Abstract
Proper expression of the dev operon is important for normal development of Myxococcus xanthus. When starved, these bacteria coordinate their gliding movements to build mounds that become fruiting bodies as some cells differentiate into spores. Mutations in the devTRS genes impair sporulation. Expression of the operon occurs within nascent fruiting bodies and depends in part on C signaling. Here, we report that expression of the dev operon, like that of several other C-signal-dependent genes, is subject to combinatorial control by the transcription factors MrpC2 and FruA. A DNA fragment upstream of the dev promoter was bound by a protein in an extract containing MrpC2, protecting the region spanning positions −77 to −54. Mutations in this region impaired binding of purified MrpC2 and abolished developmental expression of reporter fusions. The association of MrpC2 and/or its longer form, MrpC, with the dev promoter region depended on FruA in vivo, based on chromatin immunoprecipitation analysis, and purified FruA appeared to bind cooperatively with MrpC2 to DNA just upstream of the dev promoter in vitro. We conclude that cooperative binding of the two proteins to this promoter-proximal site is crucial for dev expression. 5′ deletion analysis implied a second upstream positive regulatory site, which corresponded to a site of weak cooperative binding of MrpC2 and FruA and boosted dev expression 24 h into development. This site is unique among the C-signal-dependent genes studied so far. Deletion of this site in the M. xanthus chromosome did not impair sporulation under laboratory conditions.
INTRODUCTION
Myxococcus xanthus is a Gram-negative bacterium that undergoes multicellular development (1). Upon starvation on a solid surface, cells coordinate their movements to build mounds that contain thousands of cells. Within these nascent fruiting bodies, some cells differentiate from rods to ovoid spores. Other cells lyse during the developmental process or remain outside fruiting bodies as peripheral rods. The spores remain viable during prolonged starvation and resist environmental insults. Under favorable conditions, spores germinate, producing rod-shaped cells capable of growth and division.
The developmental process of M. xanthus provides an attractive model to study signaling and gene regulatory mechanisms (1). Here, we focus on regulation of the dev operon in response to extracellular C signaling. The dev locus was identified by two transposon insertions that created reporter fusions induced during development (2, 3). Expression from the fusions was reduced in a csgA mutant incapable of C signaling (4). The transposon insertions in the dev locus prevented darkening of nascent fruiting bodies and reduced sporulation >100-fold (3, 5). The sporulation defect can be accounted for by the observation that a dev mutant fails to express the Ω7536 locus (6), the site of the exo operon, whose products are necessary for spore formation (7). How the products of the dev operon regulate the expression of the exo operon is unknown. The dev operon includes eight genes plus at least two repeats of the downstream CRISPR (clustered regularly interspaced short palindromic repeats) (8). The eight genes include a short upstream gene, three genes implicated in development (devTRS) (3, 8, 9), and four cas (CRISPR-associated) genes that typically form small interfering RNAs to inhibit the expression of plasmid and bacteriophage genes (10).
Expression of the dev operon and other C-signal-dependent genes is localized to nascent fruiting bodies (11). Numerous end-to-end contacts between cells in the outer domain of a nascent fruiting body have been proposed to facilitate C signaling, triggering the expression of the dev operon and then the exo operon and leading to spore formation (6, 11–14). Efficient C signaling requires that cells move into alignment (15–17), and sporulation demands a higher level of C signaling than does nascent fruiting body formation (18–20). Hence, regulation of the dev operon in response to C signaling appears to be a key step that couples the movement of cells into nascent fruiting bodies with differentiation into spores.
The mechanism of C signaling is partly understood. It involves CsgA (21), a 25-kDa protein (p25) that appears to associate with the inner membrane during cell growth (22) but with the outer membrane during development (23). Considerable evidence supports a model in which p25 is cleaved to a 17-kDa form (p17) that appears to be the C signal (19, 23–25). Starvation initiates a RelA-dependent proteolytic cascade in which FtsHD-dependent degradation of PopD (where FtsHD represents FtsH important for development) releases PopC for secretion (26) and PopC-dependent cleavage of p25 forms p17 at the cell surface (27). However, a p17 receptor has not been identified. The similarity of CsgA to short-chain alcohol dehydrogenases and evidence that NAD(P)+ binding is essential for activity suggested that CsgA is an enzyme that generates a C signal (28). A related protein, SocA, which, when overexpressed, can substitute for CsgA (29), oxidized lipid substrates, but the products were unstable and failed to rescue the development of a csgA mutant (30). If CsgA is an enzyme, its substrate and product remain to be identified. Interestingly, the outer membrane porin Oar is required for C signaling and has been proposed to be the channel for the export of the C signal (31).
Cellular responses to C signaling involve FruA, which is similar to the response regulators of two-component signal transduction systems (32, 33). The putative histidine protein kinase that would phosphorylate FruA in response to C signaling has not been identified. Some evidence suggests that FruA might function as a pseudoresponse regulator without being phosphorylated (34). In any case, FruA has a C-terminal DNA-binding domain that was shown previously to bind to promoter regions of FruA-dependent genes in vitro (35–37). Mapping of the dev operon promoter revealed an upstream sequence centered at position −91 with similarity to a site recognized by the FruA DNA-binding domain (8). It was shown that the region between positions −101 and −75 upstream of the dev promoter can be bound by the FruA DNA-binding domain in vitro and that mutations in this region impair dev expression in vivo (38). Hence, FruA appears to activate dev transcription by binding to an upstream cis-regulatory element.
The expression of several C-signal-dependent genes has been shown to be under the combinatorial control of FruA and MrpC2 (34, 39–41). MrpC2 is an N-terminally truncated form of MrpC (42), a protein similar to transcription factors in the cyclic AMP receptor protein (CRP) family (43). A protein serine/threonine kinase cascade negatively regulates mrpC expression during growth, apparently by phosphorylating MrpC (44), which reduces its DNA-binding activity (45) and perhaps its ability to positively autoregulate (43). MrpC2 may be produced by utilization of an alternative translation start codon, or it may be derived from MrpC by proteolytic cleavage. MrpC2 appears to activate the transcription of fruA (42). MrpC2 appeared to have higher DNA-binding activity than MrpC (45), but the two forms of the protein had similar DNA-binding activity for the fruA promoter region in a recent study (62). Since MrpC2 cannot be phosphorylated, it might play an important role in escaping negative regulation by the protein kinase cascade during development (45).
MrpC2 binds cooperatively with FruA to the promoter regions of several C-signal-dependent genes in vitro (34, 39–41). Mutations in the cooperative binding sites affect promoter activity in vivo. In each case, binding of the two proteins cooperatively to a site located immediately upstream of the promoter appears to activate transcription. However, insertion mutations in these genes cause, at most, mild defects in fruiting body formation (34, 39–41). In contrast, insertions in the dev operon impair sporulation dramatically (3, 5). To determine whether a key developmental operon is regulated by a mechanism similar to that of other C-signal-dependent genes, we undertook the investigation reported here. We show that MrpC2 and FruA appear to bind cooperatively just upstream of the dev promoter. Mutations in the MrpC2-binding site abolished promoter activity, as did mutations in the FruA-binding site described previously (38). In addition, we identify a second cis-regulatory element located further upstream of the dev promoter, where MrpC2 and FruA appear to bind with weak cooperativity and enhance the expression of the dev operon after 24 h of development.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
