Abstract
The structure of broken DNA ends is a critical determinant of the pathway used for DNA double strand break (DSB) repair. Here, we develop an approach, hairpin capture of DNA end structures (HCoDES), which elucidates chromosomal DNA end structures at single nucleotide resolution. HCoDES defines structures of physiologic DSBs generated by the RAG endonuclease, as well as those generated by nucleases widely used for genome editing. Analysis of G1-phase cells deficient in H2AX or 53BP1 reveals DNA ends that are frequently resected to form long single-stranded overhangs that can be repaired by mutagenic pathways. In addition to 3′ overhangs, many of these DNA ends unexpectedly form long 5′ single-stranded overhangs. The divergence in DNA end structures resolved by HCoDES suggests that H2AX and 53BP1 may have distinct activities in end protection. Thus, the high-resolution end structures obtained by HCoDES identify new features of DNA end processing during DSB repair.
Introduction
The structure of broken DNA ends plays a pivotal role in directing DNA double strand break (DSB) repair by either homologous recombination (HR) or non-homologous end joining (NHEJ) (Ciccia and Elledge, 2010; Huertas, 2010; Symington and Gautier, 2011). HR uses the sister chromatid as a template for precise DSB repair in S and G2 phase cells. Initiation of HR requires that broken DNA ends be resected to form long 3′ single-stranded DNA (ssDNA) overhangs. In mammalian cells, the Mre11 and CtIP nucleases initiate resection and, along with other nucleases, generate long 3′ ssDNA overhangs, which are bound by the RPA complex, initiating DSB repair by the HR pathway (Huertas, 2010; Makharashvili et al., 2014; Sartori et al., 2007; Symington and Gautier, 2011; Wang et al., 2014).
In contrast to HR, NHEJ directly ligates broken DNA ends during all phases of the cell cycle and is the main DSB repair pathway in G1 (Lieber, 2010). Extensive resection of DNA ends prior to NHEJ could generate potentially harmful chromosomal deletions. Moreover, formation of ssDNA overhangs prevents efficient repair by NHEJ and, in G1-phase cells, may allow DNA ends to access mutagenic repair pathways that have been implicated in the formation of chromosomal deletions and translocations (Gostissa et al., 2011; Huertas, 2010). Thus, mechanisms must be in place to limit DNA end resection prior to NHEJ-mediated repair. The core NHEJ factors include DNA ligase IV, XRCC4 and the Ku70/80 heterodimer (Lieber, 2010). Ku70/80 binds double stranded, but not single stranded, DNA with high affinity, possibly explaining why NHEJ does not efficiently repair ends with significant ssDNA overhangs (Blier et al., 1993; Foster et al., 2011; Grundy et al., 2014). DNA Ligase IV ligates broken DNA ends, while XRCC4 binds to DNA Ligase IV and stimulates its activity (Lieber, 2010).
In addition to genotoxic DSBs, NHEJ is also required to repair physiologic DSBs generated during processes such as V(D)J recombination, the reaction that assembles antigen receptor genes in developing lymphocytes (Helmink and Sleckman, 2012). V(D)J recombination is initiated when the RAG endonuclease introduces DNA DSBs at a pair of recombining gene segments (Bassing et al., 2002; Fugmann et al., 2000). RAG cleavage forms pairs of hairpin-sealed coding DNA ends and blunt signal ends, which are processed and joined by NHEJ to form a coding joint and signal joint, respectively (Fugmann et al., 2000; Helmink and Sleckman, 2012). Coding joint formation also depends on the Artemis endonuclease, which must open hairpin-sealed coding ends before they can be joined by core NHEJ factors (Ma et al., 2002). Like Artemis, Mre11 can open hairpin-sealed DNA ends and functions in the repair of RAG DSBs as part of the Mre11-Rad50-Nbs1 (MRN) complex (Helmink et al., 2009; Paull and Gellert, 1998). However, neither Mre11 nor other cellular nucleases can efficiently open hairpin-sealed coding ends in Artemis-deficient cells (Rooney et al., 2002). Thus, protective pathways are in place to limit nucleolytic activity at RAG DSBs.
An important component of this DNA end protective pathway is the histone variant H2AX, as demonstrated by the rapid opening and subsequent resection of hairpin-sealed coding ends in lymphocytes deficient in Artemis and H2AX (Helmink et al., 2011). The protective function of H2AX relies on its phosphorylation by the ATM kinase to form γ-H2AX in chromatin flanking RAG DSBs (Chen et al., 2000; Helmink et al., 2011; Savic et al., 2009). γ-H2AX concentrates repair factors, such as 53BP1, to DSB sites (Celeste et al., 2003; Fernandez-Capetillo et al., 2004; Lukas et al., 2004). In this regard, 53BP1 is also required to limit nuclease activity at RAG DSBs (Difilippantonio et al., 2008; Tubbs et al., 2014). Resected coding ends in H2AX or 53BP1-deficient lymphocytes are not joined efficiently by core NHEJ factors (Helmink et al., 2011; Tubbs et al., 2014). Moreover, the coding joints that do form can harbor significant deletions (Helmink et al., 2011). Similar to resection during HR in S and G2 phase cells, resection of coding ends in G1-phase lymphocytes deficient in H2AX or 53BP1 requires the CtIP nuclease (Helmink et al., 2011; Tubbs et al., 2014). Thus, H2AX and 53BP1 prevent the nucleolytic machinery of the HR pathway from functioning at RAG DSBs in G1-phase lymphocytes. When this protection fails, DNA ends can be resected and joined through pathways that promote genome instability. However, the structural features of the resected DNA ends that channel DSBs to mutagenic repair pathways are not known.
Current approaches for assessing DNA end structures are low resolution and provide little information about precise structures. For example, deposition of ssDNA-binding proteins, such as RPA or Rad51, at DSBs can be used to judge whether an end has a significant ssDNA overhang, while the loss of restriction enzyme sites near a targeted DSB can provide a low resolution map for the average extent of resection (Zhou et al., 2013). These approaches cannot distinguish between 5′ and 3′ ssDNA overhangs, which may have a differential ability to access aberrant repair pathways, nor can they determine individual DNA end structures and sequence features that may guide DNA end processing and repair outcome. We now describe a novel approach, termed Hairpin Capture of DNA End Structures (HCoDES), which elucidates chromosomal broken DNA end structures with single nucleotide resolution. Using this approach, we identified the spectrum of DNA end structures at RAG DSBs in G1-phase lymphocytes lacking either H2AX or 53BP1. In addition, we show that HCoDES can be used to reveal broken DNA end structures generated by other nucleases, such as Cas9 and zinc finger nucleases, which are frequently used for genome editing (Kim and Kim, 2014). Our findings establish HCoDES as an important new approach for elucidating the structure of broken DNA ends at DSBs generated in vivo by a variety of nucleases throughout the genome.
Results
Hairpin capture of DNA end structures (HCoDES)
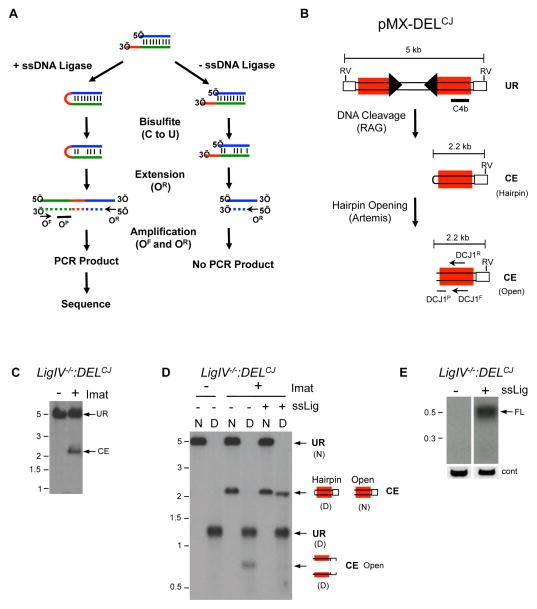
The general scheme for HCoDES is shown in Figure 1A. In brief, we reasoned that nearly any open-ended double stranded DNA break present in genomic DNA could be converted, by treatment with a ssDNA ligase, into a hairpin, which would capture the precise positions of the 5′ and 3′ end of each strand. The resultant hairpins could be amplified by PCR using complementary primers for a given sequence near the DSB if base pairing within the hairpin is first disrupted by chemical conversion with bisulfite. Using this approach, PCR products would contain all of the sequence information for resolving structures of individual DSB DNA ends (Figures 1A).
Figure 1. Ligation and amplification of coding ends.
