Abstract
Bacillus anthracis, the causative agent of anthrax, replicates as chains of vegetative cells by regulating the separation of septal peptidoglycan. Surface (S)-layer proteins and associated proteins (BSLs) function as chain length determinants and bind to the secondary cell wall polysaccharide (SCWP). In this study, we identified the B. anthracis lcpD mutant, which displays increased chain length and S-layer assembly defects due to diminished SCWP attachment to peptidoglycan. In contrast, the B. anthracis lcpB3 variant displayed reduced cell size and chain length, which could be attributed to increased deposition of BSLs. In other bacteria, LytR-CpsA-Psr (LCP) proteins attach wall teichoic acid (WTA) and polysaccharide capsule to peptidoglycan. B. anthracis does not synthesize these polymers, yet its genome encodes six LCP homologues, which, when expressed in S. aureus, promote WTA attachment. We propose a model whereby B. anthracis LCPs promote attachment of SCWP precursors to discrete locations in the peptidoglycan, enabling BSL assembly and regulated separation of septal peptidoglycan.
INTRODUCTION
Bacillus anthracis, the causative agent of anthrax, is a spore-forming, Gram-positive bacterium that germinates in host infected tissues and replicates as elongated chains of vegetative forms (1, 2). This unique growth pattern appears to be caused by the regulated separation of septal peptidoglycan, which generates bacterial chains whose mere size precludes clearance by host phagocytes (3–5). The genetic determinants for B. anthracis chain formation are conserved among pathogenic species of the Bacillus cereus group and, as demonstrated with B. cereus G9241, likely contribute to disease pathogenesis (6, 7). Hallmarks of pathogenic Bacillus species are virulence plasmids, pXO-1 and pXO-2 in B. anthracis (8, 9), providing for toxin production and capsulation (10, 11) as well as the chromosomally encoded surface (S)-layer locus (12). Two S-layer proteins of B. anthracis, Sap and EA1, are endowed with S-layer homology (SLH) domains, which retain these proteins in the bacterial envelope by binding to the secondary cell wall polysaccharide (SCWP) (13–15). S-layer protein crystallization domains, responsible for the spontaneous assembly of these polypeptides into a paracrystalline array (16), form a two-dimensional lattice that can be thought of as bacterial integument (17–19).
The structural genes of S-layer proteins, sap and eag, are flanked by genes encoding determinants for S-layer protein secretion (secA2 and slaP) (12) or pyruvylation (csaB) (14) as well as acetylation of the SCWP (patA1/2 and patB1/2) (20). CsaB-mediated pyruvylation of the terminal N-acetylmannosamine (ManNAc) of the SCWP (21), with the repeat structure [→4)-β-ManNAc-(1→4)-β-GlcNAc-(1→6)-α-GlcNAc-(1→] (22), is a prerequisite for the assembly of S-layer proteins and 22 B. anthracis S-layer-associated proteins (BSLs) (14). PatAB1/2-mediated acetylation of SCWP molecules affects the assembly of EA1 and of some but not all BSLs (20). Recent studies have begun to identify genes for SCWP synthesis, which, unlike pyruvyl or acetyl modifications of SCWP, appears to be essential for B. anthracis growth (23, 24). Nevertheless, most of the genetic determinants for SCWP synthesis, including genes whose products catalyze attachment of the polysaccharide to the peptidoglycan scaffold of B. anthracis, remain unknown (19).
LytR-CpsA-Psr (LCP) enzymes are found in many Gram-positive bacteria, with many species possessing more than one lcp gene (25). LCP proteins typically encompass a short N-terminal cytoplasmic domain, followed by 1 to 5 transmembrane helices and a C-terminal LCP domain of approximately 150 residues, which is thought to be exposed on the trans side of the membrane (25) (Fig. 1). Mutations in Streptococcus agalactiae cpsA and Streptococcus pneumoniae cps2A, the first lcp genes to be characterized, abrogate capsule synthesis, a phenotype initially attributed to transcriptional regulation (26–28). Bacillus subtilis harbors three lcp genes, designated tagTUV, which are located within a 50-kb region on the chromosome that also contains genes for the biosynthesis of teichuronic acid (TU), minor teichoic acid (TA), and wall teichoic acid (WTA) (29). Expression of at least one lcp gene—tagT, tagU, or tagV—is essential for B. subtilis growth and teichoic acid synthesis, which prompted Kawai and colleagues to propose a model whereby LCP proteins catalyze transfer of bactoprenol phosphate-linked capsular polysaccharide or teichoic acid precursors to peptidoglycan (29). In agreement with this model, the crystal structure of recombinant Cps2A and TagT revealed polyprenyl lipids bound to the active site as well as intrinsic pyrophosphatase activity for both proteins (29, 30). The genome of Staphylococcus aureus contains three lcp genes, designated lcpA, lcpB, and lcpC (31, 32). Staphylococcal mutants lacking all three lcp genes (Δlcp), although viable, display growth defects, are unable to attach either WTA or capsular polysaccharide to peptidoglycan, and release small amounts of these polymers into the extracellular medium (32–34). In Actinomyces oris, depletion of sortase A prevents bacterial growth and this phenotype can be suppressed by transposon insertions in a gene cluster encoding a LytR-CpsA-Psr (LCP)-like protein as well as with GspA (AcaA), a glycosylated surface protein substrate of sortase (35). Of note, GspA accumulated in the membrane of the sortase A mutant but not in wild-type Actinomyces (35). Thus, an LCP-like protein appears to promote the glycosylation of the GspA surface proteins in A. oris (35).
FIG 1.
Phylogenic analysis of Bacillus anthracis LytR-CpsA-Psr (LCP) proteins. (A) Predicted domains of B. anthracis LCP proteins. The transmembrane (TM) and LCP domains were predicted using the TMHMM Server v. 2.0 and Clustal W programs. (B) Phylogenetic tree revealing the relatedness of lcp genes from Staphylococcus aureus (blue), Bacillus subtilis (black), and Bacillus anthracis (red). The tree was calculated with Clustal W. B. anthracis lcp gene products were designated LcpB1 (BAS1830), LcpB2 (BAS0572), LcpB3 (BAS0746), LcpB4 (BAS3381), LcpC (BAS5115), and LcpD (BAS5047). (C) Alignment of the LytR-CpsA-Psr domains of LCP proteins from S. aureus, B. subtilis, and B. anthracis using Clustal W. Residues printed in red are conserved in all members of the LytR-CpsA-Psr protein family. Residues in blue are conserved among LcpB proteins of B. anthracis. The symbols denote identical residues in all sequences (*), a highly conserved column (:), and a weakly conserved column (.).
B. anthracis lacks the genes for ribitol phosphate (RboP) teichoic acid synthesis and elaborates poly-d-γ-glutamic acid capsule, which is tethered via amide bonds to bacterial peptidoglycan (36, 37). Screening transposon mutants of B. anthracis for chain length phenotypes, we identified the lcpD mutant, which displays a chain length phenotype and assembly defect for the S-layer protein Sap. B. anthracis encodes six LCP homologues (the products of lcpB1, lcpB2, lcpB3, lcpB4, lcpC, and lcpD), whose functions were validated by heterologous expression, measuring WTA synthesis in the S. aureus Δlcp mutant. We also report reduced cell size and diminished chain length in the lcpB3 variant, a phenotype that may be attributable to the altered deposition of two murein hydrolases, BslU and BslR, in the envelope of B. anthracis.
MATERIALS AND METHODS
Bacterial growth and reagents.
