Abstract
Levels of 2-oxoglutarate (2-OG) reflect nitrogen status in many bacteria. In heterocystous cyanobacteria, a spike in the 2-OG level occurs shortly after the removal of combined nitrogen from cultures and is an integral part of the induction of heterocyst differentiation. In this work, deletion of one of the two annotated trpE genes in Anabaena sp. strain PCC 7120 resulted in a spike in the 2-OG level and subsequent differentiation of a wild-type pattern of heterocysts when filaments of the mutant were transferred from growth on ammonia to growth on nitrate. In contrast, 2-OG levels were unaffected in the wild type, which did not differentiate under the same conditions. An inverted-repeat sequence located upstream of trpE bound a central regulator of differentiation, HetR, in vitro and was necessary for HetR-dependent transcription of a reporter fusion and complementation of the mutant phenotype in vivo. Functional complementation of the mutant phenotype with the addition of tryptophan suggested that levels of tryptophan, rather than the demonstrated anthranilate synthase activity of TrpE, mediated the developmental response of the wild type to nitrate. A model is presented for the observed increase in 2-OG in the trpE mutant.
INTRODUCTION
The Krebs cycle intermediate 2-oxoglutarate (2-OG) is an intracellular indicator of nitrogen limitation in many bacteria (reviewed in reference 1). It is the carbon skeleton for assimilation of ammonia in bacteria that primarily use the GS-GOGAT system, and therefore, the concentration of 2-OG rises when ammonia is not available. Sensors of 2-OG include the ubiquitous PII superfamily of proteins and the CRP family protein NtcA, which in cyanobacteria functionally replaces the NtrB-NtrC system of enteric bacteria (reviewed in reference 2). Direct interaction with 2-OG increases the affinity of the NtcA transcriptional regulator for DNA targets and is necessary for initiation of transcription (3) and its interaction with PII frees PipX (4), which is thought to stabilize the active form of NtcA (5). NtcA regulates the expression of genes required for transformation of inorganic forms of nitrogen such as nitrate, nitrite, and dinitrogen into ammonia, as well as the developmental program of heterocystous cyanobacteria.
Anabaena sp. strain PCC 7120 is a filamentous cyanobacterium that differentiates nitrogen-fixing heterocysts when a source of combined nitrogen is scarce (reviewed in references 6 and 7). During growth with a combined nitrogen source, usually ammonia or nitrate under laboratory conditions, Anabaena filaments are comprised exclusively of photosynthetic vegetative cells. When combined nitrogen is absent, however, a periodic pattern of morphologically distinct, nitrogen-fixing heterocyst cells is formed at approximately every 10th cell in the filament. The cascade of events involved in heterocyst differentiation can be divided into four phases: induction of differentiation, pattern formation, commitment to differentiation, and heterocyst maturation (8).
The induction phase begins with the depletion of environmental nitrogen, which causes a transient increase in the intracellular concentration of 2-OG (9, 10). This increase is necessary for the induction of differentiation and initiates a transcriptional cascade that leads to upregulation of the master regulator of differentiation, hetR, after about 30 min (11–13). HetR is a transcriptional regulator that interacts directly with DNA to modulate the expression of genes. The HetR regulon includes hetR itself, as well as a gene encoding an inhibitor of HetR activity, patS (12, 14). It is through the interaction of HetR and PatS that the spatial patterning of differentiating cells is determined. Patterned cells then proceed through commitment and maturation to produce a microoxic environment in which the nitrogenase complex functions. Thus, the developmental cascade that leads to heterocyst differentiation begins with an elevation of 2-OG levels in response to the availability of combined nitrogen. While much is known about the events following induction, comparatively little is known about the contributions of nitrogenous metabolites, such as amino acids, to this critical first step.
Previous work with amino acid analogs has suggested that tryptophan metabolism may contribute to heterocyst differentiation. The addition of dl-7-azatryptophan (AZAT), a tryptophan analog, to cultures of Anabaena sp. strain CA (15), Anabaena cylindrica (16, 17), and Anabaena variabilis (18) resulted in the formation of heterocysts even in the presence of ammonia or nitrate. Cultures of Anabaena sp. strain CA incubated with AZAT for as little as 1 to 2 h yielded functional heterocysts, but without a continued source of AZAT, additional heterocysts were not produced in successive rounds of cell division (15). AZAT was shown to be a potent inhibitor of anthranilate synthase, which is encoded by the trpE and trpG genes and catalyzes the first committed step of tryptophan biosynthesis, yet the functions of glutamate or glutamine synthase were not severely impaired (15, 19). Furthermore, the addition of AZAT to continuous cultures of Anabaena sp. strain CA resulted in a transient increase in the concentration of glutamine (20), which serves as the source of an amino group for the synthesis of anthranilate. Despite these findings supporting the promotion of differentiation by inhibition of anthranilate synthase activity, the effects of AZAT on development have, with one notable exception (21), been attributed largely to its potential to disrupt protein function in general, because unlike AZAT, tryptophan analogs that are not known to be incorporated into proteins had no effect on heterocyst differentiation (9, 15, 19).
Here, we show that one of the two trpE genes from Anabaena sp. strain PCC 7120 encodes a protein with anthranilate synthase activity that was necessary for regulation of the induction of differentiation. In its absence, a transient spike in 2-OG levels in the presence of nitrate led to the formation of a pattern of heterocysts that is observed only in the absence of fixed nitrogen in the wild type. Consistent with its role in differentiation, transcription of the trpE gene appeared to be directly regulated by HetR.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Anabaena sp. strain PCC 7120 (wild type) and its derivatives were routinely cultured in BG-11 medium as previously described (22). Medium for growth of Anabaena on 6 mM ammonia as the nitrogen source (BG-11–NH4) was prepared as previously described (23). To induce heterocyst differentiation, cultures grown to exponential phase (optical density at 750 nm [OD750] of 0.3 to 0.7) were washed three times with medium containing 17.6 mM nitrate (BG-11; for a step down from growth on BG-11–NH4) or medium lacking combined nitrogen (BG-110; for a step down from growth on BG-11). To determine heterocyst percentages, 500 cells were counted. All results are expressed as the average of three replicates. Error bars represent 1 standard deviation. dl-Tryptophan and the tryptophan analog AZAT (Sigma) were added to Anabaena cultures at concentrations of 100 and 10 μM, respectively. Transcription from the copper-inducible petE promoter was induced by the addition of copper to a final concentration of 2 μM (24). Plasmids were introduced into Anabaena strains by conjugation from Escherichia coli as previously described (25). Growth of E. coli, concentrations of antibiotics, and conditions for photomicroscopy of Anabaena were as previously described (22, 26). Fluorescence from green fluorescent protein (GFP) was imaged by confocal microscopy with an excitation wavelength of 488 nm and an emission wavelength of 510 nm as previously described (27). E. coli strains derived from JA221 were grown in M9 (ammonium chloride/salts) minimal medium with 2% Casamino Acids and supplemented with 0.1 μg/ml of thiamine and 1 μM l-tryptophan as needed (28). Transcription from the arabinose-inducible araBAD promoter was induced by the addition of 0.2% l-arabinose (29).
Plasmid construction.