(A) HCoDES schematic. Nucleotides in the 3′ ssDNA overhang (red line), top strand (blue line) and bottom strand (green line) are indicated. The relative positions of the forward (OF) and reverse (OR) oligonucleotides used for amplification and the oligonucleotide probe (OP) used to detect PCR products are shown. The newly synthesized strand (dashed line) from the first extension is shown. (B) Schematic of the pMX-DELCJ retroviral recombination substrate. The LTRs (open rectangles) and recombination signals (filled triangles) are shown. Also shown are: the positions of the EcoRV sites (RV) and probe (C4b) used for Southern blotting, the sizes of the fragments for unrearranged (UR) pMX-DELCJ and the pMX-DELCJ coding end (CE), and the relative positions of the DCJ1F and DCJ1R oligonucleotides used for HCoDES PCR and DCJ1P oligonucleotide used as a probe are shown. (C) Southern blot of EcoRV-digested LigIV−\−:DELCJ abl pre-B cell genomic DNA using the C4b probe. Cells were treated (+) or not treated (−) with imatinib (imat) to induce RAG DSBs. (D) Native (N) and Denaturing (D) Southern blot analysis of EcoRV-digested genomic DNA from LigIV−\−:DELCJ abl pre-B cells using the C4b probe. Genomic DNA was either treated (+) or not (−) with the ssDNA ligase prior to digestion with EcoRV. (E) Southern blot of HCoDES PCR amplification of ligated (+) and non-ligated (−) genomic DNA from LigIV−\−:DELCJ abl pre-B cells treated with imatinib using the DCJ1R and DCJ1F oligonucleotides for primary PCR, the Miseq-DCJ1R and Miseq-DCJ1F oligonucleotides for secondary PCR and probed with the DCJ1P oligonucleotide. The product size for a full length (FL) coding end is shown, as are the MW markers in kb. Control (cont) PCR using the CR and CF oligonucleotides is also shown. Also see related Figures S1 and S2.
We took a stepwise approach to explore the feasibility and optimization of the HCoDES technique. To test the efficiency of hairpin formation, KpnI-digested genomic DNA (4 nucleotide 3′ ssDNA overhangs) was treated with a thermostable form of T4 RNA ligase that can ligate ssDNA (hereafter referred to as ssDNA ligase) (Figure S1A) (Blondal et al., 2005). To improve ligation efficiency, DNA was ligated at 60°C in the presence of betaine, which lowers the melting temperature of DNA, allowing for single strand formation at the DNA end. Under denaturing Southern blot conditions, non-ligated KpnI fragments dissociate into single strands that migrate at a lower molecular weight than the native duplex (Figure S1B). However, following ssDNA ligation, if the top and bottom strands are covalently linked, they cannot dissociate after denaturation (Figure S1B). Denaturing Southern blot analysis revealed that the majority of the KpnI digested DNA ends were converted to hairpins upon ligation with the ssDNA ligase (Figure S1B). Analysis of DNA digested with ScaI (blunt ends) and AvrII (4 base pair 5′ overhang) yielded similar results (data not shown).
Next, we tested whether hairpin-sealed DNA ends could be amplified for sequencing. Bisulfite treatment of DNA converts cytosine to uracil, creating U-G mismatches that destabilize the duplex, and permits PCR amplification of hairpin-sealed DNA ends (Figure 1A, OF and OR oligonucleotides) (Laird et al., 2004). Forward (KpnF) and reverse (KpnR) oligonucleotides positioned 150 base pairs downstream of a KpnI site were designed to anneal to bisulfite-converted DNA (Figure S1C). Primary PCR amplification using these oligonucleotides generates a 0.3 kb product (Figure S1D). The Miseq sequencing adapters were then added in a secondary PCR using the Miseq-KpnF and Miseq-KpnR oligonucleotides, and the resulting 0.4 kb PCR product was subjected to Miseq deep sequencing (data not shown). Analysis of 98,320 sequences from pooled triplicate reactions revealed that 98% correspond to the predicted sequence for a 4 nucleotide 3′ overhang generated by KpnI digestion (Figure S1E and data not shown). Similar results were obtained from analysis of genomic DNA digested with ScaI (blunt) and AvrII (4 nucleotide 5′ overhang) (data not shown). We conclude that broken chromosomal DNA ends can be efficiently ligated, amplified, and sequenced to reveal DNA end structures at single nucleotide resolution.
Chromosomal coding end structures
RAG DSBs can be induced in pre-B cell lines transformed by the v-abl kinase, hereafter referred to as abl pre-B cells (Bredemeyer et al., 2006). Treatment of these cells with the abl kinase inhibitor imatinib leads to G1 cell cycle arrest, induction of RAG, and the initiation of V(D)J recombination at the endogenous immunoglobulin light chain kappa (Igk) locus and chromosomally integrated retroviral recombination substrates, such as pMX-DELCJ (Figure 1B and S2A) (Bredemeyer et al., 2006; Muljo and Schlissel, 2003). Induction of RAG in DNA ligase IV-deficient abl pre-B cells with single pMX-DELCJ integrants (LigIV−/−:DELCJ) results in the generation of coding ends at the Igk locus (Jk coding ends) and at pMX-DELCJ. These coding ends are not repaired due to the deficiency of DNA Ligase IV (Figure 1C and S2B).
Denaturing Southern blot analysis reveals that, in DNA Ligase IV-deficient abl pre-B cells, most hairpin-sealed pMX-DELCJ coding ends have been opened in vivo by Artemis (Figure 1D). Ligation of these coding ends with the ssDNA ligase in vitro readily converts them to hairpin-sealed DNA ends (Figure 1D). Moreover, the ssDNA ligase renders Jk1 coding ends in DNA Ligase IV-deficient abl pre-B cells resistant to treatment with exonucleases in vitro, indicative of hairpin formation (Figure S2C). Ligated and non-ligated genomic DNA from LigIV−/−:DELCJ abl pre-B cells was bisulfite treated and amplified with the appropriate primary and secondary oligonucleotides to amplify and sequence Jk1 and pMX-DELCJ coding ends (Table S1). The resulting PCR products were then deep sequenced (Figure 1E and S2D, FL).
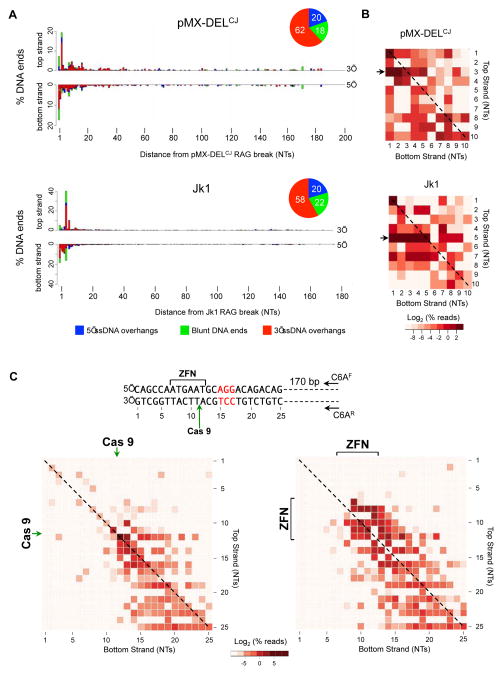
For all experiments, triplicate PCR products from two independent cell lines of each genotype were sequenced to comparable read depths using Miseq massively parallel sequencing. To identify the full spectrum of DNA end structures from sequencing data, we developed an analysis pipeline composed of open-source software and custom programs (Figure S3A). This pipeline was applied to sequence data from LigIV−/−:DELCJ abl pre-B cells to determine the 5′ and 3′ termini of each coding end and whether these DNA ends formed 5′ (blue) or 3′ (red) ssDNA overhangs or were blunt (green) (Figures 2A, 3 and S4). Most pMX-DELCJ and Jk1 coding ends are resected less than 15 bases on either the top or bottom strand. More than half of these coding ends contain 3′ overhangs with the remainder being blunt or having 5′ overhangs (Figures 2A, 3 and S4).
Figure 2. HCoDES analysis of RAG, Cas9 and ZFN DNA breaks.
(A) Barplots indicating the percentage of pMX-DELCJ (330,683 sequences analyzed) and Jk1 (315,274 sequences analyzed) coding ends with top strand (5′) and bottom strand (3′) termini at specific nucleotide (NT) distances from the RAG DSBs in LigIV−/−:DELCJ (ALig4CJ1) abl pre-B cells. Termini that are part of DSBs with 3′ (red) or 5′ (blue) ssDNA overhangs or are blunt (green) are indicated. Pie charts indicate the percentage of coding ends with 3′ (red) or 5′ (blue) ssDNA overhangs or are blunt (green). (B) Heatmap of frequencies of coding end structures with specific 5′ and 3′ termini within 10 base pairs of the RAG DSB (see Figure S3B). Arrows indicate coding end structures with the most frequent 5′ termini for Jk1 and pMX-DELCJ. Diagonal line indicates blunt coding ends. Color scale represents the log2 of the percent of total DNA ends. (C) Spacer sequence (bracket) where Eb:ZFN cleaves, PAM sequence (red nucleotides) and the site for Cas9 cleavage (green arrow) are shown. Nucleotide numbering is arbitrary. Oligonucleotides (CGAF and CGAR) used for HCoDES amplification of Eb:ZFN and Cas9 DSBs are indicated. Heat maps for DNA end structures generated after transient expression of Cas9 and the Eb:gRNA in LigIV−/− (L4B25-3.41) abl pre-B cells (left) or doxycycline induction of Eb:ZFN in LigIV−/−(BLig4ZF11) abl pre-B cells (right). The Cas9 cleavage site (green arrow) and the Eb:ZFN spacer region (ZFN, bracket) where cleavage occurs are shown. The nucleotide numbering corresponds to the sequence numbering. Blunt DNA ends are marked with a dashed line. Color scale represents the log2 of the percent of total DNA ends. Also see related figures S3, S4, S5 and S6.
