Abstract
Bacterial endospores can remain dormant for decades yet can respond to nutrients, germinate, and resume growth within minutes. An essential step in the germination process is degradation of the spore cortex peptidoglycan wall, and the SleB protein in Bacillus species plays a key role in this process. Stable incorporation of SleB into the spore requires the YpeB protein, and some evidence suggests that the two proteins interact within the dormant spore. Early during germination, YpeB is proteolytically processed to a stable fragment. In this work, the primary sites of YpeB cleavage were identified in Bacillus anthracis, and it was shown that the stable products are comprised of the C-terminal domain of YpeB. Modification of the predominant YpeB cleavage sites reduced proteolysis, but cleavage at other sites still resulted in loss of full-length YpeB. A B. anthracis strain lacking the HtrC protease did not generate the same stable YpeB products. In B. anthracis and Bacillus subtilis htrC mutants, YpeB was partially stabilized during germination but was still degraded at a reduced rate by other, unidentified proteases. Purified HtrC cleaved YpeB to a fragment similar to that observed in vivo, and this cleavage was stimulated by Mn2+ or Ca2+ ions. A lack of HtrC did not stabilize YpeB or SleB during spore formation in the absence of the partner protein, indicating other proteases are involved in their degradation during sporulation.
INTRODUCTION
Endospores produced by members of Gram-positive genera, such as Bacillus and Clostridium, possess extreme resistance properties and can remain in a fully dormant state for years. The dormant state and resistance properties are dependent on the maintenance of the spore core (cytoplasm) in a relatively dehydrated state, and this in turn depends on the intact state of the inner spore membrane and the cortex peptidoglycan (PG) wall surrounding that membrane (1). Upon exposure to nutrient germinants, spores begin to release low-molecular-weight solutes, including a large depot of Ca2+-dipicolinic acid (Ca2+-DPA), and take up water (2). Degradation of the cortex PG by germination-specific lytic enzymes (GSLEs) is required for full expansion of the membrane, full hydration of the core, and resumption of metabolism (3–6). As GSLEs hydrolyze the cortex PG before new protein synthesis can occur, they must be produced during spore formation and held stable and inactive in the dormant spore until germination is triggered (7).
Bacillus species possess two major, partially redundant GSLEs: CwlJ and SleB (7). CwlJ is produced in the mother cell of the developing sporangium (8), is associated with the spore coats on the outer surface of the cortex (9–12), and becomes active when exposed to a high concentration of Ca2+-DPA—normally when that solute is released from the germinating spore (11, 13, 14). SleB is produced within the developing forespore (15, 16) and is located interior to the cortex in the dormant spore, most likely in close association with the inner spore membrane (10, 17). The mechanisms by which SleB is held inactive during spore dormancy and released to become active during germination are unclear.
A potential factor in the regulation of SleB activity is YpeB, which is encoded in an operon with sleB and possesses a transmembrane anchor sequence that should also localize it to the outer surface of the inner spore membrane (15, 18, 19). SleB and YpeB exhibit codependence for their stable incorporation into the dormant spore (10, 18, 20). In the absence of their partner protein, both SleB and YpeB are produced and rapidly degraded during spore formation (18). It has also been observed that YpeB is proteolytically processed during spore germination (10), and it has been suggested that this processing could be involved in the initiation of SleB activity during germination (20).
The current study examined the cleavage sites generating the stable YpeB products during germination and identified HtrC as the protease responsible for these cleavage events. The resulting cleavage products were shown to contain the C terminus and PepSY domains of YpeB. Strains lacking HtrC or with mutations at the YpeB cleavage sites were constructed to evaluate the role of YpeB processing in SleB activation. In the absence of HtrC, YpeB was degraded in a more nonspecific manner during germination, and the activity of SleB was not significantly affected.
MATERIALS AND METHODS
Strains, culture conditions, and spore preparation.
Escherichia coli strains used for plasmid propagation were grown at 37°C in LB medium with 500 μg/ml erythromycin or 20 μg/ml kanamycin. Strains used for protein overexpression were grown in LB with 30 μg/ml chloramphenicol and 50 μg/ml ampicillin. Bacillus anthracis strains were derived from the Sterne strain 34F2 and were grown on brain heart infusion (BHI) (Difco) agar plates with 5 μg/ml erythromycin or 20 μg/ml kanamycin where appropriate. B. anthracis strains maintaining pBKJ236 derivatives extrachromosomally were grown at 25°C, while those in which the plasmid was integrated into the chromosome were grown at 37°C. Bacillus subtilis strains were derived from PS832, a prototrophic laboratory derivative of strain 168, and were grown on 2× SG medium (21) with 0.5 μg/ml erythromycin and 12.5 μg/ml lincomycin (macrolide-lincosamide-streptogramin B [MLS] resistance) and/or 10 μg/ml tetracycline when appropriate. Spores were prepared in modified G (22) broth for B. anthracis or 2× SG broth (21) for B. subtilis with appropriate antibiotics. After 72 h of incubation at 37°C, spores were harvested and washed in deionized water for several days, and any remaining vegetative cells were heat killed at 65°C for 25 min. B. anthracis spores were further purified by centrifugation through a 50% sodium diatrizoate (Sigma) layer as described previously (23). Where indicated, B. anthracis spores were decoated as previously described (4, 18). All spores used in this work were 99% free of vegetative cells and were stored in deionized water at 4°C until analysis.
Mutant strain construction.
All the strains and plasmids used are listed in Table 1, and the primer sequences used for plasmid construction are listed in Table 2. Construction of DPBa127, a B. anthracis strain expressing YpeB-His6, was published previously (18). In this strain, the plasmid pDPV424 encoding YpeB-His6 was integrated into the ΔypeB chromosome. Strains in which the YpeB T202-S203 or A168-S169 cleavage sites were changed to E202-L203 (YpeBT202E/S203L) or E168-L169 (YpeBA168E/S169L) were created by site-directed mutagenesis of ypeB using overlap extension PCR (24). The PCR products were subsequently cloned into pDPV424 through restriction-free cloning (25), generating plasmids encoding YpeBT202E/S203L-His6 (pDPV447) and YpeBA168E/S169L-His6 (pDPV454). To construct a strain in which both cleavage sites were mutated (YpeBA168E/S169L, T202E/S203L-His6), pDPV447 was used as the template for overlap extension PCR, and the PCR product was cloned into pDPV447, producing pDPV455. Plasmids pDPV447, pDPV454, and pDPV455 were screened for the gain or loss of a restriction site in ypeB that was designed as part of the mutagenic primers and were also verified by DNA sequencing. Plasmids were introduced into the ΔypeB strain of B. anthracis (18) by conjugation, as in the initial steps of the markerless gene replacement procedure (26), and chromosomal integrants were selected by shifting the temperature to 42°C. PCR amplification and sequencing verified plasmid integration within the 500-bp homologous region downstream of ΔypeB in the chromosome and confirmed the presence of the desired cleavage site mutations.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype/phenotypea | Constructionb | Source or reference |
|---|---|---|---|
| E. coli | |||
| DPVE13 | BL21(λDE3) pLysS (Cmr) | Novagen | |
| DPVE501 | pDPV458 (His6-MBP-HtrC45–391 Ampr) Cmr | pDPV458→DPVE13 | This study |
| B. anthracis | |||
| Sterne 34F2 | pXO1+ pXO2− | P. Hanna | |
| DPBa38 | ΔsleB | 19 | |
| DPBa89 | ΔypeB | 18 | |
| DPBa113 | ΔypeB::pDPV416 (ypeB+ Err) | 18 | |
| DPBa127 | ΔypeB::pDPV424 (YpeB-His6 Err) | 18 | |
| DPBa157 | ΔypeB::pDPV447 (YpeBT202E/S203L-His6 Err) | pDPV447→DPBa89 | This study |
| DPBa167 | ΔypeB::pDPV454 (YpeBA168E/S169L-His6 Err) | pDPV454→DPBa89 | This study |
| DPBa168 | ΔypeB::pDPV455 (YpeBA168E/S169L, T202E/S203L-His6 Err) | pDPV455→DPBa89 | This study |
| DPBa178 | ΔhtrC | pDPV460→34F2 | This study |
| DPBa182 | ΔhtrC::pDPV459 (htrC+ Err) | pDPV459→DPBa178 | This study |
| DPBa187 | ΔsleB ΔhtrC | pDPV460→DPBa38 | This study |
| DPBa188 | ΔypeB ΔhtrC | pDPV460→DPBa89 | This study |
| B. subtilis | |||
| PS832 | Prototrophic derivative of strain 168 | P. Setlow | |
| FB111 | ΔcwlJ::Tetr | 11 | |
| DPVB668 | ΔhtrC::MLSr | PCR→PS832 | This study |
| DPVB669 | ΔcwlJ::Tetr ΔhtrC::MLSr | FB111→DPVB668 | This study |
| Plasmids | |||
| pBKJ236 | Err ori(Ts) | 26 | |
| pSS4332 | Kanr, I-SceI, AmCyan | 27 | |
| pDEST-HisMBP-T | His6-MBP, Ampr Cmr | F. Schubot | |
| pDPV416 | ypeB+ | 18 | |
| pDPV424 | YpeB-His6 | 18 | |
| pDPV447 | YpeBT202E/S203L-His6 | pBKJ236::ΔsleB ypeBT202E/S203L-His6 | This study |
| pDPV454 | YpeBA168E/S169L-His6 | pBKJ236::ΔsleB ypeBA168E/S169L-His6 | This study |
| pDPV455 | YpeBA168E/S169L, T202E/S203L-His6 | pBKJ236::ΔsleB ypeBA168E/S169L, T202E/S203L-His6 | This study |
| pDPV458 | His6-MBP-HtrC45–391 | pDEST-HisMBP-T::htrC45–391 | This study |
| pDPV459 | htrC+ | pBKJ236::htrC | This study |
| pDPV460 | ΔhtrC | pBKJ236::ΔhtrC | This study |
Cmr, chloramphenicol resistance; Ampr, ampicillin resistance; Err, erythromycin resistance; Tetr, tetracycline resistance; MLSr, macrolide-lincosamide-streptogramin B resistance; ori(Ts), temperature-sensitive origin of replication; Kanr, kanamycin resistance.
