Abstract
Emerging evidence has indicated that alcohol consumption is an established risk factor for breast cancer. Deregulation of RNA polymerase III (Pol III) transcription enhances cellular Pol III gene production, leading to an increase in translational capacity to promote cell transformation and tumor formation. We have reported that alcohol intake increases Pol III gene transcription to promote cell transformation and tumor formation in vitro and in vivo. Studies revealed that tumor suppressors, pRb, p53, PTEN and Maf1 repress the transcription of Pol III genes. BRCA1 is a tumor suppressor and its mutation is tightly related to breast cancer development. However, it is not clear whether BRCA1 expression affects alcohol-induced transcription of Pol III genes. At the present studies, we report that restoring BRCA1 in HCC 1937 cells, which is a BRCA1 deficient cell line, represses Pol III gene transcription. Expressing mutant or truncated BRCA1 in these cells does not affect the ability of repression on Pol III genes. Our analysis has demonstrated that alcohol induces Pol III gene transcription. More importantly, overexpression of BRCA1 in estrogen receptor positive (ER+) breast cancer cells (MCF-7) decreases the induction of tRNALeu and 5S rRNA genes by alcohol, whereas reduction of BRCA1 by its siRNA slightly increases the transcription of the class of genes. This suggests that BRCA1 is associated with alcohol-induced deregulation of Pol III genes. These studies for the first time demonstrate the role of BRCA1 in induction of Pol III genes by alcohol and uncover a novel mechanism of alcohol-associated breast cancer.
Keywords: BRCA1, Alcohol, Pol III genes, Breast cancer
1. Introduction
Studies have indicated that RNA polymerase III-dependent (Pol III) transcriptional products are elevated in both transformed and tumor cells suggesting that they play a crucial role in tumorigenesis (White, 2001; Winter et al, 2007). Consistent with this idea, enhanced Pol III transcription is required for oncogenic transformation (Zhang et al, 2013; Johnson et al. 2008, Zhong et al, 2013). RNA Pol III transcribes a variety of untranslated RNAs, including tRNAs, 5S rRNAs, 7SL RNA, 7SK RNA and U6 RNA (Ullu et al. 1984; Dieci et al. 2007; Raha et al. 2010), while tRNA and 5S rRNA control the translational and growth capacity of cells (White, 2001; Goodfellow et al. 2006). Oncogenic proteins, such as Ras, c-Jun, and c-Myc, stimulate RNA Pol III gene transcription (Zhong et al, 2004; Johnson et al. 2008); whereas tumor suppressors, such as pRb, p53, PTEN and Maf1 repress transcription of this class of genes (White, 2004; Johnson et al. 2008; Woiwode et al. 2008). The ability of these oncogenic proteins and tumor suppressor to alter Pol III transcription results from their capacity to regulate the TFIIIB complex. The TFIIIB complex consists of TATA box-binding protein (TBP) and its associated factors, Brf1 and Bdp1. TFIIIB, together with TFIIIC and RNA Pol III, are required to transcribe tRNA genes, whereas TFIIIB, together with TFIIIA, TFIIIC and RNA Pol III, are required to transcribe 5S rRNA genes. Our studies have demonstrated that alcohol intake causes changes in Pol III gene transcription to promote cell transformation and tumor formation (Zhong et al, 2011; Zhang et al, 2013)
Alcohol consumption is most consistently associated with breast cancer risk (Hamajima et al. 2002; MacMehon, 2006; Petri, et al. 2004; Singletary et al. 2001). This association involves the estrogen receptor (ER), which is over-expressed in approximately 80% of breast cancer cases (Deandrea et al. 2008; Suzuki et al. 2008). Alcohol is known to promote mammary tumorigenesis (Singletary et al. 1995, 1991; Watabiki et al. 2000). Cancer cells have a consistent cytological feature of nucleolar hypertrophy, rRNAs are synthesized by RNA polymerase (Pol) I and III within this nucleolar compartment. Pathologists have been using enlarged nucleoli as a strong diagnostic indicator of cell transformation and neoplasia. This indicates that transformation in situ is tightly linked to the deregulation of RNA Pol I and III genes, because the size of the nucleolus reflects the levels of rRNA synthesis (White, 2004). Although alcohol-associated breast cancer is widely studied, the molecular mechanism remains to be addressed. To explore the role of alcohol in Pol III gene transcription, we treated normal and breast cancer cell lines with ethanol. Our results indicate that ethanol-induced tRNA and 5S rRNA transcription in breast cell lines is correlated with ER expression. These studies demonstrated that ERα may mediate the regulation of ethanol-induced Pol III gene transcription and that alcohol induces deregulation of Pol III gene transcription via ERα.