B. anthracis strain Sterne 34F2 (38) and its variants were grown in brain heart infusion (BHI) broth or agar at 37°C. S. aureus Newman (39) and its variants were grown in tryptic soy broth (TSB) or on tryptic soy agar (TSA) at 37°C. Escherichia coli K1077 (40) was grown in Luria-Bertani broth (LB) or agar at 37°C. Where necessary, ampicillin (Amp; 100 μg/ml), chloramphenicol (Cm; 10 μg/ml), spectinomycin (Sp; 200 μg/ml), erythromycin (Erm; 10 μg/ml), or kanamycin (Kan; 20 μg/ml) was added to cultures. S. aureus WTA synthesis was inhibited with tunicamycin (Tun; 1 μg/ml) (14). B. anthracis strains were sporulated in modified G medium (modG) (41). Spore preparations were heat treated at 68°C for 2 h to kill vegetative bacilli and then stored at 4°C. Spore titers were determined by plating aliquots of serially diluted spore preparations on BHI agar; CFU were enumerated after overnight incubation at 37°C. Spores were germinated by inoculation into BHI and grown at 37°C. To examine growth, overnight cultures were normalized to an A600 of 5 and diluted 1:100 into 100 μl of fresh medium (BHI or TSB) with appropriate antibiotics, and growth at 37°C was monitored every 30 min for 12 h in a Synergy HT plate reader (BioTek) by recording the A600.
Bacterial strains and plasmids.
Bacterial strains and plasmids utilized in this study are listed in Table S1 in the supplemental material. The alleles lcpB2::apha3 and lcpD::aad9 were obtained by transposon mutagenesis with Bursa aurealis (42). The alleles were transduced into wild-type B. anthacis Sterne with bacteriophage CP-51 (9). The location of insertional lesions was determined by sequence analyses of inverse PCR products of the transposon/chromosome junction using the primers Martn-F and SpecR (42). Deletion of lcpB1, lcpB3, lcpB4, and lcpC was achieved by allelic replacement with the temperature-sensitive vector pLM4 (43). Briefly, 1-kbp flanking DNA sequences upstream and downstream of target genes were ligated to generate 2 kbp of recombinant DNA that was cloned into pLM4. All cloning steps were performed in E. coli K1077 (with dam and dcm mutations), recombinant plasmids were electroporated into B. anthracis (44), and allelic replacement was induced via changes in incubation temperature (43). Mutant alleles were verified by PCR using chromosomal DNA as a template and primers flanking the cloning sites. S. aureus MSSA1112 and its isogenic variant lacking lcpA, lcpB, and lcpC (Δlcp) have been described previously (31). For complementation studies, B. anthracis lcp genes were cloned into pWWW412 and expressed via the constitutive hprK promoter of S. aureus (45). All lcp genes were amplified from B. anthracis Sterne chromosomal DNA by PCR using AccuPrime Pfx DNA polymerase (Invitrogen). PCR products were ligated into pWWW412, and recombinant plasmids were analyzed by DNA sequencing. Plasmid plcpASa was described earlier (32). The names and sequences of primers are listed in Table S2 in the supplemental material.
Microscopy, chain length measurements, and immunofluorescence.
To analyze chain lengths, B. anthracis spores were germinated by suspension in BHI broth at 37°C. At defined time points, aliquots of cells were removed and fixed with 4% formalin, and images were captured with a charge-coupled-device (CCD) camera on an Olympus IX81 microscope using 20× or 40× objectives. Chain lengths of bacilli were measured from acquired differential interference contrast (DIC) images using ImageJ and converted to lengths (in micrometers) using reference images of an objective micrometer (4). Statistical significance of chain length distributions between different strains was analyzed using one-way analysis of variance (ANOVA) (4).
For fluorescence microscopy, germinated spores of B. anthracis strains were fixed in 4% formalin at either 3 or 8 h postgermination. After a washing to remove fixative, samples were incubated with rabbit antisera raised against purified Sap, EA1, BslR, or BslU, and bound antibody was detected with DyLight594-conjugated anti-rabbit secondary antibody. Cells were counterstained with boron dipyrromethene (BODIPY)-vancomycin. A Leica SP5 STED-CW Superresolution laser scanning confocal microscope with a 63× objective was used to analyze the fluorescence of B. anthracis cells (12).
Preparation of murein sacculi and analysis of secondary cell wall polymers.
Purification of SCWP from B. anthracis Sterne and its variants followed a previously described protocol (14, 20). Briefly, B. anthracis strains were grown overnight at 37°C on BHI agar plates, scraped and suspended in 4% SDS, and boiled for 30 min. Bacteria were mechanically lysed with 0.1-mm glass beads and murein sacculi sedimented by centrifugation. The pellet was suspended in 100 mM Tris-HCl (pH 7.5) and incubated for 4 h at 37°C with 10 μg/ml of DNase and 10 μg/ml of RNase supplemented with 20 mM MgSO4. Samples were then incubated for 16 h at 37°C with 10 μM trypsin supplemented with 10 mM CaCl2. Enzymes were inactivated by boiling for 30 min in 1% SDS. Murein sacculi were washed sequentially with water, 100 mM Tris-HCl (pH 8.0), water, 0.1 M EDTA (pH 8.0), water, and acetone and then washed twice with water. Murein sacculi were suspended in 5 ml of water, and 25 ml of 48% hydrofluoric acid (HF) was added. Samples were incubated overnight on ice with shaking. Acid-extracted murein sacculi were sedimented and SCWP in the supernatant fractions was precipitated with ice-cold ethanol. The polysaccharide was sedimented by centrifugation, washed extensively with ice-cold ethanol, and suspended in 50 mM phosphate buffer for size exclusion high-performance liquid chromatography (SEC-HPLC) analysis.
S. aureus WTA was extracted following NaOH treatment of murein sacculi prepared from staphylococci and analyzed by polyacrylamide gel electrophoresis (PAGE) as described earlier (32).
RESULTS
Bacillus anthracis LCP genes.
A library of B. anthracis Sterne mutants with insertional lesions was screened for chain length variants (12). This search identified a variant with Bursa aurealis insertion in BAS5047, which displays increased chain length of vegetative forms compared to its wild-type parent (see Table S1 in the supplemental material). The bas5047-encoded protein product is a homologue of B. subtilis TagV (TagVBs), a member of the LytR-CpsA-Psr (LCP) family in Gram-positive bacteria (29) (Fig. 1). This family of proteins is involved in anchoring WTA and anionic polysaccharides to peptidoglycan. This was a surprising result, as B. anthracis lacks the genes for WTA synthesis and is not known to elaborate anionic polysaccharides (36).
Amino acid sequences of S. aureus LcpASa, LcpBSa and LcpCSa were used as queries for BLAST searches, which identified six LCP homologues in the genome of B. anthracis Sterne: BAS5047, BAS0572, BAS0746, BAS1830, BAS3381, and BAS5115. When queried for the presence of hydrophobic and functional domains, a conserved topology was observed with the N-terminal end inside the cell and the C-terminal end outside, resulting in the surface display of the LCP domain (Fig. 1A). The Clustal W program was used to perform a multiple-sequence alignment and derive a dendrogram comparing LCPs of B. anthracis, B. subtilis, and S. aureus (Fig. 1B and C). B. anthracis and B. subtilis LCPs segregated into three groups, one corresponding to S. aureus LcpBSa (B family), one corresponding to S. aureus LcpCSa (C family), and one without a corresponding S. aureus LCP (D family). S. aureus LcpASa (A family) did not segregate with LCPs from B. anthracis and B. subtilis (Fig. 1B). Amino acid conservation across all LCPs occurs almost exclusively within the LCP domains, and families B, C, and D are distinguished by key amino acid identities (Fig. 1C). From this analysis, we derived the following nomenclature for B. anthracis LCPs: LcpB1Ba (BAS1820), LcpB2Ba (BAS0572), LcpB3Ba (BAS0746), LcpB4Ba (BAS3381), LcpCBa (BAS5115), and LcpDBa (BAS5047). The data suggest further that B. anthracis lcpB genes likely arose via gene duplication events.