Strains were constructed by conjugation or electroporation for B. anthracis or transformation for B. subtilis. The designation preceding the arrow is the plasmid or source of the donor DNA, while the designation following the arrow is the recipient strain.
TABLE 2.
Primer sequences
| Plasmid or strain constructed | Primer name | Sequence (5′ to 3′)a |
|---|---|---|
| pDPV447 | 593 | CTTCGGACCTACCTTTGAATTAGCACAAAAAAATAAAAAAGGTGG |
| 594 | CCACCTTTTTTATTTTTTTGTGCTAATTCAAAGGTAGGTCCGAAG | |
| 603 | GTCTTCTTTACATAAAAGCGAGCCTTTTACAAAACATAACC | |
| 604 | GCTTTACGTTCTTCCATAACTTTCACATCTGGATTG | |
| pDPV454 | 605 | GTTGAGATGGCACTCGAGTTAAATCGTGATCCTGCCG |
| 606 | CGGCAGGATCACGATTTAACTCGAGTGCCATCTCAAC | |
| 603, 604 | ||
| pDPV455 | 603, 604, 605, 606 | |
| pDPV458 | 612 | GTGGAGAACCTGTACTTCCAGGGTTCAAATGATACGGGCGC |
| 613 | CCACTTTGTACAAGAAAGTTGCATTTTAATACTTTTGAATGCCTAGTTTAAC | |
| pDPV459 | 614 | CGCGGATCCGATATTGAGGTCGAGTCATTTG |
| 615 | CGCCTGCAGCAATGACATGCGTATCATCAG | |
| pDPV460 | 616 | CGCAGATCTAAAGGACATATTTGTTCACCTATC |
| 617 | CGCAGATCTAAGTATTAAAAACAGGAGGGCCTAC | |
| DPVB668 | 607 | ACAAGTACGCAGGTGCTGGC |
| 608 | CGATTATGTCTTTTGCGCAGTCGGCCCTCACGTTCGTAATCCACC | |
| 609 | GAGGGTTGCCAGAGTTAAAGGATCCCTCCGCAGACCAATTAGGC | |
| 610 | GATACACCGATTGACGTACG |
Restriction sites are underlined, TEV cleavage site regions of pDEST-HisMBP-T are italicized and in boldface, and attR2.1 regions of pDEST-HisMBP-T are italicized and underlined.
To create a B. anthracis ΔhtrC strain, htrC and approximately 500 bp flanking each side of the gene were PCR amplified from chromosomal DNA. The PCR product was inserted into the vector pBKJ236 (26) by digesting with the restriction enzymes BamHI and PstI and ligating the DNA to create pDPV459. Inverse PCR of the plasmid using primers with BglII restriction sites at the 3′ ends resulted in a linear PCR product with the majority of htrC deleted, leaving only the first five and last two codons of the gene. Subsequent BglII digestion and ligation of the PCR product produced pDPV460. This plasmid containing the htrC deletion was introduced into B. anthracis using the markerless gene replacement strategy as previously described (26), except that plasmid pSS4332 (27) was used for I-SceI expression in place of pBKJ223. Gene deletion was verified by PCR amplification and sequencing. Complementation of the ΔhtrC mutation was achieved by introduction and chromosomal integration of pDPV459. To create ΔsleB ΔhtrC and ΔypeB ΔhtrC double-deletion strains, pDPV460 was used to introduce the htrC deletion into ΔsleB (DPBa38 [19]) and ΔypeB (DPBa89 [18]) strains of B. anthracis using the markerless gene replacement strategy (26) with pSS4332 (27). Gene deletions were verified by PCR amplification and sequencing.
To create a B. subtilis ΔhtrC strain, approximately 1,000 bp flanking each side of the gene was PCR amplified from chromosomal DNA and linked to the ermC MLS antibiotic resistance gene cassette by long-flanking homology PCR (28). The PCR product was transformed into B. subtilis PS832 with selection for MLS resistance to produce DPVB668, in which all but the first seven and last eight codons of the htrC coding sequence were deleted and replaced by the MLS cassette. Chromosomal DNA from FB111 (11) was transformed into B. subtilis DPVB668 with selection for tetracycline resistance to produce DPVB669.
Spore germination assays.
The rate of germination and outgrowth of spores in liquid BHI (B. anthracis) or 2× YT (16 g tryptone, 10 g yeast extract, and 5 g NaCl per liter) (B. subtilis) was assessed by monitoring the change in optical density at 600 nm (OD) as described previously (18). For germination efficiency assays, spores at an OD of 0.2 were heat activated at 70°C for 20 min and quenched on ice. The heat-activated spores were serially diluted in deionized water, plated on BHI or 2× SG medium without antibiotics, and incubated at 37°C overnight. Colonies were counted to determine the number of CFU per OD unit. Statistical analyses of spore germination rates were performed using unpaired, two-tailed Student t tests with unequal variance.
Preparation and analysis of spore fractions.
B. anthracis sporangia samples were collected at hours 2 through 6 of sporulation from strains grown in modified G broth, as described previously (18). To prepare germinated spores for Western blot analysis, a suspension of dormant spores at a concentration of 5 OD units per ml in 10 mM Tris-HCl, pH 7.0, were heat activated at 70°C for 30 min (B. anthracis) or 75°C for 30 min (B. subtilis) and cooled on ice for 10 min. Chloramphenicol was added to a final concentration of 10 μg/ml to inhibit protein synthesis, and 7.5 OD units of spores were removed for the dormant-spore sample. The spore suspension was briefly prewarmed before the spores were germinated at 37°C with shaking at 250 rpm by the addition of 10 or 100 mM l-alanine with 1 mM inosine (B. anthracis) or 10 mM l-valine (B. subtilis). Samples equivalent to 7.5 OD units based on the starting OD were collected at various times after the addition of germinants and centrifuged at 15,800 × g for 2 min. The resulting pellets were flash frozen in liquid N2 and lyophilized.
Dried spores or sporangia were pulverized with 100 mg of 0.1-mm glass beads in a dental amalgamator (Wig-L-Bug; Dentsply) at 4,200 rpm for 20 pulses of 30 s each. Samples were put on ice for at least 30 s between pulses. Proteins were extracted from the broken material with 75 μl (dormant and germinated spores) or 100 μl (sporangia) of 1× sample loading buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.05% bromophenol blue). Following heating at 100°C for 5 min, extracts were centrifuged at 15,800 × g for 1 min, and the supernatants were analyzed by Western blotting. In some cases, proteins were extracted in sample loading buffer without bromophenol blue and quantified using amino acid analyses (29).
