Abstract
Uptake of hepatitis C virus (HCV) into hepatocytes is an orchestrated process, involving numerous host factors, virion-associated lipoproteins and a growing number of cell-associated factors. Several of these factors likely contribute to the hepatotropism and limited host range of this virus. Discerning the minimal set of human-specific factors required for viral uptake into non-human cells has facilitated the development of small animal models with inheritable HCV susceptibility. This review summarizes current knowledge of host factors required for HCV entry, the molecular mechanisms underlying HCV entry into hepatocytes, and aspects of viral entry contributing to HCV host tropism.
INTRODUCTION
At least 150 million people worldwide are currently infected with hepatitis C virus (HCV), although this value is most likely an underestimation, as almost twice as many individuals as diagnosed may carry the virus,. HCV causes persistent infection in 70–80% of those who are exposed. While the acute disease is usually asymptomatic, chronic carriers frequently develop fibrosis, cirrhosis and in some cases, hepatocellular carcinoma, especially if left untreated. The treatment for HCV has evolved rapidly in recent years. Previous therapies based on a combination of pegylated interferon (IFN) alpha and ribavirin were only partially effective and poorly tolerated by patients. However, new direct-acting antivirals (DAAs) and, most recently, all-oral IFN-free regimens have been proven highly effective across all HCV genotypes and in almost all patient populations. Yet the high costs, logistical challenges of mass deployment and the risk of drug resistance associated with even the newest DAAs are considerable obstacles that must be overcome if global levels of HCV are to be reduced. A vaccine, which would prevent infection or delay the onset of pathogenesis during a chronic infection, still does not exist. Additionally, development of other effective therapies has been delayed by the lack of suitable cell culture systems and animal models. While hepaciviruses similar to HCV have been found in a variety of species, including dogs, horses and outbred mice, HCV has a much more limited host range. While some studies have provided evidence for transient and intermittent viremia in more exotic species such as tree shrews, robust infection has only been described in humans and experimentally infected chimpanzees. Although this narrow host range of HCV is not fully understood, it can partially be explained, as described in this review, by differences in the sequences of host factors essential for viral entry.
HCV is an enveloped, positive-strand RNA virus of the genus hepacivirus in the Flaviviridae family. The 9.6-kb RNA genome is flanked by 5’ and 3’ non-translated regions and contains two open reading frames, with one encoding the entire HCV polyprotein and the other producing a single gene product, the so-called “mini core” (reviewed in (Moradpour et al., 2007)). The HCV polyprotein is processed by host-and virus-encoded proteases, releasing ten mature proteins: the structural proteins which include the core protein and the envelope glycoproteins E1 and E2; the viroporin p7; and the non-structural (NS) proteins NS2, NS3, NS4A, NS4B, NS5A and NS5B. The viral envelope proteins E1 and E2 are critically important for viral uptake into hepatocytes.
HCV entry is a highly coordinated process, requiring in addition to E1 and E2 a plethora of host cell factors, some of which contribute to the hepato- and human tropism of HCV. Here, we aim to provide a concise overview of the mechanism of HCV entry, its impact on hepato- and human tropism of HCV, and its subsequent implications for HCV animal model development.
HCV PARTICLE
The known viral components of the HCV virion are genomic RNA, the core protein and the envelope glycoproteins E1 and E2. These glycoproteins are believed to form a non-covalent heterodimer, stabilized by disulfide bridges, and help mediate receptor binding and entry into hepatocytes. As detailed below, HCV is internalized through receptor-mediated endocytosis. The viral membrane must first fuse with the endosomal membrane, thus allowing the RNA genome to gain access to the cytoplasm where it is translated (see review by (Paul et al., 2014) in this issue). This process is triggered through acidification of the endosomal compartment, which is thought to result in a conformational change of the viral envelope (Figure 1). Based primarily on computational analysis, E2 was predicted to be a class II fusion protein with a highly extended conformation of three predominantly β-sheet domains (Krey et al., 2010). However, recent crystal structures of the core HCV E2 domain demonstrate that it has a compact globular structure distinctly different from the extended, multidomain envelope-protein structures of related pesti- and flaviviruses (Khan et al., 2014; Kong et al., 2013). This suggests that HCV may follow a different fusion process involving E1, which has been predicted to contain a putative fusion peptide (Flint et al., 1999; Takikawa et al., 2000). In addition, recent studies suggest E1 may have a critical role in modulating HCV binding to receptors (Douam et al., 2014)..