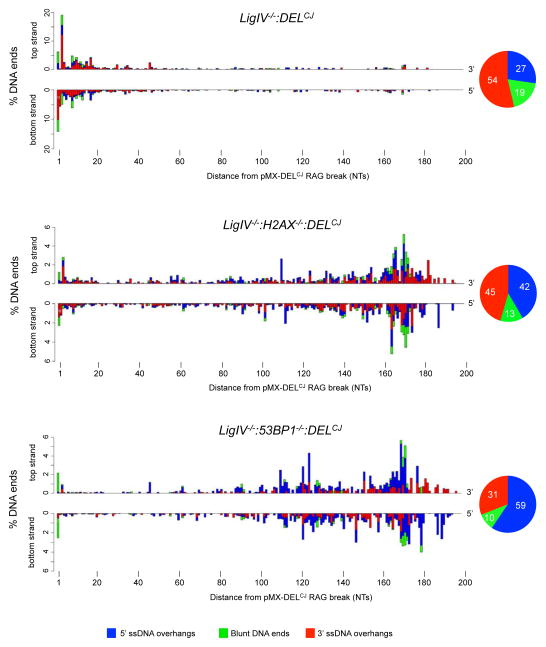
Figure 3. Coding end structures in H2AX- and 53BP1-deficient cells.
Barplots indicating the percentage of pMX-DELCJ coding ends with top strand (5′) and bottom strand (3′) termini at specific nucleotide (NT) distances from the RAG DSB in LigIV−/−:DELCJ (128,122 sequences analyzed), LigIV−/−:H2AX−/−:DELCJ (72,923 sequences analyzed), and LigIV−/−:53BP1−/−:DELCJ (337,917 sequences analyzed) abl pre-B cells as described in Figure 2A. The LigIV−/−:DELCJ (BLig4CJ7), LigIV−/−:H2AX−/−:DELCJ (LH8CJ148), and LigIV−/−:53BP1−/−:DELCJ (B53BP1Lig4CJ6) abl pre-B cells were used for analysis. Also see related figures S4 and S6.
HCoDES reveals a bias for coding ends with 5′ termini at nucleotide 3 (20%) of pMX-DELCJ and nucleotide 5 (40%) of Jk1 (Figure 2A). To assess the frequency of DNA end structures with specific 3′ and 5′ termini we generated heat maps, as schematized in Figure S3B. These analyses revealed that pMX-DELCJ coding ends with 5′ termini at nucleotide 3 primarily have 1 or 2 nucleotide 3′ ssDNA overhangs (Figure 2B, pMX-DELCJ arrow). At Jk1 coding ends, the abundant 5′ termini at nucleotide 5 primarily form 1–4 nucleotide 3′ ssDNA overhangs (Figure 2B, Jk1 arrow). These differences in pMX-DELCJ and Jk1 coding end structures may, in part, reflect the activity of Artemis in opening hairpin-sealed coding ends at distinct RAG DSBs in vivo (see Discussion) (Lu et al., 2007; Ma et al., 2002). Thus, HCoDES reveals that coding ends are minimally resected after RAG cleavage and demonstrates a bias for formation of short 3′ overhangs with preferred 5′ termini.
Structure of DSBs generated by genome editing nucleases
DNA DSBs generated by the Cas9 nuclease and a zinc finger nuclease (ZFN), two commonly used genome-editing enzymes, were analyzed by HCoDES (Kim and Kim, 2014). Cas9 forms a blunt DNA end three base pairs from the protospacer adjacent motif (PAM) flanking the guide RNA complementary sequence (Kim and Kim, 2014). Cas9 and a guide RNA (Eb:gRNA) to a region on chromosome 6 were expressed transiently in LigIV−/− abl pre-B cells arrested in G1 with imatinib, leading to robust generation of Cas9-induced DSBs (Figure S5A and S5B). HCoDES analyses using oligonucleotides ~180 base pairs from the target DSB site identified the most abundant DNA end structure (~20%) as the one expected for Cas9 cleavage 3 base pairs from the PAM sequence, with 5′ and 3′ termini at nucleotide 12 (arbitrary numbering) (Figure 2C and S5C). Most of the remaining DNA ends have 5′ and 3′ termini between positions 12 and 25 (Figure 2C). Although these DNA ends could reflect distinct Cas9 cleavage sites, they could also reflect resection of DNA ends generated at the expected Cas9 cleavage site. Notably, some blunt DNA ends terminate at nucleotide 11, and some with 3 nucleotide 5′ overhangs terminate at nucleotide 10 (Figure 2C). These DNA ends are longer than expected, and thus could not be generated by resection of DSBs generated by Cas9 cleavage 3 base pairs from the PAM.
To determine the structure of DNA ends generated by ZFNs, we used a previously described pair of zinc finger - Fok1 nuclease fusions (collectively referred to as Eb:ZFN) that introduce a DSB at a location similar to that of the Cas9 and Eb:gRNA (Figure 2C) (Tubbs et al., 2014). Fok1 cleavage generates 4 nucleotide 5′ ssDNA overhangs, and this cleavage could occur in three different registers in the six base-pair region between the ZFN binding sites (Figure 2C) (Kim and Kim, 2014). Efficient generation of DSBs by Eb:ZFN was induced using doxycycline in LigIV−/− abl pre-B cells arrested in G1 (Figure S5D). HCoDES analyses of these DNA ends reveal that many have 5′ and 3′ termini within the 6 base pair spacer region (Figures 2C and S5E). DNA ends shorter than expected and those without 4 base pair 5′ ssDNA overhangs could be generated by resection of appropriately cleaved DNA ends. In contrast to the analysis of Cas9 cleavage, we do not find Eb:ZFN DSBs with termini longer than expected, suggesting that cleavage by the Eb:ZFN is restricted primarily to the spacer region.
H2AX and 53BP1 prevent the generation of 3′ and 5′ ssDNA overhangs
Coding ends in abl pre-B cells deficient for DNA Ligase IV and either H2AX (LigIV−/−:H2AX−/−:DELCJ) or 53BP1 (LigIV−/−:53BP1−/−:DELCJ) are extensively resected (Figure S6A) (Helmink et al., 2011; Tubbs et al., 2014). HCoDES was carried out on pairs of independently derived LigIV−/−:H2AX−/−:DELCJ and LigIV−/−:53BP1−/−:DELCJ abl pre-B cells. Coding end PCR products were heterogeneous in size, with most being smaller than expected for full-length pMX-DELCJ or Jk1 coding ends (Figure S6B and data not shown). Indeed, pMX-DELCJ coding ends in LigIV−/−:H2AX−/−:DELCJ and LigIV−/−:53BP1−/−:DELCJ abl pre-B cells are substantially resected on both strands (Figures 3 and S4). The frequency distribution of 5′ and 3′ termini is not uniform. Moreover, as compared to LigIV−/−:DELCJ abl pre-B cells, the fraction of pMX-DELCJ coding ends with 3′ ssDNA overhangs (red) is decreased, while the fraction with 5′ ssDNA overhangs (blue) is increased in both LigIV−/−:H2AX−/−:DELCJ and LigIV−/−:53BP1−/−:DELCJ abl pre-B cells, although this difference is more pronounced in LigIV−/−:53BP1−/−:DELCJ abl pre-B cells (Figures 3 and S4). Analysis of Jk1 coding ends in 53BP1 and H2AX-deficient cells (Figure S4) and Eb:ZFN-induced DSBs in 53BP1-deficient cells (Figure S6C, S6D and S6E) yielded similar results.
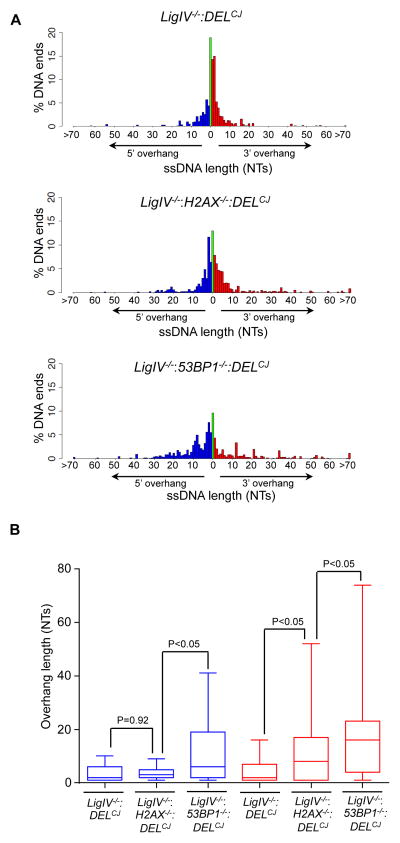
Similar to end resection in S/G2-phase cells, aberrant resection of coding ends in G1-phase abl pre-B cells deficient in either H2AX or 53BP1 generates DNA ends with long 3′ ssDNA overhangs (Figure 4). These 3′ overhangs were significantly longer in cells deficient in 53BP1 as compared to those deficient in H2AX (Figure 4). In contrast, the 5′ ssDNA overhangs are of similar size in LigIV−/−:DELCJ and LigIV−/−:H2AX−/−:DELCJ abl pre-B cells, but are significantly longer in LigIV−/−:53BP1−/−:DELCJ abl pre-B cells (Figure 4). These differences in DNA end structures suggest that H2AX and 53BP1 may have distinct activities in modulating DNA end resection in G1-phase cells (see Discussion).