To extract and purify His6-tagged YpeB cleavage products for N-terminal sequencing, 250 OD units of DPBa127 or DPBa157 spores in a final volume of 100 ml 10 mM Tris-HCl, pH 7.0, were heat activated and germinated with 100 mM l-alanine and 1 mM inosine as described above. The spores were germinated for approximately 1 h or until ≥90% of the spores had germinated, as determined by phase-contrast microscopy. The germinated spores were collected by centrifugation at 10,000 × g for 10 min at 4°C and washed with 5 ml buffer A (50 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5% glycerol, 30 mM imidazole). Following another centrifugation, the pellet was resuspended in 5 ml buffer A, split between 4 microcentrifuge tubes, and centrifuged at 15,800 × g for 2 min. The pellets were frozen at −80°C, lyophilized, and broken with glass beads as described above. Proteins were extracted by resuspending and combining samples in a total of 5 ml buffer B (500 mM NaCl, 50 mM Tris-HCl, pH 7.5, 30 mM imidazole, 8 M urea) and incubating at 4°C with slow rocking for at least 2 h. The sample was centrifuged at 6,800 × g for 10 min at 4°C, and the supernatant containing soluble extracted proteins was filtered and loaded on a 1-ml Ni-Sepharose HisTrap HP affinity column (GE Healthcare) equilibrated with buffer B. YpeB cleavage products were eluted with buffer C (500 mM NaCl, 50 mM Tris-HCl, pH 7.5, 500 mM imidazole, 8 M urea). To precipitate the affinity-purified cleavage products, deoxycholate and trichloroacetic acid (TCA) (0.05% and 20% final concentrations, respectively) were added to the pooled elution fractions, which were then incubated on ice overnight. The mixture was centrifuged at 15,000 × g for 10 min at 4°C, and the pellet was washed 3 times with 1 ml ice-cold 100% acetone. The pellet was dried and mixed with 25 μl 1× sample loading buffer, and additional 0.5 M Tris-HCl, pH 6.8, was added until the pH was ≥5.
The entire precipitated sample was run on an SDS-PAGE gel that had first been incubated overnight in 1× Tris-glycine SDS running buffer with 0.1 mM sodium thioglycolate to scavenge free radicals. The proteins were transferred to an Immobilon-PSQ polyvinylidene difluoride (PVDF) membrane (Millipore) in CAPS transfer buffer [10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS), pH 11.0, 10% methanol], and the membrane was briefly washed with deionized water before being stained with 0.1% Coomassie brilliant blue R-250 (Jersey Lab Supply) for 30 s. The membrane was destained repeatedly with 50% methanol until bands were visible. The bands corresponding to YpeB cleavage products were cut from the membrane, washed with deionized water, dried, and stored at −80°C. N-terminal sequencing via automated Edman degradation was performed on the extracted bands using an ABI 494 Protein Sequencer (Tufts University Analytical Core Facility).
Western blot analysis.
Polyclonal antibodies raised in rabbits against B. anthracis SleB and YpeB (18) and monoclonal mouse anti-His (C-terminal) antibodies (Invitrogen) were used for detection of SleB, YpeB, and derivatives in Western blots. Anti-SleB and anti-YpeB antibodies were used at 1:1,000 and 1:3,000 dilutions, respectively, while anti-His antibodies were used at a 1:5,000 dilution. Horseradish peroxidase (HRP)-conjugated secondary goat anti-rabbit antibodies (Bio-Rad) were used at a 1:5,000 (catalog number 166-2408) or 1:200,000 (catalog number 170-6515) dilution, while HRP-conjugated goat anti-mouse antibodies (PerkinElmer) were used at a 1:5,000 dilution. The Western blots utilized Amersham Hybond-P (PVDF) membranes (GE Healthcare), and antibody detection was carried out using colorimetric (BM Blue POD Substrate, Precipitating; Roche) or chemiluminescent (Amersham ECL Prime Western Blotting Detection Reagent; GE Healthcare) substrates.
HtrC and YpeB purification and assay.
The B. anthracis htrC coding sequence, lacking the first 44 codons containing the predicted transmembrane anchor, was PCR amplified and introduced into the entry vector pDonR201 (Invitrogen) and then the destination vector pDEST-HisMBP-T (a modified version of pDEST-HisMBP [30] containing a tobacco etch virus [TEV] cleavage site). The resulting plasmid, pDPV458, encoding an N-terminal His6-tagged maltose binding protein (MBP), a TEV cleavage site, and HtrC lacking its signal sequence/membrane anchor, was verified by DNA sequencing. The overexpression strain DPVE501 was cultured at 37°C until the OD reached ∼0.8, at which point isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.7 mM, the culture temperature was reduced to 12°C, and incubation was continued for 16 h.
Cells were harvested by centrifugation, resuspended in buffer D (50 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5% glycerol, 25 mM imidazole), and lysed using sonication. Soluble and insoluble protein fractions were separated by centrifugation at 117,000 × g for 1 h. Soluble His6-MBP-HtrC45–391 was purified using a Ni-Sepharose HisTrap HP affinity column equilibrated with buffer D. Protein was eluted with a step gradient of buffer D containing 100 mM increasing concentrations of imidazole, and fractions containing His6-MBP-HtrC45–391 (at 100 mM imidizole) were dialyzed in buffer D. His6-MBP-HtrC45–391 at a concentration of ∼1.5 mg/ml was incubated with 0.5 mg/ml His6-tagged TEV (S219V) protease (31) at 15°C for 16 h. Cleavage was verified by SDS-PAGE analysis. HtrC45–391 was separated from His6-TEV, His6-MBP, and undigested protein using a Ni-Sepharose HisTrap HP affinity column. Fractions containing HtrC45–391 were dialyzed in buffer D, flash frozen, and stored at −80°C. YpeB21–446 overexpression and purification were performed as described previously (18).
The protease activity of HtrC was assayed in 50 mM Tris-HCl, pH 8.0, with 1 μM HtrC45–391 and 6 μM YpeB21–446 at 37°C for 4 h, unless otherwise indicated. The reaction was terminated by adding 2× SDS-PAGE sample loading buffer to a final concentration of 1× and incubating at 100°C for 5 min. Proteins were separated using SDS-PAGE and stained with Coomassie brilliant blue.