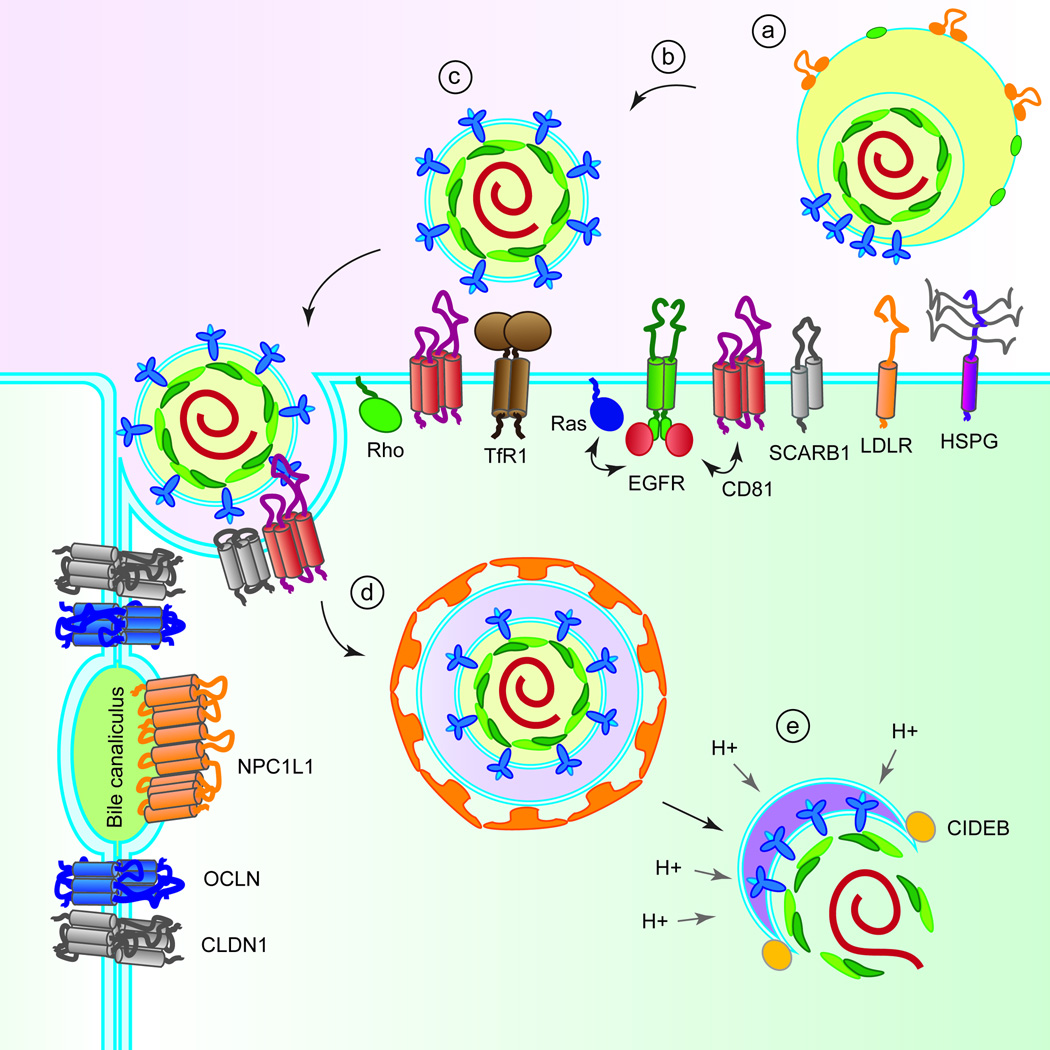
Figure 1.
Putative mechanism of HCV entry. (a) Lipoviral HCV particles attach to HSPG, LDLR and SCARB1. (b) Interaction with SCARB1 induces conformational changes in HCV E2, leading to binding of E2 to CD81. (c) CD81 binding activates signalling pathways via EGFR, Ras and Rho GTPases, triggering lateral membrane diffusion of HCV. The exact roles of TfR1, NPC1L1, CLDN1 and OCLN are not defined but they are engaged in steps post viral attachment and HCV binding to CD81. (d) Interaction of the HCV E2-CD81/CLDN1-complex initiates clathrin-mediated endocytosis. (e) Promoted by an interaction with CIDEB, the viral envelope and the endosomal membrane undergo fusion in a low pH environment.
Based on EM studies, it is thought that HCV particles have a diameter of 40–75 nm. In contrast to related enveloped RNA viruses, HCV particles contain electron-dense cores and lack obvious symmetry or surface features (reviewed in (Lindenbach and Rice, 2013)). The morphology and low buoyant density of HCV particles is due to their interaction with host lipoproteins. Biochemical analyses of HCV particles derived from patient sera revealed an association with the lipoprotein components apolipoprotein A–I (apoA-I), apoB-48, apoB-100, apoC-I and apoE (reviewed in (Bartenschlager et al., 2011)). Similarly, cell culture-derived HCV particles interact with apoE and apoC-I and have a high lipid and cholesterol content. These observations have given rise to the concept of an HCV lipoviroparticle (LVP), which can engage high and low-density lipoprotein receptors during viral uptake into hepatocytes. In addition, association with lipoproteins may also help the virus escape neutralizing antibodies.