Strains and plasmids used in this study are listed in Table 1. To construct plasmids containing different segments of dev DNA, pPV1004 was used as the template for PCR with upstream primers containing a HindIII site and downstream primer LK1344 containing a BamHI site (see Table 2 for primers). Each PCR product was cloned by using pCR2.1-TOPO and Escherichia coli strain TOP10 as described by the manufacturer. To create mutations, pPV695 served as the template, using the QuikChange site-directed mutagenesis kit (Stratagene) and pairs of primers. Cloned PCR products and genes subjected to site-directed mutagenesis were verified by DNA sequencing. To construct pREG1727 derivatives, each derivative of pCR2.1-TOPO or pPV695 was digested with HindIII and BamHI, and the DNA insert was gel purified and cloned into HindIII-BamHI-digested pREG1727 by using E. coli strain DH5α and standard methods (46). The plasmid used to delete the distal upstream site of MrpC2 and FruA binding was constructed by using overlap extension PCR (47) to synthesize a DNA fragment of ∼1 kbp spanning the region but lacking the site. In the first step, primers SS1 and SS2 and primers SS3 and SS4 were used in separate PCRs with pPV1515 as the template. In the second step, the two PCR products served as the template with primers SS1RII and SS4RII. The resulting ∼1-kbp fragment was digested with EcoRI and BamHI, and the DNA insert was gel purified and cloned into EcoRI-BamHI-digested pBJ113 to create pSS3. The insert was verified by DNA sequencing.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm with DE3, a λ prophage carrying the T7 RNA polymerase gene | Novagen |
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 59 |
| SMhisMrpC2 | BL21(DE3) containing pET16b/His10-MrpC2 | 34 |
| SMFruAhis | BL21(DE3) containing pET11km/FruA-His6 | 34 |
| TOP10 | λ− F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80 lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| M. xanthus | ||
| DK1622 | Wild type | 60 |
| DK5285 | fruA::Tn5 lac Ω4491 | 5 |
| DK11209 | ΔdevS | 8 |
| MAC1 | ΔdevS attB::pAC01 (pREG1727 with a 696-bp HindIII-BamHI fragment from pTB1) | This study |
| MAC2A | ΔdevS attB::pAC02A (pREG1727 with a 696-bp HindIII-BamHI fragment from pTB2A) | This study |
| MAC2B | ΔdevS attB::pAC02B (pREG1727 with a 696-bp HindIII-BamHI fragment from pTB2B) | This study |
| MAC5 | ΔdevS attB::pAC05 (pREG1727 with a 696-bp HindIII-BamHI fragment from pTB5) | This study |
| MLK299 | ΔdevS attB::pLK0299 (pREG1727 with a 300-bp HindIII-BamHI fragment from pAC2) | This study |
| MLK324 | ΔdevS attB::pLK0324 (pREG1727 with a 325-bp HindIII-BamHI fragment from pAC1) | This study |
| MPV35N | ΔdevS attB::pPV035N (pREG1727 with a 1,515-bp HindIII-BamHI fragment from pPV35N) | 8 |
| MPV184 | ΔdevS attB::pPV0184 (pREG1727 with a 185-bp HindIII-BamHI fragment from pPV184) | 8 |
| MPV391 | ΔdevS attB::pPV0391 (pREG1727 with a 392-bp HindIII-BamHI fragment from pPV391) | This study |
| MPV605 | ΔdevS attB::pPV0605 (pREG1727 with a 606-bp HindIII-BamHI fragment from pPV695) | This study |
| MPV695 | ΔdevS attB::pPV0695 (pREG1727 with a 696-bp HindIII-BamHI fragment from pPV695) | 38 |
| MPV1004 | ΔdevS attB::pPV01004 (pREG1727 with a 1,005-bp HindIII-BamHI fragment from pPV1004) | 8 |
| MSS1 | A distal upstream site of MrpC2 and FruA binding at positions −254 to −228 was deleted using pSS3 | This study |
| MTB3 | ΔdevS attB::pTB03 (pREG1727 with a 696-bp HindIII-BamHI fragment from pTB3) | This study |
| MTB4 | ΔdevS attB::pTB04 (pREG1727 with a 696-bp HindIII-BamHI fragment from pTB4) | This study |
| MTB6 | ΔdevS attB::pTB06 (pREG1727 with a 696-bp HindIII-BamHI fragment from pTB6) | This study |
| Plasmids | ||
| pAC1 | pCR2.1-TOPO with dev DNA spanning positions −254 to +71 generated by PCR using LK2489 and LK1344 | This study |
| pAC2 | pCR2.1-TOPO with dev DNA spanning positions −229 to +71 generated by PCR using LK2497 and LK1344 | This study |
| pBJ113 | Apr Kmr galK | 11 |
| pCR2.1-TOPO | Apr Kmr lacZα | Invitrogen |
| pET16b/His10-MrpC2 | pET16b with a gene encoding His10-MrpC2 under the control of a T7 RNA polymerase promoter | 45 |
| pET11km/FruA-His6 | pET11km with a gene encoding FruA-His6 under the control of a T7 RNA polymerase promoter | S. Inouye |
| pPV35N | pPV1515 with a TTGACG-to-GGTCAT mutation spanning positions −38 to −33 | 8 |
| pPV184 | pCR2.1-TOPO with dev DNA spanning positions −114 to +71 | 8 |
| pPV391 | pCR2.1-TOPO with dev DNA spanning positions −321 to +71 generated by PCR using Cover 7 and LK1344 | This study |
| pPV605 | pCR2.1-TOPO with dev DNA spanning positions −535 to +71 generated by PCR using Cover 6 and LK1344 | This study |
| pPV695 | pCR2.1-TOPO with dev DNA spanning positions −114 to +581 | 38 |
| pPV1004 | pCR2.1-TOPO with dev DNA spanning positions −934 to +71 generated by PCR using Cover 4 and LK1344 | 8 |
| pPV1515 | pCR2.1-TOPO with dev DNA spanning positions −934 to +581 | 8 |
| pREG1727 | Apr Kmr P1-inc attP′lacZ | 61 |
| pSS3 | pBJ113 with a ∼1-kbp region of M. xanthus DNA spanning a deletion of positions −254 to −228 upstream of the dev transcriptional start site | This study |
| pTB1 | pPV695 with M1 mutation using LK2381 and LK2382 | This study |
| pTB2A | pPV695 with M2A mutation using LK2491 and LK2492 | This study |
| pTB2B | pPV695 with M2B mutation using LK2493 and LK2494 | This study |
| pTB3 | pPV695 with M3 mutation using LK2385 and LK2386 | This study |
| pTB4 | pPV695 with M3 mutation using LK2387 and LK2388 | This study |
| pTB5 | pPV695 with M3 mutation using LK2389 and LK2390 | This study |
| pTB6 | pPV695 with M3 mutation using LK2391 and LK2392 | This study |
Where possible, the plasmid description is given in parentheses after the strain description.
TABLE 2.
Primers used in this study
| Primer | Sequencea | Descriptionb or reference |
|---|---|---|
| Cover 6 | CCCAAGCTTAAGGAGAGACCTTCTTGTGGCG | Positions −535 forward with HindIII site |
| Cover 7 | CCCAAGCTTCGCCGTTGGCTCGGATGCGGAC | Positions −321 forward with HindIII site |
| LK1298 | CGAGGACCAGCGCTCGTC | Position −19 reverse |
| LK1331 | CCAAGCTTGCTCACGTTGCAGACGGGG | Position −114 forward with HindIII site |
| LK1344 | GGCGGATCCACCTCGTACTTCGACTTCCG | Position +71 reverse with BamHI site |
| LK1861 | CCTTGAGCGCGATGGAGATA | 56 |
| LK1862 | CTCGGCGGCCTCATCGAC | 56 |
| LK2381 | GGGGCAATACAGTTGGACAAGTGAGACGATTG | M1 mutation |
| LK2382 | CAATCGTCTCACTTGTCCAACTGTATTGCCCC | M1 mutation |
| LK2385 | GGTTCAAAGTGATCATCTTGCATGCATCAG | M3 mutation |
| LK2386 | CTGATGCATGCAAGATGATCACTTTGAACC | M3 mutation |
| LK2387 | CAAAGTGAGACGAGGTACGTCATCAGCGAACG | M4 mutation |
| LK2388 | CGTTCGCTGATGACGTACCTCGTCTCACTTTG | M4 mutation |
| LK2389 | GACGATTGCATGACGACTAGAACGTTGACGAG | M5 mutation |
| LK2390 | CTCGTCAACGTTCTAGTCGTCATGCAATCGTC | M5 mutation |
| LK2391 | GCATGCATCAGCTCCATTTGACGAGCGCTG | M6 mutation |
| LK2392 | CAGCGCTCGTCAAATGGAGCTGATGCATGC | M6 mutation |
| LK2475 | GTCTGCAACGTGAGCGCG | Position −100 reverse |
| LK2476 | CACGACGGCGCCGTTGG | Position −329 forward |
| LK2481 | CGTGCTGCCTGTTCCTGG | Position −202 forward |
| LK2482 | GCCGCTGGCCCAGGAAC | Position −176 reverse |
| LK2489 | CAAGCTTCGCAACGAGCTGCGCACG | Position −254 forward with HindIII site |
| LK2491 | CAATACAGGGTTCACCTTGAGACGATTGC | M2A mutation |
| LK2492 | GCAATCGTCTCAAGGTGAACCCTGTATTG | M2A mutation |
| LK2493 | CAATACAGGGTTCACCTGTCGACGATTGCATGCATCAGCG | M2B mutation |
| LK2494 | CGCTGATGCATGCAATCGTCGACAGGTGAACCCTGTATTG | M2B mutation |
| LK2497 | CAAGCTTCACATCTCATCCTCTATGCC | Position −229 forward with HindIII site |
| SS1 | GCGAATTCGCGCCAACGCCTCGCAGC | Position +292 reverse with EcoRI site |
| SS2 | GCGCGCGCATCTCATCCTCTATGCC | Position −227 forward with overlap |
| SS3 | TGAGATGCGCGCGCCGGAGTTGCTC | Position −255 reverse with overlap |
| SS4 | GCGGATCCGGACTGGACGCCTCACTTC | Position −762 forward with BamHI site |
| SS1RII | GCGAATTCCACAGCAACAAGGACTTCCAAG | Position +272 reverse with EcoRI site |
| SS4RII | ATAGGATCCGTTGATTGACCTGCGGGGCAAG | Position −639 forward with BamHI site |
| ΔSite 1F | CACTTCCGCGCCGTGCTCTAC | Position −371 forward |
| ΔSite 1R | GAACCCTGTATTGCCCCAAAGG | Position −70 reverse |
Restriction sites are underlined. Mutations are italicized. Overlaps are in boldface type.
The position is relative to the start site of dev transcription, and the orientation (forward or reverse) is relative to the direction of dev transcription.
Growth and development.
E. coli DH5α strains containing plasmids were grown at 37°C in Luria-Bertani (LB) medium containing 50 μg/ml of either ampicillin or kanamycin sulfate. M. xanthus strains were grown at 32°C in CTT medium (1% Casitone, 10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4-K2HPO4, 8 mM MgSO4 [final pH 7.6]) (48) or on CTT agar (1.5%) plates. When required, 40 μg of kanamycin sulfate per ml was added. Fruiting body development was performed on TPM agar (1.5%) plates (10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4-K2HPO4, 8 mM MgSO4 [final pH 7.6]), as described previously (2), except for experiments in which spores were measured, in which case a submerged culture (49) was used, as described previously (50). Measurement of sonication-resistant spores, and of mature spores that were heat and sonication resistant and capable of germination, was performed as described previously (50).