BRCA1 (breast cancer susceptibility gene 1) is a human tumor suppressor (Jump et al, 1998 and 2004), called by the synonym breast cancer type 1 susceptibility protein. BRCA1 is normally expressed in the cells of breast and other tissues, where it repairs damaged DNA (Check W, 2006). If BRCA1 itself is mutated, damaged DNA is not repaired properly, and this increases the risk for breast cancer (Friedenson B, 2007). Certain variations of the BRCA1 gene lead to an increased risk for breast cancer. Women with an abnormal BRCA1 gene have up to an 80% risk of developing breast cancer (Jump et al, 2012; Kuznetsov et al, 2008).
Accumulation of Pol III gene transcripts around the nucleolus is especially evident in transformed cells (Wang et al. 2003). Consistent with the idea that a high translational capacity is necessary for rapid growth and proliferation of tumor cells, Pol III gene transcripts have been found to be increased in ovarian tumors (Winter et al, 2007). Furthermore, expression of the Pol III gene, BC200, was elevated in breast squamous cell carcinoma tissues (Chen et al. 1997). Our recent studies using both cell culture models and animal models have revealed that alcohol increases transcription of Pol III genes (Zhong et al, 2011, Zhang et al, 2013). This induction in mice fed with ethanol is associated with tumor development (Zhong et al. 2011). This implies that alcohol-induced deregulation of Pol III genes may play a critical role in tumor development. To explore the role of BRCA1 in Pol III gene transcription, we restored BRCA1 expression construct in BRCA1 deficient cell line, HCC1937, to determine Pol III gene activity. The results indicate that BRCA1 expression in HCC1937 cells represses Pol III gene transcription. Further analysis reveals that ethanol treatment of MCF-7 cells, an ER+ (estrogen receptor positive) breast cancer cell line, increases Pol III gene transcription. Overexpression of BRCA1 reduces ethanol-induced tRNALeu and 5S rRNA transcription in the cells. Repealing function of BRCA1 by mutation or truncating enhances Pol III gene transcription. These studies demonstrate, for the first time, that BRCA1 mediates alcohol-induced deregulation of Pol III genes. These novel findings provide a potential approach of treatment for alcohol-associated breast cancer patients.
2. Materials and methods
2.1. Reagents and antibodies
HCC 1937 and MCF-7 cell lines were from ATCC. Cell culture medium DMEM/F12, Lipofectamine 2000, TRizol reagent and OPTI-MEM were from Invitrogen. Antibodies against BRCA1 and β-actin were obtained from Santa Cruz Biotech. The sequences of primers of Pol III genes were described previously (Zhong et al, 2011). BRCA1 siRNAs were from Dharmacon Inc. The kit of RNase protection assay was from Ambion, Inc. The reagent of real time PCR was from Bio-Rad Biotech.
2.2. RNase protection assay
HCC 1937 cells were transfected with the pArg-maxi expression plasmid, a Pol III gene, for 48 h (Zhong et al, 2004). RNase protection assays (RPAs) were carried out as described in the protocol of Ambion, Inc. Briefly, HCC 1937 cells were harvested after transfection 48 h. The cell pellets were resuspended with 1 ml of TRIzol reagent (Invitrogen) and incubated at room temperature for 10 min. Chloroform was added (200 μl), and the mixture was incubated at room temperature for 3 min. Suspensions were centrifuged at 14,000 rpm for 20 min at 4°C.
The supernatant was mixed with an equal volume of isopropanol, incubated on dry ice for 15 min, and centrifuged at 14,000 rpm for 20 min at 4°C. The pellet was rinsed with 70% ethanol and resuspended in 20 μl of water. For the preparation of probe, the pArg-maxi gene plasmid was digested with XbaI, and labeled RNA probe was synthesized using the Maxi-script T7 kit (Ambion) with [32P] CTP. RNA (1 μg) was hybridized with the labeled RNA probe and incubated at 42°C overnight. The samples were digested by RNase A/T1 at 37°C for 30 min and precipitated. The samples were incubated on dry ice for 15 min and centrifuged at 14,000 rpm for another 15 min. The pellets were resuspended in gel loading buffer (Ambion) and separated on 8% acrylamide-8 M urea denaturing gels. The bands were visualized by autoradiography, and the RNA products were quantified by a phosphorimager.