Mutations in lcpD and lcpB3 impact Bacillus anthracis chain length.
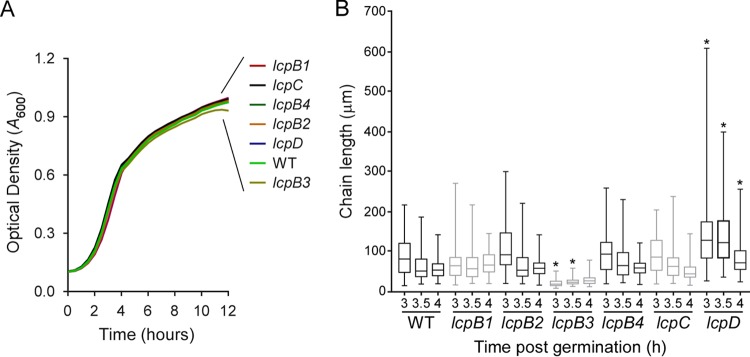
Screening transposon mutants by DNA sequencing, we identified a B. anthracis Sterne variant with a Bursa aurealis insertion in lcpB2. Allelic replacement was used to generate B. anthracis Sterne variants with deletions of BAS1820 (lcpB1), BAS0746 (lcpB3), BAS3381 (lcpB4), and BAS5115 (lcpC). By measuring increased absorbance (A600) of cultures incubated at 37°C, it was determined that B. anthracis variants lacking any one of the six lcp genes grew at a rate similar to that of their wild-type parent (Fig. 2A). Further, following incubation in sporulation medium, quantification of the abundance of heat-resistant spores showed that B. anthracis variants lacking any one of the six lcp genes sporulated with an efficiency similar to that of the wild type (data not shown). To assess chain formation, equal numbers of spores were diluted into fresh broth for germination of vegetative forms and synchronized growth. At timed intervals (3, 3.5, and 4 h), culture aliquots were fixed, images of 100 chains of vegetative bacilli were captured by DIC microcopy, and their lengths were quantified (Fig. 2B). The chain lengths of lcpB1, lcpB2, lcpB4, or lcpC mutant bacilli were not significantly different from those of wild-type B. anthracis. In contrast, disruption of lcpD resulted in increased chain lengths at each observation period, whereas disruption of lcpB3 produced shortened chains (Fig. 2B). The increased chain length phenotype of the lcpD mutant was caused by the insertional lesion in lcpD, as transformation of the mutant with a plasmid carrying wild-type lcpD reversed the phenotype to wild-type levels (Fig. 3A). Of note, the width and volume of individual lcpD cells did not differ from those of wild-type bacilli (Fig. 3C to E).
FIG 2.
Bacillus anthracis lcpD and lcpB3 mutants display chain length phenotypes. (A) Stationary-phase cultures of wild-type (WT) B. anthracis Sterne and mutants with mutational lesions in lcpB1, lcpB2, lcpB3, lcpB4, lcpC, and lcpD were normalized to an A600 of 5, inoculated 1:100 into 100 μl of fresh brain heart infusion broth, and incubated at 37°C with agitation while recording the A600 in 30-min intervals over 12 h. (B) Spores from wild-type (WT) or lcp mutant B. anthracis were germinated, aliquots of culture with synchronized growth were fixed following 3, 3.5, and 4 h of incubation, and DIC microscopy images were acquired. The lengths of 100 chains were measured using ImageJ software, and results are presented in a box-and-whisker plot. Data were analyzed with ANOVA; an asterisk denotes statistically significant differences (P < 0.05) comparing WT versus mutant at the same time point.
FIG 3.
The lcpB3 mutation diminishes cell size and chain length in Bacillus anthracis. Shown is a complementation analysis of B. anthracis chain length phenotypes, comparing the wild type (WT) and lcpD mutant and plasmids without (vector) or with wild-type lcpD (plcpD) (A) as well as WT and lcpB3 mutant and plasmid without or with wild-type lcpB3 (plcpB3) (B). (C) Spores of WT, lcpD, or lcpB3 mutant strains with or without complementing plasmid were germinated and incubated for 3.5 h, and culture aliquots were fixed for microscopy. The left side shows bright field microscopy images, whereas the right side shows fluorescence of bacteria stained with BODIPY-vancomycin. Scale bar = 10 μm. (D and E) The length and width of individual B. anthracis cells (n > 200) were measured using ImageJ and are displayed as box-and-whisker plots for cell length (D) or volume (E). Data were analyzed with ANOVA for significant differences compared to the WT unless specified as follows: **, P < 0.0001; *, P < 0.05; ns, not significant.
Plasmid-borne lcpB3 (plcpB3) increased the otherwise shortened chain length of the lcpB3 mutant to a level that exceeded even the chain length of wild-type bacilli (Fig. 3B). Microscopy images suggested that individual cells lacking lcpB3, unlike lcpD mutant bacilli, are smaller than the vegetative forms of wild-type bacilli (Fig. 3C). Using fluorescence microscopy and BODIPY-vancomycin staining of cell septa, we quantified the lengths of >200 bacilli, which revealed a reduced cell length for the lcpB3 mutant compared to wild-type B. anthracis (Fig. 3C and D). Further, when measuring cell widths and deriving volume, it was found that lcpB3 vegetative forms were smaller than wild-type B. anthracis (Fig. 3E). Both phenotypes, diminished cell length and volume, were restored when plcpB3 was transformed into the lcpB3 mutant strain (Fig. 3B). Thus, the lcpB3 mutation diminishes B. anthracis cell size and chain length, whereas the lcpD mutation increases the chain length of its vegetative forms.
Bacillus anthracis LCP enzymes attach teichoic acid to Staphylococcus aureus peptidoglycan.
The S. aureus Δlcp mutant lacks LCP enzymes and cannot attach ribitol phosphate (RboP) WTA to peptidoglycan, a phenotype that is restored by plasmid-borne expression of any one of its three lcp genes, lcpASa, lcpBSa, or lcpCSa (32). We asked whether the B. anthracis lcp genes can complement the WTA attachment phenotype of the S. aureus Δlcp mutant. Plasmids expressing any one of the six B. anthracis LCP enzymes or the lcpASa control were transformed into S. aureus MSSA1112 Δlcp. The murein sacculi from each strain were isolated and treated with dilute sodium hydroxide, which releases RboP polymers via alkaline hydrolysis from peptidoglycan. Extracts were separated by PAGE and stained with alcian blue and silver. As controls, alkaline hydrolysis liberated RboP from wild-type S. aureus but not from tunicamycin-treated cultures, in which TagO-mediated WTA synthesis was inhibited (Fig. 4A). As expected, alkaline hydrolysis did not release RboP from S. aureus Δlcp mutant murein sacculi unless the strain carried a complementing plasmid (plcpSa) (Fig. 4A). Plasmids harboring B. anthracis lcpB2 (plcpB2Ba), lcpC (plcpCBa), or lcpD (plcpDBa) restored S. aureus Δlcp mutant WTA assembly to the same level as plcpSa (Fig. 4A). Plasmids harboring B. anthracis lcpB3 (plcpB3Ba) or lcpB4 (plcpB4Ba) supported attachment of reduced amounts of WTA, whereas lcpB1 (plcpB1Ba) failed to anchor detectable levels of RboP polymer (Fig. 4A). Because plasmid plcpB1Ba restored the chain length defect of the lcpD mutant (data not shown), we surmise that failure to restore WTA assembly in S. aureus is not merely due to a lack of lcpB1Ba expression.