RESULTS
B. anthracis YpeB is proteolytically processed during spore germination.
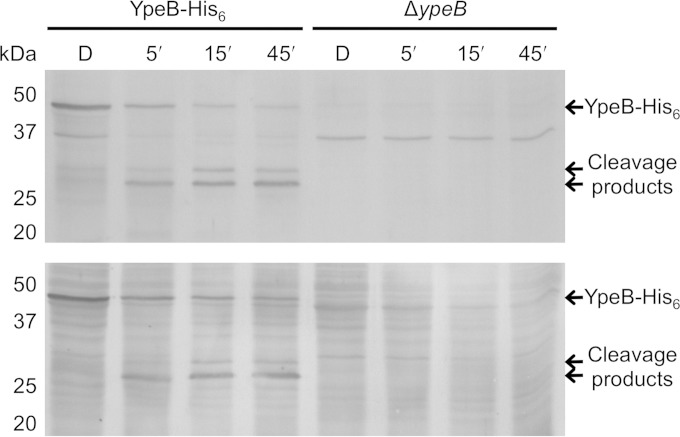
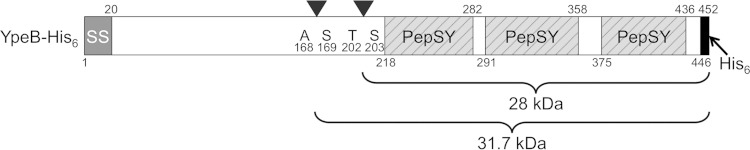
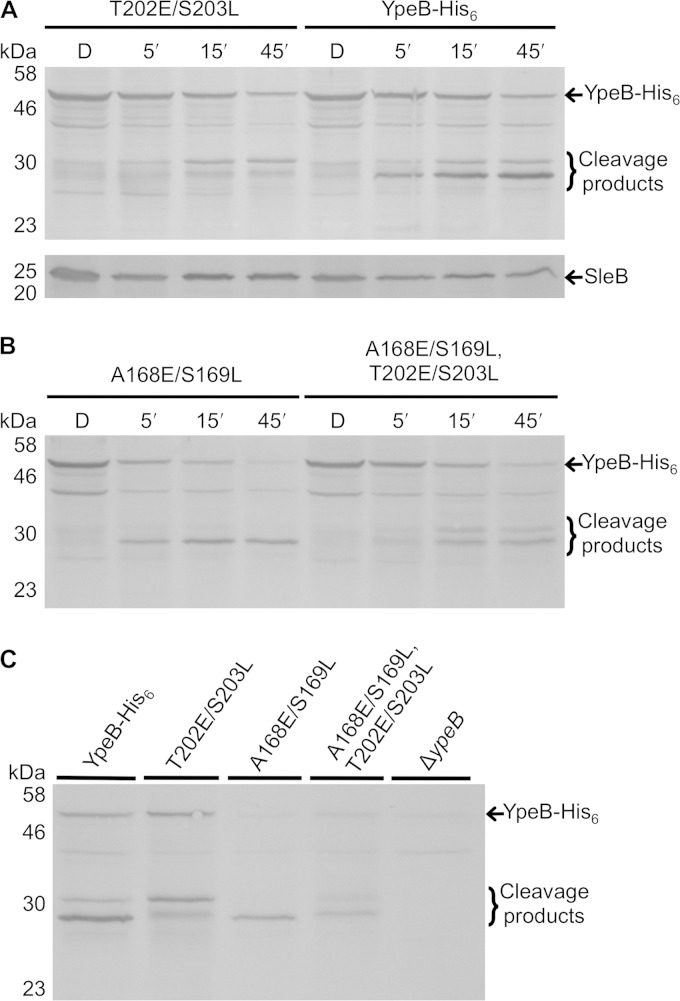
A previous study indicated that YpeB of B. subtilis is proteolytically cleaved during spore germination (10). A B. anthracis ypeB deletion mutant was previously complemented with a gene expressing YpeB with a C-terminal His6 tag (18). This His6-tagged protein accumulated in spores to an abundance equivalent to that of the wild type (WT), complemented fully for stabilization of SleB in the dormant spore, and allowed a wild-type germination rate (18). Spores of this B. anthracis YpeB-His6 strain (DPBa127) were germinated and extracted at various times for Western blotting to examine YpeB processing. By the 45-min time point, the spores had lost at least 50% of their initial OD, and examination by phase-contrast microscopy indicated that >95% of the spores in each preparation had germinated. Full-length 51-kDa YpeB-His6 diminished during germination, with the appearance of an abundant, stable, ∼28-kDa product and a minor ∼32-kDa product (Fig. 1, top). The same banding pattern was observed during germination of wild-type B. anthracis spores (not shown). Western blotting of the same YpeB-His6 germinating spore fragments using anti-His6 antibodies indicated that the 28- and 32-kDa YpeB fragments possessed the His6 tag (Fig. 1, bottom) and were therefore C-terminal portions of YpeB. The abundant 28-kDa YpeB-His6 fragment was purified from germinated spores using metal affinity chromatography, and the N-terminal sequence was determined to be SAQKN. The sequence indicated cleavage between residues T202 and S203 of YpeB (Fig. 2). In an effort to block this specific cleavage event, the ypeB-His6 allele was mutagenized to change residues T202-S203 to E202-L203. The altered YpeBT202E/S203L-His6 protein accumulated to levels equivalent to those of YpeB-His6 in the spore and complemented for SleB stabilization and for the germination rate (Fig. 3A and 4). YpeBT202E/S203L-His6 protein was not cleaved to the 28-kDa fragment during germination, indicating that the mutation at the cleavage site succeeded in blocking proteolysis, but the stability of full-length YpeB during germination was not increased; rather, the abundance of the 32-kDa product increased (Fig. 3A and C). In addition, a small amount of a new ∼29-kDa cleavage product, slightly larger than the original 28-kDa fragment, was observed (Fig. 3A and C). The 32-kDa product was purified from germinated YpeBT202E/S203L-His6 spores, and the N-terminal sequence was determined to be SNRDP, indicating processing between residues A168 and S169 (Fig. 2). Once again, the amino acids flanking this cleavage site were changed by site-specific mutagenesis, allowing the expression of YpeBA168E/S169L-His6 and YpeBA168E/S169L, T202E/S203L-His6. Both of these proteins accumulated to normal levels in the spore (Fig. 3B), and both allowed normal spore germination (Fig. 4). Western blotting of germinating spore samples indicated that the A168E/S169L change prevented proteolysis at that site (Fig. 3B and C). YpeB-A168E/S169L-His6 was instead processed to the dominant 28-kDa fragment (Fig. 3B and C). Stable cleavage products were not as strongly detected during germination of YpeBA168E/S169L, T202E/S203L-His6 spores, but the full-length protein still diminished during germination (Fig. 3B and C). Two less stable products of ∼29 and ∼33 kDa were produced (Fig. 3B and C).
FIG 1.
YpeB is cleaved during germination of B. anthracis spores. Dormant (D) DPBa127 (YpeB-His6) and DPBa89 (ΔypeB) spores were germinated with 100 mM l-alanine and 1 mM inosine at 37°C, and samples were collected at 5 (5′), 15 (15′), and 45 (45′) min. The extracted proteins were probed with anti-YpeB (top) and anti-His6 (bottom) antibodies. The predicted molecular mass of YpeB-His6 is 51 kDa. The positions of the molecular mass marker proteins (not shown) are indicated on the left.
FIG 2.
Locations of major B. anthracis YpeB cleavage sites. The scale drawing of the domain architecture of YpeB shows the N-terminal signal sequence (SS), the C-terminal PepSY domains, and the YpeB cleavage sites (arrowheads) with predicted molecular masses of the C-terminal cleavage products. The residue numbers designate the amino acid positions of domain boundaries and the positions of YpeB cleavage during spore germination.
FIG 3.
YpeB cleavage during germination of B. anthracis mutant spores. Dormant (D) spores were germinated with 100 mM l-alanine and 1 mM inosine at 37°C, and samples were collected at 5 (5′), 15 (15′), and 45 (45′) min. (A) DPBa157 (T202E/S203L) and DPBa127 (YpeB-His6) sample extracts probed with anti-YpeB (top) and anti-SleB (bottom) antibodies. (B) DPBa167 (A168E/S169L) and DPBa168 (A168E/S169L, T202E/S203L) sample extracts were probed with anti-YpeB antibodies. (C) Extracts from 45-min-germinated spores from each strain were probed with anti-YpeB antibodies. The predicted molecular mass of YpeB-His6 and derivatives is approximately 51 kDa. The positions of the molecular mass marker proteins (not shown) are indicated on the left.
FIG 4.
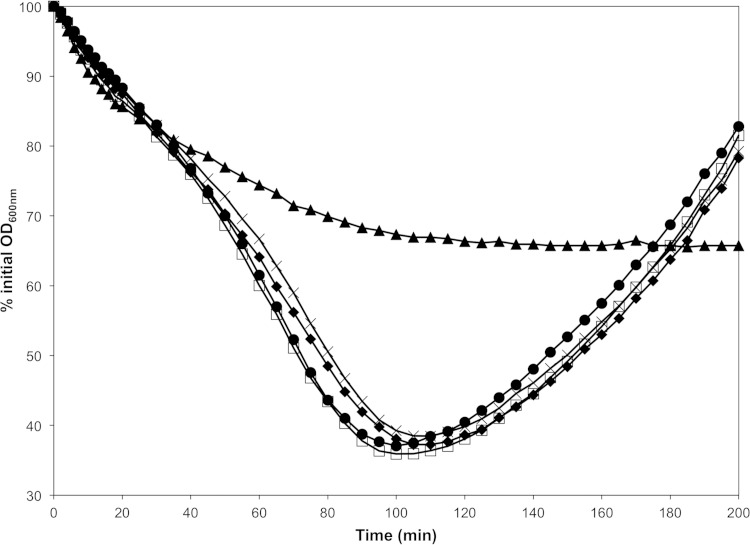
Altered YpeB proteolysis does not slow spore germination and outgrowth. Decoated ΔypeB (▲), YpeBT202E/S203L-His6 (□), YpeBA168E/S169L-His6 (◆), YpeBA168E/S169L, T202E/S203L-His6 (×), and YpeB-His6 (●) B. anthracis spores were heat activated and germinated in BHI medium at 37°C; germination and outgrowth were tracked as changes in OD. Decoating of spores eliminates CwlJ activity, and thus, completion of germination is dependent on YpeB for maintenance of SleB activity in the spores (18). The data shown are averages of results from three independent spore preparations; error bars are omitted for clarity. Germination of YpeBT202E/S203L-His6, YpeBA168E/S169L-His6, and YpeBA168E/S169L, T202E/S203L-His6 spores is not significantly different (P > 0.06) at any time point. Likewise, germination of YpeB-His6 and YpeBT202E/S203L-His6 spores is not significantly different (P > 0.07) at any time point. Focusing on stage 2 of germination, from 45 to 95 min (18), YpeBA168E/S169L-His6 and YpeB-His6 spores do not significantly differ (P > 0.07), whereas YpeBA168E/S169L, T202E/S203L-His6 and YpeB-His6 spores are not significantly different (P > 0.05) except from 75 to 80 min (P < 0.05).