2.3 RNA isolation and RT-qPCR
MCF-7 cells were cultured in 10% FBS/DMEM-F12 medium and transfected with BRCA1 siRNAs or mismatch (mm) RNA for 48 h. The cells were starved in FBS-free medium for 3–4 h and then treated with 25 mM ethanol for 1 h. The total RNA was isolated from the cells using single step extraction method TRIzol reagent (Invitrogen). The RNA samples were quantified and reverse-transcribed in a 20 μl reaction containing 1 x RT (reverse transcription) buffer. After first-strand cDNA synthesis, the cDNAs were diluted in DNase-free water and real time qPCR (RT-qPCR) were performed with specific primers as described before (Zhong et al, 2011) and PCR reagent kits (Bio-Rad Biotech) in the ABI prism 7700 Sequence Detection System. 5S rRNA and precursor of tRNALeu transcripts were measured by RT-qPCT as described previously (Zhong et al. 2011).
2.4. SDS-PAGE and immunoblot analysis
MCF-7 cells were transfected with BRCA1 expression construct or siRNAs for 48 h. The cells were incubated with 25 mM ethanol for 1 h as described above. Cells were collected with lysis buffer and sonicated. The suspensions were centrifuged to save the supernatants. Protein concentrations were determined by the Bradford method. Lysates (50 μg protein) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred from the SDS-PAGE gel to Hybond-P membrane and immunoblot analysis were performed with specific antibodies. Membranes were probed with either antibodies against BRCA1 or β-actin. Bound primary antibody was visualized using horseradish peroxidase-conjugated secondary antibody (Vector Laboratories) and enhancing chemiluminscence reagents (Santa Cruz Biotech).
3. Results
3.1. BRCA1 expression represses Pol III gene transcription in HCC 1937 cells
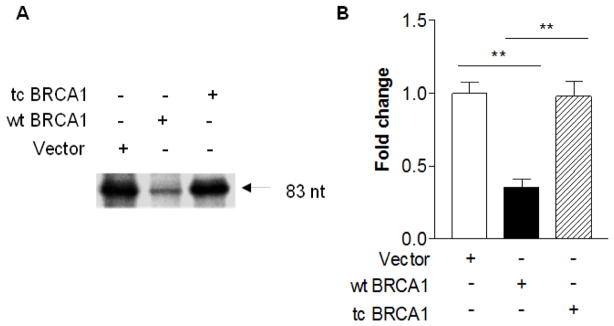
Studies have demonstrated that oncoproteins, c-Jun, c-Fos and Myc, stimulate Pol III gene transcription. In contrast, tumor suppressors, pRb, p53, PTEN and Maf1, repress the activity of the class of genes. BRCA1 is a tumor suppressor and its mutation is tightly related to breast cancer. However, it remains to be established whether BRCA1 modulates Pol III gene transcription. To investigate the role of BRCA1 in Pol III gene transcription, we transfected HCC 1937 cells with a wild type (wt) BRCA1 expression construct plus pArg Maxi gene plasmid for 48 h to perform RPAs. The results indicate that restoring BRCA1 in HCC 1937 cells decreases pArg Maxi gene transcription, compared to the cells transfected with control vector construct (Fig. 1). It suggests that BRCA1 may modulate Pol III gene transcription. Next, we further estimated the role of BRCA1 in Pol III genes. HCC 1937 cells were transfected with the construct of dominant negative mutant (mt) BRCA1. The result indicates that the transcription levels of pArg Maxi gene is not reduced by expressing DNM BRCA1, compared to expressing wt BRCA1 (Fig. 2). Further analysis reveals that expressing truncated BRCA1 could not repress pArg Maxi gene (Fig. 3). This shows that BRCA1 by mutation or truncated destroys its ability to modulate Pol III gene transcription.
Fig. 1. RNA Pol III transcription is modulated by BRCA1.
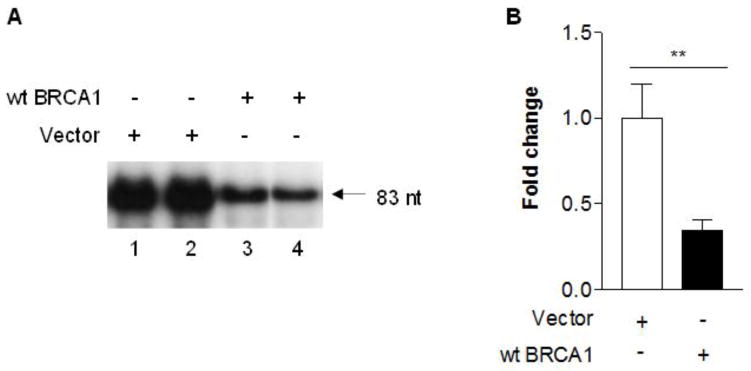
HCC 1937 cells, a BRCA1 deficient line, were transfected with vector (lane 1–2) or wild type (wt) (lane 3–4) BRCA1 expression construct plus a tRNA gene reporter plasmid, pArg Maxi gene for 48 h. RNAs ware isolated from the cells. RNase protection assay was carried out to determine the transcription levels of pArg Maxi gene (83nt). An example of autoradiogram is shown in (A). Quantification of 3 independent experiments of transfection with the constructs is showed (B). **: p<0.01.