HCV ENTRY FACTORS
Numerous host factors have been implicated in the uptake of HCV into human hepatocytes, including glycosaminoglycans (GAGs) present on heparan sulfate proteoglycans (HSPGs), low-density-lipoprotein receptor (LDLR) (Agnello et al., 1999), CD81 (Pileri et al., 1998), scavenger receptor class B member 1 (SCARB1) (Scarselli et al., 2002), the tight junction proteins claudin-1 (CLDN1) (Evans et al., 2007) and occludin (OCLN) (Liu et al., 2009; Ploss et al., 2009), the receptor tyrosine kinases epidermal growth factor receptor (EGFR) and ephrin receptor A2 (EphA2) (Lupberger et al., 2011), the cholesterol transporter Niemann-Pick C1-like 1 (NPC1L1) (Sainz et al., 2012), transferring receptor 1 (TfR1) (Martin and Uprichard, 2013) and the cell death-inducing DFFA-like effector b (CIDEB) (Wu et al., 2014) (Figure 1).
In addition, the C-type lectins liver/lymph node-specific intercellular adhesion molecule-3-grabbing integrin (L-SIGN, (Cormier et al., 2004)), expressed on liver sinusoidal endothelial cells, and the dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN), present on dendritic cells and liver resident macrophages, can bind to HCV E2 (Gardner et al., 2003; Lozach et al., 2003; Pohlmann et al., 2003). Conceivably, C-type lectins on sinusoidal cells may be able to capture HCV in the liver, but it is unclear whether this can lead to productive infection of hepatocytes in trans or whether sequestered HCV is subjected to lysosomal degradation.
CD81 was the first molecule to be identified as an HCV receptor, directly binding the soluble form of HCV E2 (Pileri et al., 1998). CD81 is a ubiquitously expressed tetraspanin, containing a small extracellular and a large extracellular loop (LEL). The LEL seems to play an important role in mediating the interaction of CD81 with E2, as soluble recombinant human CD81 LEL can inhibit infections with cell culture-derived HCV (HCVcc) and lentiviral pseudoparticles bearing HCV glycoproteins (HCVpp) (Zhang et al., 2004). Time course studies with CD81-specific antibodies, which block HCV infection, suggest that CD81 mediates a post-binding event in HCV entry (Bertaux and Dragic, 2006). Consistent with this, the CD81-binding region of E2 is obstructed by its hypervariable region 1 (HVR1), suggesting that native E2 undergoes a conformational change before its engagement with CD81 (Bankwitz et al., 2010).
SCARB1 was identified as an HCV receptor by its ability to bind recombinant HCV E2 (Scarselli et al., 2002). SCARB1 is highly expressed in the liver and can bind highdensity lipoproteins (HDLs), very-low-density lipoproteins (VLDLs) and oxidized forms of LDLs, mediating the uptake of cholesterol from these lipoproteins by hepatocytes. SCARB1 is a glycoprotein with cytoplasmic N- and C-terminal domains separated by a large extracellular domain. Recent studies show that SCARB1 has multiple stepwise roles during HCV entry. First, the initial attachment of HCV particles to SCARB1 is mediated by virus-associated lipoproteins, such as apoE, which is present on HCV LVPs. Second, the lipid transfer function of SCARB1 may facilitate the exposure of CD81 binding sites on HCV E2 and transfer of the virus particle to CD81.. Finally, direct interactions between SCARB1 and the HVR1 of E2 enhance infectivity of the particle post-attachment (Bartosch et al., 2005). Together, these studies suggest that HCV entry results from a complex interplay between lipoproteins, SCARB1, and HCV envelope glycoproteins.
CLDN1 is a member of the claudin family of tight junction proteins and is expressed in many epithelial tissues, including the liver. CLDN1 was identified as an essential HCV entry factor through expression cloning of a cDNA that made HEK293 cells permissive to HCVpp entry (Evans et al., 2007). Recent studies suggest that HCV E1 but not E2 may directly interact with CLDN1 (Douam et al., 2014). During the HCV entry process, CLDN1 interacts with CD81, forming a complex critical for virus internalization. Other members of the claudin family, specifically CLDN6 and CLDN9, can also mediate HCV entry (Zheng et al., 2007). However, usage of different claudins appears to be dependent on the HCV isolate (Haid et al., 2014) but less in human hepatocytes (Fofana et al., 2013).
Using a similar cDNA expression cloning strategy, another tight junction protein, OCLN, was identified as a missing host factor needed to facilitate HCV uptake into mouse and hamster cell lines expressing human CD81, CLDN1 and OCLN (Ploss et al., 2009). Gain- and loss-of-function studies also established the critical role of OCLN for HCV entry into human cells. OCLN is a four-transmembrane protein expressed in the tight junctions of polarized cells. To date, there is no experimental evidence demonstrating a direct interaction of OCLN with the HCV glycoproteins. Although the exact role of OCLN in viral uptake is not completely understood, it appears to function at a post-attachment step in HCV entry (Sourisseau et al., 2013b).
NPC1L1 is a 13-transmembrane-domain cell surface cholesterol-sensing receptor expressed on the apical surface of intestinal enterocytes and human hepatocytes. NPC1L1 is responsible for cellular cholesterol absorption and whole-body cholesterol homeostasis. Ezetimibe is a 2-azetidinone–class drug that has been shown to be a direct inhibitor of NPC1L1 internalization. An ezetimibe-mediated inhibition assay suggested that NPC1L1 reduces HCV entry at a post-binding, pre-fusion step (Sainz et al., 2012). Given their high expression in hepatocytes, NPC1L1, CLDN1 and SCARB1 may largely define the hepatotropism of HCV at the level of entry.