FIG 4.
Bacillus anthracis LCP enzymes attach teichoic acid to Staphylococcus aureus peptidoglycan. (A) Purified murein sacculi from S. aureus MSSA1112 (WT with empty vector) grown in the absence (lane 1) or presence (lane 2) of tunicamycin (tun) were treated with dilute sodium hydroxide and the released wall teichoic acids (WTA) were separated by PAGE and stained with alcian blue and silver. Lanes 3 to 10, WTA extracts of murein sacculi from the S. aureus Δlcp mutant carrying either empty plasmid (vector), control plasmid with S. aureus lcpA (plcpASa), or plasmids with any one of the six lcp genes of B. anthracis: plcpB1, plcpB2, plcpB3, plcpB4, plcpC, and plcpD. The bottom portion shows an immunoblot for staphylococcal sortase (SrtA) using bacterial lysate samples for the corresponding WTA extracts. (B) Growth of the S. aureus Δlcp mutant carrying vector, plcpASa, or plasmids with the six lcp genes of B. anthracis was monitored by first normalizing stationary-phase cultures to an A600 of 5, next diluting these cultures (1:100) into fresh brain heart infusion broth, and then recording the A600 during the 12-h incubation at 37°C with rotation.
Compared to wild-type S. aureus, the Δlcp mutant strain displays a growth defect, presumably because bactoprenol, a carrier required for peptidoglycan synthesis, is sequestered in the WTA synthesis pathway (32). In agreement with this model, transformation of the S. aureus Δlcp mutant with plcpSa but not empty vector promoted growth of the mutant strain (32) (Fig. 4B). Similar to the case with plcpSa, transformation of the Δlcp mutant with plcpB2Ba, plcpCBa, or plcpDBa also promoted growth, whereas plcpB3Ba, plcpB4Ba, or plcpB1Ba did not (Fig. 4B). Together these data indicate that lcpB2, lcpB3, lcpB4, lcpC, and lcpD encode functional LCP enzymes that can catalyze transfer of bactoprenol-linked RboP to peptidoglycan in a heterologous host.
LcpD promotes SCWP attachment to Bacillus anthracis peptidoglycan.
Similar to S. aureus WTA, the SCWP of B. anthracis can be released from murein sacculi via hydrofluoric acid (HF) treatment (14, 20). HF-extracted SCWP can be purified and quantified by size exclusion high-performance liquid chromatography (SEC-HPLC), resulting in a homogenous absorbance peak at 206 nm (20) (Fig. 5A). To quantify the abundance of SCWP attached to peptidoglycan, equal amounts of murein sacculi from wild-type B. anthracis Sterne and its lcpD mutant carrying either vector alone or plasmid-encoded lcpD (plcpD) were extracted with HF and analyzed by SEC-HPLC. As determined by quantifying the absorbance peak at 206 nm, significantly less SCWP was extracted by HF from the envelope of the lcpD mutant than from that of wild-type bacilli (Fig. 5A and B). In contrast, plasmid-borne expression of wild-type lcpD in the lcpD mutant resulted in an increase in HF-extractable SCWP to a level that exceeded even that of wild-type bacilli (Fig. 5A and B). These data suggest that plasmid-encoded LcpD is overproduced (a phenomenon described before [46]) and that the LcpD enzyme promotes the attachment of SCWP to B. anthracis peptidoglycan.
FIG 5.
The lcpD mutation diminishes the attachment of secondary cell wall polysaccharide (SCWP) to Bacillus anthracis peptidoglycan. (A) Equal amounts of purified murein sacculi from B. anthracis Sterne (wild type [WT] with empty vector) or its lcpD mutant carrying empty vector or complementing plasmid (plcpD) were treated with hydrofluoric acid, released SCWP was separated by SEC-HPLC, and absorbance was recorded at 206 nm (mAu). (B) SEC-HPLC peak sizes were measured (mAu2×106) in triplicate from independent samples, and the average peak size and standard deviation were calculated. Comparison of SCWP abundance between strains with the two-tailed Student's t test was used to analyze statistically significant differences (*, P < 0.05).
Assembly of S-layer and S-layer-associated proteins in Bacillus anthracis lcp mutants.
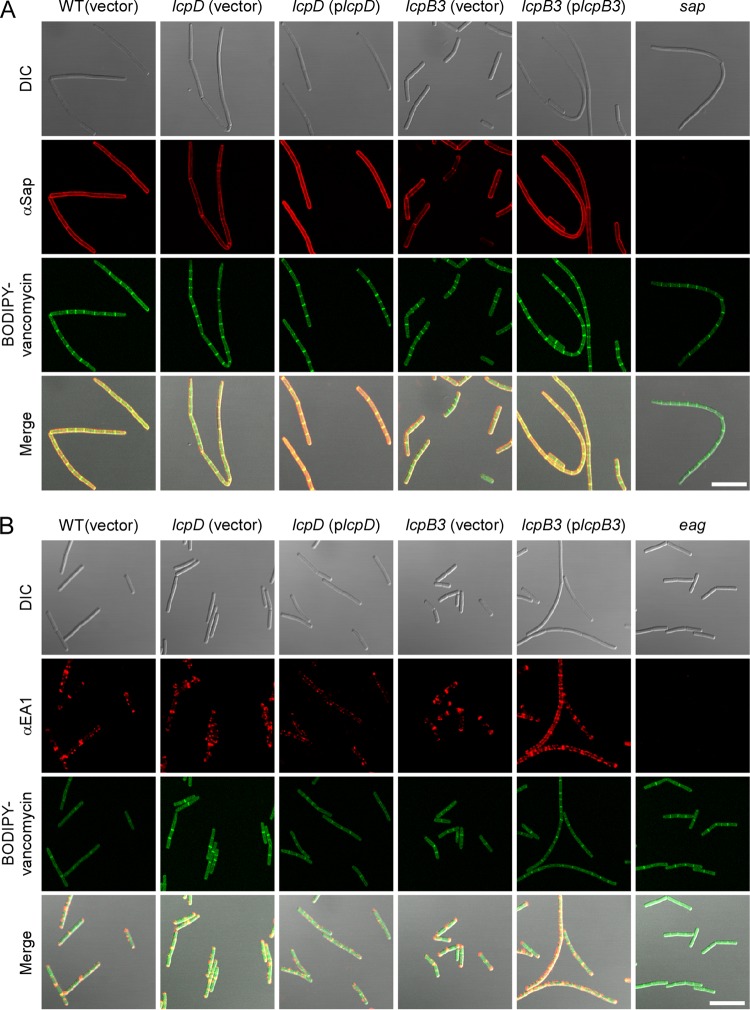
B. anthracis chain length is impacted by genes that affect SCWP synthesis (gneZ) (23) or modification (csaB, patA1/2, and patB1/2) (14, 20) as well as the S-layer (sap) and S-layer-associated proteins that are anchored to it (bslO) or that promote the secretion of their precursors (secA2 and slaP) (4, 5, 12). We asked whether lcp mutations, which appear to affect SCWP attachment to peptidoglycan, also impact the assembly of S-layer and S-layer-associated proteins. Spores from individual B. anthracis lcp mutants were inoculated into broth and incubated. At timed intervals, culture aliquots were examined for the deposition of Sap, EA1, BslR, and BslU on the surface of vegetative cells by immunofluorescence microscopy. The lcpB3 and lcpD mutants displayed S-layer assembly phenotypes, and our data analysis was limited to these strains. Compared to the case with wild-type bacilli, the abundance of Sap, the predominant S-layer protein of B. anthracis, was diminished in both lcpD and lcpB3 variants (Fig. 6A). This defect was rescued for each of these mutants by plasmid-borne expression of the corresponding wild-type lcp gene (Fig. 6A). As a control, sap mutant bacilli did not generate immunofluorescent signals during staining with Sap antiserum, demonstrating the specificity of these antibodies (Fig. 6A). The abundance and localized deposition of the EA1 S-layer protein at the septa and cell poles were not affected in the lcpD and lcpB3 mutants (Fig. 6B). BODIPY-vancomycin labeling was also unchanged, suggesting that the loss of lcp genes or their overexpression did not alter early cell division events.