ChIP.
M. xanthus strains DK1622 and DK5285 were used for chromatin immunoprecipitation (ChIP) as described previously (34, 37, 41). The primers used for PCR of the dev promoter region were LK1298 and LK1331, and those used for PCR of the rpoC coding region were LK1861 and LK1862.
Preparation of proteins.
DNA-binding proteins partially purified (ammonium sulfate [AS] fraction) from 12-h-developing M. xanthus DZF1 cells, as described previously (42), were a gift from Sumiko Inouye. Recombinant His10-MrpC2 (34) and FruA-His6 (45) were expressed in E. coli BL21(DE3) and purified as described previously.
EMSAs and footprinting.
For electrophoretic mobility shift assays (EMSAs), 32P-labeled DNA fragments from the dev promoter region were generated by PCR using the wild-type or mutant plasmid as the template and primers labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England BioLabs). The DNA fragment was purified after 15% PAGE (46). EMSAs were performed as described previously (37), except that binding reaction mixtures were incubated at 25°C for 15 min. For footprinting, a 32P-labeled DNA probe was synthesized by PCR after labeling primer LK1331 as described above, and probes were purified after 5% PAGE (46). Footprint analysis of gel slices using 1,10-phenanthroline-copper ion was performed as described previously (51).
Construction of M. xanthus strains and determination of lacZ expression during development.
Strains containing a plasmid integrated at the Mx8 phage attachment site, attB, were constructed by electroporation (52). Transformants were selected on CTT agar plates containing kanamycin sulfate and screened on TPM agar plates containing 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside per ml, in order to avoid rare transformants with unusual developmental lacZ expression (53). Three transformants were typically chosen for further analysis, and β-galactosidase activity was measured as described previously (2). Strains containing a deletion of the distal upstream site of cooperative MrpC2 and FruA binding were constructed by electroporating pSS3 into M. xanthus, selecting a kanamycin-resistant transformant, growing the transformant without selection, and then selecting galactose-resistant clones as described previously (54). Three clones in which the wild-type allele had been replaced with the deletion by homologous recombination were identified by colony PCR with primers ΔSite 1F and ΔSite 1R and verified by sequencing of the PCR product. The three clones were collectively designated MSS1.
RESULTS
Binding of a protein in a fraction from developing M. xanthus cells to the dev promoter region.
A preparation containing DNA-binding proteins that was partially purified from 12-h-developing M. xanthus cells was shown previously to contain MrpC2 (34, 42). The preparation, called the AS fraction, was incubated with a 32P-labeled DNA probe spanning positions −114 to −19 of the dev promoter region, and EMSAs revealed a single shifted complex (Fig. 1A). This finding suggested that a protein in the AS fraction binds to the dev promoter region. To characterize the position of binding, the protein-DNA complex and the unbound DNA probe (as a control) were excised from the gel and subjected to footprinting by treatment with 1,10-phenanthroline-copper followed by electrophoresis on a DNA sequencing gel. The complex showed protection from positions −77 to −54 (Fig. 1B). Interestingly, this region is adjacent to a site spanning positions −101 to −75 that is bound by FruA (38). Since FruA and MrpC2 bind cooperatively to adjacent sites in the promoter regions of several M. xanthus genes (34, 39–41), we hypothesized that MrpC2 in the AS fraction was binding to the region spanning positions −77 to −54.
FIG 1.
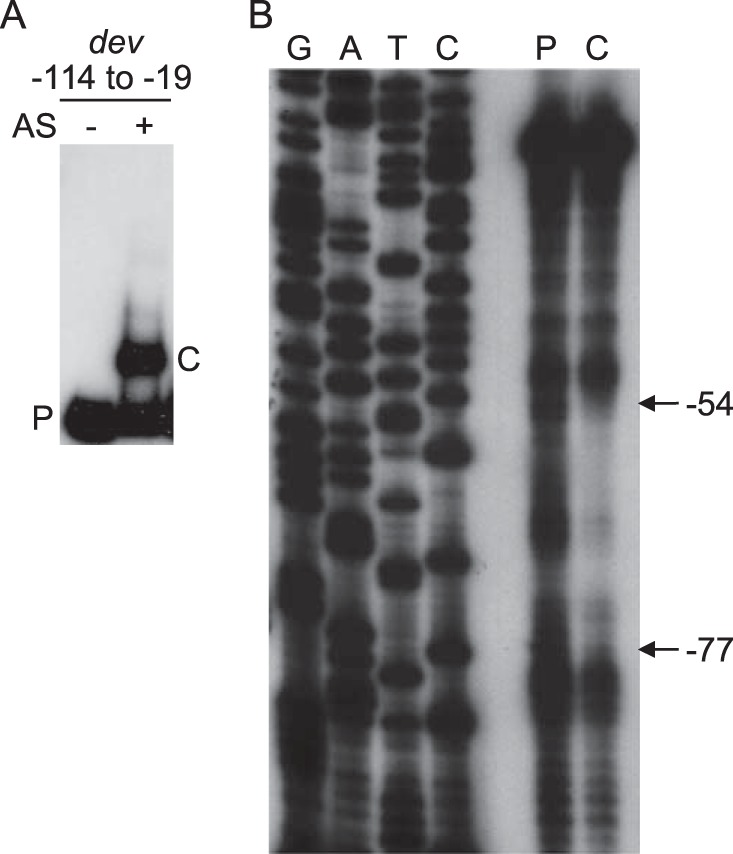
Binding of a protein in a fraction from developing M. xanthus cells to the dev promoter region. (A) EMSAs. A 32P-labeled DNA probe (2 nM) spanning positions −114 to −19 of the dev promoter region was incubated alone or with proteins in the AS fraction (0.7 μg/μl) and subjected to EMSAs. The unbound probe (P) and the shifted complex (C) produced by the AS fraction are indicated. (B) Footprinting. The unbound probe (P) and the shifted complex (C) from a gel like that shown in panel A were subjected to footprint analysis with 1,10-phenanthroline-copper. Lanes G, A, T, and C show sequence ladders generated by primer LK1331.
MrpC and FruA associate with the dev promoter region in vivo.
To test whether MrpC and FruA associate with the dev promoter region in developing M. xanthus cells, we performed ChIP assays with polyclonal antibodies to MrpC, which also recognize MrpC2 (45), and polyclonal antibodies to FruA. Cells that had been developing for 18 h, when both transcription factors are known to be expressed, were subjected to ChIP with affinity-purified anti-MrpC immunoglobulin G (IgG) (or IgG from a nonimmunized rabbit as a control) or anti-FruA serum (or preimmune serum from the same rabbit as a control). The ChIP DNA was analyzed by PCR with primers designed to amplify the dev promoter region (positions −119 to −19) or, as a control, the rpoC coding region (positions +1780 to +1905) (37). PCR analysis showed enrichment of the dev promoter region by ChIP with anti-MrpC compared to control IgG (Fig. 2A, lanes 5 and 6) and with anti-FruA compared to control preimmune serum (Fig. 2B, lanes 5 and 6). Similar results were observed in two additional experiments. As expected, PCR analysis with primers designed to amplify the rpoC coding region showed no enrichment by ChIP with anti-MrpC or anti-FruA compared to the controls. We conclude that MrpC and/or MrpC2 and FruA associate with the dev promoter 18 h into development.
FIG 2.
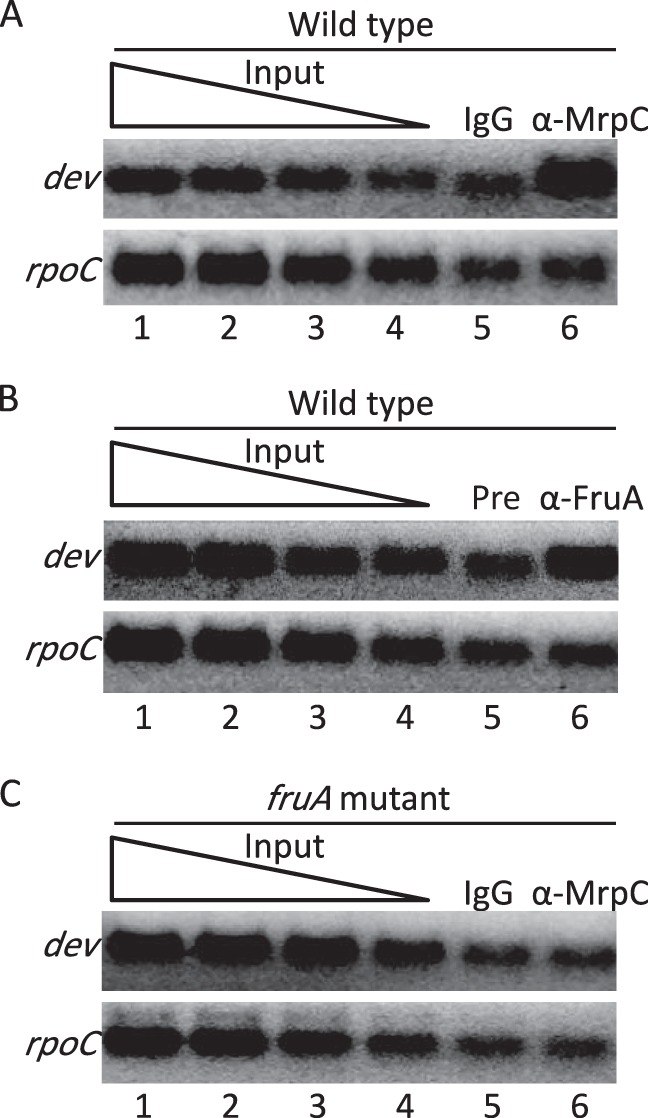
Association of MrpC and FruA with the dev promoter region in developing M. xanthus cells. Eighteen hours into development, cells were subjected to ChIP analysis. DNA was amplified with primers for the dev promoter region spanning positions −114 to −19 or for the rpoC coding region spanning positions +1780 to +1905 as a control. A 2-fold dilution series of input DNA (lanes 1 to 4 in each panel) was used as a template in parallel PCRs to show that the PCR conditions allow detection of differences in DNA concentrations for each primer set. (A) Wild-type strain DK1622 with affinity-purified IgG antibodies against MrpC (α-MrpC) or, as a control, with total IgG (IgG) from nonimmunized rabbits. (B) Wild-type strain DK1622 with antiserum against FruA (α-FruA) or, as a control, preimmune antiserum (Pre). (C) fruA mutant strain DK5285 with antibodies as described above for panel A.
