression induces the derepression of another subtelomeric gene, YFR057w. However, silencing at the hidden mating type loci is maintained, as seen by the comparable level of the HMLα transcript. ACT1 was used as the normalization control. Error bars indicate SD (n = 3).
DISCUSSION
In this article, we provide a mechanistic understanding of how Hsp90 homeostasis regulates Sir2 function in the cell. We provide compelling evidence that Hsp90 regulates the transcription of SIR2 under heat stress and thereby controls the cellular abundance of Sir2 protein. Previous work in our laboratory had demonstrated that heat shock treatment as well as Hsp90 overexpression caused a drastic reduction in the endogenous level of Sir2 (5). The reduced pool of Sir2 was functionally active, but its limiting quantity was insufficient to establish silencing across all 32 telomeres. However, it was adequate to silence hidden mating type loci. Findings from this work help in understanding how Hsp90 overexpression results in a reduced pool of Sir2 protein.
Molecular players involved in transcriptional regulation of SIR2 gene have remained elusive. Here, we report for the first time the identification of a transcriptional repressor that regulates SIR2 gene transcription. We provide several lines of evidence that unequivocally establish Cup9 as the transcriptional repressor of SIR2 gene expression.
First, in the Δcup9 background, neither heat shock nor overexpression of Hsp82 had any effect on SIR2 transcription. Second, Cup9 overexpression caused a drastic reduction in SIR2 transcription, resulting in the derepression of subtelomeric genes. Such an effect was independent of heat stress. Third, bioinformatics analysis predicted a Cup9 binding sequence within (−419 to −399) upstream regions of SIR2. Finally, a chromatin immunoprecipitation assay further demonstrated that endogenously expressed Cup9 was recruited at the 5′ end on the yeast SIR2 promoter under the Hsp82 overexpression condition.
Our current data show that under heat stress, Cup9 expression is induced. Under such conditions, Cup9 binds to the SIR2UAS, leading to the transcriptional downregulation of SIR2 and thereby causing derepression of subtelomeric genes. This was further supported by a reporter gene analysis using various deletion constructs of SIR2UAS. The effect of Hsp82 on transcriptional downregulation of the reporter gene was abolished when the Cup9 binding region was deleted from SIR2UAS. Currently, it is not known how Hsp82 regulates Cup9 expression. Previously, it was observed that copper stress causes transcriptional upregulation of CUP9 when cells are grown on lactose medium (13). Our study demonstrates that expression of CUP9 is also upregulated during heat shock treatment. It was previously reported that high temperature (37°C) strengthens mating and telomere silencing (38). Our results corroborate this finding, since CUP9 transcription remained unaltered between 30°C and 37°C. However, at 39°C, there was a nearly 2.5-fold increase in the CUP9 transcript. It has been observed that expression of HSP90 increases severalfold not only during heat shock but also in response to several other environmental stresses (39–41). Thus, it will be interesting to investigate whether upregulation of Cup9 is a specific or general stress response phenomenon.
Transcriptional regulation of Sir2 in yeast in response to environmental stimuli has never been identified before. However, in a human cell line, it has been demonstrated that SIRT1p associates with HIC1 (hypermethylated in cancer 1) and the complex binds to the SIRT1 promoter to repress its own transcription (42). Epigenetic silencing of HIC1 through hypermethylation of the HIC1 promoter has been associated with aging. Reduction in HIC1 expression results in SIRT1 upregulation, which results in excessive deacetylation and deactivation of p53 function and thus increases the cancer risk in mammals. Any such feedback inhibition of SIR2 expression by Sir2 itself in yeast is not known. To the best of our knowledge, Cup9 is the only repressor identified so far that regulates SIR2 gene expression in yeast.