FIG 6.
Deposition of S-layer proteins in the envelope of wild-type and lcpB3 and lcpD mutant Bacillus anthracis. Spores from wild-type B. anthracis Sterne (WT with empty vector) or lcpB3 or lcpD mutant strains containing empty vector or complementing plasmid were germinated in BHI broth and incubated for 3 h (A) or 8 h (B), fixed in formalin, and stained with polyclonal anti-Sap (A) or anti-EA1 (B) rabbit antibodies. Antibody binding and bacterial architecture were revealed with DyLight594-conjugated anti-rabbit secondary antibody and BODIPY-vancomycin, respectively. Differential interference contrast (DIC) and fluorescence microscopy images were acquired. Data sets were merged to reveal the location of the proteins in relation to the cell septa (BODIPY-vancomycin). As controls, sap and eag mutants were also stained with their respective antisera. Scale bar = 10 μm.
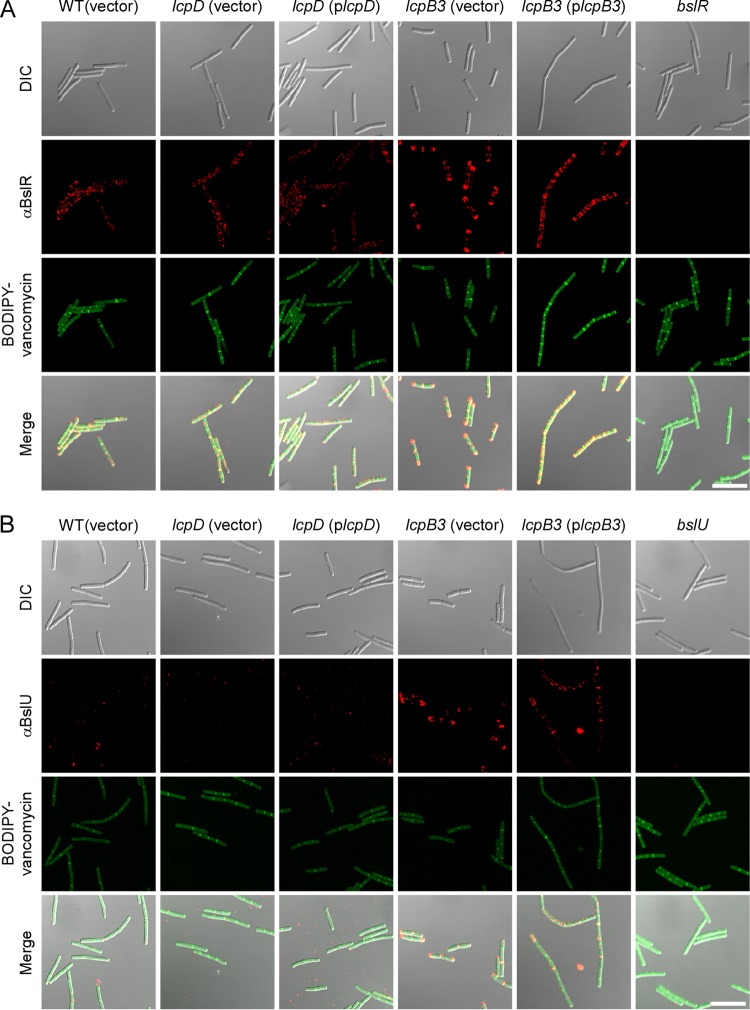
BslR and BslU, two S-layer-associated proteins with predicted murein hydrolase activity (N-acetylmuramoyl-l-alanine-amidase domains), attach to the SCWP and the envelope of B. anthracis in a manner requiring N-terminal S-layer homology domains, similar to Sap and EA1 (7). Unlike the two highly abundant S-layer proteins, BSLs are deposited with lesser abundance and at discrete locations in the envelope of B. anthracis (5). Immunofluorescence microscopy of wild-type bacteria revealed that BslR localizes in uneven patches throughout the envelope, whereas BslU is deposited in a few discrete puncta (Fig. 7). The lcpD mutation did not affect the distribution or abundance of BslR and BslU, as these proteins were detected in similar abundances and locations in the envelope of the lcpD mutant (Fig. 7). In contrast, in the lcpB3 mutant, BslR- and BslU-specific staining was increased in the lcpB3 mutant, where these proteins appear to accumulate at the most distal poles of chains formed from vegetative cells (Fig. 7). Plasmid-borne expression of wild-type lcpB3 (plcpB3) somewhat improved the distribution of these proteins along the elongated chains of vegetative forms; however, the intensity of BslU and BslR staining remained increased compared to that for wild-type bacilli (Fig. 7). We surmise that the phenotypes of B. anthracis lcpB3 (plcpB3) cells are caused by the overexpression of lcpB3 from the constitutive hrpK promoter in the high-copy-number plasmid pWWW412 (see Table S1 in the supplemental material). As noted above, lcpB3 overexpression also resulted in elongated chains of vegetative bacilli. We presume that the increased abundance of BslU and BslR near the poles and cell septa of the B. anthracis lcpB3 mutant may be responsible for the reduced chain length phenotype of this variant. On the other hand, the reduced abundance of Sap is likely responsible for the increased chain length phenotype of the B. anthracis lcpD mutant (5).
FIG 7.
Deposition of S-layer-associated proteins in the envelope of wild-type and lcpB3 and lcpD mutant Bacillus anthracis. Spores from wild-type B. anthracis Sterne (WT with empty vector) or lcpB3 or lcpD mutant strains containing empty vector or complementing plasmid were germinated in BHI broth, incubated for 8 h, fixed in formalin, and stained with polyclonal anti-BslR (A) or anti-BslU (B) rabbit antibodies. Antibody binding and bacterial architecture were revealed with DyLight594-conjugated anti-rabbit secondary antibody and BODIPY-vancomycin, respectively. Differential interference contrast (DIC) and fluorescence microscopy images were acquired. Data sets were merged to reveal the location of the proteins in relation to the cell septa (BODIPY-vancomycin). As controls, bslR and bslU mutants were also stained with their respective antisera. Scale bar = 10 μm.