Fig. 2. Expression of a mutational BRCA1 does not affect the Pol III gene.
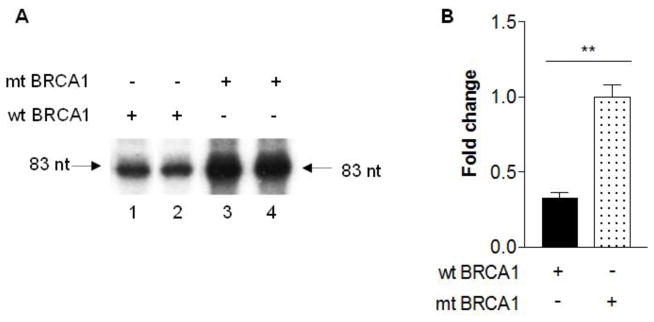
HCC 1937 cells were transfected with wt BRCA1 (lane 1–2) or a dominant negative mutant (mt) BRCA1 (lane 3–4) expression construct plus pArg Maxi gene as described in Fig. 1. RNase protection assay was carried out to determine the transcription levels of pArg Maxi gene (83nt). An example of autoradiogram is shown in (A). Quantification of 3 independent experiments of transfection with the constructs is showed (B). **: p<0.01.
Fig. 3. Expression of a truncated BRCA1 does not repress the activity of Pol III genes.
HCC 1937 cells were transfected with wt BRCA1 or a truncated BRCA1 expression construct plus pArg Maxi gene as described in Fig. 1. RNase protection assay was carried out to determine the transcription levels of pArg Maxi gene (83nt). An example of autoradiogram is shown in (A). Quantification of 3 independent experiments of transfection with the constructs is showed (B). Truncated BRCA1 lost its function repressing Pol III gene transcription. **: p<0.01.
3.2. Alteration of BRCA1 affects alcohol-induced Pol III gene transcription
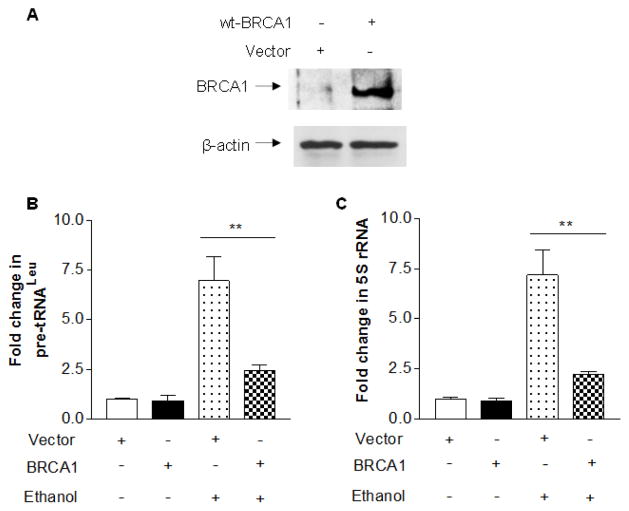
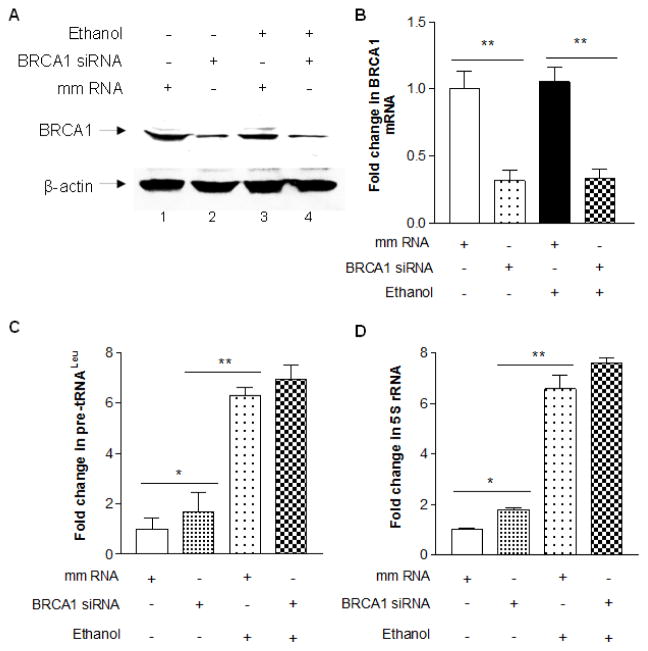
Alcohol consumption is associated with development of human breast cancer (Wang et al, 2012; Wong et al, 2012). Recent, we have demonstrated that ethanol treatment increases Pol III gene transcription and cell transformation in breast cells (Zhang et al, 2013). Therefore, we further investigate whether BRCA1 expression affects the induction of Pol III genes by alcohol. Our recent studies have demonstrated that ethanol dramatically enhances transcription of pre-tRNAleu and 5S rRNA in MCF-7 cells (Zhang et al, 2013). To determine changes in alcohol-induced transcription of Pol III genes by BRCA1, MCF-7 cells were transfected with wt-BRCA1 expression construct for 48 h and treated with 25 mM ethanol. The results reveal that cellular levels of BRCA1 protein is increased in the cells transfected with its expression construct, compared to the cells with vector alone (Fig. 4A). Overexpression of BRCA1 decreases the induction of pre-tRNAleu and 5S rRNA by ethanol (Fig. 4B and 4C). This implies that BRCA1 mediates alcohol-induced Pol III gene transcription, whereas ethanol treatment does not obviously change cellular level of BRCA1 (Fig. 5A, Lane 1 and Lane 3). To confirm the observation of repressing the induction of Pol III genes by BRCA1, we further determine whether reduction of BRCA1 by its siRNA is able to affect alcohol-induced Pol III gene transcription. The result shows that BRCA1 siRNA efficiently decreases the cellular level of its protein and mRNA (Fig. 5A and 5B). Next, we estimated the