The strains and plasmids used in this study are listed in Table 1. The primers used in this study are listed in Table S1 in the supplemental material. The integrity of all PCR-derived constructs was verified by sequencing.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| Anabaena sp. | ||
| PCC 7120 | Wild type | Institut Pasteur Culture Collection |
| UHM101 | ΔpatA | 49 |
| UHM103 | ΔhetR | 22 |
| UHM335 | ΔtrpE | This study |
| E. coli JA221 | ΔtrpE, tryptophan auxotroph | 45 |
| Plasmids | ||
| pAM504 | Shuttle vector for replication in E. coli and Anabaena; Kmr Nmr | 30 |
| pAM1956 | Shuttle vector pAM504 with promoterless gfp | 33 |
| pRL277 | Suicide vector; Spr Smr | 12 |
| pRR159 | pAM504 with PhetN-hetN-YFP | 27 |
| pSW7848 | Source of araC-ParaBAD | 32 |
| pET28b+hetR | Expression vector for purifying polyhistidine epitope-tagged HetR; Kmr | 35 |
| pPJAV123 | pAM504 with NdeI site removed | 50 |
| pPJAV149 | pRL277 used to make UHM335 | This study |
| pPJAV153 | Shuttle vector pAM504 for creating copper-inducible C-terminal YFP translational fusions | 50 |
| pPJAV297 | pAM504 with PpetE-trpE translationally fused to YFP | This study |
| pPJAV360 | pAM504 with araC-ParaBAD | This study |
| pPJAV370 | pAM504 with ParaBAD-trpE | This study |
| pPJAV381 | pAM1956 with PtrpE | This study |
| pPJAV382 | pAM1956 with PtrpE with mutated HetR binding site | This study |
| pAHB112 | pRL277 with PtrpE-trpE | This study |
| pAHB113 | pRL277 with PtrpE-trpE with mutated HetR binding site | This study |
Km, kanamycin; Nm, neomycin; Sp, spectinomycin; Sm, streptomycin.
Plasmid pPJAV149 is a suicide vector based on pRL277 (12) that was used to cleanly delete all but the first 30 and last 30 nucleotides of the coding region of alr3233, one of the two trpE genes. Regions upstream and downstream of trpE were amplified by PCR from chromosomal DNA with primer sets alr3233-up-F and alr3233-up-R and alr3233-dn-F and alr3233-dn-R, respectively. The up- and downstream fragments were fused together by overlap extension PCR (31) and cloned into the SmaI site of pBlueScript SK+ (Stratagene). The fused product was cloned as a BamHI-SacI fragment into the BglII-SacI sites in pRL277.
Plasmid pPJAV297 is a mobilizable shuttle vector based on pAM504 carrying PpetE-trpE translationally fused to yfp. The coding region of trpE was amplified by PCR from chromosomal DNA with primers trpE-NdeI-F and trpE-BglII-R and cloned into the SmaI site of pBlueScript SK+. The trpE coding region was moved as an NdeI-BglII fragment into the NdeI-BamHI sites of pPJAV153 (50).
Plasmid pPJAV360 is a mobilizable shuttle vector based on pAM504 (30) carrying araC-ParaBAD with the BamHI site removed from ParaBAD. Fragments up- and downstream of the BamHI site in ParaBAD were amplified by PCR from pSW7848 (32) with primers araC-XhoI-F and araC-dBam-OEX-R and primers araC-dBam-OEX-F and PmrBAD-BaS-SacI-R, respectively. The up- and downstream fragments were fused together by overlap extension PCR and cloned as an XhoI-SacI fragment into the SalI-SacI sites of pAM504.
Plasmid pPJAV370 is a mobilizable shuttle vector based on pAM504 carrying ParaBAD-trpE. The coding region of trpE was amplified by PCR from chromosomal DNA with primers trpE-BamHI-F and trpE-SacI-R and cloned as a BamHI-SacI fragment into the same sites in pPJAV360.
Plasmid pPJAV381 is a mobilizable shuttle vector based on pAM1956 (33) carrying the trpE promoter region transcriptionally fused to gfp. A fragment containing the trpE promoter region was amplified by PCR from chromosomal DNA with primers PtrpE-XhoI-F and PtrpE-OEX-R. The product was cloned into the SmaI site of pAM1956, and directionality was verified by PCR.
Plasmid pPJAV382 is a mobilizable shuttle vector based on pAM1956 carrying the trpE promoter region transcriptionally fused to gfp with transversions at each of the 17 nucleotides comprising the HetR binding core (from 5′-GATGGGTTACACCCCTC-3′ to 5′-TCGTTTGGCACAAAAGA-3′). The trpE promoter region containing a mutated HetR binding site was amplified by PCR from pAHB113 with primers PtrpE-XhoI-F and PtrpE-OEX-R. The product was cloned into the SmaI site of pAM1956, and directionality was verified by PCR.
Plasmid pAHB112 is a suicide vector based on pRL277 used to integrate the native trpE promoter and coding region into the trpE locus in UHM335 as a single recombinant. The trpE promoter and coding region were amplified by PCR from chromosomal DNA with primers PtrpE-BamHI-F and trpE-SacI-R and cloned as a BamHI-SacI fragment into the BglII-SacI sites in pRL277.
Plasmid pAHB113 is a suicide vector based on pRL277 used to integrate the trpE promoter and coding region with transversions at each of the 17 nucleotides comprising the HetR binding core (as above) into the trpE locus in UHM335 as a single recombinant. Fragments up- and downstream of the HetR binding site were amplified by PCR from chromosomal DNA with primers PtrpE-BamHI-F and PtrpE-Rmut-OEX-R and primers PtrpE-Rmut-OEX-F and trpE-SacI-R, respectively. The up- and downstream fragments were fused together by overlap extension PCR and cloned as a BamHI-SacI fragment into the BglII-SacI sites in pRL277.
Strain construction.
The trpE gene, except for the first 30 and last 30 nucleotides of the coding region, was cleanly deleted from chromosomal DNA by allelic replacement as previously described (34) with plasmid pPJAV149 to create UHM335. The resultant strain was verified by PCR with primers trpE-up-out and trpE-down-out, which anneal outside the region of DNA used to make the deletion. The correct insertion of all single recombinants at the trpE locus was verified with primers trpE-out-F and trpE-int-R. The integrity of the HetR binding site in the trpE promoter region of all of the single recombinants tested was verified by sequencing.
Purification of recombinant HetR and DNA binding assays.
Recombinant HetR was overexpressed from pET28b+hetR and purified from BL21(DE3) cells as previously described (35). Electrophoretic mobility shift assays (EMSAs) were performed essentially as before (35), except that binding was assessed with 6% gel retardation polyacrylamide gels (Invitrogen), rather than agarose gels, according to the manufacturer's instructions. The 29-bp DNA binding fragments for hetP, hetPmut, trpE, and trpEmut were prepared by annealing primers R-bind 29mer top and R-bind 29mer btm, primers R-bsmut 29mer top and R-bsmut 29mer btm, primers trpE-110 and trpE-110-C, and primers trpE-Rmutfull and trpE-Rmutfull-C, respectively, as previously described (35). The larger DNA fragments of lacZ (268 bp), hetP (124 bp), trpE (155 bp), and trpE with a mutated HetR binding site (155 bp) that were utilized for binding assays were amplified from E. coli DH5α (Life Technologies) chromosomal DNA, Anabaena chromosomal DNA, or pAHB113 (as described above), respectively, by PCR with primers K12-PlacZ-F and K12-PlacZ-R, primers 233-F3-EcoRI and 233-R1-BamHI, and primers trpE-155-F and trpE-155-R, respectively.
Acetylene reduction assays, 2-OG quantification, and fluorescence quantification.
Acetylene reduction assays (22, 36), quantification of intracellular 2-OG pools (9, 37), measurement of total protein (38), and quantification and normalization of green and red fluorescence to report transcriptional levels (8) were performed as previously described.