DISCUSSION
In this study, we characterized lcp genes as determinants of B. anthracis cell size and chain length, a unique growth pattern that involves assembly of the SCWP and attached S-layer and BSL proteins (19). Earlier work had identified LCP proteins as catalysts of WTA and capsular polysaccharide attachment to peptidoglycan (29), specifically the formation of HF-sensitive phosphodiester bonds between the C-6 hydroxyl of N-acetylmuramic acid and WTA or capsular polysaccharide (47–49). Of note, recent work implicated an LCP-like protein from A. orsi in the glycosylation of a sortase-anchored surface protein, although the chemical nature of the glycosyl modification is still unknown (35). S. aureus Newman variants lacking all three lcp genes (Δlcp) cannot attach either WTA or the type 5 capsule, a polysaccharide with the repeat structure [→4)-β-ManNAc-(1→4)-α-FucNAc(OAc)-(1→3)-β-FucNAc-(1→]n (50), to peptidoglycan (32, 33). Staphylococcal WTA synthesis requires tagO and tagA, whose products promote synthesis of murein linkage units (GlcNAc-ManNAc) on bactoprenol carriers (49, 51, 52). The tagO murein linkage pathway is dispensable for the synthesis of capsular polysaccharide, which also involves bactoprenol as the lipid carrier (33). When examined with single and double lcp mutants, S. aureus LcpASa, LcpBSa, and LcpCSa have been found to exert substrate preferences for WTA or capsular polysaccharide. Nevertheless, each enzyme displays functional redundancy and catalyzes attachment of both WTA and polysaccharide to peptidoglycan (33). Testing all six lcp genes of B. anthracis, we showed in this study for the first time that heterologous expression of LCPs, specifically encoded by lcp genes from B. anthracis (which does not synthesize RboP), restores WTA synthesis in the S. aureus Δlcp mutant. Full complementation and growth restoration were achieved with plasmids harboring lcpB2Ba, lcpCBa, and lcpDBa, whereas lcpB3Ba and lcpB4Ba plasmids achieved partial complementation. Only the lcpB1Ba plasmid failed to complement. As lcpB1Ba may not be expressed in staphylococci, it seems plausible that all LCP enzymes of B. anthracis are capable of catalyzing WTA attachment in S. aureus. We showed further that lcpD is required for efficient attachment of SCWP to B. anthracis peptidoglycan. Together, these observations suggest that LCP enzymes recognize bactoprenol-(PO4)2-linked amino sugars but not RboP as the substrate for transfer to peptidoglycan. Lipid-linked LCP substrates include the GlcNAc-ManNAc moiety of murein linkage units, the ManNAc-FucNAc-FucNAc repeats of capsular polysaccharide, and the ManNAc-GlcNAc-GlcNAc repeats of SCWP.
Mutations in B. anthracis lcpD or lcpB3 are associated with cell size and chain length phenotypes that have their underpinnings in SCWP assembly defects. In this regard, B. anthracis more closely resembles B. subtilis, in which mutations in individual lcp genes (tagT, tagU, and tagV) display discrete defects in teichoic acid attachment to peptidoglycan and in which the LCP enzymes TagT, TagU, and TagV assemble along MreB scaffolds (29, 53), cytoskeletal elements that inform assembly and disassembly of peptidoglycan and thereby determine bacterial cell shape (54, 55). The phenotypic differences between bacilli and staphylococci may be explained through differences in cell structure and cytokinesis. S. aureus forms spherical cells that replicate without MreB by forming cross wall structures for cytokinesis, peptidoglycan synthesis, and cell separation (56, 57). Rod-shaped bacilli, on the other hand, require MreB proteins for maintenance of cell shape and discrete mechanisms that support peptidoglycan and teichoic acid synthesis at the cylindrical and polar portions of their envelope (29). In agreement with these models, mreB, tagTUV, and tagO mutations each abrogate the rod-shaped character of B. subtilis (29). We presume that mutations in the corresponding genes of B. anthracis would cause similar phenotypes, as occurs when TagO is inhibited by tunicamycin (14).
Why has B. anthracis evolved six LCP enzymes when B. subtilis makes do with three? Unlike B. subtilis (58), B. anthracis, although unable to synthesize RboP, appears to require tagO for growth (14). The tagO gene is part of a 50-kb genomic region of B. subtilis that harbors virtually all genes required for WTA synthesis, including tagTUV (29). On the B. anthracis chromosome, lcpD (BAS5047) is also located in close proximity to tagO (BAS5050) and gneY (BAS5048) yet at a distance from gneZ (BAS5117) (23); of note, only gneZ is required for SCWP synthesis and is located within the sps gene cluster, which has been proposed to support SCWP synthesis in B. cereus group members (23, 24). Thus, the chromosomal region of B. anthracis associated with SCWP synthesis, which is currently thought to encompass the tagO and the sps gene clusters, harbors only two of the six lcp genes, lcpD and lcpC (BAS5115). The remaining genes—lcpB1, lcpB2, lcpB3, and lcpB4—are distributed elsewhere in the chromosome and are not associated with genes that are known to contribute to the synthesis of murein linkage units or polysaccharides. We presume that the expanded repertoire of B. anthracis LCP enzymes may attach different types of secondary cell wall polymers to peptidoglycan. To explore this possibility, future work could seek to identify peptidoglycan-attached polymers in variants that express only one of the six LCP enzymes.
Interestingly, lcpB3 mutant bacilli form smaller cells as well as shortened chains and, conversely, plasmid overexpression of lcpB3 results in chains that are longer than wild type. These phenotypes are unique to lcpB3 and suggest a direct contribution to the bacterial cell cycle compared, for example, to the small cell phenotype observed upon overexpression of the virulence gene set (59). Both the lcpD and lcpB3 mutants assembled reduced amounts of Sap, the predominant S-layer protein, in their envelope, although the spatial distribution of Sap was not affected. A clear difference between the lcpD and lcpB3 mutants was observed with BslU and BslR, two predicted murein hydrolases with SLH domains (60). In wild-type bacilli, BslR is found in small patches along the cylindrical surface, whereas BslU appears to be deposited as small puncta in the immediate vicinity of septal peptidoglycan. This distribution was not affected in lcpD bacilli. In contrast, the deposition of BslR and BslU into the envelope of lcpB3 mutant bacilli was increased and both proteins accumulated at the poles of the most distal cells within lcpB3 chains. BslR and BslU are members of the family of B. anthracis N-acetylmuramoyl-l-alanine amidases with S-layer homology domains, which exist neither in B. subtilis nor in S. aureus (7). Plasmid pXO-2, which has been lost in the B. anthracis Sterne strain, encodes AmiA, one of the seven members of this family of amidases, while the remaining six genes are located on the chromosome (60). Future work must reveal the contributions of BslR and BslU toward regulated chain length of B. anthracis and the specific requirements for the unique distribution pattern of these BSLs in the bacterial envelope.
Supplementary Material
ACKNOWLEDGMENTS
We thank Derek Elli for assistance and members of our laboratory for discussion and comments on the manuscript.
M.L.Z. and J.M.L. are trainees of the Medical Scientist Training Program at the University of Chicago and are supported by a National Institutes of Health (NIH) training grant (GM07281). J.M.L. is the recipient of NIH Ruth L. Kirschstein National Research Service award 1F30AI110036-01. Y.G.Y.C. is the recipient of American Heart Association award 13POST16980091. This research was supported by grant AI069227 from the National Institute of Allergy and Infectious Diseases, Infectious Disease Branch (to O.S. and D.M.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02364-14.