Identification of candidate B. anthracis proteases that might cleave YpeB.
A variety of data sources were used to identify a protease, here called HtrC, as a candidate to be involved in YpeB cleavage. Recent studies of B. anthracis and B. subtilis spore membrane proteomes indicated that this protease might be associated with the membrane that YpeB is embedded in (Y. Chen and D. L. Popham, unpublished data). A transcriptome study had indicated that htrC (BAS5314) was expressed during the latter stages of B. anthracis sporulation (32). In B. subtilis, the apparent ortholog (htrC, also known as yycK) is expressed during sporulation under the control of σG, which would place it in the forespore compartment (33), along with sleB-ypeB expression (16). Studies of HtrC and paralogs in other species have indicated that these proteases cross and remain associated with the outer surface of the cytoplasmic membrane due to an uncleaved signal sequence/membrane anchor (34). HtrC expressed in the developing spore would therefore be expected to remain on the outer surface of the inner spore membrane, in the same location as YpeB. While multiple proteases in each species possess some properties consistent with a role in YpeB processing, the HtrC proteins of B. anthracis and B. subtilis were considered the strongest candidates.
Strains lacking HtrC are altered in YpeB proteolysis.
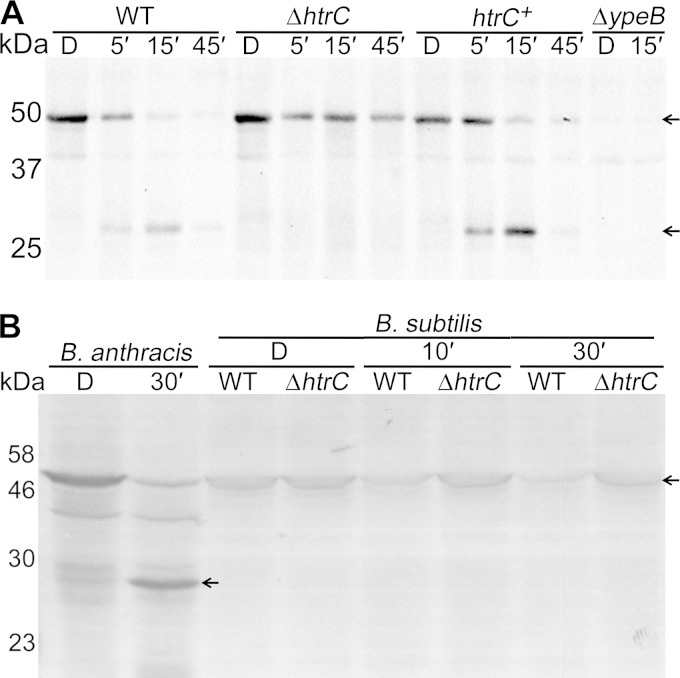
B. anthracis and B. subtilis strains carrying null mutations in htrC were constructed, and YpeB cleavage was examined by Western blotting using antisera against B. anthracis YpeB (18). The B. anthracis ΔhtrC mutant lost full-length YpeB during germination more slowly than the wild type and did not accumulate the specific 27- and 31-kDa cleavage products (Fig. 5A). (These cleavage products are slightly smaller than those in Fig. 1 because they lack the His6 tag.) In some cases, Western blots exhibited a heavier background in the 30- to 50-kDa range for germinating ΔhtrC spores (Fig. 5A), suggesting that YpeB might be degraded to nonspecific products, but this has not been further confirmed. While the antibodies did not recognize the cleavage products of B. subtilis YpeB, the htrC mutant maintained increased amounts of full-length YpeB during germination relative to the wild-type strain (Fig. 5B).
FIG 5.
HtrC cleaves YpeB during germination of B. anthracis and B. subtilis spores. Spores were prepared, germinated, and analyzed by Western blotting with B. anthracis anti-YpeB antibodies as described in Materials and Methods. The positions of YpeB (∼51 kDa) and YpeB cleavage products (∼27 kDa) are indicated by arrows. (A) B. anthracis spores were dormant (D) or were collected 5 (5′), 15 (15′), and 45 (45′) min after addition of germinants. The strains were DPBa2 (WT), DPBa178 (ΔhtrC), DPBa182 (htrC+), and DPBa89 (ΔypeB). (B) B. anthracis and B. subtilis spores were dormant (D) or were collected 10 (10′) and 30 (30′) min after addition of germinants. The strains were DPBa2 (B. anthracis), PS832 (B. subtilis WT), and DPVB668 (B. subtilis ΔhtrC). The predicted molecular masses of YpeB proteins from B. anthracis and B. subtilis are 50 and 51 kDa, respectively. The positions of the molecular mass marker proteins (not shown) are indicated on the left.
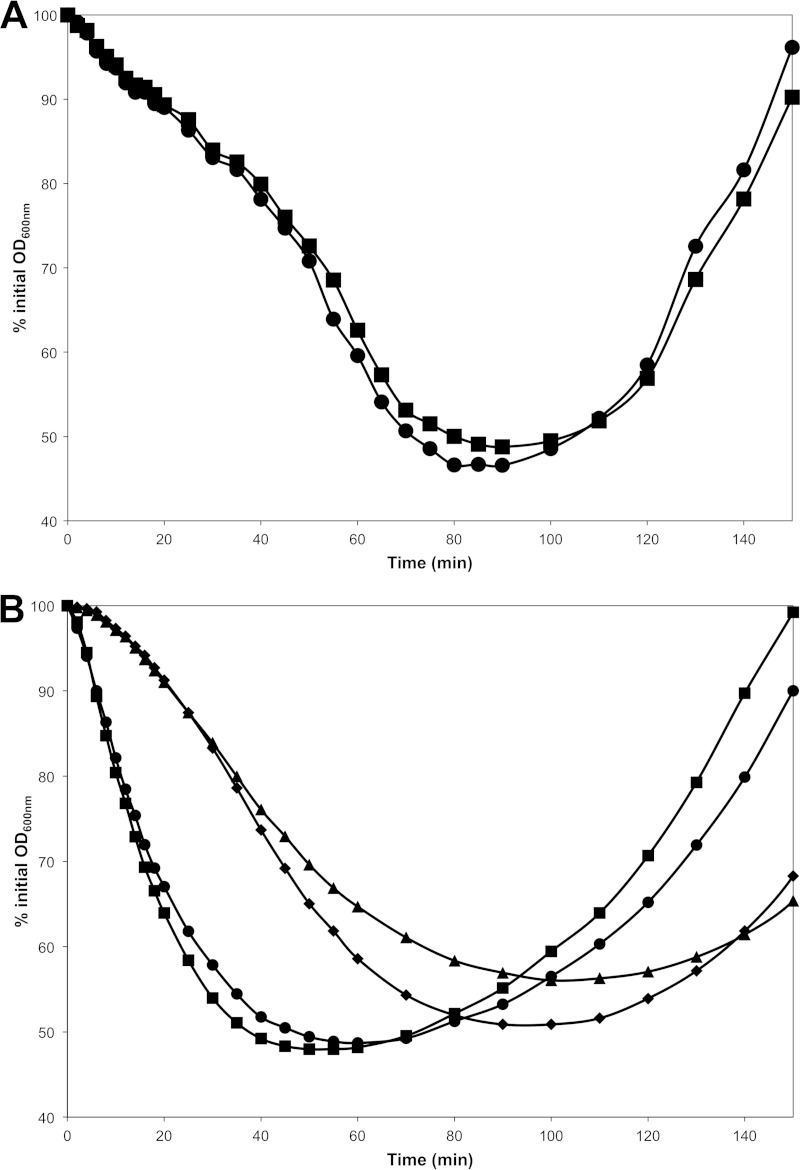
The germination of htrC mutant spores was examined to determine if slowed degradation of YpeB resulted in decreased SleB activity. This was done in B. anthracis decoated spores, which have lost CwlJ activity, and in a B. subtilis ΔcwlJ background in order to render the function of SleB more easily observed (13, 35). In both Bacillus species, the germination rates of ΔhtrC spores were not significantly different from those of the wild-type spores (Fig. 6A and B), and microscopic examination revealed no clear differences between the germinating spores of these strains (data not shown). In both species, the colony-forming efficiency of ΔhtrC spores lacking CwlJ activity was also not different from that of the corresponding wild-type spores (Table 3).