Figure 4. Length of ssDNA overhangs in H2AX- and 53BP1-deficient cells.
(A) Percentage of coding ends with specific length of 3′ ssDNA (red) and 5′ ssDNA (blue) overhangs and blunt DNA ends (green) in LigIV−/−:DELCJ (ALig4CJ1), LigIV−/−:H2AX−/−:DELCJ (LH8CJ148), and LigIV−/−:53BP1−/−:DELCJ (B53BP1Lig4CJ6) abl pre-B cells. (B) Box and whisker plots showing the range of 3′ (red) and 5′ (blue) ssDNA overhang lengths (Wilcoxon signed rank test). The median (midline), the boundries of the box mark the 25th and 75th percentiles and the whiskers, which mark the 10th and 90th percentiles are shown.
Resected coding ends bind RPA
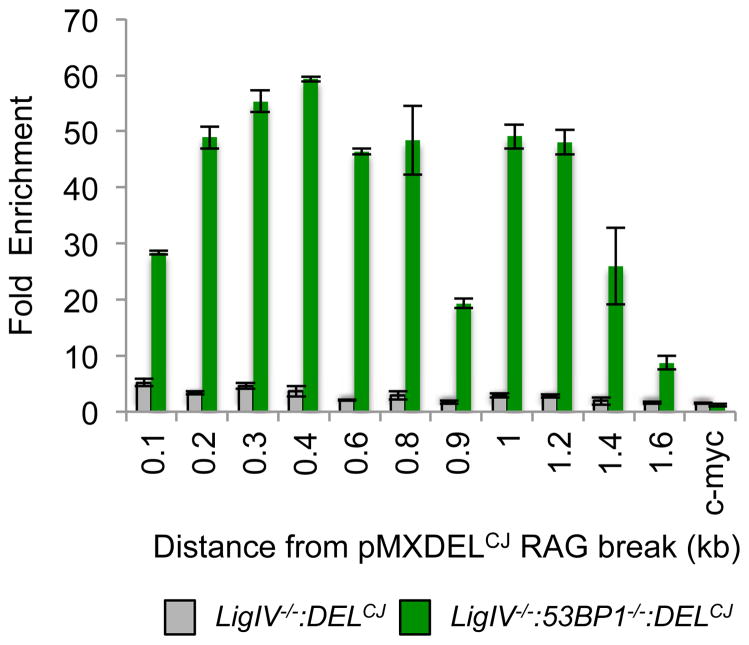
RPA binding to ssDNA is the initial step of HR in S/G2-phase cells. Heterotrimeric RPA requires a minimum of 26 bases of ssDNA for binding in vitro (Cai et al., 2007). In this regard, many of the coding ends in LigIV−/−:53BP1−/−:DELCJ abl pre-B cells have ssDNA overhangs (5′ and 3′) 26 bases or longer, whereas very few DNA ends have ssDNA overhangs this long in LigIV−/−:DELCJ cells (Figure 4). To determine whether RPA binds to these resected coding ends in G1-phase cells, we performed chromatin immunoprecipitation (ChIP) using an antibody to the RPA32 subunit and amplified DNA with primer pairs at increasing distances from the site of RAG cleavage in pMX-DELCJ (Figure 5). Minimal binding of RPA32 was detected at pMX-DELCJ coding ends generated in LigIV−/−:DELCJ abl pre-B cells (Figure 5). However, in LigIV−/−:53BP1−/−:DELCJ abl pre-B cells, where RAG DSBs are resected to form long ssDNA overhangs, we observed robust RPA32 binding up to 1.4 kb from the site of the RAG DSBs (Figure 5). We conclude that coding ends processed to generate ssDNA overhangs can bind RPA in G1-phase abl pre-B cells.
Figure 5. RPA binds coding ends with ssDNA overhangs in G1-phase cells.
ChIP for RPA32 binding at increasing distances (kb) from the pMX-DELCJ coding end in LigIV−/−:DELCJ (BLig4CJ7) and LigIV−/−:53BP1−/−:DELCJ (B53BP1Lig4CJ6) abl pre-B cells. Y axis shows fold enrichment compared to RPA32 binding to c-myc. Bars represent mean values from at least two independent PCR reactions, +/- SEM. Similar results were obtained with two independent LigIV−/−:DELCJ and LigIV−/−:53BP1−/−:DELCJ abl pre-B cell lines.
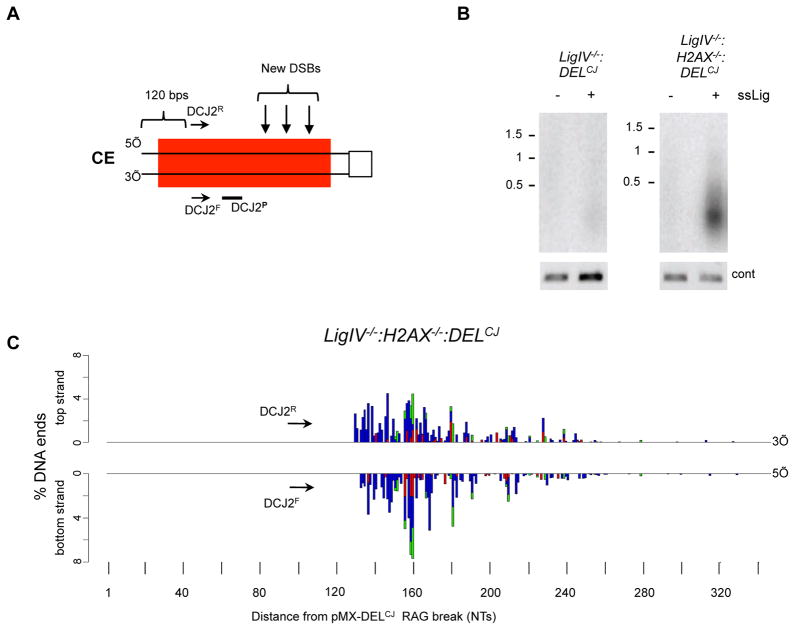
De novo DSBs can be generated during DNA end resection
DNA resection is carried out both by exonucleases and endonucleases, which could potentially generate DSBs as part of the resection process. To investigate this possibility, we modified the HCoDES approach to detect DSBs generated during resection (Figure 6A). To this end, primary PCR oligonucleotides (DCJ2F and DCJ2R) were used that amplify de novo secondary breaks generated as part of the resection process, downstream of the initiating RAG DSB in pMX-DELCJ (Figure 6A). Indeed, PCR products were observed when amplifying ligated genomic DNA from LigIV−/−:H2AX−/−:DELCJ abl pre-B cells, but not LigIV−/−:DELCJ abl pre-B cells (Figure 6B). Sequence analysis revealed that these products are heterogeneous in size and end structure, with 5′ and 3′ termini detected up to 320 base pairs from the site of the RAG DSB (Figure 6C). We conclude that the resection of coding ends in G1-phase lymphocytes involves endonuclease activity that is able to generate de novo DNA DSBs several hundred base pairs from the site of the RAG DSB. This activity would lead to the formation of small double-stranded DNA fragments.
Figure 6. Generation of DSBs during coding end resection.
(A) Schematic of pMX-DELCJ coding end (CE) as described in Figure 1B showing the relative position of the DCJ2R and DCJ2F oligonucleotides used to amplify DSBs generated downstream of the coding end and the DCJ2P oligonucleotide probe. (B) Southern blot of PCR amplified pMX-DELCJ coding ends from LigIV−/−:DELCJ (BLig4CJ7) and LigIV−/−:H2AX−/−:DELCJ (LH8CJ148) abl pre-B cells using DCJ2R and DCJ2F and DCJ2P as a probe. Genomic DNA was treated (+) or not (−) with the ssDNA ligase prior to PCR amplification. Control PCR was performed as in Figure 1E. (C) Barplots from HCoDES analysis using the DCJ2R and DCJ2F oligonucleotides indicating the percentage of de novo pMX-DELCJ DSBs with top strand (3′) and bottom strand (5′) termini at specific nucleotide (NT) distances from the RAG DSB in LigIV−/−:H2AX−/−:DELCJ (LH8CJ148) abl pre-B cells. 50,437 sequences were analyzed.