Dietary restriction is proven to be an environmental factor that increases longevity from yeast to mammals (43, 44). NAD+-induced histone deacetylase activity of Sir2 is required for increased longevity during starvation, and this effect was not observed in a sir2 mutant strain (45). It had been demonstrated previously that under normal conditions, Cup9 is a short-lived protein, having approximately 5 min as its half-life (46). The presence of imported di- or tripeptides causes the activation of E3 ubiquitin ligase Ubr1 and accelerates the Ubr1-dependent ubiquitylation of Cup9 (46). Thus, under normal conditions, due to the unstable nature of Cup9, Sir2 can be transcribed optimally during glucose starvation. However, under heat shock conditions (39°C), an elevated level of Cup9 leads to repression of SIR2 transcription, thereby mimicking sir2 knockdown in yeast, which is inherited for many generations. Thus, it is tempting to predict that Sir2 may not influence longevity under heat stress conditions. This hypothesis is supported by the following lines of evidence. First of all, the Δsir2 mutant strain does not show life span extension in yeast, and upon heat treatment, the steady-state level of Sir2 decreased drastically and remained almost undetectable for up to 6 days in our Western blot analysis. Second, deletion of cup9, a condition that increases Sir2 abundance, yielded an extended life span under cold conditions (47). Another study performed with Caenorhabditis elegans (48) also documented that decreasing the temperature progressively lengthened the life span of worms. It will be interesting to explore whether such Hsp90-induced regulation of mammalian sirtuins also results in their suboptimal activity.
Our work reveals that transient heat shock results in heritability of derepressed subtelomeric chromatin. Previously, it had been reported that telomere structure regulates the heritability of silenced subtelomeres (49). An elongated telomere track leads to the increased inheritance of the silenced subtelomeric state and is independent of yeast chromatin assembly factor 1 (yCAF-1). In another study, it was reported that prolonged exposure to heat stress (37°C) as well as Hsp82 overexpression led to telomere shortening in wild-type cells (32, 33). Thus, it is important to understand whether transient heat shock causes any change in telomere structure and can thereby influence telomere silencing. Our study shows that transient heat shock leads to shortening of the telomere length, which is gradually restored to the wild-type length (at the end of the 6th day). The restoration of telomere length might be one of the factors for the reappearance of the telomere position effect, since TPE is not reestablished before the telomere length returns to normal. On the other hand, the reappearance of Sir2p is also likely to be the reason behind restoration of wild-type-like silencing at the telomere, as the timing of Sir2 reappearance and that of the reestablishment of TPE coincide very closely. Thus, it is possible that transient heat shock-mediated derepression as well as reestablishment of TPE is multifactorial and a period of 7 to 8 days is required for the full reestablishment of subtelomeric silencing.
Although the mechanism behind heritable repression of SIR2 is not clear at present, this work has unraveled a cryptic pathway of SIR2 regulation that is induced under heat stress (39°C) or under a condition in which Hsp90 is overexpressed in cells. Our work showed that a short period of heat shock rendered the cells sir2 knockdown cells for more than 90 generations. Our finding on derepression of the HMLα transcript upon transient heat shock implies a likely defect in yeast mating behavior which has tremendous implications for yeast physiology. This observation is also important in a broader context, as Sir2 is one of the epigenetic factors that establishes silencing at subtelomeric regions in many eukaryotes. Sir2-mediated telomere silencing plays a major role in mutually exclusive expression of virulent multigene family in protozoan parasites such as Plasmodium and trypanosomes (50, 51). Such a mechanism controls antigenic variation and thereby causes evasion of the host immune system (52). In light of our findings, it will be interesting to explore whether exposure to febrile temperature (around 39°C) as a natural consequence of Plasmodium infection has any correlation with derepression of subtelomeric virulent genes as a consequence of poorer Sir2 activity.
ACKNOWLEDGMENTS
The work was supported by grants from the Council of Scientific and Industrial Research, Government of India [37(1549)/12/EMR-II], and the Department of Biotechnology (India) [BT-BRB-TF-3-2013] to S.B. S.L. was supported by a senior research fellowship from the University Grants Commission, Government of India.
We thank Arthur Lustig (Tulane University) for the telomeric probe. We thank Meenu Babu and Meera Babu for critically reading the manuscript.
We declare that we have no conflicts of interest.