Association of MrpC and/or MrpC2 with the dev promoter region requires FruA.
It was shown previously that the association of MrpC and/or MrpC2 with the fmgA (34), fmgBC (40), and fmgE (41) promoter regions depends on FruA. This appears to be due to cooperative binding of the two proteins just upstream of the promoters, based on in vitro DNA-binding studies. To test whether the association of MrpC and/or MrpC2 with the dev promoter region requires FruA, we performed ChIP-PCR analysis of a fruA mutant at 18 h of development. PCR analysis showed no enrichment of the dev promoter region by ChIP with anti-MrpC compared to control IgG (Fig. 2C, lanes 5 and 6). Similar results were observed in two additional experiments. Likewise, there was no enrichment of the rpoC coding region compared to the control, as expected. Taken together, the results of our ChIP-PCR analysis show that the association of MrpC and/or MrpC2 with the dev promoter region 18 h into development requires FruA. These results suggest that FruA binds cooperatively with MrpC and/or MrpC2 to the dev promoter region.
MrpC2 and FruA bind to the dev promoter region in vitro.
Purified His10-MrpC2 and FruA-His6 were tested for binding to a 32P-labeled DNA fragment spanning positions −114 to −19 of the dev promoter region in EMSAs. His10-MrpC2 produced two shifted complexes, one more abundant and faster migrating than the other (Fig. 3, lane 2). The slower-migrating complex presumably has more than one His10-MrpC2 bound. FruA-His6 produced a single shifted complex (Fig. 3, lane 3). Together, the two proteins produced a single shifted complex that migrated more slowly and was more abundant (lane 4) than the complex produced by FruA-His6 alone or the faster-migrating complex produced by His10-MrpC2 alone. This pattern was shown previously to be indicative of cooperative binding (34).
FIG 3.
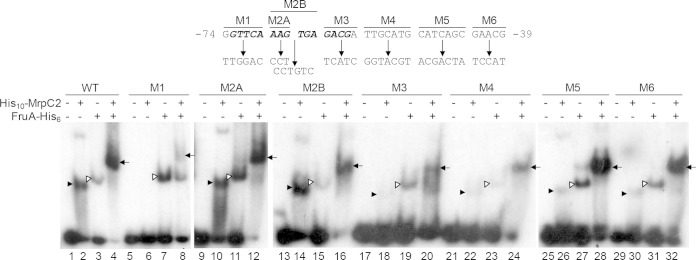
Binding of MrpC2 and FruA to the dev promoter region. (A) Mutations made in the dev promoter region between positions −74 and −39. The name of each multiple-base-pair transversion is shown at the top. Boldface italic type indicates the complement of the sequence CGTCN8AAC, which best matches the MrpC2 consensus binding sequence TGTYN8RAC (62). (B) EMSAs with 32P-labeled dev DNA (2 nM) spanning positions −114 to −19 and His10-MrpC2 (0.13 μM), FruA-His6 (3 μM), or both His10-MrpC2 (0.13 μM) and FruA-His6 (3 μM), as indicated. The probe DNA had the wild-type (WT) sequence or the indicated mutation. Filled arrowheads point to shifted complexes produced by His10-MrpC2 alone, open arrowheads point to complexes produced by FruA-His6 alone, and arrows point to complexes indicative of cooperative binding of the two proteins.
To test the hypothesis mentioned above, that MrpC2 binds to the region spanning positions −77 to −54 of the dev promoter region, we made several mutations between positions −74 and −39 (Fig. 3) and measured the effects on the binding of His10-MrpC2 and FruA-His6 in EMSAs. The mutations were multiple-base-pair transversions that dramatically altered the sequence. Mutations M1 and M3 have changes at the ends of a sequence (CGTCN8AAC, on the opposite strand of the one shown in bold italic type in Fig. 3) very similar to a consensus sequence for MrpC binding (TGTYN8RAC). The consensus sequence was identified by bioinformatic analysis of sequences bound by MrpC, as determined by chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) of M. xanthus that had formed nascent fruiting bodies (62). Mutations M2A and M2B have changes in the middle (i.e., the nonspecific N8 part) of the sequence matching the consensus. Mutation M1 eliminated detectable binding of His10-MrpC2 (Fig. 3, lane 6), and mutations M3 (lane 18), M4 (lane 22), M5 (lane 26), and M6 (lane 30) greatly diminished binding. These results demonstrate that His10-MrpC2 binds to the sequence between positions −74 and −39 in the dev promoter region. Mutations M2A (Fig. 3, lane 10) and M2B (lane 14) did not impair His10-MrpC2 binding, nor did mutations M2A (lane 12) and M2B (lane 16) impair the formation of the slower-migrating complex indicative of cooperative binding of His10-MrpC2 and FruA-His6. In contrast, mutations M1 (Fig. 3, lane 8) and M3 (lane 20) reduced the formation of the slower-migrating complex. The ability of mutation M1 to form a small amount of this complex suggests that His10-MrpC2 can bind in the presence of FruA-His6, even though His10-MrpC2 alone did not bind detectably (Fig. 3, lane 6). Interestingly, mutations M4 (Fig. 3, lane 24), M5 (lane 28), and M6 (lane 32) formed considerable amounts of the slower-migrating complex, leaving little of the probe unbound and very little of the probe bound by just one of the proteins, as if cooperative binding occurred but not to the extent observed for the wild-type DNA sequence (Fig. 3, lane 4). Altogether, our results indicate that FruA binds cooperatively with MrpC and/or MrpC2 to sequences located immediately upstream of the dev promoter, with MrpC and/or MrpC2 being proximal to the promoter and MrpC2 very likely accounting for the binding of a protein in the AS fraction to the dev promoter region (Fig. 1).
Mutations in the MrpC2-binding site abolish dev expression.
To test the effects of the mutations described above on dev expression in vivo, we constructed M. xanthus strains with mutant promoter regions transcriptionally fused to the E. coli lacZ reporter gene and measured β-galactosidase specific activity during development. As shown previously (38), the wild-type dev promoter region spanning positions −114 to +581 fused to lacZ and integrated ectopically in a devS-null mutant (to relieve negative autoregulation) resulted in increasing expression by 12 h into development that reached ∼1,000 units by 48 h (Fig. 4). Likewise, the corresponding promoter region with the M2B mutation, which did not impair His10-MrpC2 binding in vitro (Fig. 3, lane 14), resulted in similar developmental lacZ expression (Fig. 4). In contrast, the M1, M3, M4, M5, and M6 mutations that impaired His10-MrpC2 binding in vitro (Fig. 3) abolished dev expression in vivo (Fig. 4). Although these mutations appeared to allow cooperative binding of His10-MrpC2 and FruA-His6 in vitro to some extent (Fig. 3), the binding in developing cells was apparently insufficient to activate transcription from the dev promoter (Fig. 4). As a negative control, a mutation in the −35 region of the dev promoter abolished dev expression (Fig. 4), as expected (8). We conclude that high-affinity, cooperative binding of MrpC and/or MrpC2 with FruA to a site immediately upstream of the dev promoter is crucial to activate transcription of the dev operon during development.
FIG 4.
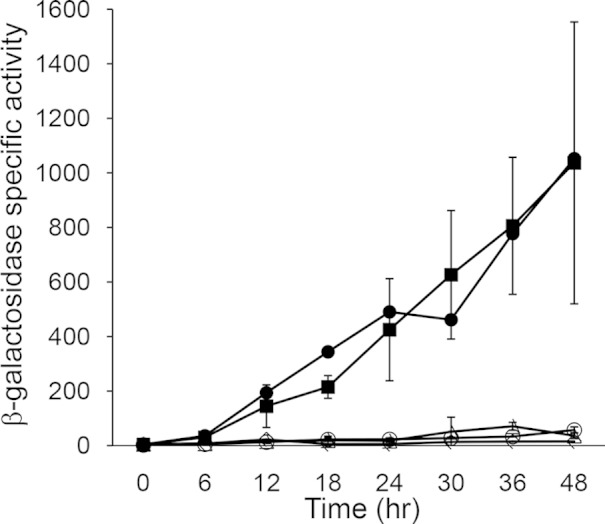
Effects of mutations in the MrpC2-binding site on developmental dev-lacZ expression. β-Galactosidase specific activity during development was measured for lacZ fused to dev spanning positions −114 to +581. Fragments were inserted into pREG1727 to create lacZ fusions, and the resulting plasmids were transformed into M. xanthus ΔdevS mutant strain DK11209. Expression from transformants bearing the wild-type promoter (●) or the M2B (■), M5 (△), or M3 (○) mutant promoter region was measured, as was that from the M1, M4, and M6 mutant promoter regions, which is not shown since it was indistinguishable from the negative control. As a negative control, expression from a fragment spanning positions −934 to +581 with a mutation in the −35 region of the dev promoter that abolishes activity (◇) was likewise measured. The units of activity are nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. For the mutant promoter regions, points show average values for three independent transformants, and error bars represent 1 standard deviation from the mean. Expression from the wild-type promoter and the negative control was measured once and was consistent with results reported previously (8, 38).