RESULTS
HetR binds to an inverted DNA repeat in the trpE promoter region.
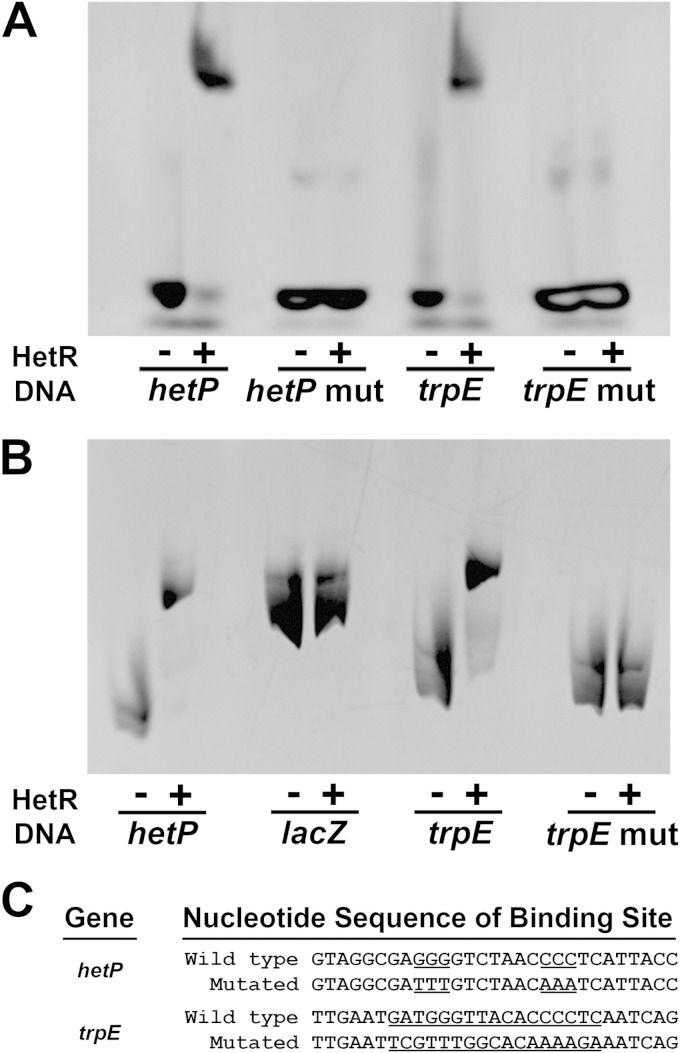
As part of a study designed to define additional HetR binding sites in the Anabaena genome, an inverted repeat in the promoter region of the gene alr3233, which is annotated as one of two trpE genes in the genome, was identified (8). To test for DNA-protein interaction, an EMSA was conducted with HetR and a 29-bp DNA fragment containing the potential HetR binding site derived from the trpE promoter region (Fig. 1A). A shifted species, indicative of HetR binding, was present for both the previously published hetP positive control 29-bp DNA fragment and the trpE binding site (26). Because the hetP and trpE binding sites migrated the same distance in the assay and near-complete shifting was observed at the HetR concentration utilized, we posit that HetR binds to these two sites similarly in vitro.
FIG 1.
EMSAs displaying the specific interaction of HetR with the binding site in the trpE promoter. The HetR concentration was 25 μM (A) or 2.5 μM (B), and the DNA concentration was 2 μM (A) or 0.3 μM (B). The DNA lengths are 29 bp (A), 124 bp (B; hetP), 268 bp (B; lacZ), and 155 bp (B; trpE and trpEmut). Nucleotide sequence comparison of the 29-bp DNA fragments utilized in EMSAs in this study (C). Transversion mutations of selected nucleotides are underlined. DNA sequences are presented in the 5′-to-3′ direction.
The sequence specificity of HetR has been tested for the hetP and hepA binding sites with the mutation of 6 and 17 nucleotides, respectively, present in the inverted-repeat-containing binding cores (8, 26). Mutations introduced into the hetP and hepA binding sites both abrogated binding in vitro, and mutations in the hepA binding site also abolished heterocyst-specific transcription from the hepA promoter. To test the sequence specificity of the trpE binding site, the binding of HetR to a 29-bp DNA fragment with transversions at each of the 17 bp comprising the trpE binding site core was assessed by EMSA (Fig. 1A). Even though the inverted-repeat structure of the binding site core was maintained, no shift was evident from either the mutated hetP or trpE binding sites, indicating that HetR did not interact with these DNA fragments in vitro. When larger DNA fragments were assessed for binding to determine whether HetR interacted with additional sequences outside the 29 bp utilized above for binding, only DNA with an intact HetR binding site produced a shifted species (Fig. 1B). We conclude that HetR binds specifically to the trpE binding site in a manner similar to that in which it binds to the hetP binding site.
A ΔtrpE mutant differentiates heterocysts under nitrogen-replete conditions.
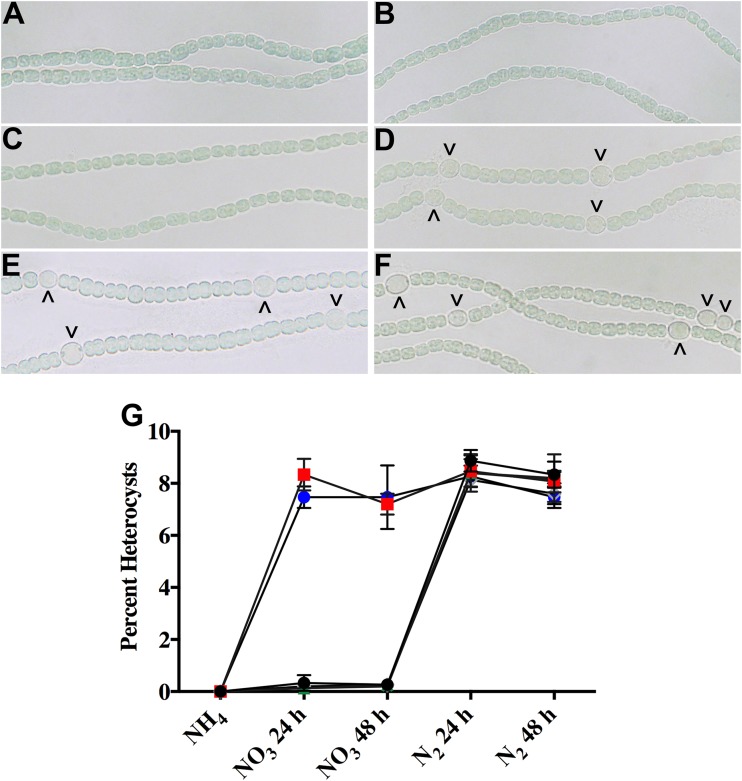
Many genes with a HetR binding site in their promoter regions (hetR [39], patS, pknE [40], hetP [26], hetZ [41], hepA [39], and alr1000 [8]) have been shown to contribute to heterocyst development or function. To determine whether the trpE gene (alr3233) is involved in heterocyst differentiation, the trpE coding region was cleanly deleted from the Anabaena chromosome. alr3233 is one of only two genes predicted to encode a TrpE protein in Anabaena, and we found that the ΔtrpE mutant did not require supplementation of the medium with tryptophan for survival. Growth of the wild type and the ΔtrpE mutant on ammonia, the preferred nitrogen source and the most repressive condition for heterocyst differentiation, resulted in filaments containing exclusively vegetative cells (Fig. 2A, B, and G). A step down from ammonia to growth on nitrate resulted in the development of a wild-type pattern of heterocysts (∼8%) in the ΔtrpE mutant, whereas the wild type remained largely undifferentiated under the same conditions (Fig. 2C, D, and G). The removal of combined nitrogen from nitrate-grown cultures resulted in the creation of a wild-type pattern of heterocysts (∼8%) in the wild type and maintenance of this pattern in the ΔtrpE mutant (Fig. 2E to G). The heterocysts produced by the ΔtrpE mutant during growth on nitrate were not capable of aerobic nitrogen fixation (see Fig. S1 in the supplemental material). However, 2 h after the removal of combined nitrogen, the heterocysts that formed in the presence of nitrate fixed nitrogen under aerobic conditions. A C-terminal translational fusion of TrpE to yfp, expressed from the copper-inducible petE promoter and induced with 2 μM copper, complemented the ΔtrpE mutant (Fig. 2G). Though the TrpE-YFP translational fusion complemented the ΔtrpE mutant, fluorescence localized to puncta. These fluorescent puncta are quite similar to those observed with Clp proteases and their various substrates tagged with GFP, suggesting that TrpE-YFP may be transported to discrete positions in the cell for degradation (see Fig. S2 in the supplemental material) (42–44). Complementation indicated that the formation of heterocysts in the presence of nitrate by the mutant was not due to a polar mutation. Taken together, these data suggest that the trpE gene product is involved in preventing differentiation during growth on nitrate.