REFERENCES
- 1.De Ruijter AJ, Van Gennip AH, Caron HN, Kemp S, Van Kuilenburg AB. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.North BJ, Verdin E. 2004. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol 5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imai S, Armstrong CM, Kaeberlein M, Guarente L. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 4.Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. 2007. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell 27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laskar S, Bhattacharyya MK, Shankar R, Bhattacharyya S. 2011. HSP90 controls SIR2 mediated gene silencing. PLoS One 6:e23406. doi: 10.1371/journal.pone.0023406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol 22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmen AA, Milne L, Grunstein M. 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J Biol Chem 277:4778–4781. doi: 10.1074/jbc.M110532200. [DOI] [PubMed] [Google Scholar]
- 8.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80:583–592. doi: 10.1016/0092-8674(95)90512-X. [DOI] [PubMed] [Google Scholar]
- 9.Hickman MA, Froyd C, Rusche LN. 2011. Reinventing heterochromatin in budding yeasts: Sir2 and the origin recognition complex take center stage. Eukaryot Cell 10:1183–1192. doi: 10.1128/EC.05123-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang CL, Landry J, Sternglanz R. 2008. A yeast Sir2 mutant temperature sensitive for silencing. Genetics 180:1955–1962. doi: 10.1534/genetics.108.094516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1. J Biol Chem 277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 12.Gallo CM, Smith DL Jr, Smith JS. 2004. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol 24:1301–1312. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knight SA, Tamai KT, Kosman DJ, Thiele DJ. 1994. Identification and analysis of a Saccharomyces cerevisiae copper homeostasis gene encoding a homeodomain protein. Mol Cell Biol 14:7792–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Protchenko O, Shakoury-Elizeh M, Keane P, Storey J, Androphy R, Philpott CC. 2008. Role of PUG1 in inducible porphyrin and heme transport in Saccharomyces cerevisiae. Eukaryot Cell 7:859–871. doi: 10.1128/EC.00414-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurent A, Bihan R, Omilli F, Deschamps S, Pellerin I. 2008. PBX proteins: much more than Hox cofactors. Int J Dev Biol 52:9–20. doi: 10.1387/ijdb.072304al. [DOI] [PubMed] [Google Scholar]
- 17.Astell CR, Jonasson LA, Smith M. 1981. The sequence of the DNAs coding for the mating-type loci of Saccharomyces cerevisiae. Cell 10:15–23. [DOI] [PubMed] [Google Scholar]
- 18.Xia Z, Turner GC, Hwang CS, Byrd C, Varshavsky A. 2008. Amino acids induce peptide uptake via accelerated degradation of CUP9, the transcriptional repressor of the PTR2 peptide transporter. J Biol Chem 283:28958–28968. doi: 10.1074/jbc.M803980200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai H, Hauser M, Naider F, Becker JM. 2007. Differential regulation and substrate preferences in two peptide transporters of Saccharomyces cerevisiae. Eukaryot Cell 6:1805–1813. doi: 10.1128/EC.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai H, Kauffman S, Naider F, Becker JM. 2006. Genomewide screen reveals a wide regulatory network for di/tripeptide utilization in Saccharomyces cerevisiae. Genetics 172:1459–1476. doi: 10.1534/genetics.105.053041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, Luga V, Przulj V, Robinson M, Suzuki H, Hayashizaki Y, Jurisica I, Wrana JL. 2005. High-throughput mapping of a dynamic signaling network in mammalian cells. Science 307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 22.Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. 2012. Quantitative analysis of Hsp90-client interactions reveals principles of substrate recognition. Cell 150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Picard D. 2006. Chaperoning steroid hormone action. Trends Endocrinol Metab 17:229–235. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Nathan DF, Vos MH, Lindquist S. 1997. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci U S A 94:12949–12956. doi: 10.1073/pnas.94.24.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picard D. 2002. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci 59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frydman J, Hohfeld J. 1997. Chaperones get in touch: the Hip-Hop connection. Trends Endocrinol Metab 22:87–92. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y, Mosser DD, Morimoto RI. 1998. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev 12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voellmy R. 2004. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones 9:122–133. doi: 10.1379/CSC-14R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longtine MS, Mckenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Ppringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. doi:. [DOI] [PubMed] [Google Scholar]