Localization of a second positive cis-regulatory element.
A series of 5′ deletions with upstream ends located at position −934, −535, or −114 and the same downstream end at position +834 fused to lacZ showed a graded loss of dev promoter activity in previous work (8). These results implied that additional positive cis-regulatory elements lie upstream of position −114. However, other results suggested that upstream and downstream regulatory elements functionally interact (8). To minimize such interactions and to further localize upstream regulatory elements, a series of 5′ deletions with upstream ends at position −934, −535, −321, or −114 and the same downstream end at position +71 fused to lacZ was constructed. These dev-lacZ fusions were integrated ectopically into M. xanthus with a devS-null mutation, and developmental lacZ expression was measured. Surprisingly, the fusions with upstream ends at position −535 or −321 exhibited higher expression levels during development than the fusion with its upstream end at position −934 (Fig. 5). This suggests that a negative regulatory element lies between positions −934 and −535. The expression level from the fusion with its upstream end at position −114 was lower than the expression level from the fusions with upstream ends at position −934 or −535 (Fig. 5), as observed previously for fusions with their downstream end at position +834 (8). The much higher expression level from the fusion with its upstream end at position −321 than from the fusion with its upstream end at position −114 (Fig. 5) suggests that a strong positive cis-regulatory element lies between positions −321 and −114. We further characterized this regulatory element, as described below.
FIG 5.
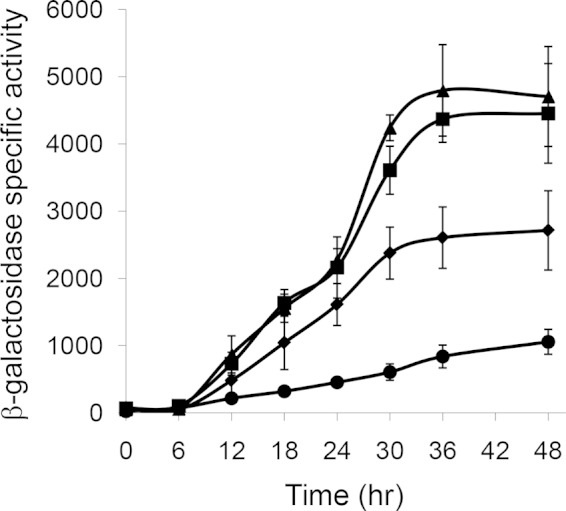
Effects of 5′ deletions on developmental dev-lacZ expression. Fragments spanning positions −934 (◆), −535 (■), −321 (▲), or −114 (●) to +71 were inserted into pREG1727, the resulting plasmids were transformed into M. xanthus ΔdevS mutant strain DK11209, and β-galactosidase specific activity during development was determined for three independent transformants. The units of activity are nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. Points show average values, and error bars represent 1 standard deviation from the mean.
MrpC2 and FruA bind to a second site upstream of the dev promoter.
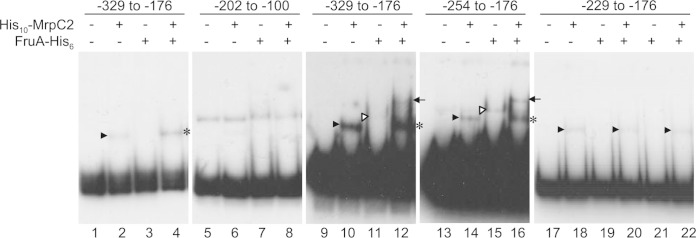
Since MrpC and/or MrpC2 binds cooperatively with FruA immediately upstream of the dev promoter to activate transcription, as described above, and since these transcription factors bind in the vicinity of other promoters in various arrangements to regulate their transcription (34, 39–41), we tested whether His10-MrpC2 and FruA-His6 bind to two overlapping DNA probes spanning positions −329 to −176 and positions −202 to −100 upstream of the dev promoter, which covers the region corresponding to the second positive cis-regulatory element mentioned above plus a few base pairs on each end in case a binding site is close to an end. His10-MrpC2 produced a single shifted complex with the probe spanning positions −329 to −176 (Fig. 6, lane 2) but not with the probe spanning positions −202 to −100 (Fig. 6, compare lanes 5 and 6) (note that this probe was contaminated with a species migrating more slowly, but there was no difference when His10-MrpC2 was added). FruA-His6 did not bind detectably to either probe (Fig. 6, lanes 3 and 7); however, in combination with His10-MrpC2, FruA-His6 appeared to enhance the formation of a shifted complex with the probe spanning positions −329 to −176, although little if any of the complex migrated more slowly than the complex produced by His10-MrpC2 alone (Fig. 6, compare lanes 2 and 4). Because these results suggested that there might be weak cooperative binding of the two proteins to the probe spanning positions −329 to −176, we repeated the experiment with slightly more concentrated probe and slightly less FruA-His6. Under these conditions, FruA-His6 alone produced a shifted complex that was barely detectable (Fig. 6, lane 11), and in combination with His10-MrpC2, the two proteins produced two shifted complexes (Fig. 6, lane 12). The amount of the two shifted complexes was slightly larger than expected from the binding of the two proteins individually, indicative of weak cooperative binding. A probe spanning positions −254 to −176 exhibited a similar pattern of binding (Fig. 6, lanes 13 to 16). In contrast, a probe spanning residues −229 to −176 showed no detectable binding by FruA-His6 and no indication of cooperative binding, although it was bound by His10-MrpC2 (Fig. 6, lanes 17 to 22). Because we were unable to prepare this probe at a high concentration, we cannot be certain whether FruA-His6 alone is able to bind, but it did not enhance complex formation in combination with His10-MrpC2, as we had observed with the probe spanning positions −329 to −176 at a lower concentration (Fig. 6, compare lanes 4 and 22). Therefore, we conclude that DNA between positions −254 and −229 in the dev promoter region is necessary for the weak cooperative binding of FruA and MrpC2 to a site located between positions −254 and −176, with MrpC2 binding being proximal to the promoter.
FIG 6.
Binding of MrpC2 and FruA to a second site in the dev promoter region. Shown are results of EMSAs with 32P-labeled dev DNA spanning the indicated positions with His10-MrpC2 (1 μM) and/or FruA-His6, as indicated. The probe DNA concentration was 2 nM (lanes 1 to 8 and 17 to 22) or 6 nM (lanes 9 to 12). The FruA-His6 concentration was 3 μM (lanes 3, 4, 7, 8, and 19 to 22) or 1.5 μM (lanes 11, 12, 15, and 16). Filled arrowheads point to shifted complexes produced by His10-MrpC2 alone, open arrowheads point to complexes produced by FruA-His6 alone, arrows point to complexes indicative of cooperative binding of the two proteins, and asterisks indicate FruA-His6-dependent enhancement of complexes that comigrate with those produced by His10-MrpC2 alone.
The second site of MrpC2 and FruA binding is important for dev expression.
To test whether DNA between positions −254 and −229 contributes to dev transcription in vivo, 5′ deletions with upstream ends at position −254 or −229 and the same downstream end at position +71 fused to lacZ were constructed and integrated ectopically into M. xanthus with a devS-null mutation, and developmental lacZ expression was measured. The expression level from the fusion with its upstream end at position −254 was only slightly lower than the expression level from the fusion with its upstream end at position −321, which was measured as a control (Fig. 7). Interestingly, the expression level from the fusion with its upstream end at position −229 increased similarly during the first 24 h of development, then stopped increasing until 36 h, and finally rose slightly by 48 h, reaching about half the level observed for the fusion with its upstream end at position −254. As a control, expression from the fusion with its upstream end at position −114 was measured in parallel. The expression level increased less rapidly during the first 24 h but continued to rise later in development, reaching a level by 48 h slightly below that observed for the fusion with its upstream end at position −229. Taken together, these results demonstrate that the region between positions −254 and −229 of the dev promoter region, which is necessary for weak cooperative binding of MrpC2 and FruA in vitro (Fig. 6), is important for the normal pattern of dev expression in vivo, particularly after 24 h of development (Fig. 7). Additional cis-regulatory elements between positions −114 and −229 appear to boost dev expression during the first 24 h of development, and elements between positions −254 and −321 appear to boost expression later during development.
FIG 7.