changes of Pol III genes by BRCA1 siRNA. The results indicate that reduction of BRCA1 by the siRNA slightly increases the induction of pre-tRNAleu and 5S rRNA by ethanol in MCF-7 cells, compared to mm (mismatch) RNA (Fig. 5C and 5D). These results of MCF-7 cells are consistent with ones in HCC 1937 cells. This shows that BRCA1 negatively modulates Pol III gene transcription and the overexpression of BRCA1 decreases the induction of Pol III genes by alcohol.
Fig. 4. Overexpression of BRCA1 represses the induction of Pol III genes by alcohol.
MCF-7 cells were transfected with vector or wt BRCA1 expression construct as described in Fig. 1. The resultant cell lysates and total RNA from these cells was extracted. Immunoblot analysis was carried out to determine protein level of BRCA1 (A). RT-qPCR was performed to measure the levels of pre-tRNALeu (B), 5S rRNA (C) transcription. The fold change was calculated by normalizing to the amount of GAPDH. The bars represent Mean ± SE of at least three independent determinations of transfection with the constructs. The results indicate that overexpression of BRCA1 decreases the induction of Pol III gene transcription. **: p<0.01.
Fig. 5. Reduction of BRCA1 increases Pol III gene transcription.
MCF-7 cells were transfected with mismatch RNA (mm RNA, as a control) or BRCA1 siRNA for 48 h. The resultant cell lysates and total RNAs from these cells was extracted. Immunoblot analysis was carried out to determine protein level of BRCA1 (A). RT-qPCR was performed to measure the levels of BRCA1 mRNA (B), pre-tRNALeu (C), 5S rRNA (D) transcription. The fold change was calculated by normalizing to the amount of GAPDH. The bars represent Mean ± SE of at least three independent determinations of transfection with mm RNA or BRCA1 siRNA. The results indicate that repression of BRCA1 by its siRNA increases the induction of Pol III gene transcription, compared to mm RNA. **: p<0.01.
Discussion
BRCA1 status is tightly associated with breast cancer. In this study, we used both RPA and RT-qPCR approaches to determine whether alteration of BRCA1 expression affects Pol III gene transcription. These results indicate that restoring BRCA1 expression represses pArg Maxi gene transcription in HCC 1937 cells. Alcohol dramatically increases transcription of pre-tRNAleu and 5S rRNA in MCF-7 cells. Overexpressing BRCA1 markedly inhibits the induction of Pol III genes by alcohol. In contrast, reduction of BRCA1 expression by its siRNA slightly enhances the induction of Pol III genes. These studies for the first time demonstrate that BRCA1 modulates Pol III gene transcription induced by alcohol. The alteration of cellular level of BRCA1 affects alcohol-induced deregulation of Pol III genes in breast cancer cells.