Discussion
HCoDES is a novel approach that can elucidate a broad spectrum of diverse chromosomal DNA end structures at single nucleotide resolution. We have used HCoDES to examine the structure of DSBs generated by three distinct nucleases (RAG, Cas9, and a ZFN) in the presence or absence of two proteins that function to protect DNA ends from resection in G1-phase cells (H2AX and 53BP1). The HCoDES analyses reveal important new insights into how broken DNA ends are processed in G1-phase cells and how this processing may promote mutagenic DSB repair and genome instability. In addition, these analyses establish the utility of HCoDES as an approach that can be broadly applied to elucidate diverse broken DNA end structures generated throughout the genome.
The HCoDES assay incorporates several salient features to distinguish it from other existing measures of DNA end structure. The first step is a ligation of the top and bottom strands of broken DNA ends, which requires the presence of a 5′ phosphate and a 3′ hydroxyl group. The HCoDES ligation conditions have been optimized to ensure that ligation proceeds efficiently when these criteria are met, even at a blunt DNA end where ligation of the top strand to the bottom strand of the same DNA end is an unfavorable reaction. It is conceivable that, at a small fraction of DNA ends, these groups may be removed or have adducts that prevent ligation. However, such DNA ends are nucleolytically processed in vivo to generate a 5′ phosphate and 3′ hydroxyl group. Thus, HCoDES should reveal the complete spectrum of DNA ends generated during the resection of chromosomal DSBs. Importantly, the assay is able to detect these broken DNA ends when they are present at a frequency of one or two per cell.
HCoDES can determine the full spectrum of DNA end structures captured in PCR products up to 500 base pairs in length, which is the current limit for robust paired end sequencing. Undoubtedly, as sequencing technologies improve, larger HCoDES PCR products will be amenable to analysis. Here, we used forward and reverse oligonucleotides to a complementary region approximately 200 base pairs from the DNA cleavage sites, allowing us to determine all of the DNA end structures within this region. However, HCoDES is not restricted to analyses of DSB-proximal end structures and can be used to study hyper-resected ends by designing forward and reverse oligonucleotide pairs to complementary regions at greater distances from the cleavage site. Moreover, staggering the forward and reverse primers to non-complementary regions would permit the detection of DNA ends with longer 5′ or 3′ ssDNA overhangs. Thus, the HCoDES approach provides a flexible platform to assess the full spectrum of DNA end structures generated in vivo during chromosomal DSB repair.
Several aspects of our study support the ability of HCoDES to accurately assign chromosomal DNA end structures. First, Jk1 and pMX-DELCJ coding ends generated in DNA Ligase IV-deficient abl pre-B cells exhibit minimal resection (<15 base pairs) on the top and bottom strands, similar to the spectrum of nucleotide loss observed in normal coding joints (Gauss and Lieber, 1996). Second, Artemis opens hairpin-sealed coding ends in vitro by cleaving after the first to the fourth nucleotide 3′ of the hairpin coding end, with a bias for cleaving after the second nucleotide (Lu et al., 2007; Ma et al., 2002). In this regard, we find that pMX-DELCJ coding ends have 5′ termini at the third nucleotide, consistent with Artemis cleavage after the second nucleotide. The 5′ termini of Jk1 coding ends are preferentially located at the fifth position, suggesting that Artemis opens these hairpins after the fourth nucleotide, which is in agreement with previous analyses of Jk1 ligation mediated PCR products (Schlissel, 1998). Third, cleavage by Cas9 generates a significant fraction of blunt DNA ends three base pairs from the PAM sequence, as would be expected for precise Cas9 cleavage (Kim and Kim, 2014). Many of the DNA ends that do not have this structure could have been generated by nucleolytic resection of precisely generated Cas9 DSBs. Indeed, this type of DSB processing prior to NHEJ forms the basis for Cas9-mediated gene inactivation. However, the generation of broken DNA ends that are longer than expected suggests that in vivo Cas9 may, on occasion, cleave at locations other than 3 base pairs from the PAM. Fourth, the majority of the Eb:ZFN-induced DSBs occur within the six base pair spacer region between the two zinc finger binding sites, which is the expected site for cleavage (Kim and Kim, 2014). Many of the DNA ends do not have 4 base pair 5′ ssDNA overhangs, and a significant fraction are shorter and outside of the spacer region, both of which could reflect resection of precise Eb:ZFN DSBs.
In the absence of H2AX or 53BP1, coding ends generated by RAG cleavage are significantly resected in G1-phase cells (Helmink et al., 2011; Tubbs et al., 2014). This is not unique to RAG DSBs, as Eb:ZFN-induced DSBs are also significantly resected in 53BP1-deficient cells (Tubbs et al., 2014). HCoDES reveals that this resection is not uniformly processive; rather, the 5′ and 3′ termini of these DNA ends exhibit some regional preferences. A bias toward specific 5′ or 3′ termini could be due to the targeting of endonuclease activities, as well as exonuclease processivity, which may be affected by several factors. For example, nucleosome positioning and histone modifications that lead to changes in chromatin structure could impede nuclease accessibility and processivity. Indeed, proteins with chromatin modifying activities have been implicated in the regulation of DNA end resection in yeast (Chen et al., 2012; Costelloe et al., 2012). Secondary structures formed by exposed ssDNA could also promote biases for specific 5′ and 3′ termini. This includes the formation of hairpin structures from the annealing of complementary regions on ssDNA, which could inhibit exonuclease processivity, leading to an increased frequency of DNA ends with 5′ and 3′ termini at locations where hairpins form. Moreover, cleavage by Mre11 or Artemis at the single to double strand transition of the hairpin loop could also promote specific 5′ or 3′ termini (Ma et al., 2002; Paull and Gellert, 1998). In addition, CtIP has endonuclease activity that targets 5′ flaps and the 5′ base of hairpin structures (Makharashvili et al., 2014; Wang et al., 2014). These endonuclease activities may be restricted once ssDNA overhangs bind RPA, which antagonizes the formation of ssDNA secondary structures in yeast (Deng et al., 2014).
Using a modified version of the HCoDES approach, we show that de novo DSBs can be generated during coding end resection. Although the nucleases that form these DSBs are not known, CtIP and Mre11 could generate DSBs as part of the resection process by forming closely staggered nicks downstream of a broken DNA end. Moreover, at single stranded gap intermediates, the exposed ssDNA may form hairpins that could be cleaved by Mre11, Artemis, or CtIP, forming a DSB. These DSBs would release small double stranded DNA fragments that could be an additional source of genome instability if they were inserted during the repair process at DSB sites elsewhere in the genome.
The resection of coding ends in G1-phase lymphocytes deficient for H2AX or 53BP1 produces DNA ends with ssDNA overhangs of substantial length. Indeed, some of these DNA ends can bind to RPA in G1-phase cells. Surprisingly, in addition to 3′ ssDNA overhangs, this resection also leads to the formation of a significant number of DNA ends with long 5′ ssDNA overhangs, especially in cells deficient in 53BP1. In S- and G2-phase cells, broken DNA ends with 5′ ssDNA overhangs would not be repaired by HR, because they would be unable to prime DNA synthesis. However, DNA ends with 5′ ssDNA overhangs could be repaired by mutagenic pathways that use small regions of homology (Gostissa et al., 2011; Huertas, 2010). For this to occur, the resection polarity (5′ or 3′) at the two DNA ends would have to be the same to permit base pairing, although it is possible that a strand invasion mechanism could promote the joining of DNA ends with dissimilar (5′ and 3′) ssDNA overhangs.
The retention of 53BP1 by γ-H2AX at broken DNA ends should inhibit resection. However, the increase in DNA ends with long 5′ ssDNA overhangs in cells deficient in 53BP1 as compared to H2AX suggests that H2AX has additional functions in modulating resection in G1-phase cells. In this regard, γ-H2AX retains Mre11 at DSB sites and could potentially affect the association and function of other nucleases at broken DNA ends (Celeste et al., 2003; Kobayashi et al., 2002; Lukas et al., 2004). Mre11 has 3′ to 5′ exonuclease activity that could generate 5′ ssDNA overhangs, and perhaps this activity is regulated by 53BP1 at DSBs in G1-phase cells.
Loss of 53BP1 rescues the HR-mediated DSB repair defect in Brca1-deficient cells, presumably by permitting resection (Bunting et al., 2010; Cao et al., 2009). However, the rescue is only partial, and these cells still exhibit DSB repair defects and genome instability. In this regard, if deficiency of 53BP1 in Brca1-deficient cells leads to the generation of both 3′ and 5′ ssDNA overhangs in S and G2 phase cells, as we observe in G1-phase cells, only DNA ends with 3′ ssDNA overhangs would be repaired by HR. In contrast, those DNA ends with 5′ ssDNA overhangs would persist unrepaired or be resolved by mutagenic repair pathways that promote genome instability.