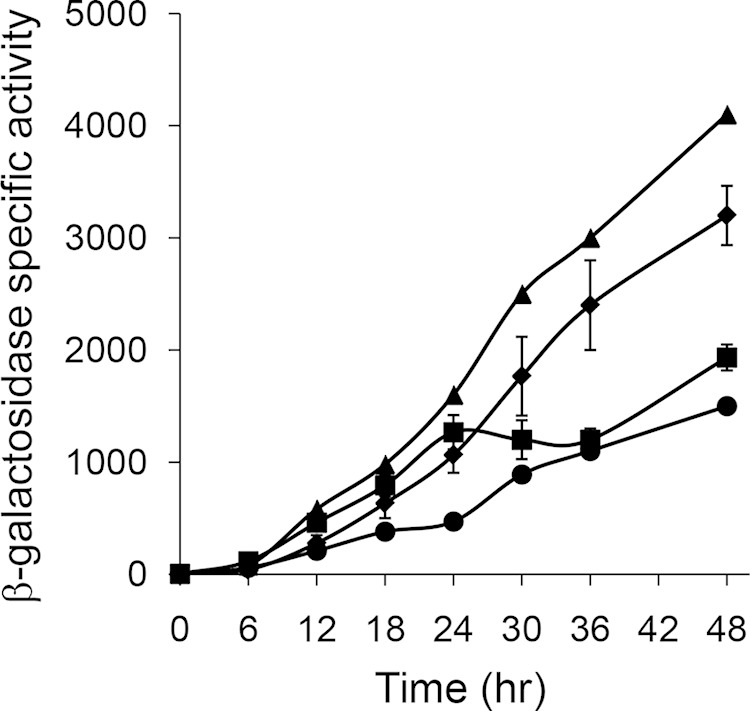
Effects of 5′ deletions to position −254 or −229 on developmental dev-lacZ expression. Fragments spanning positions −321 (▲), −254 (◆), −229 (■), or −114 (●) to +71 were inserted into pREG1727, the resulting plasmids were transformed into M. xanthus ΔdevS mutant strain DK11209, and β-galactosidase specific activity during development was determined. The units of activity are nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. For the 5′ deletions to positions −254 and −229, points show average values for three independent transformants, and error bars represent 1 standard deviation from the mean. Expression from the 5′ deletions to positions −321 and −114 was measured once and was consistent with the results shown in Fig. 5.
Deletion of the distal upstream site of MrpC2 and FruA binding does not impair spore formation.
The period between 24 and 30 h poststarvation is crucial for commitment to sporulation during M. xanthus development (50). Since the distal upstream site of MrpC2 and FruA binding, located at least in part between positions −254 and −229, boosts dev expression at 24 to 36 h of development (Fig. 7), we hypothesized that this cis-regulatory element is important for commitment to sporulation. To test this hypothesis, we deleted DNA between positions −254 and −228 in the M. xanthus chromosome, which also creates a small in-frame deletion in the upstream gene MXAN_7267. We derived three such strains from wild-type strain DK1622 and measured sonication-resistant spore formation at 24, 30, 36, 48, and 72 h poststarvation, with strain DK1622 as a control. We observed no significant difference between the deletion and control strains, and the results were very similar to those described previously for strain DK1622 (50). There was also no significant difference in the number of mature spores (i.e., heat- and sonication-resistant spores capable of germination) at 72 h poststarvation. We conclude that the distal upstream site of MrpC2 and FruA binding is not required for sporulation. Together with our results shown in Fig. 7, this suggests that a boost in dev expression at 24 to 36 h is not important for commitment to sporulation, at least under laboratory conditions.
DISCUSSION
Our results show that the regulation of a key developmental operon is under combinatorial control of MrpC2 and FruA, which appear to bind cooperatively to two sites upstream of the dev promoter and contribute to positive regulation. Below, we discuss the implications of our findings related to this novel arrangement of MrpC2- and FruA-binding sites.
Cooperative binding of MrpC2 and FruA immediately upstream of the dev promoter is crucial for expression.
A common theme among several C-signal-dependent genes studied previously is that MrpC2 and FruA bind cooperatively just upstream of the promoter (34, 39–41) (Fig. 8). At this location, either protein may bind proximal to the promoter. Immediately upstream of the dev promoter, MrpC2 and FruA bind cooperatively, with MrpC2 being proximal to the promoter, as in the fmgA promoter region. Cooperative binding to the dev promoter region is supported by our in vivo and in vitro data. We found that the association of MrpC and/or MrpC2 with the dev promoter region in vivo requires FruA by performing ChIP-PCR analysis of a fruA mutant (Fig. 2C). We did not perform a similar analysis of an mrpC mutant since fruA is not transcribed in such a mutant (42). Our evidence for the cooperative binding of MrpC2 and FruA in vitro is based on the migration and abundance of shifted complexes produced in EMSAs (Fig. 3, lanes 2 to 4). This pattern of shifted complexes was initially observed for the fmgA promoter region, and cooperative binding was demonstrated by using DNase I footprinting (34). A similar pattern of shifted complexes in EMSAs provided evidence for cooperative binding just upstream of the fmgBC, fmgD, and fmgE promoters (39–41). In these cases, FruA binds proximal to the promoter (Fig. 8).
FIG 8.
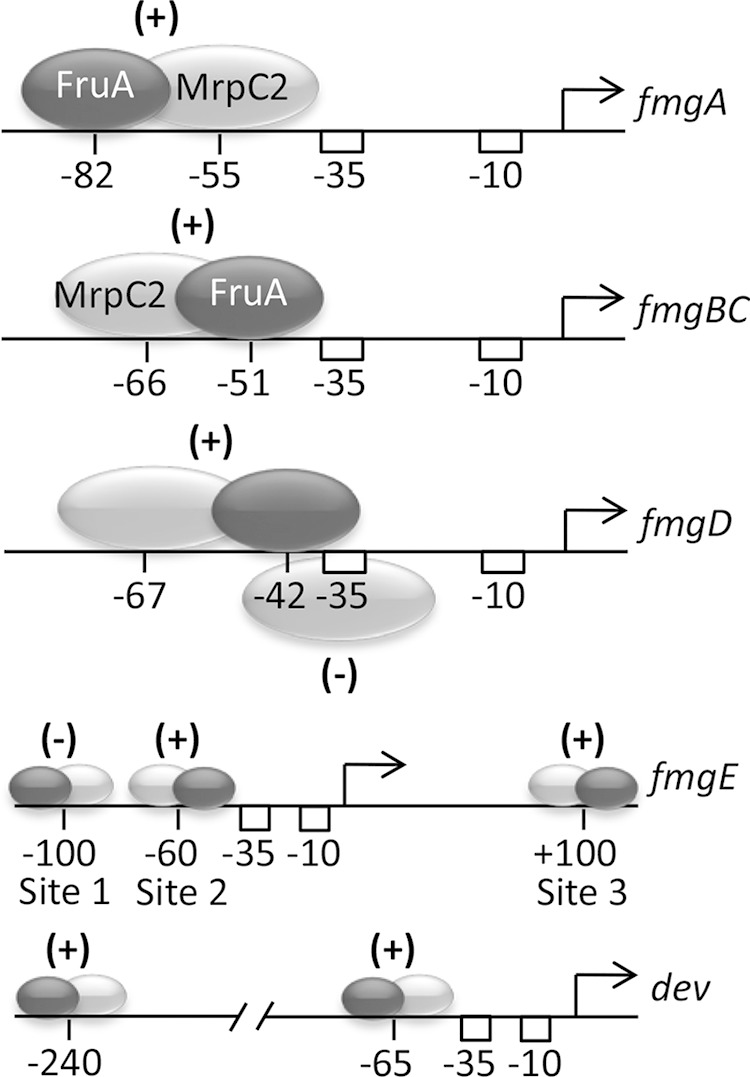
Arrangements of MrpC2- and FruA-binding sites in promoter regions of C-signal-dependent genes. The approximate positions of His10-MrpC2 (light gray ovals) and FruA-His6 (dark gray ovals) binding in vitro are shown, based on footprinting and the effects of mutations in DNA probes in EMSAs for fmgA (34, 37) and dev (38) (Fig. 1 and 3) or just on the effects of mutations in DNA probes in EMSAs for fmgBC, fmgD, and fmgE (39–41). (+) and (−) indicate positive and negative effects on promoter activity, respectively, as inferred from expression of lacZ fusions in promoter regions with mutations in transcription factor-binding sites (38, 53, 56–58) (Fig. 4). A larger region is shown for the fmgE and dev promoter regions.
Mutations that reduce or eliminate binding in vitro of MrpC2 or FruA to their sites near the dev promoter impair or abolish promoter activity in vivo. This was shown previously for the FruA DNA-binding domain and mutations in the region between positions −101 and −75 (38) and was shown here for MrpC2 and mutations in the region between positions −74 and −39 (Fig. 3 and 4). We conclude that cooperative binding of MrpC2 and FruA immediately upstream of the dev promoter is crucial for expression. Presumably, the transcription factors recruit RNA polymerase to the promoter and/or facilitate a subsequent step in transcription initiation.
Interestingly, mutations M1 and M3, which strongly impair MrpC2 binding and cooperative binding in vitro, have changes at the ends of a sequence very similar to a consensus sequence for MrpC binding, whereas mutations M2A and M2B, which did not impair MrpC2 or cooperative binding, have changes in the middle (i.e., the nonspecific N8 part) of the sequence matching the consensus (Fig. 3). Mutations M4, M5, and M6 had less of an impact on MrpC2 and cooperative binding than did mutations M1 and M3 (Fig. 3), yet all five mutations abolished dev expression in vivo (Fig. 4). A plausible explanation for this finding is that the binding reaction conditions in vitro (i.e., purified proteins and DNA) do not fully reflect the conditions present in vivo (e.g., many potential binding sites competing for binding of the proteins).