FIG 2.
Phenotype of the ΔtrpE mutant (UHM335) under various growth conditions. Bright-field micrographs of the wild type (A, C, E) and UHM335 (B, D, F) during growth on ammonia (NH4) (A, B), on nitrate (NO2) (C, D), and without a source of combined nitrogen (N2) (E, F). Carets indicate heterocysts. The percentage of cells that were heterocysts in individual cultures was tracked over time as the source of nitrogen was varied (G). Cultures were initially grown with ammonia, and percentages of heterocysts were recorded. Ammonia was then replaced with nitrate, and heterocyst percentages were determined after 24 and 48 h. Nitrate was then removed from the culture medium, and heterocyst percentages were determined after 24 and 48 h of growth with dinitrogen as the only source of nitrogen. Average heterocyst percentages were determined from counts of 500 cells in three cultures of the wild type (black), UHM335 (red), the wild type with a plasmid containing PpetE-trpE-YFP (green), UHM335 (asterisks) with a plasmid containing PpetE-trpE-YFP, UHM335 with the trpE gene (open diamonds), and UHM335 with the trpE gene containing the mutated HetR binding site (blue). Error bars represent standard deviations.
The trpE gene requires an intact HetR binding site for proper expression.
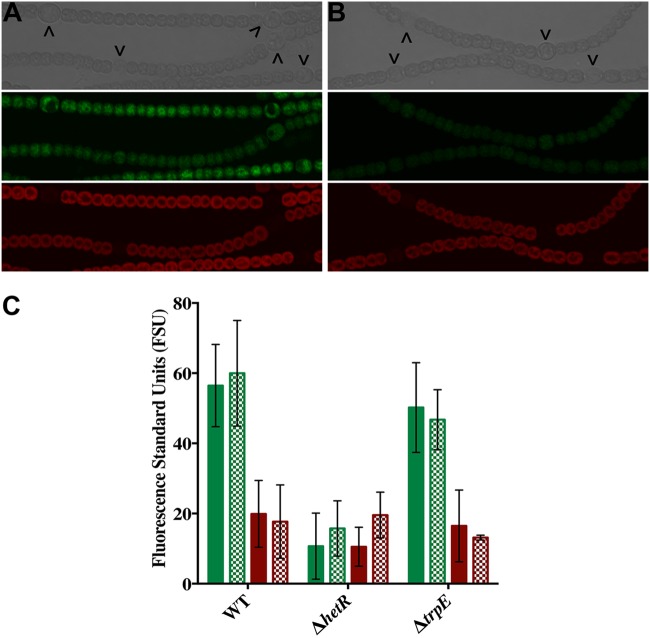
Recent studies have shown that an intact HetR binding site is required for proper expression of the hetZ and hepA genes (8, 41). To determine the involvement of the HetR binding site in trpE expression, transcriptional fusions of the gene for GFP (gfp) with the trpE promoter region (PtrpE) containing either a native or a mutated HetR binding site were constructed and fluorescence was assessed in three different strain backgrounds: the wild type, a ΔhetR mutant, and a ΔtrpE mutant. In all of the backgrounds tested, fluorescence from transcription of both PtrpE fusions was present in all cells in the presence or absence of combined nitrogen (Fig. 3A and B and data not shown). To assess the contribution of HetR to trpE transcription at the population level, GFP fluorescence was quantified and used to compare transcriptional reporter activities across genetic backgrounds. Fluorescence from the PtrpE-gfp fusion was roughly equivalent in the wild type and the ΔtrpE mutant but was decreased in the ΔhetR mutant in the presence or absence of combined nitrogen (Fig. 3C). When the same mutation in the trpE binding site that abrogated HetR binding in vitro was introduced into the PtrpE-gfp fusion, fluorescence from PtrpE(mut)-gfp in the wild type and the ΔtrpE mutant was diminished to the level produced in the ΔhetR mutant strain (Fig. 3A to C). Fluorescence in the ΔhetR mutant strain was unaffected by a native or mutated HetR binding site in PtrpE. These results indicate that HetR is necessary for wild-type levels of trpE transcription in vivo and likely acts via direct binding to the HetR binding site in the promoter region of trpE.
FIG 3.
The HetR binding site present in the trpE promoter is required for proper expression. Shown are bright-field (top), GFP fluorescence (middle), and chlorophyll autofluorescence (bottom) micrographs of the wild type (WT) bearing PtrpE-gfp in pPJAV381 (A) or PtrpE(mut)-gfp with a mutated HetR binding site in pPJAV382 (B). Fluorescence was quantified in the wild type and the ΔhetR (UHM103) and ΔtrpE (UHM335) mutants bearing pPJAV381 (green bars) or pPJAV382 (red bars) grown in the presence of nitrate (solid bars) or 24 h after the removal of combined nitrogen (hatched bars). All fluorescence measurements were taken at an OD750 of 0.1. Error bars represent standard deviations.
If transcription of the trpE gene is activated by the direct interaction of HetR with its binding site in the trpE promoter and this activation is required for correctly timed differentiation, mutation of the HetR binding site in the trpE promoter in the genome should yield a phenotype similar to that of the ΔtrpE mutant. To determine whether the HetR binding site in the trpE promoter has biological relevance, trpE with a native or mutated HetR binding site was reintroduced into the ΔtrpE mutant and complementation was assessed. Introduction of trpE with the native HetR binding site into its promoter complemented the ΔtrpE mutant, yielding a strain that failed to develop heterocysts when grown in nitrate (Fig. 2G). In contrast, introduction of trpE with a mutated HetR binding site did not complement the ΔtrpE mutant; the strain differentiated a wild-type pattern of heterocysts when grown in nitrate, indicating that an intact HetR binding site in the trpE promoter region is necessary for complementation of the ΔtrpE mutant. We infer that mutation of the binding site interferes with HetR-mediated transcriptional activation by abrogating direct interaction with this site.
alr3233 encodes an anthranilate synthase.