REFERENCES
- 1.Koch R. 1876. Die Ätiologie der Milzbrand-Krankheit, begründet auf die Entwicklungsgeschichte des Bacillus anthracis. Beitr Biol Pflanzen 2:277–310. [Google Scholar]
- 2.Mock M, Fouet A. 2001. Anthrax. Annu Rev Microbiol 55:647–671. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- 3.Ruthel G, Ribot WJ, Bavari S, Hoover T. 2004. Time-lapse confocal imaging of development of Bacillus anthracis in macrophages. J Infect Dis 189:1313–1316. doi: 10.1086/382656. [DOI] [PubMed] [Google Scholar]
- 4.Anderson VJ, Kern JW, McCool JW, Schneewind O, Missiakas DM. 2011. The SLH domain protein BslO is a determinant of Bacillus anthracis chain length. Mol Microbiol 81:192–205. doi: 10.1111/j.1365-2958.2011.07688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kern VJ, Kern JW, Theriot JA, Schneewind O, Missiakas DM. 2012. Surface (S)-layer proteins Sap and EA1 govern the binding of the S-layer associated protein BslO at the cell septa of Bacillus anthracis. J Bacteriol 194:3833–3840. doi: 10.1128/JB.00402-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang YT, Oh SY, Hendrickx AP, Lunderberg JM, Schneewind O. 2013. Bacillus cereus G9241 S-layer assembly contributes to the pathogenesis of anthrax-like disease in mice. J Bacteriol 195:596–605. doi: 10.1128/JB.02005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kern JW, Schneewind O. 2008. BslA, a pXO1-encoded adhesin of Bacillus anthracis. Mol Microbiol 68:504–515. doi: 10.1111/j.1365-2958.2008.06169.x. [DOI] [PubMed] [Google Scholar]
- 8.Okinaka RT, Cloud K, Hampton O, Hoffmaster AR, Hill KK, Keim P, Koehler TM, Lamke G, Kumano S, Mahillon J, Manter D, Martinez Y, Ricke D, Svensson R, Jackson PJ. 1999. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J Bacteriol 181:6509–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green BD, Battisti L, Koehler TM, Thorne CB, Ivins BE. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect Immun 49:291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collier RJ, Young JA. 2003. Anthrax toxin. Annu Rev Cell Dev Biol 19:45–70. doi: 10.1146/annurev.cellbio.19.111301.140655. [DOI] [PubMed] [Google Scholar]
- 11.Candela T, Fouet A. 2006. Poly-gamma-glutamate in bacteria. Mol Microbiol 60:1091–1098. doi: 10.1111/j.1365-2958.2006.05179.x. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen-Mau S-M, Oh SY, Kern V, Missiakas D, Schneewind O. 2012. Secretion genes as determinants of Bacillus anthracis chain length. J Bacteriol 194:3841–3850. doi: 10.1128/JB.00384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mesnage S, Fontaine T, Mignot T, Delepierre M, Mock M, Fouet A. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J 19:4473–4484. doi: 10.1093/emboj/19.17.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kern J, Ryan C, Faull K, Schneewind O. 2010. Bacillus anthracis surface-layer proteins assemble by binding to the secondary cell wall polysaccharide in a manner that requires csaB and tagO. J Mol Biol 401:757–775. doi: 10.1016/j.jmb.2010.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kern JW, Wilton R, Zhang R, Binkowski A, Joachimiak A, Schneewind O. 2011. Structure of the SLH domains from Bacillus anthracis surface array protein. J Biol Chem 286:26042–26049. doi: 10.1074/jbc.M111.248070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baranova E, Fronzes R, Garcia-Pino A, Van Gerven N, Papapostolou D, Péhau-Arnaudet G, Pardon E, Steyaert J, Howorka S, Remaut H. 2012. SbsB structure and lattice reconstruction unveil Ca2+ triggered S-layer assembly. Nature 487:119–122. [DOI] [PubMed] [Google Scholar]
- 17.Mesnage S, Tosi-Couture E, Mock M, Gounon P, Fouet A. 1997. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol Microbiol 23:1147–1155. doi: 10.1046/j.1365-2958.1997.2941659.x. [DOI] [PubMed] [Google Scholar]
- 18.Couture-Tosi E, Delacroix H, Mignot T, Mesnage S, Chami M, Fouet A, Mosser G. 2002. Structural analysis and evidence for dynamic emergence of Bacillus anthracis S-layer networks. J Bacteriol 184:6448–6456. doi: 10.1128/JB.184.23.6448-6456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fagan RP, Fairweather NF. 2014. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol 12:211–222. doi: 10.1038/nrmicro3213. [DOI] [PubMed] [Google Scholar]
- 20.Lunderberg JM, Nguyen-Mau SM, Richter GS, Wang YT, Dworkin J, Missiakas DM, Schneewind O. 2013. Bacillus anthracis acetyltransferases PatA1 and PatA2 modify the secondary cell wall polysaccharide and affect the assembly of S-layer proteins. J Bacteriol 195:977–989. doi: 10.1128/JB.01274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forsberg LS, Abshire TG, Friedlander A, Quinn CP, Kannenberg EL, Carlson RW. 2012. Localization and structural analysis of a conserved pyruvylated epitope in Bacillus anthracis secondary cell wall polysaccharides and characterization of the galactose deficient wall polysaccharide from avirulent B. anthracis CDC 684. Glycobiology 22:1103–1117. doi: 10.1093/glycob/cws080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhury B, Leoff C, Saile E, Wilkins P, Quinn CP, Kannenberg EL, Carlson RW. 2006. The structure of the major cell wall polysaccharide of Bacillus anthracis is species specific. J Biol Chem 281:27932–27941. doi: 10.1074/jbc.M605768200. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y-T, Missiakas D, Schneewind O. 9 June 2014. GneZ, a UDP-GlcNAc 2-epimerase, is required for S-layer assembly and vegetative growth of Bacillus anthracis. J Bacteriol doi: 10.1128/JB.01829-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuch R, Pelzek AJ, Raz A, Euler CW, Ryan PA, Winer BY, Farnsworth A, Bhaskaran SS, Stebbins CE, Xu Y, Clifford A, Bearss DJ, Vankayalapati H, Goldberg AR, Fischetti VA. 2013. Use of a bacteriophage lysin to identify a novel target for antimicrobial development. PLoS One 8:e60754. doi: 10.1371/journal.pone.0060754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubscher J, Luthy L, Berger-Bachi B, Stutzmann Meier P. 2008. Phylogenetic distribution and membrane topology of the LytR-CpsA-Psr protein family. BMC Genomics 9:617. doi: 10.1186/1471-2164-9-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bender MH, Cartee RT, Yother J. 2003. Positive correlation between tyrosine phosphorylation of CpsD and capsular polysaccharide production in Streptococcus pneumoniae. J Bacteriol 185:6057–6066. doi: 10.1128/JB.185.20.6057-6066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cieslewicz MJ, Kasper DL, Wang Y, Wessels MR. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J Biol Chem 276:139–146. doi: 10.1074/jbc.M005702200. [DOI] [PubMed] [Google Scholar]
- 28.Hanson BR, Runft DL, Streeter C, Kumar A, Carion TW, Neely MN. 2012. Functional analysis of the CpsA protein of Streptococcus agalactiae. J Bacteriol 194:1668–1678. doi: 10.1128/JB.06373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, Lewis RJ, Vollmer W, Daniel RA, Errington J. 2011. A widespread family of bacterial cell wall assembly proteins. EMBO J 30:4931–4941. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eberhardt A, Hoyland CN, Vollmer D, Bisle S, Cleverley RM, Johnsborg O, Håvarstein LS, Lewis RJ, Vollmer W. 2012. Attachment of capsular polysaccharide to the cell wall in Streptococcus pneumoniae. Microb Drug Resist 18:240–255. doi: 10.1089/mdr.2011.0232. [DOI] [PubMed] [Google Scholar]