FIG 6.
Loss of HtrC does not alter the germination rates of B. anthracis and B. subtilis spores. Spores were heat activated and germinated in rich medium as described in Materials and Methods; germination and outgrowth were tracked as changes in OD600. The data shown are averages of results from three independent spore preparations; error bars are omitted for clarity. (A) B. anthracis wild-type (DPBa2; ■) and ΔhtrC (DPBa178; ●) spores were decoated and germinated in BHI broth. The two strains were not statistically different from one another (P > 0.3) at any time point. (B) B. subtilis wild-type (PS832; ■), ΔcwlJ (FB111; ◆), ΔhtrC (DPVB668; ●), and ΔcwlJ ΔhtrC (DPVB669; ▲) spores were germinated in 2× YT broth. The wild-type and ΔhtrC strains were not significantly different, and the ΔcwlJ and ΔcwlJ ΔhtrC strains were not significantly different (P > 0.2) at any time point. The wild-type and ΔcwlJ strains were significantly different from 30 to 60 min, and the ΔhtrC and ΔcwlJ ΔhtrC strains were significantly different from 12 to 50 min (P < 0.05).
TABLE 3.
Germination efficiencies of ΔhtrC B. anthracis and B. subtilis spores
| Species | Strain | Genotype | No. of CFU/OD unit/mla |
|---|---|---|---|
| B. anthracis | DPBa2 | WT | 6.7 × 107 |
| DPBa178 | ΔhtrC | 6.0 × 107 | |
| DPBa182 | htrC+ | 6.3 × 107 | |
| B. subtilis | PS832 | WT | 1.7 × 108 |
| FB111 | ΔcwlJ | 3.0 × 108 | |
| DPVB668 | ΔhtrC | 1.9 × 108 | |
| DPVB669 | ΔcwlJ ΔhtrC | 2.0 × 108 |
Decoated spores were used for B. anthracis studies. The values are averages of three independent spore preparations. Among the strains for each species, no statistical difference in colony-forming efficiency was present.
In vitro cleavage of YpeB by HtrC.
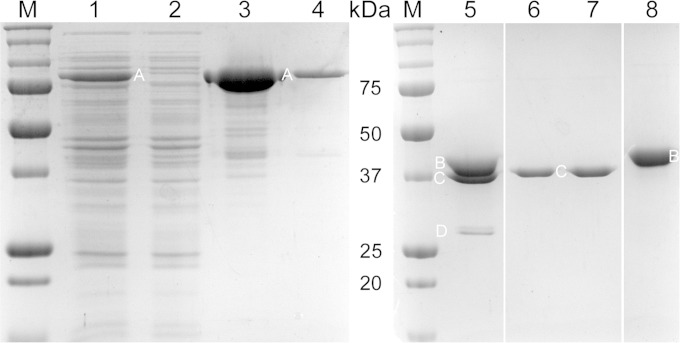
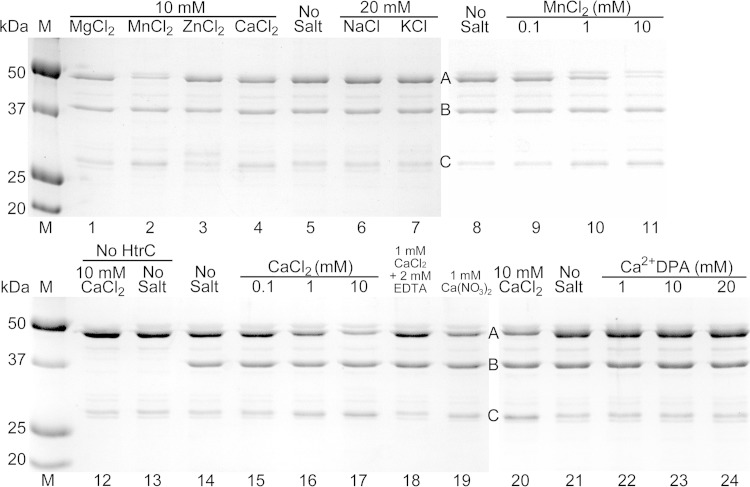
B. anthracis HtrC, lacking its signal sequence/membrane anchor, was expressed in E. coli as a His6-MBP fusion protein and was purified by metal affinity chromatography. Following purification of His6-MBP-HtrC45–391, the His6-MBP domain was removed using TEV protease, and HtrC45–391 was purified (Fig. 7). The pure protease exhibited little activity against a generic substrate (fluorescently labeled casein) (data not shown) and, initially, very little activity against purified B. anthracis YpeB (Fig. 8, lanes 13 and 14). Assay attempts under a wide variety of conditions revealed that HtrC-catalyzed cleavage of YpeB to produce an ∼27-kDa fragment, as observed in vivo, was greatly stimulated by the presence of Mn2+ (Fig. 8, lanes 8 to 11) and, to a lesser degree, by Ca2+ (Fig. 8, lanes 14 to 17) but not by Mg2+, Zn2+, Na+, or K+ (Fig. 8, lanes 1 to 10). Relatively high concentrations of Mn2+ or Ca2+ were required to achieve maximum HtrC activity (Fig. 8, lanes 8 to 11 and 14 to 17). Metal-stimulated HtrC activity was inhibited by EDTA, a chelator of divalent cations (Fig. 8, lanes 14, 16, and 18). The presence of an equimolar concentration of DPA, another strong chelator, blocked the positive effect of Ca2+ on HtrC (Fig. 8, lanes 20 to 24). HtrC action on YpeB was found to be maximal at pH 7 to 9 (data not shown).
FIG 7.
Purification of B. anthracis HtrC. A His6-MBP-HtrC45–391 fusion protein was overexpressed in E. coli, purified by metal affinity chromatography, cleaved with TEV protease, and separated from the His6-MBP tag and His6-TEV using a second metal affinity column. Samples were soluble protein extracts of induced (lane 1) and uninduced (lane 2) cells, eluate fractions from the metal affinity column containing His6-MBP-HtrC45–391 (A) (lanes 3 and 4), His6-MBP-HtrC45–391 digested with His6-TEV protease (D) (lane 5), flowthrough fractions from the second metal affinity column containing HtrC45–391 (C) (lanes 6 and 7), and eluate from the second metal affinity column containing His6-MBP (B) (lane 8). Lanes 1 to 4 and 5 to 8 are from two different gels; the intervening lanes between lanes 5 and 6 and lanes 7 and 8 were removed for clarity. The masses of standard proteins (M) are indicated in the center.
FIG 8.
In vitro cleavage of YpeB by HtrC. Purified YpeB21–446 (A) (6 μM) was incubated for 4 h with purified HtrC45–391 (B) (1 μM) unless otherwise indicated, with the indicated small-molecule additions. Reactions were terminated using SDS and heat. Samples were separated by SDS-PAGE, and proteins, including the YpeB cleavage product (C), were detected by staining with Coomassie brilliant blue. Protein concentrations were 12 μM YpeB21–446 and 2 μM HtrC45–391 in lanes 20 to 24. Lanes 1 to 7, 8 to 11, 12 to 19, and 20 to 24 are from four different gels. The masses of standard proteins (M) are indicated on the left.
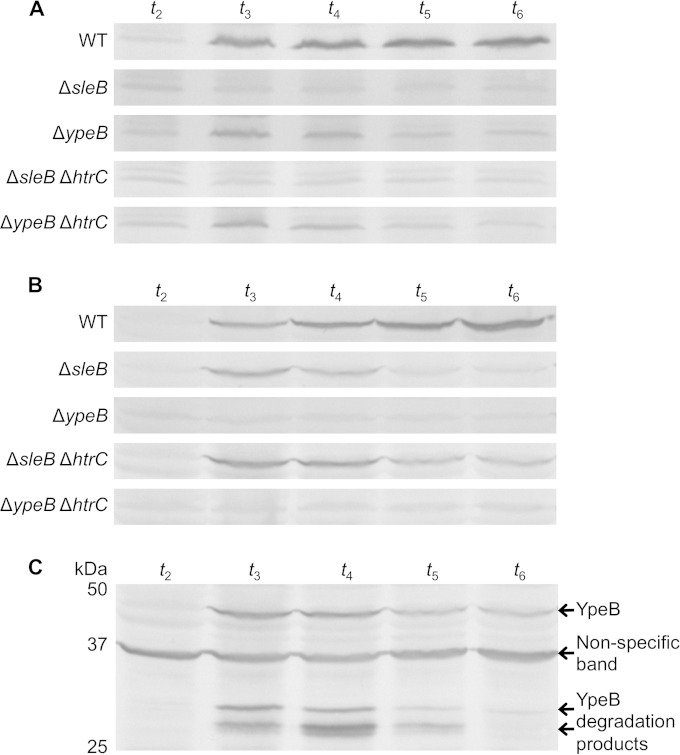
HtrC is not an important factor in SleB or YpeB degradation during spore formation.