On the basis of amino acid homology, alr3233 is predicted to encode an anthranilate synthase (TrpE) that catalyzes the first committed step of tryptophan biosynthesis; TrpE converts chorismate to anthranilate. To determine whether the protein product of alr3233 is an anthranilate synthase, alr3233 was expressed in E. coli anthranilate synthase mutant strain JA221 (45), a tryptophan auxotroph with a mutation in the trpE gene, and complementation was assessed. Strain JA221 with the empty expression vector as a negative control was unable to grow without the addition of exogenous tryptophan (see Fig. S3 in the supplemental material). In contrast, the strain with the same vector containing alr3233 grew in the presence or absence of tryptophan, which is indicative of complementation of the trpE mutation by alr3233. We conclude that alr3233 encodes an anthranilate synthase capable of catalyzing the same reaction as E. coli TrpE.
AZAT induces heterocyst differentiation under nitrogen-replete conditions.
The exogenous addition of AZAT to cultures of heterocystous cyanobacteria has been shown to induce differentiation in the presence of ammonia or nitrate. To determine the effect of AZAT on wild-type Anabaena and the ΔtrpE mutant, various concentrations of AZAT were added to ammonia- or nitrate-grown cultures and heterocyst formation was assessed. The addition of AZAT to the wild type or the ΔtrpE mutant during growth on ammonia did not result in the production of heterocysts (see Fig. S4 in the supplemental material). In contrast, AZAT addition to a final concentration of 10 μM and incubation for 24 h were sufficient to produce a normal pattern of heterocysts in the wild type when it was grown on nitrate. Interestingly, the addition of tryptophan did not affect the differentiation of the wild type but complemented the ΔtrpE mutant such that it no longer formed a wild-type pattern of heterocysts during growth on nitrate. Ammonia at the same concentration had no effect on the phenotype of the ΔtrpE mutant (see Fig. S4). Differentiation of heterocysts by nitrate-grown wild-type Anabaena in response to AZAT resembled the phenotype of the ΔtrpE mutant, and the mutation in trpE was complemented by the addition of tryptophan.
2-OG levels increase in response to nitrate in the ΔtrpE mutant.
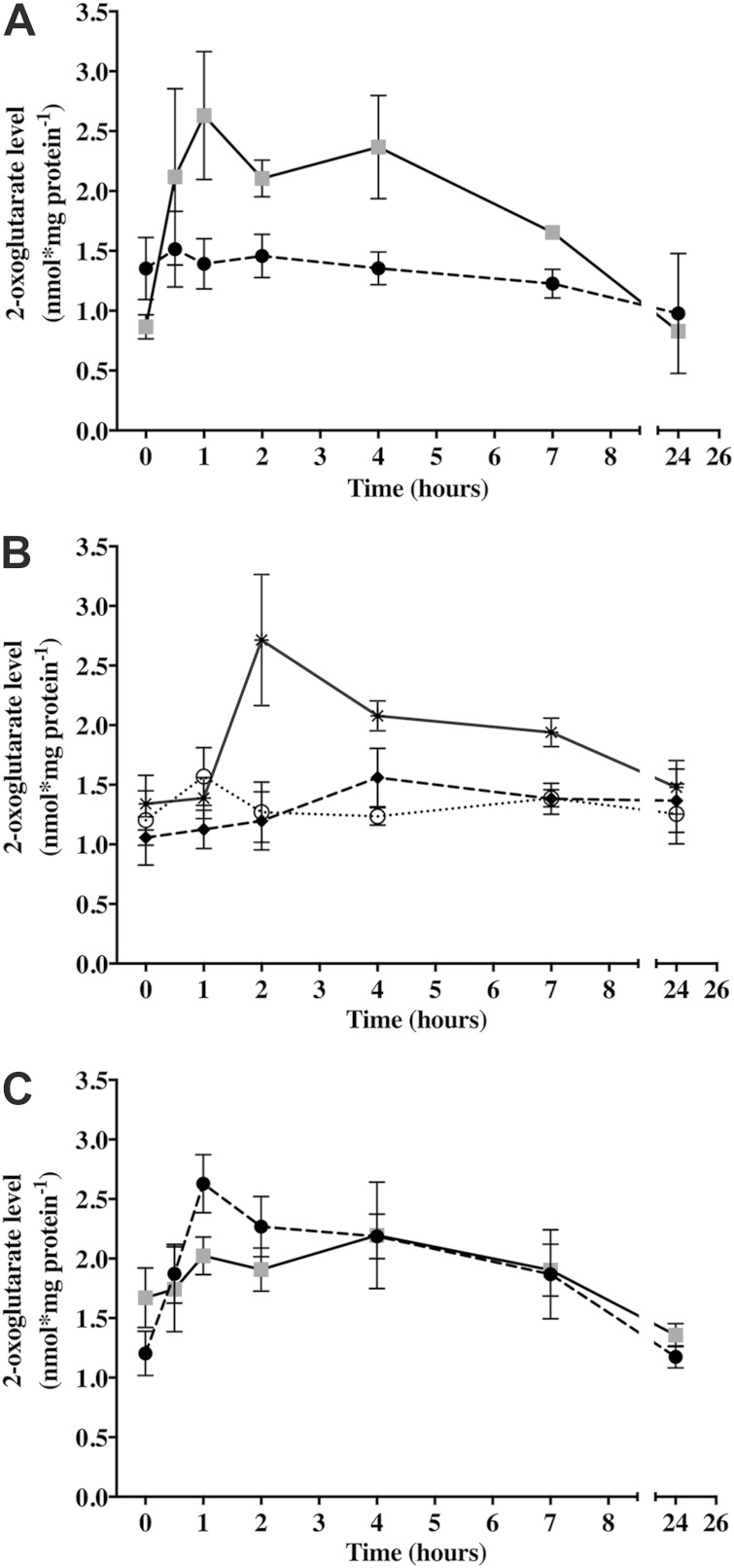
Previous work has shown that when combined nitrogen was removed from nitrogen-replete Anabaena cultures, the intracellular pool of 2-OG quickly increased from 1.15 ± 0.27 nmol/mg protein to a maximum of 2.51 ± 0.2 nmol/mg protein after 1 h (9). 2-OG returned to prestarvation levels 16 to 24 h after the concentration spike. In addition, the exogenous addition of 2-OG was sufficient to induce heterocyst differentiation in the presence of nitrate but not ammonia in an Anabaena strain expressing an E. coli kgtP 2-OG permease to permit efficient uptake of 2-OG (10). These results provide strong support for 2-OG as the intracellular nitrogen starvation signal that facilitates the induction of differentiation. Consistent with the earlier work, the wild type did not display fluctuations in 2-OG levels in our experiments when grown in ammonia or nitrate or stepped down from ammonia to nitrate (Fig. 4A and B), but removal of combined nitrogen from nitrate-grown wild-type cultures resulted in a transient spike in 2-OG, with a maximum of 2.63 ± 0.42 nmol/mg protein, that occurred 1 h following nitrogen removal (Fig. 4C). To determine whether differentiation following the addition of AZAT resulted in altered 2-OG levels, indicative of a nitrogen starvation response, the intracellular pool of 2-OG was measured over time under various conditions. When 10 μM AZAT was exogenously added to the wild type grown in nitrate, the differentiation of a normal pattern of heterocysts within 24 h was observed (see Fig. S4 in the supplemental material), a phenotype similar to the differentiation that resulted from the addition of exogenous 2-OG to the Anabaena strain expressing the kgtP 2-OG permease (10). In addition, an increase in the 2-OG concentration to 2.71 ± 0.95 nmol/mg protein was observed 2 h after the addition of 10 μM AZAT (Fig. 4B). 2-OG returned to the basal level of 1.48 ± 0.39 nmol/mg protein by 24 h postaddition. To control for the presence of exogenous amino acids, dl-tryptophan was added to nitrate-grown wild-type cultures to a concentration of 100 μM, but neither a 2-OG spike nor heterocyst differentiation resulted (Fig. 4B; see Fig. S4). We infer that the increase in 2-OG resulting from the addition of AZAT, which led to heterocyst differentiation, was due to a nitrogen starvation response similar to that seen after the removal of combined nitrogen from wild-type Anabaena cultures.