Our analyses establish the power of the HCoDES approach for studying the processing of broken DNA ends during chromosomal DSB repair in vivo. We have shown that HCoDES can be used to analyze DNA end structures generated by different nucleases at a variety of genomic locations. Moreover, we have shown that the high-resolution DNA end structures from HCoDES analyses can reveal potential differences in the function of proteins (H2AX and 53BP1) implicated in protecting DNA ends in G1-phase cells. By analogy, HCoDES can be used to investigate the function of the many other catalytic and non-catalytic proteins required for DNA end resection. In addition, as HCoDES involves ligation of the top and bottom strand and bisulfite treatment, it provides strand-specific information about the CpG methylation status of broken DNA ends. Thus, subsequent HCoDES analyses may provide a mechanistic connection between CpG methylation and DNA end processing. Finally, although our analyses have focused on assessing the structure of chromosomal DNA ends generated in vivo, the HCoDES approach could also be used to reveal precise DNA end structures generated by different classes of genome editing enzymes in vitro and during resection in vitro using purified proteins and protein complexes involved in DNA end processing.
Experimental Procedures
Single stranded DNA ligation and bisulfite treatment
Genomic DNA was isolated as previously described (Blondal et al., 2005). For ssDNA ligation, 10 μg of genomic DNA was ligated at 60°C for 16 hrs in a total volume of 50 μL of 2.5 mM MnCl2, 1M betaine, 0.05 mM ATP, 1x CircLigase buffer and 200 units CircLigase (epicenter biotechnology). 2 μg of ligated genomic DNA was bisulfite treated using the EZ DNA Methylation-Gold Kit (Zymo Research) following the manufactures instructions. Briefly, 130 μL of prepared CT conversion reagent was mixed thoroughly with 20ul of 100 ng/μL ligated genomic DNA. This solution was then incubated at 98°C for 10 min followed by 64°C for 2.5 hrs and then stored at 4°C up to 20 hours. Bisulphite converted DNA was then purified and eluted into 60 ul of H2O following the manufactures instructions.
PCR amplification
For amplification of bisulfite treated genomic DNA, 6 μL of the eluted bisulphite treated DNA solution was amplified in a 50 μL reaction volume of 50 units of Maxima Hot Start Taq DNA polymerase (Fermentase), 1x Maxima Hot Start reaction buffer, 3mM MgCl2, 5mM dNTPs, 0.3 μM forward primer and 0.3 μM reverse primer. The oligonucleotides used for the primary PCR reaction are listed in Table S1 and have omitted the “MiSeq-“ prefix. The primary PCR cycling condition was 95° for 4 min followed by 26 cycles of 95° for 30 sec, 54° for 45 sec, 72° for 1 min 30 sec, followed by a final extension at 72° for 5 min. Appropriate MiSeq sequencing adaptors were then added by nested PCR. For this, the primary PCR primers were removed by adding 1 μL (20 units) of Exonuclease I to the primary PCR reaction (total volume of 51 μL) and incubated at 37° for 45 minutes. Exonuclease I was heat inactivated at 80° for 30 minutes. To this reaction was added, 2.5 units (0.5 μL) of maxima hotstart taq DNA polymerase (Fermentase), 0.75 μL DMSO, 1 μL of each 15 μM stock of forward and reverse oligonucleotides that have indexed sequencing adaptors for sequencing on the MiSeq sequencing platform (0.28 μM final concentration). The PCR cycling condition for addition of the MiSeq sequencing adaptors was 95° for 4 min followed by 9 cycles of 95° for 30 sec, 54° for 45 sec, 72° 1 min 30 sec, followed by a final extension at 72° for 5 min. All nested PCR reactions were done with the oligonucleotides with the same name as that described for the primary PCR, but have the “MiSeq-“ prefix (see Table S1). For sequencing the single stranded ligase treated KpnI control, PCR was done with oligonucleotides KpnIR and KpnIF followed by a nested PCR with oligonucleotides MiSeq-KpnIR and MiSeq-KpnIF (Table S1). The cycling parameters for the loading control PCRs was the same as for the primary PCR reaction, except the extension time was 1 minute and it was done for 35 cycles.
Purification of PCR products and sequencing
PCR products were excised from a 1.5% TBE low melting point agarose gel (Invitrogen). DNA from excised gel slices was purified using a Qiagen Gel Extraction kit (Qiagen) following the manufacturer’s instructions, including the optional steps of isopropanol addition and the extra QG buffer wash. PCR products were eluted in 15 μL to 20 μL of Qiagen elution buffer and quantitated using the Qubit dsDNA HS Assay Kit (Life Technologies). Each purified sample was then run on a 2100 Bioanalyzer (Agilent Technologies) to assess removal of primer dimer, determine molarity and evaluate the expected size distribution of end products. Samples were then sequenced on the Illumina MiSeq Desktop Sequencer using the 2 × 250 bp kit and 40% Phi-X DNA.
Sequence processing
For sequence analysis of amplified bisulphite treated DNA, paired end reads from the MiSeq Desktop Sequencer were first demultiplexed by the sample indexes allowing for a maximum 1 base pair mismatch in the index. The adaptor sequences were clipped from reads 1 and 2 using FLEXBAR version 2.4 (parameters: -m 10 -u 5 -ae RIGHT -r -aa -at 0.5) (Dodt et al., 2012). The paired reads were then assembled using FLASH-1.2.7 software (parameters: -m 10 -M 100 -x 0.25) (Magoc and Salzberg, 2011). The assembly parameters were adjusted based on the read length distribution of a particular sample. When a sample was dominated by short reads that resulted in 100% sequence overlap between read 1 and 2, ~50 bases from the end of the reads was removed using PrinSeq in order to the prevent mis-assembly (Schmieder and Edwards, 2011). The trimmed reads were assembled again by FLASH. Individual assembled reads (contigs) were aligned pairwise to the reference sequence using the MUSCLE aligner (parameters: default) as a scheduled batch job using The Genome Institute clusters (Edgar, 2004). Alignments were subjected to four filtering steps with customized Perl scripts: (1) Remove alignments with >5bp deletions on either strand (mis-assembled sequences due to poor sequence quality); (2) Remove alignments without >10 bps of continuous reference sequence (primer dimers); (3) Remove alignments where assembled sequences appear to be longer than the full length coding end (usually mis-assembled reads, small deletions near coding end or insertions); (4) Remove alignments for products smaller than 50 base pairs. The remaining alignments were used to identify the position of the 5′ and 3′ ends of a DSB. The end structures from triplicate PCR reactions were then pooled and DNA end structures with single reads were removed. As a convention, nucleotides at the ligation junction that could be assigned to either the 5′ or 3′ end were assigned to the 5′ end except for the analysis of Cas9, Eb:ZFN and KpnI DNA ends, where assignment to generate blunt, 4 nucleotide 5′ overhangs and 4 nucleotide 3′ overhangs, were made, respectively.
Additional experimental details can all be found in Supplemental methods.
Supplementary Material
Figure S1, related to Figure 1. Analysis of KpnI-digested DNA. (A) Schematic of ligation of KpnI-digested genomic DNA and Southern blotting strategy for panel B. (B) Native (N) or denaturing (D) Southern blot analysis of KpnI-digested DNA that was ligated (+) or not (−) with the ssDNA ligase (ssLig), digested with EcoRV and probed with the C4b probe. Molecular weight markers are shown in kilobases. (C) Schematic for amplification of a ligated KpnI site in pMX-DELCJ using the KpnR and KpnF oligonucleotides. (D) PCR amplification of KpnI site after genomic DNA was treated (+) or not treated (−) with the ssDNA ligase (ssLig). Control (cont) PCR is shown as are MW markers in kilobases (kb). (E) Sequence schematic of the ligation, bisulfite treatment and amplification of a KpnI digested DNA end. Nucleotides derived from the top strand (blue), bottom strand (green) and 4 base 3′ ssDNA overhang (red) are shown.
Figure S2, related to Figures 1 and 2. Ligation of Igk coding ends. (A) Schematic of the Igk locus with Vk and Jk gene segments. EcoRI and SacI restriction enzyme sites and JkIII probe used for Southern blotting are shown. Fragments generated by RAG cleavage at the four Jk gene segments are shown. The relative position of the Jk1R and Jk1F oligonucleotides used to amplify Jk1 coding ends are also shown. (B) Southern blot of EcoRI and SacI digested genomic DNA from LigIV−/−:DELCJ abl pre-B cells not treated (−) or treated (+) with imatinib (imat). Hybridizing bands from the unrearranged (UR) and Jk coding ends (CE) are indicated. (C) Southern blot analysis as described in panel B of genomic DNA from imatinib-treated LigIV−/−:DELCJ abl pre-B cells treated (+) or not (−) with ssDNA ligase (ssLig) followed by Exonuclease I and Lambda Exonuclease I (Exo) treatment (+) or no treatment (−). (D) Southern blot using oligonucleotide Jk1P as a probe for products of HCoDES PCR amplification of Jk1 coding ends using the Jk1R and Jk1F oligonucleotides for primary PCR and Miseq- Jk1R and Miseq-Jk1F oligonucleotides for secondary PCR. Control PCR for these DNA samples shown in figure 1E. Genomic DNA from LigIV−/−:DELCJ abl pre-B cells treated (+) or not (−) with ssDNA ligase (ssLig) was analyzed.