To determine if HtrC is involved in the degradation of SleB and/or YpeB in the absence of the partner protein during spore formation, ΔsleB ΔhtrC and ΔypeB ΔhtrC double mutants were constructed in B. anthracis. As demonstrated previously (18), in a ΔsleB mutant, YpeB failed to accumulate during spore formation, and in a ΔypeB mutant, SleB did not accumulate (Fig. 9A and B). The additional deletion of htrC from these strains did not alter the degradation of YpeB or SleB, respectively, during sporulation (Fig. 9), indicating that other proteases are likely active in their degradation during spore formation. Additionally, the production of YpeB during ΔsleB ΔhtrC spore formation coincided with the appearance of YpeB-specific degradation products (Fig. 9C), similar to those seen during sporulation of the ΔsleB strain (18) and to specific cleavage products seen during spore germination. This finding indicates that, at least during sporulation, other proteases are capable of producing similar cleavage products.
FIG 9.
SleB and YpeB production during sporulation. WT, ΔsleB, ΔypeB, ΔsleB ΔhtrC, and ΔypeB ΔhtrC strains of B. anthracis were grown in modified G broth at 37°C, and sporangia collected during sporulation were subjected to Western blot analysis. t2 through t6 indicate the number of hours since the initiation of sporulation. (A) Sporangia probed with anti-SleB antibodies. (B) Sporangia probed with anti-YpeB antibodies. (C) Sporangia from the ΔsleB ΔhtrC strain probed with anti-YpeB antibodies. The positions of the molecular mass marker proteins (not shown) are indicated on the left.
DISCUSSION
B. anthracis YpeB is processed to a relatively stable fragment during germination, as in B. subtilis (10). The ultimate N termini of the stable YpeB fragments were determined to be in a region between an undefined N-terminal YpeB domain and the C-terminal region containing three putative PepSY domains. This C-terminal fragment persists well into the germination process. The N-terminal cleavage product was observed neither in Western blots of germinating spore extracts nor in in vitro reactions using purified proteins. While this might be due to poor recognition of the domain by the antiserum and/or poor staining of the domain, it raises the possibility that the domain is subject to more rapid degradation to smaller fragments. Interestingly, the YpeB N-terminal domain was recently shown to exert an inhibitory effect on SleB activity in vitro (20). Processing of YpeB could play a role in the activation of SleB during germination.
Deletion of htrC was found to result in the disappearance of specific YpeB degradation products in both species, though the amount of full-length YpeB still decreased during germination. This indicates that multiple proteases are active in this region of the spore during germination. The degradation of YpeB during germination may be a multistep process, carried out by multiple proteases, with the ultimate stable products being produced by HtrC. However, initial cleavage of YpeB by another protease is apparently not a requirement for HtrC action, as the action of HtrC was reproducible in vitro using purified proteins.
The inability to fully block YpeB degradation during germination precludes demonstration of a role for YpeB instability in SleB activation during germination. Even when YpeB degradation was slowed by the loss of HtrC, the spore germination rate was not significantly affected. Under the experimental conditions tested, germination was dependent on SleB activity (11, 19). A previous study indicated that a 30% decrease in SleB abundance resulted in an observable germination defect (18), so the delay in YpeB degradation observed here must not have inhibited SleB to this degree. This may indicate that YpeB degradation is not required for SleB activation and that YpeB degradation is merely the natural disposal of the protein following the breakage of spore dormancy. Alternatively, the slowed kinetics of YpeB degradation may not have been sufficient to affect the outward assays of germination progression. Unfortunately, strains in which N-terminal regions of YpeB were deleted, including the identified HtrC cleavage sites and potential upstream cleavage sites, did not retain stable SleB in the dormant spore (18) and thus could not be used to determine what effect a noncleavable YpeB has on SleB activity during germination.
Alteration of the dominant HtrC cleavage sites in YpeB resulted in the appearance of alternative dominant in vivo cleavage sites in the same region of the protein. Several pieces of evidence suggest that the C-terminal domain of YpeB is relatively protease resistant while the N-terminal domain is more sensitive, and the region linking the domains may be especially sensitive. The N-terminal domain was not observed in either in vivo or in vitro experiments where the C-terminal domain could be detected. In the absence of SleB, YpeB is degraded during spore formation, producing dominant cleavage products similar in size to those produced during germination (18). Similar YpeB cleavage products were produced during sporulation of a ΔsleB ΔhtrC mutant, indicating that another protease(s) targets the same region between the YpeB domains. These experiments also revealed that proteases other than HtrC must be involved in the degradation of both YpeB and SleB when produced in the absence of SleB or YpeB, respectively, during spore formation. The use of multiple protease-deficient strains to block SleB and YpeB degradation during sporulation, and potentially during germination, is challenging, as multiple proteases present in the intermembrane space of the developing spore are necessary for proper spore formation (36, 37).
B. anthracis HtrC, expressed and purified from E. coli, cleaved YpeB in vitro but with relatively poor kinetics. A 4-h digestion was required to achieve catalysis of approximately three YpeB molecules per HtrC molecule. This activity was dependent on the presence of relatively high concentrations of Mn2+ or Ca2+ ions. The high ion concentrations required suggest that their function is not directly in catalysis, which is consistent with the absence of a role for metal in catalysis by this class of serine proteases (38). Interestingly, HtrC action on YpeB coincides closely with the release of a large amount of Ca2+-DPA from the spore core during germination. The local Ca2+-DPA concentration is apparently quite high at this time, as it can activate CwlJ activity, a process that requires >20 mM Ca2+-DPA (39). While most of the Ca2+ released during germination is likely bound to DPA, and our findings indicate that the Ca2+-DPA complex is ineffective in promoting HtrC activity, it is possible that enough free Ca2+ is present to increase HtrC activity during germination. Spores also contain significant amounts of Mn2+ (40, 41), which is likely complexed with DPA and released during germination and could thus play a role in stimulating HtrC.
Chemicals that function as protease inhibitors have been shown to block spore germination at different stages, suggesting that protease activity is critical during germination (42–45). While it has not been shown that this activity is actually due to inhibition of a protease rather than another protein function or what specific germination processes are affected, these results raise the possibility that initiation of cortex hydrolysis requires a protease activity. Proteolytic processing of a clostridial GSLE, SleC, is required for induction of activity during germination, but this GSLE is unrelated to Bacillus GSLEs. Further study of the roles of proteases in spore germination, and of their potential direct effect on activation of cortex hydrolysis, may allow the improvement of spore decontamination or application methods.
ACKNOWLEDGMENTS
The research reported in this article was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R21AI088298.
The content is solely our responsibility and does not necessarily represent the official views of the National Institutes of Health.
We thank Roger Plaut and Scott Stibitz for the gift of plasmid pSS4332. Florian Schubot contributed reagents, and Jessica Blackwood provided laboratory assistance.