FIG 4.
Quantification of 2-OG levels in the wild type and the ΔtrpE mutant (UHM335) under various conditions. (A) The wild type (circles) and UHM335 (circles) were grown in BG-11 medium containing ammonia (NH4) and stepped down to growth on nitrate (NO2). (B) The wild type was grown in nitrate supplemented with 10 μM AZAT (solid line), 100 μM dl-tryptophan (dashed line), or nothing (dotted line). (C) The wild type (circles) and UHM335 (circles) were stepped down from nitrate-containing growth medium to that lacking a source of combined nitrogen. The 0-h time point corresponds to the time of the step down or the addition of amino acids. 2-OG was quantified in triplicate at the time points shown. Error bars represent standard deviations.
The ΔtrpE mutant produced a normal pattern of heterocysts following the step down from growth on ammonia to growth on nitrate (Fig. 2D). Because this phenotype resembles the differentiation that accompanies the addition of AZAT to nitrate-grown wild-type cultures, the intracellular 2-OG pool of the ΔtrpE mutant was measured following the transitions from growth on ammonia to growth on nitrate and then the removal of combined nitrogen. The basal 2-OG level was lower in the ΔtrpE mutant at 0.87 ± 0.17 nmol/mg protein than in the wild type at 1.35 ± 0.45 nmol/mg protein when cultured with ammonia (Fig. 4A). Unlike the wild type, which did not display a change in 2-OG levels after the step down from ammonia growth to nitrate growth, the intracellular 2-OG concentration within the ΔtrpE mutant increased to a maximum of 2.69 ± 0.93 nmol/mg protein before returning to the basal level within 24 h. After the removal of combined nitrogen, 2-OG levels in the wild type increased to 2.63 ± 0.42 nmol/mg protein after 1 h and returned to the preinduction level of 1.17 ± 0.16 within 24 h (Fig. 4C). In contrast, the ΔtrpE mutant reached a maximum intracellular 2-OG concentration of 2.04 ± 0.77 nmol/mg protein after 4 h and returned to the basal level of 1.36 ± 0.17 nmol/mg protein within 24 h. Taken together, these results suggest that the ΔtrpE mutation induces differentiation in medium containing nitrate in response to 2-OG, the same signal that induces differentiation of the wild type when combined nitrogen is unavailable.
DISCUSSION
Heterocystous cyanobacteria have distinct physiological responses to three different, general types of bioavailable nitrogen, ammonia, nitrate/nitrite, and dinitrogen (reviewed in reference 6). When ammonia is sufficient to support growth, it is assimilated directly through the glutamine synthetase-glutamate synthase (GS-GOGAT) system, and NtcA activity does not result in a nitrogen starvation response. Alternatively, when forms of inorganic nitrogen such as nitrate and nitrite that do not require nitrogenase for reduction to ammonia are present but ammonia itself is not, NtcA activates the transcription of genes necessary for their utilization. These include genes for the uptake of nitrate and nitrite, as well as their reduction to ammonia. In this case, intracellular levels of 2-OG do not increase substantially above those seen when ammonia is present. Levels of 2-OG were similar before and after transfer of filaments from medium containing ammonia to that containing only nitrate (Fig. 4A). Lastly, when dinitrogen is the only form of nitrogen capable of supporting growth, a transient rise in 2-OG levels is observed and NtcA initiates the induction phase of heterocyst differentiation that facilitates the fixation of dinitrogen to ammonia. The phenotype of the trpE mutant described here implicates the alr3233 gene in the differential responses of heterocystous cyanobacteria to nitrate and dinitrogen.
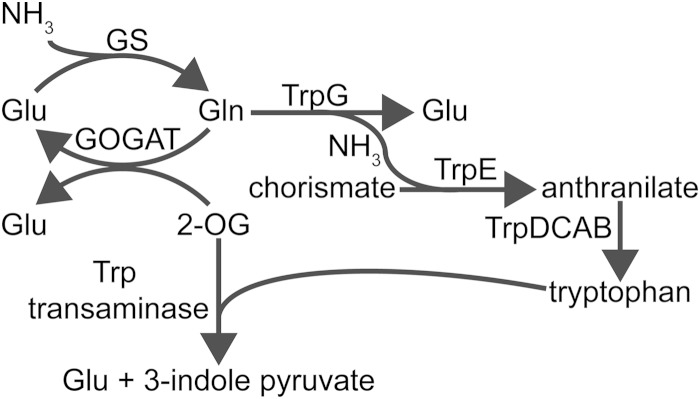
In wild-type Anabaena, the transition from growth on ammonia to growth on nitrate does not normally result in the differentiation of a wild-type pattern of heterocysts or a spike in 2-OG levels like that seen during the transition to nitrogen-depleted conditions. In contrast, the trpE mutant experiences an elevation of 2-OG and differentiates heterocysts likely as a consequence. So, how could the intact alr3233 gene, which complemented the lack of anthranilate synthase activity in an E. coli trpE mutant, prevent the spike in 2-OG that is seen in the mutant shortly after transfer to nitrate? The answer is not apparent, so we can only speculate on the basis of the potential effects of increased tryptophan and/or anthranilate synthase activity on metabolite levels. Together the trpE and trpG gene products catalyze the transfer of an amino group from glutamine to chorismate to yield glutamate and anthranilate. The glutamine consumed is therefore not available to react with 2-OG to make glutamate as part of the GS-GOGAT system, so increased anthranilate synthase activity seems to increase, not decrease, levels of 2-OG (Fig. 5). On the other hand, tryptophan, the end product of the synthesis pathway, can be used in a reaction that consumes 2-OG. Transfer of an amino group from tryptophan to 2-OG by an aminotransferase would yield glutamate and 3-indole pyruvate. At least four genes predicted to encode transaminases capable of catalyzing this reaction, one of which is located directly downstream of hetR, are found in the Anabaena genome. At first, the consumption of glutamine by anthranilate synthase and subsequent consumption of 2-OG appear to have a net zero effect on 2-OG levels. However, tryptophan-dependent consumption of 2-OG implies that, for a short period of time, higher levels of tryptophan represent increased potential for the reduction of 2-OG levels by a tryptophan transaminase. Induction of the nitrate assimilation genes, including nir and nrtA, which encode a nitrate reductase and a nitrate transporter, respectively, occurs 0.5 to 1 h after the addition of nitrate to a culture of Anabaena (46). This delay in the availability of nitrate-dependent ammonia coincides with the timing of the 2-OG spike when the trpE mutant is transferred from ammonia to nitrate, a spike in 2-OG that is absent when a functional alr3233 gene is present.
FIG 5.
Diagram of proposed interactions that regulate the induction response to nitrogen starvation in Anabaena. See the text for details. Abbreviations: NH3, ammonia; Trp, tryptophan; Glu, glutamate; Gln, glutamine; TrpG, glutamine amidotransferase; TrpE, anthranilate synthase; TrpD, anthranilate phosphoribosyltransferase; TrpC, indole-3-glycerol-phosphate synthase; TrpA and TrpB, tryptophan synthase (described in the order in which reactions occur during tryptophan biosynthesis).