Figure S3, related to Figures 2, 3, 4 and 6. Sequence analysis. (A) Schematic depicting the workflow of informatic processing of HCoDES data. (B) Heat maps show the frequency of broken DNA ends with specific 5′ and 3′ termini. The X-axis depicts nucleotide (NT) position on the bottom strand and the Y-axis shows the corresponding nucleotide on the top strand. The green dashed line marks boxes that correspond to blunt DNA ends. The boxes to the left are 3′ ssDNA overhangs and those to the right are 5′ ssDNA overhangs. Four different DNA ends structures (a–d) are shown as examples.
Figure S4, related to Figure 3. Jk1 and pMX-DELCJ coding end structures. Barplots of Jk1 coding ends in LigIV−/−:DELCJ (BLig4CJ7, 305,805 sequences analyzed), LigIV−/−:H2AX−/−:DELCJ (LH8CJ148, 157,970 sequences analyzed) and LigIV−/−:53BP1−/−:DELCJ (B53BP1Lig4CJ6, 258,051 sequences analyzed) abl pre-B cells as described in legend to Figure 3. Also shown are barplots of pMX-DELCJ coding ends in LigIV−/−:H2AX−/−:DELCJ (LH5CJ48, 76,560 sequences analyzed) and LigIV−/−:53BP1−/−:DELCJ (C5LCJ5, 416,959 sequences analyzed) as described in Figure 3.
Figure S5, related to Figure 2C. Cas9 and Eb:ZFN induced DSBs. (A) Schematic of the Southern blot strategy for analyzing Cas9 and Eb:ZFN DSBs on chromosome 6. Shown are the HindIII sites and the location of the Eb probe relative to the location of the Cas9 and Eb:ZFN DSBs. (B) LigIV−/− (L4B25-3.41) abl pre-B cells were treated with imatinib followed by transient expression of Cas9 or Cas9 and Eb:gRNA. Genomic DNA was digested with HindIII and probed with Eb. (C) Southern blot of HCoDES PCR amplification of genomic DNA from LigIV−/− abl pre-B cells after expression of Cas9 and Eb:gRNA (panel B) using the C6AF and C6AR oligonucleotides for primary PCR, Miseq-C6AF and Miseq-C6AR oligonucleotides for secondary PCR and the C6AP oligonucleotide probe. Genomic DNA was ligated (+) or not ligated (−) with the ssDNA ligase (ssLig) prior to PCR. Control PCR is as described in figure 1E. (D) Southern blot of genomic DNA from LigIV−/− (BLig4ZF11) abl pre-B cells not treated (−) or treated (+) with doxycycline (DOX) for induction of the Eb:ZFN as described in panel A. (E) HCoDES PCR of genomic DNA from LigIV−/− abl pre-B cells after induction of Eb:ZFN as described in panel C.
Figure S6, related to Figure 3. pMX-DELCJ coding end and Eb:ZFN DNA ends in H2AX- and 53BP1-deficient cells. (A) Southern blot analysis of pMX-DELCJ in LigIV−/−:H2AX−/−:DELCJ (LH5CJ48) and LigIV−/−:53BP1−/−:DELCJ (B53BP1Lig4CJ6) abl pre-B cells as described in Figure 1C. Bands due to unrearranged pMX-DELCJ (UR) and resected pMX-DELCJ coding ends are shown. (B) Southern blot of HCoDES amplification of pMX-DELCJ coding ends from panel A as described in Figure 1E. (C) Southern blot of genomic DNA from LigIV−/−:53BP1−/− (CL5ZFN12) abl pre-B cells after induction (+) or no induction (−) of Eb:ZFN with doxycycline as described in figure S7D. (D) HCoDES PCR of genomic DNA from LigIV−/−:53BP1−/− abl pre-B cells after induction of Eb:ZFN as described in figure S7E. (E) DNA end structures generated by DNA cleavage by Eb:ZFN in LigIV−/−:53BP1−/−:DELCJ abl pre-B cells as described in figure 3. Nucleotide numbering on the X axis is arbitrary. The position of the spacer region where the Eb:ZFN introduces DSBs is indicated.
Table S1. Oligonucleotides used for HCoDES. Bold text denotes complementary sequences downstream of the DSB. Red text are example index sequences. Non-bold text are MiSeq adaptor sequences. Numbers in parenthesis are the distances from the 3′ end of the oligonucleotide to the non-resected DNA end.
Table S2. Primers used for RPA ChIP. Numbers in position columns indicate distance from coding end. F: Forward; R: Reverse; NA: Not applicable.
Acknowledgments
We would like to thank Jessica Hoisington-Lopez from the Center for Genome Sciences and Systems Biology for her sequencing expertise. We would like to thank Jennifer Schmidt for her expertise with the Agilent 2100 Bioanalyzer. This work was supported by the National Institutes of Health grants AI074953 (B.P.S.), AI047829 (B.P.S.), AI079732 (E.M.O.) and AI081224 (E.M.O.) and by an American Cancer Society fellowship 250637 (Y.D.)
Footnotes
Accession number: Sequencing reads were deposited in the NCBI Gene Expression Omnibus (GEO) database (accession number GSE62534).
Author Contributions: Y.D. developed the HCoDES assay. Y.D., A.T.T. and C.P. performed HCoDES assays. B-R.C. and B-S.L. provided cells for HCoDES assay. N.D.H. and R.G. assisted with experiments. Y.Z. developed the informatic analysis pipeline. D.D. and Z.M. carried out RPA ChIP. G.M.W., E.S. and A.R. provided sequencing advice. E.M.O. and J-Y.Z. developed Cas9 cleavage assay. Y.D., A.L.B., B.P.S. and J.E.P. designed experiments and developed data analysis plan. Y.D., J.E.P., A.L.B and B.P.S wrote the manuscript.
Supplemental Information: Supplemental information can be found online and includes Supplemental Methods, six figures and two tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- Blier PR, Griffith AJ, Craft J, Hardin JA. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. 1993;268:7594–7601. [PubMed] [Google Scholar]
- Blondal T, Thorisdottir A, Unnsteinsdottir U, Hjorleifsdottir S, Aevarsson A, Ernstsson S, Fridjonsson OH, Skirnisdottir S, Wheat JO, Hermannsdottir AG, et al. Isolation and characterization of a thermostable RNA ligase 1 from a Thermus scotoductus bacteriophage TS2126 with good single-stranded DNA ligation properties. Nucleic Acids Res. 2005;33:135–142. doi: 10.1093/nar/gki149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, Sleckman BP. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Roginskaya M, Qu Y, Yang Z, Xu Y, Zou Y. Structural characterization of human RPA sequential binding to single-stranded DNA using ssDNA as a molecular ruler. Biochemistry. 2007;46:8226–8233. doi: 10.1021/bi7004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- Chen HT, Bhandoola A, Difilippantonio MJ, Zhu J, Brown MJ, Tai X, Rogakou EP, Brotz TM, Bonner WM, Ried T, Nussenzweig A. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science. 2000;290:1962–1965. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cui D, Papusha A, Zhang X, Chu CD, Tang J, Chen K, Pan X, Ira G. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature. 2012;489:576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costelloe T, Louge R, Tomimatsu N, Mukherjee B, Martini E, Khadaroo B, Dubois K, Wiegant WW, Thierry A, Burma S, et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature. 2012;489:581–584. doi: 10.1038/nature11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:405–412. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt M, Roehr JT, Ahmed R, Dieterich C. FLEXBAR-Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms. Biology (Basel) 2012;1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Foster SS, Balestrini A, Petrini JH. Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Mol Cell Biol. 2011;31:4379–4389. doi: 10.1128/MCB.05854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- Gauss GH, Lieber MR. Mechanistic constraints on diversity in human V(D)J recombination. Mol Cell Biol. 1996;16:258–269. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol. 2011;29:319–350. doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- Grundy GJ, Moulding HA, Caldecott KW, Rulten SL. One ring to bring them all-The role of Ku in mammalian non-homologous end joining. DNA Repair (Amst) 2014;17:30–38. doi: 10.1016/j.dnarep.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Helmink BA, Bredemeyer AL, Lee BS, Huang CY, Sharma GG, Walker LM, Bednarski JJ, Lee WL, Pandita TK, Bassing CH, Sleckman BP. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J Exp Med. 2009;206:669–679. doi: 10.1084/jem.20081326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmink BA, Sleckman BP. The Response to and Repair of RAG-Mediated DNA Double-Strand Breaks. Annu Rev Immunol. 2012;30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmink BA, Tubbs AT, Dorsett Y, Bednarski JJ, Walker LM, Feng Z, Sharma GG, McKinnon PJ, Zhang J, Bassing CH, Sleckman BP. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 2011;469:245–249. doi: 10.1038/nature09585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol. 2002;12:1846–1851. doi: 10.1016/s0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- Laird CD, Pleasant ND, Clark AD, Sneeden JL, Hassan KM, Manley NC, Vary JC, Jr, Morgan T, Hansen RS, Stoger R. Hairpin-bisulfite PCR: assessing epigenetic methylation patterns on complementary strands of individual DNA molecules. Proc Natl Acad Sci U S A. 2004;101:204–209. doi: 10.1073/pnas.2536758100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Schwarz K, Lieber MR. Extent to which hairpin opening by the Artemis:DNA-PKcs complex can contribute to junctional diversity in V(D)J recombination. Nucleic Acids Res. 2007;35:6917–6923. doi: 10.1093/nar/gkm823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makharashvili N, Tubbs AT, Yang SH, Wang H, Barton O, Zhou Y, Deshpande RA, Lee JH, Lobrich M, Sleckman BP, et al. Catalytic and Noncatalytic Roles of the CtIP Endonuclease in Double-Strand Break End Resection. Mol Cell. 2014;54:1022–1033. doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo SA, Schlissel MS. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat Immunol. 2003;4:31–37. doi: 10.1038/ni870. [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, DeVido J, Foy D, Chaudhuri J, Lombard D, Alt FW. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic V, Yin B, Maas NL, Bredemeyer AL, Carpenter AC, Helmink BA, Yang-Iott KS, Sleckman BP, Bassing CH. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol Cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel MS. Structure of nonhairpin coding-end DNA breaks in cells undergoing V(D)J recombination. Mol Cell Biol. 1998;18:2029–2037. doi: 10.1128/mcb.18.4.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Tubbs AT, Dorsett Y, Chan E, Helmink B, Lee BS, Hung P, George R, Bredemeyer AL, Mittal A, Pappu RV, et al. KAP-1 Promotes Resection of Broken DNA Ends Not Protected by gamma-H2AX and 53BP1 in G1-Phase Lymphocytes. Mol Cell Biol. 2014;34:2811–2821. doi: 10.1128/MCB.00441-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li Y, Truong LN, Shi LZ, Hwang PY, He J, Do J, Cho MJ, Li H, Negrete A, et al. CtIP Maintains Stability at Common Fragile Sites and Inverted Repeats by End Resection-Independent Endonuclease Activity. Mol Cell. 2014;54:1012–1021. doi: 10.1016/j.molcel.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Caron P, Legube G, Paull TT. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2013;42:e19. doi: 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 1. Analysis of KpnI-digested DNA. (A) Schematic of ligation of KpnI-digested genomic DNA and Southern blotting strategy for panel B. (B) Native (N) or denaturing (D) Southern blot analysis of KpnI-digested DNA that was ligated (+) or not (−) with the ssDNA ligase (ssLig), digested with EcoRV and probed with the C4b probe. Molecular weight markers are shown in kilobases. (C) Schematic for amplification of a ligated KpnI site in pMX-DELCJ using the KpnR and KpnF oligonucleotides. (D) PCR amplification of KpnI site after genomic DNA was treated (+) or not treated (−) with the ssDNA ligase (ssLig). Control (cont) PCR is shown as are MW markers in kilobases (kb). (E) Sequence schematic of the ligation, bisulfite treatment and amplification of a KpnI digested DNA end. Nucleotides derived from the top strand (blue), bottom strand (green) and 4 base 3′ ssDNA overhang (red) are shown.
Figure S2, related to Figures 1 and 2. Ligation of Igk coding ends. (A) Schematic of the Igk locus with Vk and Jk gene segments. EcoRI and SacI restriction enzyme sites and JkIII probe used for Southern blotting are shown. Fragments generated by RAG cleavage at the four Jk gene segments are shown. The relative position of the Jk1R and Jk1F oligonucleotides used to amplify Jk1 coding ends are also shown. (B) Southern blot of EcoRI and SacI digested genomic DNA from LigIV−/−:DELCJ abl pre-B cells not treated (−) or treated (+) with imatinib (imat). Hybridizing bands from the unrearranged (UR) and Jk coding ends (CE) are indicated. (C) Southern blot analysis as described in panel B of genomic DNA from imatinib-treated LigIV−/−:DELCJ abl pre-B cells treated (+) or not (−) with ssDNA ligase (ssLig) followed by Exonuclease I and Lambda Exonuclease I (Exo) treatment (+) or no treatment (−). (D) Southern blot using oligonucleotide Jk1P as a probe for products of HCoDES PCR amplification of Jk1 coding ends using the Jk1R and Jk1F oligonucleotides for primary PCR and Miseq- Jk1R and Miseq-Jk1F oligonucleotides for secondary PCR. Control PCR for these DNA samples shown in figure 1E. Genomic DNA from LigIV−/−:DELCJ abl pre-B cells treated (+) or not (−) with ssDNA ligase (ssLig) was analyzed.
Figure S3, related to Figures 2, 3, 4 and 6. Sequence analysis. (A) Schematic depicting the workflow of informatic processing of HCoDES data. (B) Heat maps show the frequency of broken DNA ends with specific 5′ and 3′ termini. The X-axis depicts nucleotide (NT) position on the bottom strand and the Y-axis shows the corresponding nucleotide on the top strand. The green dashed line marks boxes that correspond to blunt DNA ends. The boxes to the left are 3′ ssDNA overhangs and those to the right are 5′ ssDNA overhangs. Four different DNA ends structures (a–d) are shown as examples.
Figure S4, related to Figure 3. Jk1 and pMX-DELCJ coding end structures. Barplots of Jk1 coding ends in LigIV−/−:DELCJ (BLig4CJ7, 305,805 sequences analyzed), LigIV−/−:H2AX−/−:DELCJ (LH8CJ148, 157,970 sequences analyzed) and LigIV−/−:53BP1−/−:DELCJ (B53BP1Lig4CJ6, 258,051 sequences analyzed) abl pre-B cells as described in legend to Figure 3. Also shown are barplots of pMX-DELCJ coding ends in LigIV−/−:H2AX−/−:DELCJ (LH5CJ48, 76,560 sequences analyzed) and LigIV−/−:53BP1−/−:DELCJ (C5LCJ5, 416,959 sequences analyzed) as described in Figure 3.
Figure S5, related to Figure 2C. Cas9 and Eb:ZFN induced DSBs. (A) Schematic of the Southern blot strategy for analyzing Cas9 and Eb:ZFN DSBs on chromosome 6. Shown are the HindIII sites and the location of the Eb probe relative to the location of the Cas9 and Eb:ZFN DSBs. (B) LigIV−/− (L4B25-3.41) abl pre-B cells were treated with imatinib followed by transient expression of Cas9 or Cas9 and Eb:gRNA. Genomic DNA was digested with HindIII and probed with Eb. (C) Southern blot of HCoDES PCR amplification of genomic DNA from LigIV−/− abl pre-B cells after expression of Cas9 and Eb:gRNA (panel B) using the C6AF and C6AR oligonucleotides for primary PCR, Miseq-C6AF and Miseq-C6AR oligonucleotides for secondary PCR and the C6AP oligonucleotide probe. Genomic DNA was ligated (+) or not ligated (−) with the ssDNA ligase (ssLig) prior to PCR. Control PCR is as described in figure 1E. (D) Southern blot of genomic DNA from LigIV−/− (BLig4ZF11) abl pre-B cells not treated (−) or treated (+) with doxycycline (DOX) for induction of the Eb:ZFN as described in panel A. (E) HCoDES PCR of genomic DNA from LigIV−/− abl pre-B cells after induction of Eb:ZFN as described in panel C.
Figure S6, related to Figure 3. pMX-DELCJ coding end and Eb:ZFN DNA ends in H2AX- and 53BP1-deficient cells. (A) Southern blot analysis of pMX-DELCJ in LigIV−/−:H2AX−/−:DELCJ (LH5CJ48) and LigIV−/−:53BP1−/−:DELCJ (B53BP1Lig4CJ6) abl pre-B cells as described in Figure 1C. Bands due to unrearranged pMX-DELCJ (UR) and resected pMX-DELCJ coding ends are shown. (B) Southern blot of HCoDES amplification of pMX-DELCJ coding ends from panel A as described in Figure 1E. (C) Southern blot of genomic DNA from LigIV−/−:53BP1−/− (CL5ZFN12) abl pre-B cells after induction (+) or no induction (−) of Eb:ZFN with doxycycline as described in figure S7D. (D) HCoDES PCR of genomic DNA from LigIV−/−:53BP1−/− abl pre-B cells after induction of Eb:ZFN as described in figure S7E. (E) DNA end structures generated by DNA cleavage by Eb:ZFN in LigIV−/−:53BP1−/−:DELCJ abl pre-B cells as described in figure 3. Nucleotide numbering on the X axis is arbitrary. The position of the spacer region where the Eb:ZFN introduces DSBs is indicated.
Table S1. Oligonucleotides used for HCoDES. Bold text denotes complementary sequences downstream of the DSB. Red text are example index sequences. Non-bold text are MiSeq adaptor sequences. Numbers in parenthesis are the distances from the 3′ end of the oligonucleotide to the non-resected DNA end.
Table S2. Primers used for RPA ChIP. Numbers in position columns indicate distance from coding end. F: Forward; R: Reverse; NA: Not applicable.