- 30.Liaw H, Lustig A. 2006. Sir3 C-terminal domain involvement in the initiation and spreading of heterochromatin. Mol Cell Biol 26:7616–7631. doi: 10.1128/MCB.01082-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao H, Moss DL, Parke C, Tatum D, Lustig AJ. 2014. The Ctf18RFC clamp loader is essential for telomere stability in telomerase-negative and mre11 mutant alleles. PLoS One 9(2):e88633. doi: 10.1371/journal.pone.0088633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grandin N, Charbonneau M. 2001. Hsp90 levels affect telomere length in yeast. Mol Genet Genomics 265:126–143. doi: 10.1007/s004380000398. [DOI] [PubMed] [Google Scholar]
- 33.Romano GH, Harari Y, Yehuda T, Podhorzer A, Rubinstein L, Shamir R, Gottlieb A, Silberberg Y, Pe'er D, Ruppin E, Sharan R, Kupiec M. 2013. Environmental stresses disrupt telomere length homeostasis. PLoS Genet 9(9):e1003721. doi: 10.1371/journal.pgen.1003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berg OG, Von Hippel PH. 1987. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J Mol Biol 193:723–750. [DOI] [PubMed] [Google Scholar]
- 35.Roider HG, Kanhere A, Manke T, Vingron M. 2007. Predicting transcription factor affinities to DNA from a biophysical model. Bioinformatics 23:134–141. doi: 10.1093/bioinformatics/btl565. [DOI] [PubMed] [Google Scholar]
- 36.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryne JC, Valen E, Tang MHE, Marstrand T, Winther O, Piedade ID, Krogh A, Lenhard B, Sandelin A. 2008. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res 36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bi X, Yu Q, Sandmeier JJ, Elizondo S. 2004. Regulation of transcriptional silencing in yeast by growth temperature. J Mol Biol 344:893–905. doi: 10.1016/j.jmb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Piper PW, Talreja K, Panaretou B, Moradas-Ferreira P, Byrne K, Praekelt UM, Meacock P, Recnacq M, Boucherie H. 1994. Induction of major heat-shock proteins of Saccharomyces cerevisiae, including plasma membrane Hsp30, by ethanol levels above a critical threshold. Microbiology 140:3031–3038. doi: 10.1099/13500872-140-11-3031. [DOI] [PubMed] [Google Scholar]
- 40.Aranda A, Querol A, Olmo MLD. 2002. Correlation between acetaldehyde and ethanol resistance and expression of HSP genes in yeast strains isolated during the biological aging of sherry wines. Arch Microbiol 177:304–312. doi: 10.1007/s00203-001-0391-1. [DOI] [PubMed] [Google Scholar]
- 41.Imai J, Yahara I. 2000. Role of HSP90 in salt stress tolerance via stabilization and regulation of calcineurin. Mol Cell Biol 20:9262–9270. doi: 10.1128/MCB.20.24.9262-9270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen WY, Wang DH, Yen RWC, Luo J, Gu W, Baylin SB. 2005. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 43.Guarente L, Picard F. 2005. Calorie restriction—the SIR2 connection. Cell 120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 44.Wu Z, Liu SQ, Huang D. 2013. Dietary restriction depends on nutrient composition to extend chronological lifespan in budding yeast Saccharomyces cerevisiae. PLoS One 8:e64448. doi: 10.1371/journal.pone.0064448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin SJ, Defossez PA, Guarente L. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 46.Byrd C, Turner GC, Varshavsky A. 1998. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J 17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Postma L, Lehrach H, Ralser M. 2009. Surviving in the cold: yeast mutants with extended hibernating lifespan are oxidant sensitive. Aging 11:957–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dancsó B, Spiro Z, Arslan MA, Nguyen MT, Papp D, Csermely P, Sti C. 2010. The heat shock connection of metabolic stress and dietary restriction. Curr Pharm Biotechnol 11:139–145. doi: 10.2174/138920110790909704. [DOI] [PubMed] [Google Scholar]
- 49.Park Y, Lustig AJ. 2000. Telomere structure regulates the heritability of repressed subtelomeric chromatin in Saccharomyces cerevisiae. Genetics 154:587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Figueiredo LM, Pirrit LA, Scherf A. 2000. Genomic organisation and chromatin structure of Plasmodium falciparum chromosome ends. Mol Biochem Parasitol 106:169–174. doi: 10.1016/S0166-6851(99)00199-1. [DOI] [PubMed] [Google Scholar]
- 51.Vanhamme L, Pays E. 1995. Control of gene expression in trypanosomes. Microbiol Rev 59:223–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scherf A, Rubio JJL, Riviere L. 2008. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol 62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]