Functional complementation of the trpE mutant by the addition of tryptophan (see Fig. S4 in the supplemental material) suggests that it is the level of tryptophan, and not anthranilate synthase activity, per se, that is necessary to prevent a similar 2-OG spike in the wild type in response to nitrate, consistent with the model above. Like tryptophan transaminase, other amino acid aminotransferases also transfer an amino group from the amino acid to 2-OG, suggesting that other amino acids would have the potential to temporarily maintain 2-OG levels during the transfer of filaments from ammonia to nitrate, as we suggest for tryptophan. Along those lines, β-2-thienyl-dl-alanine, a phenylalanine analog, has also been shown to induce the formation of heterocysts in the presence of combined nitrogen in Anabaena CA in a manner similar to that of AZAT (18).
The model presented above may account for the response to nitrate, but it does not necessarily address how filaments sense the presence of nitrate. When filaments of Anabaena are transferred from ammonia to dinitrogen, a spike in 2-OG is observed and subsequently heterocyst differentiation ensues. Neither the spike in 2-OG nor differentiation was observed when transfer was to a medium containing nitrate. Proteins such as the sensor histidine kinases NarQ and NarX sense and respond to nitrate in other bacteria, and proteins with similar functions may be important for the differential response of Anabaena to the presence of nitrate (47, 48). Alternatively, differences in 2-OG levels in response to ammonia or nitrate may exist but are beyond the level of resolution of the assay used here. In this case, small changes in 2-OG may be sufficient to elicit NtcA-mediated expression of nitrate assimilatory genes but not that of hetR or other genes involved in differentiation. Tryptophan metabolism could have a buffering effect on 2-OG levels, acting as a safety valve that prevents spikes in 2-OG that would otherwise lead to differentiation during growth on nitrate.
The alr3233 gene is one of two predicted to encode anthranilate synthase activity in Anabaena. Its mutation did not affect the growth rate, and its expression is regulated by HetR, suggesting a primarily developmental role. Regulation is likely direct, given the binding of HetR to a region of DNA upstream of trpE that contains a 17-bp inverted repeat that is necessary for binding in vitro, and the binding site is likely important for regulation of transcription in vivo, given that its mutation reduced transcription levels from a transcriptional reporter fusion and prevented complementation of the trpE mutant by a wild-type version of trpE that was integrated into the chromosome. A classification system for DNA sequences known to be bound by HetR was recently proposed (8). Category I sites are poorly defined and require large ratios of HetR to DNA to show signs of binding in vitro, category II sites are bound at an intermediate level and are characterized by a relatively well-conserved 17-bp inverted repeat, and category III sites show the most complete and consistent shifts in EMSAs and contain a 17-bp inverted repeat distinct from those in class II sites. The site upstream of trpE described here appears to be the third category III site to be described. The others occur upstream of hetP and hetZ. Induction of transcription of these two positive regulators of differentiation occurs in cells that conform to the eventual pattern of heterocysts, and their products appear to function at the end of the patterning phase or commitment phase of differentiation (26, 41). In contrast, transcription of trpE appears to be uniform along filaments and its product appears to participate in suppression of differentiation in nitrate. These different regulatory outcomes suggest that additional factors coordinate with the binding of HetR to target DNA sequences to yield distinct spatiotemporal patterns of transcription.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tina Carvalho and Marilyn Dunlap at the University of Hawaii Biological Electron Microscope Facility for assistance with confocal microscopy, Ray Dixon for his consultation on the 2-OG quantification assay, and an anonymous reviewer for noting the potential for NtcA to sense subtle fluctuations in 2-OG.
This work was supported by grant MCB-1121346 from the National Science Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02145-14.
REFERENCES
- 1.Leigh JA, Dodsworth JA. 2007. Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol 61:349–377. doi: 10.1146/annurev.micro.61.080706.093409. [DOI] [PubMed] [Google Scholar]
- 2.Herrero A, Muro-Pastor AM, Flores E. 2001. Nitrogen control in cyanobacteria. J Bacteriol 183:411–425. doi: 10.1128/JB.183.2.411-425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanigawa R, Shirokane M, Maeda SI, Omata T, Tanaka K, Takahashi H. 2002. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc Natl Acad Sci U S A 99:4251–4255. doi: 10.1073/pnas.072587199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espinosa J, Forchhammer K, Burillo S, Contreras A. 2006. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol Microbiol 61:457–469. doi: 10.1111/j.1365-2958.2006.05231.x. [DOI] [PubMed] [Google Scholar]
- 5.Llácer JL, Espinosa J, Castells MA, Contreras A, Forchhammer K, Rubio V. 2010. Structural basis for the regulation of NtcA-dependent transcription by protein PipX and PII. Proc Natl Acad Sci U S A 107:15397–15402. doi: 10.1073/pnas.1007015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang C-C, Laurent S, Sakr S, Peng L, Bedu S. 2006. Heterocyst differentiation and pattern formation in cyanobacteria: a chorus of signals. Mol Microbiol 59:367–375. doi: 10.1111/j.1365-2958.2005.04979.x. [DOI] [PubMed] [Google Scholar]
- 7.Golden J W, Yoon H-S. 2003. Heterocyst development in Anabaena. Curr Opin Microbiol 6:557–563. doi: 10.1016/j.mib.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Videau P, Ni S, Rivers OS, Ushijima B, Feldmann EA, Cozy LM, Kennedy MA, Callahan SM. 2014. Expanding the direct HetR regulon in Anabaena sp. strain PCC 7120. J Bacteriol 196:1113–1121. doi: 10.1128/JB.01372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laurent S, Chen H, Bedu S, Ziarelli F, Peng L, Zhang C-C. 2005. Nonmetabolizable analogue of 2-oxoglutarate elicits heterocyst differentiation under repressive conditions in Anabaena sp. PCC 7120. Proc Natl Acad Sci U S A 102:9907–9912. doi: 10.1073/pnas.0502337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J-H, Laurent S, Konde V, Bedu S, Zhang C-C. 2003. An increase in the level of 2-oxoglutarate promotes heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Microbiology 149:3257–3263. doi: 10.1099/mic.0.26462-0. [DOI] [PubMed] [Google Scholar]
- 11.Ehira S, Ohmori M. 2006. NrrA directly regulates expression of hetR during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 188:8520–8525. doi: 10.1128/JB.01314-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Black TA, Cai Y, Wolk CP. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol 9:77–84. doi: 10.1111/j.1365-2958.1993.tb01670.x. [DOI] [PubMed] [Google Scholar]
- 13.Valladares A, Flores E, Herrero A. 2008. Transcription activation by NtcA and 2-oxoglutarate of three genes involved in heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 190:6126–6133. doi: 10.1128/JB.00787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan Q, Huang G, Lechno-Yossef S, Wolk CP, Kaneko T, Tabata S. 2005. Clustered genes required for synthesis and deposition of envelope glycolipids in Anabaena sp. strain PCC 7120. Mol Microbiol 58:227–243. doi: 10.1111/j.1365-2958.2005.04818.x. [DOI] [PubMed] [Google Scholar]