TfR1 is ubiquitously expressed in all tissues and is the main receptor for cellular iron uptake via clathrin-mediated endocytosis. Interestingly, HCV infection can decrease TfR1 expression, and TfR1 has been found to bind HCV viral particles. Time-of-addition experiments using anti-TfR1 antibodies indicate that TfR1 may play a role in HCV uptake after the virus engages CD81 (Martin and Uprichard, 2013). However, further studies are needed to clarify the exact roles of TfR1 contributing to HCV entry.
Receptor tyrosine kinases EGFR and EphA2 were identified as HCV entry factors using a functional RNAi kinase screen. The compounds erlotinib (a specific inhibitor of EGFR) and dasatinib (a specific inhibitor of EphA2) can block HCV entry into hepatocytes (Lupberger et al., 2011). It should be noted that EGFR does not directly interact with the HCV particle, but EGFR-dependent signaling promotes the formation of CD81-CLDN1 complexes, which is required for HCV entry (Lindenbach and Rice, 2013)..
Recently, CIDEB has been implicated in HCV uptake (Wu et al., 2014). CIDEB is highly expressed in the liver and is one of the members of the cell death-inducing DFFAlike effector (CIDE) protein family. Based on an infection time-course assay and the fact that CIDEB affects uptake of HCVcc but not HCVpp, it appears that CIDEB functions in a late step of HCV entry, possibly to facilitate membrane fusion.. However, the precise mechanism by which HCV utilizes CIDEB for entry is not known.
PUTATIVE MECHANISM OF HCV ENTRY
HCV entry is a highly orchestrated process requiring tight spatial control and temporal engagement of cellular co-factors (Figure 1). GAGs, LDLR and SCARB1 are thought to mediate the initial attachment of the lipoprotein-associated HCV particles to the surface of hepatocytes. The interaction with SCARB1 is believed to aid in the dissociation of HCV-associated lipoproteins from viral particles and induce conformational changes in the HCV E2 glycoprotein, exposing the CD81 interaction domains. It is reported that TfR1 is engaged at a post-CD81 step in HCV entry, but the precise role is not known (Martin and Uprichard, 2013). Likewise, OCLN functions at a post-binding stage prior to endosomal acidification (Sourisseau et al., 2013b).
Engagement of E2 with CD81 subsequently activates signalling pathways via EGFR and H-Ras, as well as through Rho GTPases, which trigger lateral membrane diffusion of HCV–CD81 complexes to the tight junction (Lupberger et al., 2011; Zona et al., 2013). The interaction of HCV-CD81 complexed with CLDN1 is thought to induce clathrin-mediated endocytosis. Following uptake, HCV–co-receptor complexes are trafficked to early endosomes for HCV fusion. The interaction of E2 with CD81 induces fusion between the viral envelope and the endosomal membrane in a low pH environment. Following fusion, HCV genomic RNA is released from the viral nucleocapsid into the cytosol, where it is translated into viral proteins, initiating the subsequent steps in the HCV life cycle.
The entry mechanisms described above are thought to mostly apply to cell-free HCV particle transmission. In addition, HCV can spread to neighboring hepatocytes via direct cell-to-cell transmission. In contrast to cell-free HCV particle transmission, HCV may be shielded from neutralizing antibodies during cell-to-cell transmission, aiding the establishment of persistent infection. Recent studies have also demonstrated that cell-to-cell transmission may be a major transmission route for antiviral-resistant strains, thus highlighting the importance of studying this process further. HCV cell-to-cell transmission is dependent on numerous host factors also required during cell-free entry, including SCARBI, CLDN1, OCLN, EGFR, EphA2, and NPC1L1 and possibly CD81. In contrast, the yery-low-density lipoprotein (VLDL) pathway, which is required for the secretion of cell-free infectious virus, does not seem to be required for cell-to-cell transmission. In addition, TfR1 is involved in HCV cell-free entry but has a less prominent role in cell-to-cell transmission (Barretto et al., 2014).