- 31.Dengler V, Stutzmann Meier P, Heusser R, Kupferschmied P, Fazekas J, Friebe S, Staufer SB, Majcherczyk PA, Moreillon P, Berger-Bächi B, McCallum N. 2012. Deletion of hypothetical wall teichoic acid ligases in Staphylococcus aureus activates the cell wall stress response. FEMS Microbiol Lett 333:109–120. doi: 10.1111/j.1574-6968.2012.02603.x. [DOI] [PubMed] [Google Scholar]
- 32.Chan YGY, Frankel MB, Dengler V, Schneewind O, Missiakas DM. 2013. Staphylococcus aureus mutants lacking the LytR-CpsA-Psr (LCP) family of enzymes release wall teichoic acids into the extracellular medium. J Bacteriol 195:4650–4659. doi: 10.1128/JB.00544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan YG, Kim HK, Schneewind O, Missiakas D. 2014. The capsular polysaccharide of Staphylococcus aureus is attached to peptidoglycan by the LytR-CpsA-Psr (LCP) family of enzymes. J Biol Chem 289:15680–15690. doi: 10.1074/jbc.M114.567669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Over B, Heusser R, McCallum N, Schulthess B, Kupferschmied P, Gaiani JM, Sifri CD, Berger-Bächi B, Stutzmann Meier P. 2011. LytR-CpsA-Psr proteins in Staphylococcus aureus display partial functional redundancy and the deletion of all three severely impairs septum placement and cell separation. FEMS Microbiol Lett 320:142–151. doi: 10.1111/j.1574-6968.2011.02303.x. [DOI] [PubMed] [Google Scholar]
- 35.Wu C, Huang I-H, Chang C, Reardon-Robinson ME, Das A, Ton-That H. 17 September 2014. Lethality of sortase depletion in Actinomyces oris caused by excessive membrane accumulation of a surface glycoprotein. Mol Microbiol doi: 10.1111/mmi.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garufi G, Hendrickx AP, Beeri K, Kern JW, Sharma A, Richter SG, Schneewind O, Missiakas DM. 2012. Synthesis of lipoteichoic acids in Bacillus anthracis. J Bacteriol 194:4312–4321. doi: 10.1128/JB.00626-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richter GS, Anderson VJ, Garufi G, Lu L, Joachimiak A, He C, Schneewind O, Missiakas D. 2009. Capsule anchoring in Bacillus anthracis occurs by a transpeptidation mechanism that is inhibited by capsidin. Mol Microbiol 71:404–420. doi: 10.1111/j.1365-2958.2008.06533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sterne M. 1937. Avirulent anthrax vaccine. Onderstepoort J Vet Sci Animal Ind 21:41–43. [PubMed] [Google Scholar]
- 39.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. 2007. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes. J Bacteriol 190:300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fulford W, Model P. 1984. Specificity of translational regulation by two DNA-binding proteins. J Mol Biol 173:211–226. doi: 10.1016/0022-2836(84)90190-6. [DOI] [PubMed] [Google Scholar]
- 41.Kim HU, Goepfert JM. 1974. A sporulation medium for Bacillus anthracis. J Appl Bacteriol 37:265–267. doi: 10.1111/j.1365-2672.1974.tb00438.x. [DOI] [PubMed] [Google Scholar]
- 42.Tam C, Glass EM, Anderson DM, Missiakas D. 2006. Transposon mutagenesis of Bacillus anthracis strain Sterne using Bursa aurealis. Plasmid 56:74–77. doi: 10.1016/j.plasmid.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 43.Marraffini LA, Schneewind O. 2006. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Mol Microbiol 62:1402–1417. doi: 10.1111/j.1365-2958.2006.05469.x. [DOI] [PubMed] [Google Scholar]
- 44.Gaspar AH, Marraffini LA, Glass EM, DeBord KL, Ton-That H, Schneewind O. 2005. Bacillus anthracis sortase A (SrtA) anchors LPXTG motif-containing surface proteins to the cell wall envelope. J Bacteriol 187:4646–4655. doi: 10.1128/JB.187.13.4646-4655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bubeck Wardenburg J, Williams WA, Missiakas D. 2006. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci U S A 103:13831–13836. doi: 10.1073/pnas.0603072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson M, Aly KA, Chen YH, Missiakas D. 2013. Secretion of atypical protein substrates by the ESAT-6 secretion system of Staphylococcus aureus. Mol Microbiol 90:734–743. doi: 10.1111/mmi.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munoz E, Ghuysen J-M, Heymann H. 1967. Cell walls of Streptococcus pyogenes type 14. C polysaccharide-peptidoglycan and G polysaccharide-peptidoglycan complexes. Biochemistry 6:3659–3670. [DOI] [PubMed] [Google Scholar]
- 48.Yokoyama K, Miyashita T, Arakai Y, Ito E. 1986. Structure and functions of linkage unit intermediates in the biosynthesis of ribitol teichoic acids in Staphylococcus aureus H and Bacillus subtilis W23. Eur J Biochem 161:479–489. doi: 10.1111/j.1432-1033.1986.tb10469.x. [DOI] [PubMed] [Google Scholar]
- 49.Kojima N, Arakai Y, Ito E. 1985. Structure of the linkage units between ribitol teichoic acids and peptidoglycan. J Bacteriol 161:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones C. 2005. Revised structures for the capsular polysaccharides from Staphylococcus aureus types 5 and 8, components of novel glycoconjugate vaccines. Carbohydr Res 340:1097–1106. doi: 10.1016/j.carres.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 51.Xia G, Peschel A. 2008. Toward the pathway of S. aureus WTA biosynthesis. Chem Biol 15:95–96. doi: 10.1016/j.chembiol.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 52.D'Elia MA, Pereira MP, Chung YS, Zhao W, Chau A, Kenney TJ, Sulavik MC, Black TA, Brown ED. 2006. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J Bacteriol 188:4183–4189. doi: 10.1128/JB.00197-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones LJ, Carballido-López R, Errington J. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913–922. doi: 10.1016/S0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 54.Graumann PL. 2009. Dynamics of bacterial cytoskeletal elements. Cell Motil Cytoskeleton 66:909–914. doi: 10.1002/cm.20381. [DOI] [PubMed] [Google Scholar]
- 55.Domínguez-Cuevas P, Porcelli I, Daniel RA, Errington J. 2013. Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis. Mol Microbiol 89:1084–1098. doi: 10.1111/mmi.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinho MG, Kjos M, Veening JW. 2013. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol 11:601–614. doi: 10.1038/nrmicro3088. [DOI] [PubMed] [Google Scholar]
- 57.Giesbrecht P, Kersten T, Maidhof H, Wecke J. 1998. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol Mol Biol Rev 62:1371–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D'Elia MA, Henderson JA, Beveridge TJ, Heinrichs DE, Brown ED. 2009. The N-acetylmannosamine transferase catalyzes the first committed step of teichoic acid assembly in Bacillus subtilis and Staphylococcus aureus. J Bacteriol 191:4030–4034. doi: 10.1128/JB.00611-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mujtaba S, Winer BY, Jaganathan A, Patel J, Sgobba M, Schuch R, Gupta YK, Haider S, Wang R, Fischetti VA. 2013. Anthrax SET protein: a potential virulence determinant that epigenetically represses NF-kappaB activation in infected macrophages. J Biol Chem 288:23458–23472. doi: 10.1074/jbc.M113.467696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kern JW, Schneewind O. 2010. BslA, the S-layer adhesin of Bacillus anthracis, is a virulence factor for anthrax pathogenesis. Mol Microbiol 75:324–332. doi: 10.1111/j.1365-2958.2009.06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.