- 15.Bottomley PJ, van Baalen C, Tabita FR. 1980. Heterocyst differentiation and tryptophan metabolism in the cyanobacterium Anabaena sp. CA. Arch Biochem Biophys 203:204–213. doi: 10.1016/0003-9861(80)90170-8. [DOI] [PubMed] [Google Scholar]
- 16.Bothe H, Eisbrenner G. 1977. Effect of 7-azatryptophan on nitrogen-fixation and heterocyst formation in the blue-green alga Anabaena cylindrica. Biochem Physiol Pflanz 171:322–332. [Google Scholar]
- 17.Adams DG. 1992. The effect of dl-7-azatryptophanon heterocyst development in the cyanobacterium Anabaena cylindrica. Microbiology 138:355–362. [Google Scholar]
- 18.Rogerson AC. 1979. Modifiers of heterocyst repression and spacing and formation of heterocysts without nitrogenase in the cyanobacterium Anabaena variabilis. J Bacteriol 140:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CH, Van Baalen C, Tabita FR. 1987. Nitrogen starvation mediated by dl-7-azatryptophan in the cyanobacterium Anabaena sp. strain CA. J Bacteriol 169:1107–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen CH, Van Baalen C, Tabita FR. 1987. dl-7-Azatryptophan and citrulline metabolism in the cyanobacterium Anabaena sp. strain 1F. J Bacteriol 169:1114–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchison GJ, Wilcox M. 1973. Alteration in heterocyst pattern of Anabaena produced by 7-azatryptophan. Nat New Biol 246:229–233. doi: 10.1038/newbio246229a0. [DOI] [PubMed] [Google Scholar]
- 22.Borthakur PB, Orozco CC, Young-Robbins SS, Haselkorn R, Callahan SM. 2005. Inactivation of patS and hetN causes lethal levels of heterocyst differentiation in the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol 57:111–123. doi: 10.1111/j.1365-2958.2005.04678.x. [DOI] [PubMed] [Google Scholar]
- 23.Mitschke J, Vioque A, Haas F, Hess WR, Muro-Pastor AM. 2011. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc Natl Acad Sci U S A 108:20130–20135. doi: 10.1073/pnas.1112724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buikema WJ, Haselkorn R. 2001. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc Natl Acad Sci U S A 98:2729–2734. doi: 10.1073/pnas.051624898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elhai J, Wolk CP. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol 167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 26.Higa KC, Callahan SM. 2010. Ectopic expression of hetP can partially bypass the need for hetR in heterocyst differentiation by Anabaena sp. strain PCC 7120. Mol Microbiol 77:562–574. doi: 10.1111/j.1365-2958.2010.07257.x. [DOI] [PubMed] [Google Scholar]
- 27.Higa KC, Rajagopalan R, Risser DD, Rivers OS, Tom SK, Videau P, Callahan SM. 2012. The RGSGR amino acid motif of the intercellular signaling protein, HetN, is required for patterning of heterocysts in Anabaena sp. strain PCC 7120. Mol Microbiol 83:682–693. doi: 10.1111/j.1365-2958.2011.07949.x. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 29.Le Roux F, Binesse J, Saulnier D, Mazel D. 2007. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol 73:777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei T-F, Ramasubramanian R, Golden JW. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J Bacteriol 176:4473–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higuchi R, Krummel B, Saiki RK. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res 16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Val ME, Skovgaard O, Ducos-Galand M, Bland MJ, Mazel D. 2012. Genome engineering in Vibrio cholerae: a feasible approach to address biological issues. PLoS Genet 8:e1002472. doi: 10.1371/journal.pgen.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon H-S, Golden JW. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935–938. doi: 10.1126/science.282.5390.935. [DOI] [PubMed] [Google Scholar]
- 34.Callahan SM, Buikema WJ. 2001. The role of HetN in maintenance of the heterocyst pattern in Anabaena sp. PCC 7120. Mol Microbiol 40:941–950. doi: 10.1046/j.1365-2958.2001.02437.x. [DOI] [PubMed] [Google Scholar]
- 35.Feldmann EA, Ni S, Sahu ID, Mishler CH, Risser DD, Murakami JL, Tom SK, McCarrick RM, Lorigan GA, Tolbert BS, Callahan SM, Kennedy MA. 2011. Evidence for direct binding between HetR from Anabaena sp. PCC 7120 and PatS-5. Biochemistry 50:9212–9224. doi: 10.1021/bi201226e. [DOI] [PubMed] [Google Scholar]
- 36.Ernst A, Black T, Cai Y, Panoff J-M, Tiwari DN, Wolk CP. 1992. Synthesis of nitrogenase in mutants of the cyanobacterium Anabaena sp. strain PCC 7120 affected in heterocyst development or metabolism. J Bacteriol 174:6025–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reyes-Ramirez F, Little R, Dixon R. 2001. Role of Escherichia coli nitrogen regulatory genes in the nitrogen response of the Azotobacter vinelandii NifL-NifA complex. J Bacteriol 183:3076–3082. doi: 10.1128/JB.183.10.3076-3082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Risser DD, Callahan SM. 2008. HetF and PatA control levels of HetR in Anabaena sp. strain PCC 7120. J Bacteriol 190:7645–7654. doi: 10.1128/JB.01110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang X, Dong Y, Zhao J. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc Natl Acad Sci U S A 101:4848–4853. doi: 10.1073/pnas.0400429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saha SK, Golden JW. 2011. Overexpression of pknE blocks heterocyst development in Anabaena sp. strain PCC 7120. J Bacteriol 193:2619–2629. doi: 10.1128/JB.00120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du Y, Cai YA, Hou S, Xu X. 2012. Identification of the HetR recognition sequence upstream of hetZ in Anabaena sp. strain PCC 7120. J Bacteriol 194:2297–2306. doi: 10.1128/JB.00119-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simmons LA, Grossman AD, Walker GC. 2008. Clp and Lon proteases occupy distinct subcellular positions in Bacillus subtilis. J Bacteriol 190:6758–6768. doi: 10.1128/JB.00590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirstein J, Strahl H, Moliere N, Hamoen LW, Turgay K. 2008. Localization of general and regulatory proteolysis in Bacillus subtilis cells. Mol Microbiol 70:682–694. doi: 10.1111/j.1365-2958.2008.06438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. 2006. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A 103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chinault AC, Carbon J. 1979. Overlap hybridization screening: isolation and characterization of overlapping DNA fragments surrounding the leu2 gene on yeast chromosome III. Gene 5:111–126. doi: 10.1016/0378-1119(79)90097-0. [DOI] [PubMed] [Google Scholar]
- 46.Cai Y, Wolk CP. 1997. Nitrogen deprivation of Anabaena sp. strain PCC 7120 elicits rapid activation of a gene cluster that is essential for uptake and utilization of nitrate. J Bacteriol 179:258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiang RC, Cavicchioli R, Gunsalus RP. 1992. Identification and characterization of NarQ, a second nitrate sensor for nitrate-dependent gene regulation in Escherichia coli. Mol Microbiol 6:1913–1923. doi: 10.1111/j.1365-2958.1992.tb01364.x. [DOI] [PubMed] [Google Scholar]
- 48.Stewart V, Parales J Jr. 1988. Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J Bacteriol 170:1589–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orozco CC, Risser DD, Callahan SM. 2006. Epistasis analysis of four genes from Anabaena sp. strain PCC 7120 suggests a connection between PatA and PatS in heterocyst pattern formation. J Bacteriol 188:1808–1816. doi: 10.1128/JB.188.5.1808-1816.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rivers OS, Videau P, Callahan SM. 11 November 2014. Mutation of sepJ reduces the intercellular signal range of a hetN-dependent paracrine signal, but not of a patS-dependent signal, in the filamentous cyanobacterium Anabaena sp. strain PCC 7120. Mol Microbiol doi: 10.1111/mmi.12836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.