CONTRIBUTION OF HCV ENTRY TO HOST TROPISM
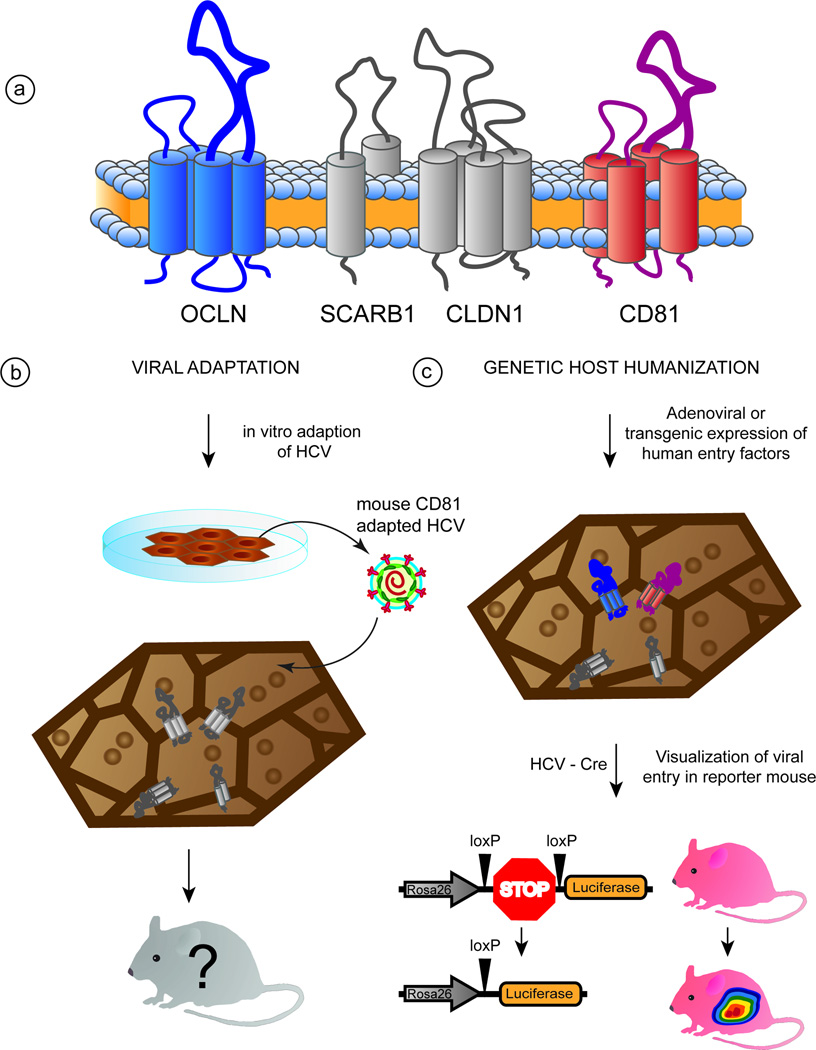
While HCV can establish chronic infections in humans, chimpanzees and, to a limited extent, tree shrews, other species, including rodents and smaller, non-human primates, appear to be resistant to infection. Although the basis of this highly restricted species tropism is not completely understood, several studies have demonstrated that it is partially defined at the level of viral entry. Previous reports showed that CD81 from various non-human primate species could facilitate HCVpp uptake into human HepG2 hepatoma cells, which lack endogenous expression of CD81(Flint et al., 2006). However, mice remained insusceptible to HCV infection, even upon transgenic expression of human CD81 (Masciopinto et al., 2002), indicating that other human-specific factors might be required to facilitate HCV entry into murine cells. Subsequent work revealed that human OCLN rendered mouse cells permissive for HCV and therefore was the missing factor needed to overcome the HCV species barrier at the level of entry (Ploss et al., 2009). While CD81, SCARB1, CLDN1 and OCLN were ultimately shown to all be required for uptake into mouse cells, CD81 and OCLN constitute the minimal set of human-specific factors needed for viral uptake into mouse or hamster cells (Ploss et al., 2009). As was the case for CD81, primate orthologues of OCLN functioned like the human counterpart, whereas canine, hamster, mouse and rat OCLN had intermediate to low activities, and guinea pig OCLN was completely nonfunctional (Michta et al., 2010). These observations can partially be explained by differences between the second extracellular loops of both CD81 and OCLN from humans and non-permissive species (Flint et al., 2006; Michta et al., 2010). In contrast, the residues within the first extracellular loop of CLDN1 needed for HCV uptake (Evans et al., 2007) are conserved between mice and humans, explaining the observation that mouse CLDN1 can support viral entry. Although it was originally demonstrated that human but not mouse SCARB1 could bind soluble HCV E2 (Scarselli et al., 2002) both in vitro (Ploss et al., 2009) and in vivo (Dorner et al., 2013; Dorner et al., 2011) infection assays demonstrated that mouse and human SCARB1 were equally functional. These findings suggest that HCV E2 binding may not be a reliable proxy for functional entry assays.
To date, the varying susceptibility of non-human cells to HCV entry is largely accounted for by differences at critical residues within orthologues of CD81, SCARB1, CLDN1 and OCLN or by insufficient expression of these proteins. However, the contribution of other human entry factors to the species tropism of HCV has yet to be determined. The human and murine orthologues of the entry factors described in this paper share a high degree of sequence similarity (Table 1). However, differences in individual residues can drastically affect viral uptake efficiency. NPC1L1 and TfR1, which are both highly expressed in human hepatocytes, were shown to be important for HCV uptake in human cells. In contrast, mouse fibroblasts expressing human CD81 and OCLN but not NPC1L1 and TfR1 can still support HCV entry (Vogt et al., 2013). Furthermore, while the murine ortholog of NPC1L1 is abundantly expressed only in the gut and not in the liver, HCV is still capable of entering into hepatocytes of mice modified to express a combination of human CD81 and OCLN (Dorner et al., 2013; Dorner et al., 2011). This suggests some level of redundancy for NPC1L1 in HCV uptake in mouse cells. It is conceivable but remains to be experimentally tested if HCV entry into mouse hepatocytes would possibly become more efficient if human TfR1 and NPC1L1 were ectopically co-expressed in the mouse liver along with human CD81 and OCLN.