Why is the expression of the dev operon and several other C-signal-dependent genes under the combinatorial control of MrpC2 and FruA? Since FruA responds to C signaling, which appears to increase as cells become aligned in nascent fruiting bodies, FruA may communicate positional information to the gene regulatory network. Since several mechanisms link MrpC and MrpC2 levels to starvation, it was proposed previously that the abundance of these proteins is responsive to nutrient conditions as development proceeds (34). In agreement with that proposal, it was found that MrpC and MrpC2 are highly sensitive to nutrient-regulated proteolysis both before and during a critical period of commitment to sporulation at ∼24 to 30 h poststarvation (50). The addition of nutrients halted the expression of the dev operon. Hence, combinatorial regulation appears to ensure that the dev operon and other C-signal-dependent genes are fully expressed, committing cells to form spores, only if cells are both starving (to allow sufficient MrpC and MrpC2 activity) and aligned within a nascent fruiting body (to allow efficient C signaling and therefore sufficient FruA activity).
Binding of MrpC2 and FruA to a distal upstream site boosts dev expression during the critical period of commitment to sporulation.
Our findings imply that the expression of the dev operon is subject to a novel regulatory strategy compared with other C-signal-dependent genes studied so far. We found that a second cis-regulatory element located further upstream of the dev promoter corresponds to a site of weak cooperative binding of MrpC2 and FruA and contributes positively to dev expression after 24 h of development. This is a more complex arrangement of cooperative binding sites than those identified in the fmgA (34) and fmgBC (40) promoter regions, where a single cooperative binding site just upstream of the promoter was found (Fig. 8). Expression from these promoters depends in part on C signaling, as does the expression of the dev operon (4), but 5′ deletion analysis indicated additional complexity in the regulation of the dev promoter (8). Two positive cis-regulatory elements were inferred, one between positions −934 and −535 and the other between positions −535 and −114, both upstream of the promoter-proximal cooperative binding site for MrpC2 and FruA discussed above. This type of complexity has not been observed for the promoter regions of other C-signal-dependent genes examined so far, although two genes that depend completely on C signaling for expression appear to have more complex regulatory strategies than those of fmgA and fmgBC. The fmgD promoter region has two MrpC2-binding sites (39) (Fig. 8). The downstream MrpC2 site overlaps the FruA site and the promoter. Mutational analysis supports a model in which the two transcription factors compete for binding, resulting in repression when two MrpC2s bind cooperatively (early in development) and in activation when FruA binds cooperatively with upstream MrpC2 (late in development, as C signaling causes the concentration of active FruA to increase). The fmgE promoter region has three sites where MrpC2 and FruA bind cooperatively (41) (Fig. 8). Site 1, centered at about position −100, mediates negative regulation, whereas site 2, immediately upstream of the promoter, and site 3, at about position +100, mediate positive regulation. The relative binding affinities support a model in which site 3 recruits MrpC2 and FruA to the promoter region, site 1 competes with site 2 for binding of the two proteins, and binding to site 2 is required to activate the promoter, which occurs only as C signaling produces a high concentration of active FruA late in development. Regulation of the dev operon might also rely on differential affinities of binding of MrpC2 and FruA to cooperative binding sites. Binding to the site immediately upstream of the promoter was stronger in vitro than binding to the distal upstream site (Fig. 3 and 6). Perhaps the proximal upstream site can be occupied at relatively low MrpC2 and FruA concentrations in vivo, accounting for expression early in development that does not depend on C signaling to increase the concentration of active FruA. We propose that the occupancy of the distal upstream site requires a higher concentration of active FruA, produced in response to efficient C signaling as cells become aligned in nascent fruiting bodies and accounting for the boost in dev expression observed after 24 h of development. Since the boost in dev expression coincides with the critical period of commitment to sporulation mentioned above (50), we hypothesized that increased dev expression is important for sporulation. However, we found that deletion of the distal upstream region of MrpC2 and FruA binding did not impair spore formation under laboratory conditions. Perhaps under other conditions, the boost in dev expression brought about by the distal upstream site is important for sporulation.
In addition to the two upstream cooperative binding sites for MrpC2 and FruA characterized here, regulation of the dev operon also involves downstream elements. A positive regulatory element located at about position +350 is bound by LadA, a LysR-type transcription factor (38). Two other positive regulatory elements and one negative regulatory element are inferred from 3′ deletion analysis, and several results suggest that downstream regulatory elements interact functionally with upstream elements (8). DNA looping was proposed to explain the apparent long-range interactions. It seems likely that DNA looping also allows MrpC2 and FruA bound to the distal upstream site to activate transcription from the dev promoter, as looping often explains how transcription factors exert effects from promoter-distal sites (55). In any case, the identification of the distal upstream site was facilitated here by performance of a 5′ deletion analysis with lacZ fusions containing very little DNA downstream of the dev promoter, to eliminate potential long-range interactions between upstream and downstream regulatory elements. The effect of DNA between positions −934 and −535 was negative for fusions ending at position +71 (Fig. 5) but positive for fusions ending at position +834 (8), supporting the notion of long-range interactions in the latter case. Although some mysteries remain about regulation of the dev operon, our results show that combinatorial regulation by MrpC2 and FruA is an important aspect.
Combinatorial regulation by MrpC2 and FruA appears to be used widely during M. xanthus development. In addition to the dev promoter and the four fmg promoters mentioned above, ChIP-seq analysis of cells that had formed nascent fruiting bodies revealed 1,608 putative MrpC-binding sites, and when 15 of these were tested for cooperative binding of MrpC2 and FruA, there was evidence for cooperative binding in 13 cases (62). These include sites near the 5′ ends of genes important for development, such as genes that code for protein kinases or transcription factors, and genes involved in signal production, spore formation, and motility.
Combinatorial regulation by MrpC2 and FruA is also versatile. Depending on the arrangement and affinities of binding sites for MrpC2 and FruA, a variety of regulatory patterns can be achieved in terms of the strength and timing of expression. Our analysis of dev regulation provides the first example of a distal upstream cooperative binding site for MrpC2 and FruA that boosts expression late in development.
ACKNOWLEDGMENTS
We are grateful to Sumiko Inouye for providing the AS fraction, protocols for protein purification, and the antibodies against MrpC. We thank Sheenu Mittal for purified proteins and technical advice.
This research was supported by NSF grants MCB-0744343 and MCB-1411272 and by Michigan State University AgBioResearch.
REFERENCES
- 1.Yang Z, Higgs P. 2014. Myxobacteria: genomics, cellular and molecular biology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 2.Kroos L, Kuspa A, Kaiser D. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol 117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 3.Thony-Meyer L, Kaiser D. 1993. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J Bacteriol 175:7450–7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroos L, Kaiser D. 1987. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev 1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- 5.Kroos L, Kuspa A, Kaiser D. 1990. Defects in fruiting body development caused by Tn5 lac insertions in Myxococcus xanthus. J Bacteriol 172:484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Licking E, Gorski L, Kaiser D. 2000. A common step for changing cell shape in fruiting body and starvation-independent sporulation of Myxococcus xanthus. J Bacteriol 182:3553–3558. doi: 10.1128/JB.182.12.3553-3558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller FD, Schink CW, Hoiczyk E, Cserti E, Higgs PI. 2012. Spore formation in Myxococcus xanthus is tied to cytoskeleton functions and polysaccharide spore coat deposition. Mol Microbiol 83:486–505. doi: 10.1111/j.1365-2958.2011.07944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viswanathan P, Murphy K, Julien B, Garza AG, Kroos L. 2007. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J Bacteriol 189:3738–3750. doi: 10.1128/JB.00187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boysen A, Ellehauge E, Julien B, Sogaard-Andersen L. 2002. The DevT protein stimulates synthesis of FruA, a signal transduction protein required for fruiting body morphogenesis in Myxococcus xanthus. J Bacteriol 184:1540–1546. doi: 10.1128/JB.184.6.1540-1546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karginov FV, Hannon GJ. 2010. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell 37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julien B, Kaiser AD, Garza A. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc Natl Acad Sci U S A 97:9098–9103. doi: 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sager B, Kaiser D. 1993. Two cell-density domains within the Myxococcus xanthus fruiting body. Proc Natl Acad Sci U S A 90:3690–3694. doi: 10.1073/pnas.90.8.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sager B, Kaiser D. 1993. Spatial restriction of cellular differentiation. Genes Dev 7:1645–1653. doi: 10.1101/gad.7.9.1645. [DOI] [PubMed] [Google Scholar]
- 14.Sager B, Kaiser D. 1994. Intercellular C-signaling and the traveling waves of Myxococcus. Genes Dev 8:2793–2804. doi: 10.1101/gad.8.23.2793. [DOI] [PubMed] [Google Scholar]