Breast cancer is the most common malignant disease in females. Individuals harboring germline mutations in the breast cancer susceptibility gene BRCA1 carry an 80% lifetime risk of developing breast cancer (Ford, et al. 1998). The protein product of the BRCA1 gene has many important cellular functions including DNA repair, cell cycle regulation and transcriptional regulation. Accordingly, deficiency in BRCA1 leads to accelerated proliferation, aberrant mitosis, increased chromosome instability and tumorigenesis (Deng CX., 2006; Mullan et al, 2006). BRCA1 transcription is regulated by diverse environmental stimuli including genotoxic agents, hypoxia and mitogenic hormone stimulation. The best-characterized stimulant of BRCA1 expression is estrogen, which induces the highest elevations in BRCA1 mRNA levels that routinely peaks just before the onset of DNA synthesis (Spillman et al, 1996; Marks et al, 1997). Studies from our lab and others have demonstrated that tumor suppressors repress Pol III gene transcription (White, 2004; Johnson et al. 2008; Woiwode et al. 2008) and deregulation of Pol III gene tightly links to cell transformation and tumor formation (Zhang et al, 2011, 2013; Zhong et al, 2011, 2013; Johnson et al 2008). Although a study showed that BRCA1 decreased Pol III gene transcription in HCC1937 cells (Veras et al, 2009), the role of BRCA1 in alcohol-induced deregulation of Pol III genes remains to be addressed. Here, this present study indicates that BRCA1 negatively mediates Pol III gene transcription and decreases the induction of the genes by alcohol. The mutational and truncated BRCA1 lost its function on Pol III genes.
Epidemiological studies have indicated alcohol consumption has consistently been associated with an increased risk for breast cancer in women (Petri et al. 2004; Singletary et al. 2001). Studies by Wang et al have demonstrated that alcohol increased MCP-1 and CRR2 expression, which promoted mammary tumor growth in alcohol-fed mice (Wang et al. 2012). Alcohol intake was associated with ER+ (estrogen receptor positive) breast cancer cases more than to ER− cases (Deandrea et al. 2008; Suzuki et al. 2008; Dumitrescu et al. 2005). A study indicated that alcohol increased ERα expression to promote breast tumor formation in mice (Wong et al 2012). However, the exact mechanism, by which alcohol promotes development of ER+ breast cancer, is still unknown. A previous study demonstrated that alcohol down-regulated the expression of BRCA1, a potent inhibitor of ERα, thereby contributing to breast cancer (Dumitrescu et al. 2005). Alcohol consumption was also shown to increase the transcriptional activity of ERα (Fan et al. 2000, Zhang et al, 2013). Recently, we reported that alcohol increases Pol III gene transcription in both normal and cancer breast cell lines. The induction in ER+ breast cancer cell lines (MCF-7 and T47D) is significantly higher (5~6 fold) than in ER− normal breast cell lines (MCF-10A, MCF-10F and MCF-12A) and ER− breast cancer cells (MAD-MB231 and SKRB-3). Ethanol increases ERα expression, resulting in an increase in products of Pol III genes (Zhang et al, 2013). These results support the idea that alcohol increases ERα expression to elevate Pol III gene transcription to bring about greater phenotypic changes. Given that alcohol down-regulates BRCA1 expression (Dumitrescu et al. 2005), this implies that BRCA1 may mediate alcohol-induced deregulation of Pol III genes. Therefore, it is critically important to determine the role of BRCA1 in alcohol-induced Pol III gene transcription. In the present studies, our studies indicate that overexpression of BRCA1 reduces the induction of Pol III genes by alcohol. Inhibiting BRCA1 function by mutation, truncation or its siRNA destroys the repression of BRCA1 on Pol III genes. Next, we will further investigate whether BRCA1 affects TFIIIB function or it interacts with subunits of TFIIIB complex to modulate Pol III gene transcription.
In summary, the present study provides evidence that BRCA1 represses Pol III gene transcription. Overexpression of BRCA1 decreases the induction of Pol III genes by alcohol. It is the first report that BRCA1 modulates RNA Pol III-dependent transcription induced by alcohol. The novel findings indicate that BRCA1 may play critical role in alcohol-associated breast cancer.
Research highlights.
Overexpression of BRCA1 represses Pol III gene transcription;
Mutant or truncated BRCA1 lost the function of modulation on Pol III genes;
Alcohol increases Pol III gene transcription;
BRCA1 represses the induction of Pol III genes elevated by alcohol
Acknowledgments
We would like to thank Drs. M. R. Stallcup (University of Southern California) for scientific discussions. This work was supported by National Institutes of Health grants AA017288, AA021114 and AA023247 to S.Z.