Table 1. Gene.
Expression of HCV entry factors in humans and mice.
| Gene | Tissue Expression | Liver expression |
Protein Identity (%) |
|||
|---|---|---|---|---|---|---|
| Human | Mouse | Human | Mouse | Human | Mouse | |
| LDLR | Ldlr | Ubiquitous | Ubiquitous | + | + | 79 |
| SCARB1 | Scarb1 | Adrenal tissue, placenta, liver | Adrenal gland,ovary, liver | + | + | 80 |
| CD81 | Cd81 | Ubiquitous | Ubiquitous | + | + | 92 |
| CLDN1 | Cldn1 | Ubiquitous | Most abundantly skin,liver | + | + | 91 |
| CLDN6 | Cldn6 | Ubiquitous | Placenta | + | Low | 87 |
| CLDN9 | Cldn9 | Heart, muscle | Pituitary, pancreas | + | Low | 97 |
| OCLN | Ocln | Tight junctions | Tight junctions | + | + | 89 |
| EGFR | Egfr | Placenta, prostate, liver | Liver,cornea,osteobla st | + | + | 88 |
| EphA2 | Epha2 | Epithelial, endothelial cells | Epithelial, endothelial cells | + | + | 92 |
| TfR1 | Tfr1 | Early erythroid cells, liver | Bone marrow, placenta, liver | Low | Low | 77 |
| NPC1L1 | Npc1l1 | Ubiquitous | Small intestine | + | − | 77 |
| CIDEB | Cideb | Liver, small intestine | Liver, small intestine, kidney | + | + | 86 |
The observation that CD81 and OCLN comprise the minimal set of human factors required to facilitate HCV uptake into rodent cell lines hinted how a mouse supporting HCV entry would need to be engineered. Indeed, it was shown that adenoviral delivery or transgenic expression of human CD81 and OCLN is sufficient to allow HCV infection of fully immunocompetent inbred mice (Dorner et al., 2013; Dorner et al., 2011). Using intergenotypic virus chimeras expressing CRE recombinase to activate a cellular reporter in vivo, HCV glycoprotein-mediated uptake of diverse HCV genotypes can be conveniently quantified by bioluminescent imaging (Figure 2). This platform has been used to genetically dissect HCV entry in vivo and to evaluate pre-clinically passive and active immunization strategies (Dorner et al., 2011). When crossed to immunodeficient backgrounds, HCV entry factor transgenic mice support the entire HCV life-cycle (Dorner et al., 2013), suggesting that immune-mediated restrictions exist post-entry and are not entirely insurmountable.
Figure 2.
Approaches to overcome species-specific blocks of HCV entry and in vivo visualization of HCV entry. (a) The second extracellular loops of OCLN (blue) and CD81 (red) are responsible for the species tropism of viral entry. (b) Genetic viral and host adaptation approaches. Mouse CD81-adapted HCV can enter mouse cell lines expressing mouse orthologues of CD81 and OCLN. (c) Transgenic or adenoviral expression of human HCV entry factors facilitates HCV entry into mouse cells in vitro and in vivo. HCV uptake can be visualized in vivo using a cellularly encoded reporter that is activated by HCV expressing Cre-recombinase (HCV-Cre).
Complementary to the genetics host adaptation approach to facilitate viral uptake into hepatocytes of non-permissive species, it was shown that the HCV envelope proteins can be adapted to use entry factor orthologues from other species. Taking advantage of the high mutational plasticity of HCV, the viral envelope proteins were adapted to utilize mouse CD81 more efficiently (Bitzegeio et al., 2010). Three adaptive mutations in HCV E1 and E2 were shown to be responsible for the observed gain of function phenotype. Interestingly, the resulting mouse CD81-adapted HCV was also capable of engaging mouse OCLN more efficiently and had a lower dependency on SCARB1. Although it remains to be demonstrated whether mouse CD81-adapted HCV is capable of infecting mouse hepatocytes in vivo, this study provides an important proof-of-concept for the feasibility of adapting HCV to engage orthologues of essential host factors from other species. Interestingly, recent studies suggest that these mutations also facilitate more efficient HCV uptake in stem cell-derived hepatocyte-like cells from pigtailed macaques (Sourisseau et al., 2013a). A suitable explanation for the broadened host range of this particular virus could be conformational changes in the virion envelope as a result of the three described mutations in E1 and E2. However, some caution is warranted as it remains to be tested whether mouse CD81-adapted HCV faithfully recapitulates viral entry.