REFERENCES
- 1.Setlow P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol 101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 2.Setlow P. 2014. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol 196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atrih A, Zöllner P, Allmaier G, Foster SJ. 1996. Structural analysis of Bacillus subtilis 168 endospore peptidoglycan and its role during differentiation. J Bacteriol 178:6173–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popham DL, Helin J, Costello CE, Setlow P. 1996. Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc Natl Acad Sci U S A 93:15405–15410. doi: 10.1073/pnas.93.26.15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekiguchi J, Akeo K, Yamamoto H, Khasanov FK, Alonso JC, Kuroda A. 1995. Nucleotide sequence and regulation of a new putative cell wall hydrolase gene, cwlD, which affects germination in Bacillus subtilis. J Bacteriol 177:5582–5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Setlow B, Melly E, Setlow P. 2001. Properties of spores of Bacillus subtilis blocked at an intermediate stage in spore germination. J Bacteriol 183:4894–4899. doi: 10.1128/JB.183.16.4894-4899.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Popham DL, Heffron JD, Lambert EA. 2012. Degradation of spore peptidoglycan during germination, p 121–142. In Abel-Santos E. (ed), Bacterial spores: current research and applications. Caister Academic Press, Norwich, United Kingdom. [Google Scholar]
- 8.Ishikawa S, Yamane K, Sekiguchi J. 1998. Regulation and characterization of a newly deduced cell wall hydrolase gene (cwlJ) which affects germination of Bacillus subtilis spores. J Bacteriol 180:1375–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagyan I, Setlow P. 2002. Localization of the cortex lytic enzyme CwlJ in spores of Bacillus subtilis. J Bacteriol 184:1219–1224. doi: 10.1128/jb.184.4.1219-1224.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chirakkal H, O'Rourke M, Atrih A, Foster SJ, Moir A. 2002. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology 148:2383–2392. [DOI] [PubMed] [Google Scholar]
- 11.Paidhungat M, Ragkousi K, Setlow P. 2001. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J Bacteriol 183:4886–4893. doi: 10.1128/JB.183.16.4886-4893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ragkousi K, Eichenberger P, van Ooij C, Setlow P. 2003. Identification of a new gene essential for germination of Bacillus subtilis spores with Ca2+-dipicolinate. J Bacteriol 185:2315–2329. doi: 10.1128/JB.185.7.2315-2329.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heffron JD, Lambert EA, Sherry N, Popham DL. 2010. Contributions of four cortex lytic enzymes to germination of Bacillus anthracis spores. J Bacteriol 192:763–770. doi: 10.1128/JB.01380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Setlow B, Peng L, Loshon CA, Li YQ, Christie G, Setlow P. 2009. Characterization of the germination of Bacillus megaterium spores lacking enzymes that degrade the spore cortex. J Appl Microbiol 107:318–328. doi: 10.1111/j.1365-2672.2009.04210.x. [DOI] [PubMed] [Google Scholar]
- 15.Boland FM, Atrih A, Chirakkal H, Foster SJ, Moir A. 2000. Complete spore-cortex hydrolysis during germination of Bacillus subtilis 168 requires SleB and YpeB. Microbiology 146:57–64. [DOI] [PubMed] [Google Scholar]
- 16.Moriyama R, Fukuoka H, Miyata S, Kudoh S, Hattori A, Kozuka S, Yasuda Y, Tochikubo K, Makino S. 1999. Expression of a germination-specific amidase, SleB, of bacilli in the forespore compartment of sporulating cells and its localization on the exterior side of the cortex in dormant spores. J Bacteriol 181:2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korza G, Setlow P. 2013. Topology and accessibility of germination proteins in the Bacillus subtilis spore inner membrane. J Bacteriol 195:1484–1491. doi: 10.1128/JB.02262-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernhards CB, Popham DL. 2014. The role of YpeB in cortex hydrolysis during germination of Bacillus anthracis spores. J Bacteriol 196:3399–3409. doi: 10.1128/JB.01899-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heffron JD, Orsburn B, Popham DL. 2009. Roles of germination-specific lytic enzymes CwlJ and SleB in Bacillus anthracis. J Bacteriol 191:2237–2247. doi: 10.1128/JB.01598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Butzin XY, Davis A, Setlow B, Korza G, Üstok FI, Christie G, Setlow P, Hao B. 2013. Activity and regulation of various forms of CwlJ, SleB, and YpeB proteins in degrading cortex peptidoglycan of spores of Bacillus species in vitro and during spore germination. J Bacteriol 195:2530–2540. doi: 10.1128/JB.00259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leighton TJ, Doi RH. 1971. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem 254:3189–3195. [PubMed] [Google Scholar]
- 22.Kim HU, Goepfert JM. 1974. A sporulation medium for Bacillus anthracis. J Appl Bacteriol 37:265–267. doi: 10.1111/j.1365-2672.1974.tb00438.x. [DOI] [PubMed] [Google Scholar]
- 23.Nicholson WL, Setlow P. 1990. Sporulation, germination, and outgrowth, p 391–450. In Harwood CR, Cutting SM (ed), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England. [Google Scholar]
- 24.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 25.van den Ent F, Löwe J. 2006. RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods 67:67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Janes BK, Stibitz S. 2006. Routine markerless gene replacement in Bacillus anthracis. Infect Immun 74:1949–1953. doi: 10.1128/IAI.74.3.1949-1953.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bishop-Lilly KA, Plaut RD, Chen PE, Akmal A, Willner KM, Butani A, Dorsey S, Mokashi V, Mateczun AJ, Chapman C, George M, Luu T, Read TD, Calendar R, Stibitz S, Sozhamannan S. 2012. Whole genome sequencing of phage resistant Bacillus anthracis mutants reveals an essential role for cell surface anchoring protein CsaB in phage AP50c adsorption. Virol J 9:246. doi: 10.1186/1743-422X-9-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wach A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259–265. doi:. [DOI] [PubMed] [Google Scholar]
- 29.González-Castro MJ, López-Hernández J, Simal-Lozano J, Oruña-Concha MJ. 1997. Determination of amino acids in green beans by derivitization with phenylisothiocyanate and high-performance liquid chromatography with ultraviolet detection. J Chrom Sci 35:181–185. doi: 10.1093/chromsci/35.4.181. [DOI] [Google Scholar]
- 30.Austin BP, Nallamsetty S, Waugh DS. 2009. Hexahistidine-tagged maltose-binding protein as a fusion partner for the production of soluble recombinant proteins in Escherichia coli. Methods Mol Biol 498:157–172. doi: 10.1007/978-1-59745-196-3_11. [DOI] [PubMed] [Google Scholar]
- 31.Kapust RB, Tözsér J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. 2001. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng 14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 32.Bergman NH, Anderson EC, Swenson EE, Niemeyer MM, Miyoshi AD, Hanna PC. 2006. Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J Bacteriol 188:6092–6100. doi: 10.1128/JB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fabret C, Hoch JA. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol 180:6375–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalbey RE, Wang P, van Dijl JM. 2012. Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol Mol Biol Rev 76:311–330. doi: 10.1128/MMBR.05019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giebel JD, Carr KA, Anderson EC, Hanna PC. 2009. The germination-specific lytic enzymes SleB, CwlJ1, and CwlJ2 each contribute to Bacillus anthracis spore germination and virulence. J Bacteriol 191:5569–5576. doi: 10.1128/JB.00408-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campo N, Rudner DZ. 2007. SpoIVB and CtpB are both forespore signals in the activation of the sporulation transcription factor σK in Bacillus subtilis. J Bacteriol 189:6021–6027. doi: 10.1128/JB.00399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mastny M, Heuck A, Kurzbauer R, Heiduk A, Boisguerin P, Volkmer R, Ehrmann M, Rodrigues CD, Rudner DZ, Clausen T. 2013. CtpB assembles a gated protease tunnel regulating cell-cell signaling during spore formation in Bacillus subtilis. Cell 155:647–658. doi: 10.1016/j.cell.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clausen T, Southan C, Ehrmann M. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell 10:443–455. doi: 10.1016/S1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- 39.Paidhungat M, Setlow P. 2000. Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J Bacteriol 182:2513–2519. doi: 10.1128/JB.182.9.2513-2519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Granger AC, Gaidamakova EK, Matrosova VY, Daly MJ, Setlow P. 2011. Effects of Mn and Fe levels on Bacillus subtilis spore resistance and effects of Mn2+, other divalent cations, orthophosphate, and dipicolinic acid on protein resistance to ionizing radiation. Appl Environ Microbiol 77:32–40. doi: 10.1128/AEM.01965-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slepecky R, Foster JW. 1959. Alterations in metal content of spores of Bacillus megaterium and the effect on some spore properties. J Bacteriol 78:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boschwitz H, Gofshtein-Gandman L, Halvorson HO, Keynan A, Milner Y. 1991. The possible involvement of trypsin-like enzymes in germination of spores of Bacillus cereus T and Bacillus subtilis 168. J Gen Microbiol 137:1145–1153. doi: 10.1099/00221287-137-5-1145. [DOI] [PubMed] [Google Scholar]
- 43.Boschwitz H, Halvorson HO, Keynan A, Milner Y. 1985. Trypsinlike enzymes from dormant and germinated spores of Bacillus cereus T and their possible involvement in germination. J Bacteriol 164:302–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boschwitz H, Milner Y, Keynan A, Halvorson HO, Troll W. 1983. Effect of inhibitors of trypsin-like proteolytic enzymes on Bacillus cereus T spore germination. J Bacteriol 153:700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cortezzo DE, Setlow B, Setlow P. 2004. Analysis of the action of compounds that inhibit the germination of spores of Bacillus species. J Appl Microbiol 96:725–741. doi: 10.1111/j.1365-2672.2004.02196.x. [DOI] [PubMed] [Google Scholar]