Abbreviation
- Pol III genes
RNA polymerase III-dependent genes
- BRCA1
breast cancer 1, early onset
- RPA
RNase protection assay
- Mm
mismatch
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Check W. BRCA: What we know now. College of American Pathologists. 2006 Retrieved 2010-08-23. [Google Scholar]
- Chen W, Böcker W, Brosius J, Tiedge H. Expression of neural BC200 RNA in human tumours. J Pathol. 1997;183:345–351. doi: 10.1002/(SICI)1096-9896(199711)183:3<345::AID-PATH930>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Deandrea S, Talamini R, Foschi R, Montella M, Dal Maso L, Falcini F, La Vecchia C, Franceschi S, Negri E. Alcohol and breast cancer risk defined by estrogen and progesterone receptor status: a case-control study. Cancer Epidemiol Biomarkers Prev. 2008;17:2025–2028. doi: 10.1158/1055-9965.EPI-08-0157. [DOI] [PubMed] [Google Scholar]
- Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006;34:1416–1426. doi: 10.1093/nar/gkl010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Dumitrescu RG, Shields PG. The etiology of alcohol-induced breast cancer. Alcohol. 2005;35:213–225. doi: 10.1016/j.alcohol.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Fan S, Meng Q, Gao B, Grossman J, Yadegari M, Goldberg ID, Rosen EM. Alcohol stimulates estrogen receptor signaling in human breast cancer cell lines. Cancer Res. 2000;60:5635–5639. [PubMed] [Google Scholar]
- Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedenson B. The BRCA1/2 pathway prevents hematologic cancers in addition to breast and ovarian cancers. BMC Cancer. 2007;7:152–162. doi: 10.1186/1471-2407-7-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow SJ, Innes F, Derblay LE, MacLellan WR, Scott PH, White RJ. Regulation of RNA polymerase III transcription during hypertrophic growth. EMBO J. 2006;25:1522–1533. doi: 10.1038/sj.emboj.7601040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamajima N, Hirose K, Tajima K, Rohan T, Calle EE, Heath CW, Jr, Coates RJ, Liff JM, Talamini R, Chantarakul N, et al. Alcohol, tobacco and breast cancer--collaborative reanalysis of individual data from 53 epidemiological studies, including 58,515 women with breast cancer and 95,067 women without the disease. Br J Cancer. 2002;87:1234–1245. doi: 10.1038/sj.bjc.6600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DL, Johnson SA. Cell biology. RNA metabolism and oncogenesis. Science. 2008;320:461–462. doi: 10.1126/science.1158680. [DOI] [PubMed] [Google Scholar]
- Johnson SA, Dubeau L, Johnson DL. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem. 2008;283:19184–19191. doi: 10.1074/jbc.M802872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jump up. Genetics. Breastcancer.org. 2012 2012-09-17. [Google Scholar]
- Jump up. Duncan JA, Reeves JR, Cooke TG. BRCA1 and BRCA2 proteins: roles in health and disease. Molecular pathology MP. 1998;51(5):237–47. doi: 10.1136/mp.51.5.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jump up. Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer science. 2004;95(11):866–871. doi: 10.1111/j.1349-7006.2004.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov SG1, Liu P, Sharan SK. Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat Med. 2008;14:875–881. doi: 10.1038/nm.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMahon B. Epidemiology and the causes of breast cancer. Int J Cancer. 2006;118:2373–2378. doi: 10.1002/ijc.21404. Review. [DOI] [PubMed] [Google Scholar]
- Marks JR1, Huper G, Vaughn JP, Davis PL, Norris J, McDonnell DP, Wiseman RW, Futreal PA, Iglehart JD. BRCA1 expression is not directly responsive to estrogen. Oncogene. 1997;14(1):115–121. doi: 10.1038/sj.onc.1200808. [DOI] [PubMed] [Google Scholar]
- Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–5863. doi: 10.1038/sj.onc.1209872. Review. [DOI] [PubMed] [Google Scholar]
- Petri A, Tjønneland M, Gamborg D, Johansen S, Høidrup TIA, Sørensen Grønbaek M. Alcohol intake, type of beverage, and risk of breast cancer in pre- and postmenopausal women. Alcohol Clin Exp Res. 2004;28:1084–1090. doi: 10.1097/01.alc.0000130812.85638.e1. [DOI] [PubMed] [Google Scholar]
- Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Snyder M. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci U S A. 2010;107:3639–3644. doi: 10.1073/pnas.0911315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singletary KM, Nelshoppen J, Wallig M. Enhancement by chronic ethanol intake of N-methyl-N-nitrosourea-induced rat mammary tumorigenesis. Carcinogenesis. 1995;16:959–964. doi: 10.1093/carcin/16.4.959. [DOI] [PubMed] [Google Scholar]