PERSPECTIVES
HCV entry is a highly orchestrated process influenced not only by cell- but also virion-associated host factors. Many of the essential host factors required for viral uptake into human cells have been identified, yielding potential targets for therapeutic interventions and providing insights into the determinants governing HCV tissue and host tropism. Defining the minimal set of human specific factors required for rendering rodent cells permissive to HCV uptake has facilitated the construction of mice with inheritable susceptibility to HCV infection. These genetically humanized animals show promise as a tractable small animal model for HCV, but additional refinements are still needed to increase the robustness of the system.
A major challenge remains to define the exact roles of the numerous entry factors in this complex and finely controlled process of viral entry. HCV pseudoparticles have undoubtedly played a critical role in the discovery of some of the entry factors and are also particularly useful for analyzing HCV entry into cells that do not readily support HCV RNA replication. However, given the distinct biophysical properties of HCV particles, it remains important to use LVPs produced in cells with an intact VLDL pathway. Many host factors involved in viral uptake have been identified both in hepatoma cells and in non-hepatic cells. However, neither faithfully recapitulates the polarized cellular architecture of hepatocytes in the liver. Primary cell culture systems using human adult primary hepatocytes, fetal hepatoblasts or even stem cell-derived hepatocyte-like cells have been developed as a more physiologically relevant platform to study viral uptake. Integrating available in vivo platforms amenable to genetic manipulations will ultimately allow us to dissect HCV uptake and other parts of the viral life-cycle in the 3D context of the liver.
ACKNOWLEDGEMENTS
The authors thank Jenna Gaska and Benjamin Winer for edits and critical discussion of the manuscript. Work in the laboratory is in part supported by grants from the National Institutes of Health (2 R01 AI079031-05A1, 1 R01 AI107301-01, 1 R56 AI106005-01), the Walter Reed Army Institute of Research, the Bill and Melinda Gates Foundation and the Grand Challenge Program of Princeton University. M.v.S. is a recipient of a fellowship from the German Research Foundation (Deutsche Forschungsgemeinschaft). We apologize to all colleagues whose work could not be cited due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci U S A. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankwitz D, Steinmann E, Bitzegeio J, Ciesek S, Friesland M, Herrmann E, Zeisel MB, Baumert TF, Keck ZY, Foung SK, et al. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J Virol. 2010;84:5751–5763. doi: 10.1128/JVI.02200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N, Sainz B, Jr, Hussain S, Uprichard SL. Determining the involvement and therapeutic implications of host cellular factors in hepatitis C virus cell-to-cell spread. J Virol. 2014;88:5050–5061. doi: 10.1128/JVI.03241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Penin F, Lohmann V, Andre P. Assembly of infectious hepatitis C virus particles. Trends in microbiology. 2011;19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Bartosch B, Verney G, Dreux M, Donot P, Morice Y, Penin F, Pawlotsky JM, Lavillette D, Cosset FL. An Interplay between Hypervariable Region 1 of the Hepatitis C Virus E2 Glycoprotein, the Scavenger Receptor BI, High-Density Lipoprotein Promotes both Enhancement of Infection and Protection against Neutralizing Antibodies. J Virol. 2005;79:8217–8229. doi: 10.1128/JVI.79.13.8217-8229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertaux C, Dragic T. Different domains of CD81 mediate distinct stages of hepatitis C virus pseudoparticle entry. J Virol. 2006;80:4940–4948. doi: 10.1128/JVI.80.10.4940-4948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitzegeio J, Bankwitz D, Hueging K, Haid S, Brohm C, Zeisel MB, Herrmann E, Iken M, Ott M, Baumert TF, et al. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog. 2010;6:e1000978. doi: 10.1371/journal.ppat.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier EG, Durso RJ, Tsamis F, Boussemart L, Manix C, Olson WC, Gardner JP, Dragic T. L-SIGN (CD209L) and DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis C virus. Proc Natl Acad Sci U S A. 2004;101:14067–14072. doi: 10.1073/pnas.0405695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M, Horwitz JA, Donovan BM, Labitt RN, Budell WC, Friling T, Vogt A, Catanese MT, Satoh T, Kawai T, et al. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature. 2013;501:237–241. doi: 10.1038/nature12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douam F, Dao Thi VL, Maurin G, Fresquet J, Mompelat D, Zeisel MB, Baumert TF, Cosset FL, Lavillette D. Critical interaction between E1 and E2 glycoproteins determines binding and fusion properties of hepatitis C virus during cell entry. Hepatology. 2014;59:776–788. doi: 10.1002/hep.26733. [DOI] [PubMed] [Google Scholar]
- Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- Flint M, Thomas JM, Maidens CM, Shotton C, Levy S, Barclay WS, McKeating JA. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J Virol. 1999;73:6782–6790. doi: 10.1128/jvi.73.8.6782-6790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint M, von Hahn T, Zhang J, Farquhar M, Jones CT, Balfe P, Rice CM, McKeating JA. Diverse CD81 proteins support hepatitis C virus infection. J Virol. 2006;80:11331–11342. doi: 10.1128/JVI.00104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fofana I, Zona L, Thumann C, Heydmann L, Durand SC, Lupberger J, Blum HE, Pessaux P, Gondeau C, Reynolds GM, et al. Functional analysis of claudin-6 and claudin-9 as entry factors for hepatitis C virus infection of human hepatocytes by using monoclonal antibodies. J Virol. 2013;87:10405–10410. doi: 10.1128/JVI.01691-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JP, Durso RJ, Arrigale RR, Donovan GP, Maddon PJ, Dragic T, Olson WC. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc Natl Acad Sci U S A. 2003;100:4498–4503. doi: 10.1073/pnas.0831128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haid S, Grethe C, Dill MT, Heim M, Kaderali L, Pietschmann T. Isolate-dependent use of claudins for cell entry by hepatitis C virus. Hepatology. 2014;59:24–34. doi: 10.1002/hep.26567. [DOI] [PubMed] [Google Scholar]
- Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Bohannon CD, Jacob J, et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature. 2014;509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey T, d'Alayer J, Kikuti CM, Saulnier A, Damier-Piolle L, Petitpas I, Johansson DX, Tawar RG, Baron B, Robert B, et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 2010;6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol. 2013;11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yang W, Shen L, Turner JR, Coyne CB, Wang T. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83:2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozach PY, Lortat-Jacob H, de Lacroix de Lavalette A, Staropoli I, Foung S, Amara A, Houles C, Fieschi F, Schwartz O, Virelizier JL, et al. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. The Journal of biological chemistry. 2003;278:20358–20366. doi: 10.1074/jbc.M301284200. [DOI] [PubMed] [Google Scholar]
- Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Uprichard SL. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci U S A. 2013;110:10777–10782. doi: 10.1073/pnas.1301764110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masciopinto F, Freer G, Burgio VL, Levy S, Galli-Stampino L, Bendinelli M, Houghton M, Abrignani S, Uematsu Y. Expression of human CD81 in transgenic mice does not confer susceptibility to hepatitis C virus infection. Virology. 2002;304:187–196. doi: 10.1006/viro.2002.1631. [DOI] [PubMed] [Google Scholar]
- Michta ML, Hopcraft SE, Narbus CM, Kratovac Z, Israelow B, Sourisseau M, Evans MJ. Species-specific regions of occludin required by hepatitis C virus for cell entry. J Virol. 2010;84:11696–11708. doi: 10.1128/JVI.01555-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Paul D, Madan V, Bartenschlager R. Hepatitis C virus RNA replication and assembly: Living on the fat of the land. Cell Host & Microbe. 2014 doi: 10.1016/j.chom.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmann S, Zhang J, Baribaud F, Chen Z, Leslie GJ, Lin G, Granelli-Piperno A, Doms RW, Rice CM, McKeating JA. Hepatitis C Virus glycoproteins Interact with DC-SIGN and DC-SIGNR. J Virol. 2003;77:4070–4080. doi: 10.1128/JVI.77.7.4070-4080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B, Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, et al. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012;18:281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO Journal. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourisseau M, Goldman O, He W, Gori JL, Kiem HP, Gouon-Evans V, Evans MJ. Hepatic cells derived from induced pluripotent stem cells of pigtail macaques support hepatitis C virus infection. Gastroenterology. 2013a;145:966–969. doi: 10.1053/j.gastro.2013.07.026. e967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourisseau M, Michta ML, Zony C, Israelow B, Hopcraft SE, Narbus CM, Parra Martin A, Evans MJ. Temporal analysis of hepatitis C virus cell entry with occludin directed blocking antibodies. PLoS Pathog. 2013b;9:e1003244. doi: 10.1371/journal.ppat.1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takikawa S, Ishii K, Aizaki H, Suzuki T, Asakura H, Matsuura Y, Miyamura T. Cell fusion activity of hepatitis C virus envelope proteins. J Virol. 2000;74:5066–5074. doi: 10.1128/jvi.74.11.5066-5074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt A, Scull MA, Friling T, Horwitz JA, Donovan BM, Dorner M, Gerold G, Labitt RN, Rice CM, Ploss A. Recapitulation of the hepatitis C virus life-cycle in engineered murine cell lines. Virology. 2013;444:1–11. doi: 10.1016/j.virol.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Lee EM, Hammack C, Robotham JM, Basu M, Lang J, Brinton MA, Tang H. Cell death-inducing DFFA-like effector b is required for hepatitis C virus entry into hepatocytes. J Virol. 2014;88:8433–8444. doi: 10.1128/JVI.00081-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Randall G, Higginbottom A, Monk P, Rice CM, McKeating JA. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J Virol. 2004;78:1448–1455. doi: 10.1128/JVI.78.3.1448-1455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng A, Yuan F, Li Y, Zhu F, Hou P, Li J, Song X, Ding M, Deng H. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol. 2007;81:12465–12471. doi: 10.1128/JVI.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zona L, Lupberger J, Sidahmed-Adrar N, Thumann C, Harris HJ, Barnes A, Florentin J, Tawar RG, Xiao F, Turek M, et al. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe. 2013;13:302–313. doi: 10.1016/j.chom.2013.02.006. [DOI] [PubMed] [Google Scholar]