- 15.Kim SK, Kaiser D. 1990. Cell motility is required for the transmission of C-factor, an intercellular signal that coordinates fruiting body morphogenesis of Myxococcus xanthus. Genes Dev 4:896–905. doi: 10.1101/gad.4.6.896. [DOI] [PubMed] [Google Scholar]
- 16.Kim SK, Kaiser D. 1990. Cell alignment required in differentiation of Myxococcus xanthus. Science 249:926–928. doi: 10.1126/science.2118274. [DOI] [PubMed] [Google Scholar]
- 17.Kroos L, Hartzell P, Stephens K, Kaiser D. 1988. A link between cell movement and gene expression argues that motility is required for cell-cell signaling during fruiting body development. Genes Dev 2:1677–1685. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- 18.Kim SK, Kaiser D. 1991. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J Bacteriol 173:1722–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruse T, Lobedanz S, Berthelsen NM, Sogaard-Andersen L. 2001. C-signal: a cell surface-associated morphogen that induces and co-ordinates multicellular fruiting body morphogenesis and sporulation in Myxococcus xanthus. Mol Microbiol 40:156–168. doi: 10.1046/j.1365-2958.2001.02365.x. [DOI] [PubMed] [Google Scholar]
- 20.Li S-F, Lee B, Shimkets LJ. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev 6:401–410. doi: 10.1101/gad.6.3.401. [DOI] [PubMed] [Google Scholar]
- 21.Shimkets LJ, Gill RE, Kaiser D. 1983. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc Natl Acad Sci U S A 80:1406–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simunovic V, Gherardini FC, Shimkets LJ. 2003. Membrane localization of motility, signaling, and polyketide synthetase proteins in Myxococcus xanthus. J Bacteriol 185:5066–5075. doi: 10.1128/JB.185.17.5066-5075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobedanz S, Sogaard-Andersen L. 2003. Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev 17:2151–2161. doi: 10.1101/gad.274203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SK, Kaiser D. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell 61:19–26. doi: 10.1016/0092-8674(90)90211-V. [DOI] [PubMed] [Google Scholar]
- 25.Kim SK, Kaiser D. 1990. Purification and properties of Myxococcus xanthus C-factor, an intercellular signaling protein. Proc Natl Acad Sci U S A 87:3635–3639. doi: 10.1073/pnas.87.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konovalova A, Lobach S, Sogaard-Andersen L. 2012. A RelA-dependent two-tiered regulated proteolysis cascade controls synthesis of a contact-dependent intercellular signal in Myxococcus xanthus. Mol Microbiol 84:260–275. doi: 10.1111/j.1365-2958.2012.08020.x. [DOI] [PubMed] [Google Scholar]
- 27.Rolbetzki A, Ammon M, Jakovljevic V, Konovalova A, Sogaard-Andersen L. 2008. Regulated secretion of a protease activates intercellular signaling during fruiting body formation in M. xanthus. Dev Cell 15:627–634. doi: 10.1016/j.devcel.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Lee B-U, Lee K, Mendez J, Shimkets L. 1995. A tactile sensory system of Myxococcus xanthus involves an extracellular NAD(P)+-containing protein. Genes Dev 9:2964–2973. doi: 10.1101/gad.9.23.2964. [DOI] [PubMed] [Google Scholar]
- 29.Lee K, Shimkets LJ. 1996. Suppression of a signaling defect during Myxococcus xanthus development. J Bacteriol 178:977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avadhani M, Geyer R, White DC, Shimkets LJ. 2006. Lysophosphatidylethanolamine is a substrate for the short-chain alcohol dehydrogenase SocA from Myxococcus xanthus. J Bacteriol 188:8543–8550. doi: 10.1128/JB.01047-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhat S, Zhu X, Patel RP, Orlando R, Shimkets LJ. 2011. Identification and localization of Myxococcus xanthus porins and lipoproteins. PLoS One 6:e27475. doi: 10.1371/journal.pone.0027475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellehauge E, Norregaard-Madsen M, Sogaard-Andersen L. 1998. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in Myxococcus xanthus development. Mol Microbiol 30:807–817. doi: 10.1046/j.1365-2958.1998.01113.x. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa M, Fujitani S, Mao X, Inouye S, Komano T. 1996. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol Microbiol 22:757–767. doi: 10.1046/j.1365-2958.1996.d01-1725.x. [DOI] [PubMed] [Google Scholar]
- 34.Mittal S, Kroos L. 2009. A combination of unusual transcription factors binds cooperatively to control Myxococcus xanthus developmental gene expression. Proc Natl Acad Sci U S A 106:1965–1970. doi: 10.1073/pnas.0808516106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ueki T, Inouye S. 2005. Identification of a gene involved in polysaccharide export as a transcription target of FruA, an essential factor for Myxococcus xanthus development. J Biol Chem 280:32279–32284. doi: 10.1074/jbc.M507191200. [DOI] [PubMed] [Google Scholar]
- 36.Ueki T, Inouye S. 2005. Activation of a development-specific gene, dofA, by FruA, an essential transcription factor for development of Myxococcus xanthus. J Bacteriol 187:8504–8506. doi: 10.1128/JB.187.24.8504-8506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoder-Himes D, Kroos L. 2006. Regulation of the Myxococcus xanthus C-signal-dependent Ω4400 promoter by the essential developmental protein FruA. J Bacteriol 188:5167–5176. doi: 10.1128/JB.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viswanathan P, Ueki T, Inouye S, Kroos L. 2007. Combinatorial regulation of genes essential for Myxococcus xanthus development involves a response regulator and a LysR-type regulator. Proc Natl Acad Sci U S A 104:7969–7974. doi: 10.1073/pnas.0701569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J, Son B, Viswanathan P, Luethy P, Kroos L. 2011. Combinatorial regulation of fmgD by MrpC2 and FruA during Myxococcus xanthus development. J Bacteriol 193:1681–1689. doi: 10.1128/JB.01541-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mittal S, Kroos L. 2009. Combinatorial regulation by a novel arrangement of FruA and MrpC2 transcription factors during Myxococcus xanthus development. J Bacteriol 191:2753–2763. doi: 10.1128/JB.01818-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Son B, Liu Y, Kroos L. 2011. Combinatorial regulation by MrpC2 and FruA involves three sites in the fmgE promoter region during Myxococcus xanthus development. J Bacteriol 193:2756–2766. doi: 10.1128/JB.00205-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueki T, Inouye S. 2003. Identification of an activator protein required for the induction of fruA, a gene essential for fruiting body development in Myxococcus xanthus. Proc Natl Acad Sci U S A 100:8782–8787. doi: 10.1073/pnas.1533026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun H, Shi W. 2001. Genetic studies of mrp, a locus essential for cellular aggregation and sporulation of Myxococcus xanthus. J Bacteriol 183:4786–4795. doi: 10.1128/JB.183.16.4786-4795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nariya H, Inouye S. 2005. Identification of a protein Ser/Thr kinase cascade that regulates essential transcriptional activators in Myxococcus xanthus development. Mol Microbiol 58:367–379. doi: 10.1111/j.1365-2958.2005.04826.x. [DOI] [PubMed] [Google Scholar]
- 45.Nariya H, Inouye S. 2006. A protein Ser/Thr kinase cascade negatively regulates the DNA-binding activity of MrpC, a smaller form of which may be necessary for the Myxococcus xanthus development. Mol Microbiol 60:1205–1217. doi: 10.1111/j.1365-2958.2006.05178.x. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 47.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 48.Hodgkin J, Kaiser D. 1977. Cell-to-cell stimulation of motility in nonmotile mutants of Myxococcus. Proc Natl Acad Sci U S A 74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuner JM, Kaiser D. 1982. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J Bacteriol 151:458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajagopalan R, Kroos L. 2014. Nutrient-regulated proteolysis of MrpC halts expression of genes important for commitment to sporulation during Myxococcus xanthus development. J Bacteriol 196:2736–2747. doi: 10.1128/JB.01692-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuwabara MD, Sigman DS. 1987. Footprinting DNA-protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry 26:7234–7238. doi: 10.1021/bi00397a006. [DOI] [PubMed] [Google Scholar]
- 52.Kashefi K, Hartzell P. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthux frzF− defect. Mol Microbiol 15:483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x. [DOI] [PubMed] [Google Scholar]
- 53.Viswanathan P, Kroos L. 2003. cis elements necessary for developmental expression of a Myxococcus xanthus gene that depends on C signaling. J Bacteriol 185:1405–1414. doi: 10.1128/JB.185.4.1405-1414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ueki T, Inouye S, Inouye M. 1996. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183:153–157. doi: 10.1016/S0378-1119(96)00546-X. [DOI] [PubMed] [Google Scholar]
- 55.Cournac A, Plumbridge J. 2013. DNA looping in prokaryotes: experimental and theoretical approaches. J Bacteriol 195:1109–1119. doi: 10.1128/JB.02038-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoder D, Kroos L. 2004. Mutational analysis of the Myxococcus xanthus Ω4400 promoter region provides insight into developmental gene regulation by C signaling. J Bacteriol 186:661–671. doi: 10.1128/JB.186.3.661-671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoder D, Kroos L. 2004. Mutational analysis of the Myxococcus xanthus Ω4499 promoter region reveals shared and unique properties in comparison with other C-signal-dependent promoters. J Bacteriol 186:3766–3776. doi: 10.1128/JB.186.12.3766-3776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Viswanathan K, Viswanathan P, Kroos L. 2006. Mutational analysis of the Myxococcus xanthus Ω4406 promoter region reveals an upstream negative regulatory element that mediates C-signal dependence. J Bacteriol 188:515–524. doi: 10.1128/JB.188.2.515-524.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 60.Kaiser D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci U S A 76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fisseha M, Gloudemans M, Gill R, Kroos L. 1996. Characterization of the regulatory region of a cell interaction-dependent gene in Myxococcus xanthus. J Bacteriol 178:2539–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson M, Son B, Kroos D, Kroos L. Transcription factor MrpC binds to promoter regions of hundreds of developmentally-regulated genes in Myxococcus xanthus. BMC Genomics, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]