- Singletary KW, McNary MQ, Odoms AM, Nelshoppen J, Wallig MA. Ethanol consumption and DMBA-induced mammary carcinogenesis in rats. Nutr Cancer. 1991;16:13–23. doi: 10.1080/01635589109514136. [DOI] [PubMed] [Google Scholar]
- Singletary KW, Gapstur SM. Alcohol and breast cancer: review of epidemiologic and experimental evidence and potential mechanisms. JAMA. 2001;286:2143–2151. doi: 10.1001/jama.286.17.2143. [DOI] [PubMed] [Google Scholar]
- Spillman MA, Bowcock AM. BRCA1 and BRCA2 mRNA levels are coordinately elevated in human breast cancer cells in response to estrogen. Oncogene. 1996;13(8):1639–1645. [PubMed] [Google Scholar]
- Suzuki R, Orsini N, Mignone L, Saji S, Wolk A. Alcohol intake and risk of breast cancer defined by estrogen and progesterone receptor status--a meta-analysis of epidemiological studies. Int J Cancer. 2008;122:1832–1841. doi: 10.1002/ijc.23184. [DOI] [PubMed] [Google Scholar]
- Ullu E, Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984;312:171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Veras I, Rosen EM, Schramm L. Inhibition of RNA polymerase III transcription by BRCA1. J Mol Biol. 2009;387:523–531. doi: 10.1016/j.jmb.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Wang C, Politz JC, Pederson T, Huang S. RNA polymerase III transcripts and the PTB protein are essential for the integrity of the perinucleolar compartment. Mol Biol Cell. 2003;14:2425–2435. doi: 10.1091/mbc.E02-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Xu M, Li F, Wang X, Bower KA, Frank JA, Lu Y, Chen G, Zhang Z, Ke Z, Shi X, Luo J. Ethanol promotes mammary tumor growth and angiogenesis: the involvement of chemoattractant factor MCP-1. Breast Cancer Res Treat. 2012;133:1037–1048. doi: 10.1007/s10549-011-1902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabiki Y, Okii Y, Tokiyasu S, Yoshimura M, Yoshida A, Akane N, Shikata N, Tsubura A. Long-term ethanol consumption in ICR mice causes mammary tumor in females and liver fibrosis in males. Alcohol Clin Exp Res. 2000;24:117S–122S. [PubMed] [Google Scholar]
- White RJ. RNA polymerase III transcription and cancer. Oncogene. 2004;23:3208–3216. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- Winter A, Sourvinos G, Allison S, Tosh K, Scott P, Spandidos D, White RJ. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumors. Proc Natl Acad Sci U S A. 2007;97:12619–12624. doi: 10.1073/pnas.230224097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woiwode A, Johnson SA, Zhong S, Zhang C, Roeder RG, Teichmann M, Johnson DL. PTEN represses RNA polymerase III-dependent transcription by targeting the TFIIIB complex. Mol Cell Biol. 2008;28:4204–4214. doi: 10.1128/MCB.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AW, Dunlap SM, Holcomb VB, Nunez NP. Alcohol Promotes Mammary Tumor Development via the Estrogen Pathway in Estrogen Receptor Alpha-Negative HER2/neu Mice. Alcohol Clin Exp Res. 2012;36:577–587. doi: 10.1111/j.1530-0277.2011.01654.x. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Jin J, Zhong Q, Yu XL, Levy D, Zhong S. ERα mediates alcohol-induced deregulation of Pol III genes in breast cancer cells. Carcinogenesis. 2013;34:28–37. doi: 10.1093/carcin/bgs316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhong Q, Evans AG, Levy D, Zhong S. Phosphorylation of histone H3 serine 28 modulates RNA polymerase III-dependent transcription. Oncogene. 2011;30:3943–3952. doi: 10.1038/onc.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Shi GG, Zhang QS, Zhang YM, Levy D, Zhong S. Role of phosphorylated histone H3 serine 10 in DEN-induced deregulation of Pol III genes and cell proliferation and transformation. Carcinogenesis. 2013;34(11):2460–2469. doi: 10.1093/carcin/bgt219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Machida K, Tsukamoto H, Johnson DL. Alcohol induces RNA polymerase III-dependent transcription through c-jun by coregulating TBP and Brf1 expression. J Biol Chem. 2011;286:2393–2401. doi: 10.1074/jbc.M110.192955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zheng C, Johnson DL. Epidermal Growth Factor enhances cellular TBP levels and induces RNA polymerase I- and III-dependent gene activity. Mol Cell Biol. 2004;24:5119–5129. doi: 10.1128/MCB.24.12.5119-5129.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]