Abstract
Eukaryotic cells express at least three unique nuclear RNA polymerases. The selective advantage provided by this enhanced complexity is a topic of fundamental interest in cell biology. It has long been known that the gene targets and transcription initiation pathways for RNA polymerases (Pols) I, II and III are distinct; however, recent genetic, biochemical and structural data suggest that even the core enzymes have evolved unique properties. Among the three eukaryotic RNA polymerases, Pol I is considered the most divergent. Transcription of the ribosomal DNA by Pol I is unmatched in its high rate of initiation, complex organization within the nucleolus and functional connection to ribosome assembly. Furthermore, ribosome synthesis is intimately linked to cell growth and proliferation. Thus, there is intense selective pressure on Pol I. This review describes key features of Pol I transcription, discusses catalytic activities of the enzyme and focuses on recent advances in understanding its unique role among eukaryotic RNA polymerases.
Introduction
Transcription is the universal process by which organisms express their genomes. Enzymes that convert genetic information encoded by DNA into RNA are called DNA-dependent RNA polymerases (RNAPs). All cells utilize multi-subunit RNAPs for transcription of their genomes. Whereas prokaryotic cells accomplish this task using a single RNA polymerase, eukaryotic cells utilize at least three specialized nuclear RNAPs: RNA polymerase I (Pol I), RNA polymerase II (Pol II) and RNA polymerase III (Pol III). Pol I transcribes ribosomal DNA (rDNA); Pol II synthesizes all messenger RNAs (mRNAs) and most regulatory non-coding RNAs; and Pol III primarily produces transfer RNAs (tRNAs) and 5S rRNA. The diversity of multisubunit RNAPs, their conservation, and subunit composition are summarized in Table 1.
Table 1.
Diversity of the Multisubunit RNA Polymerases.
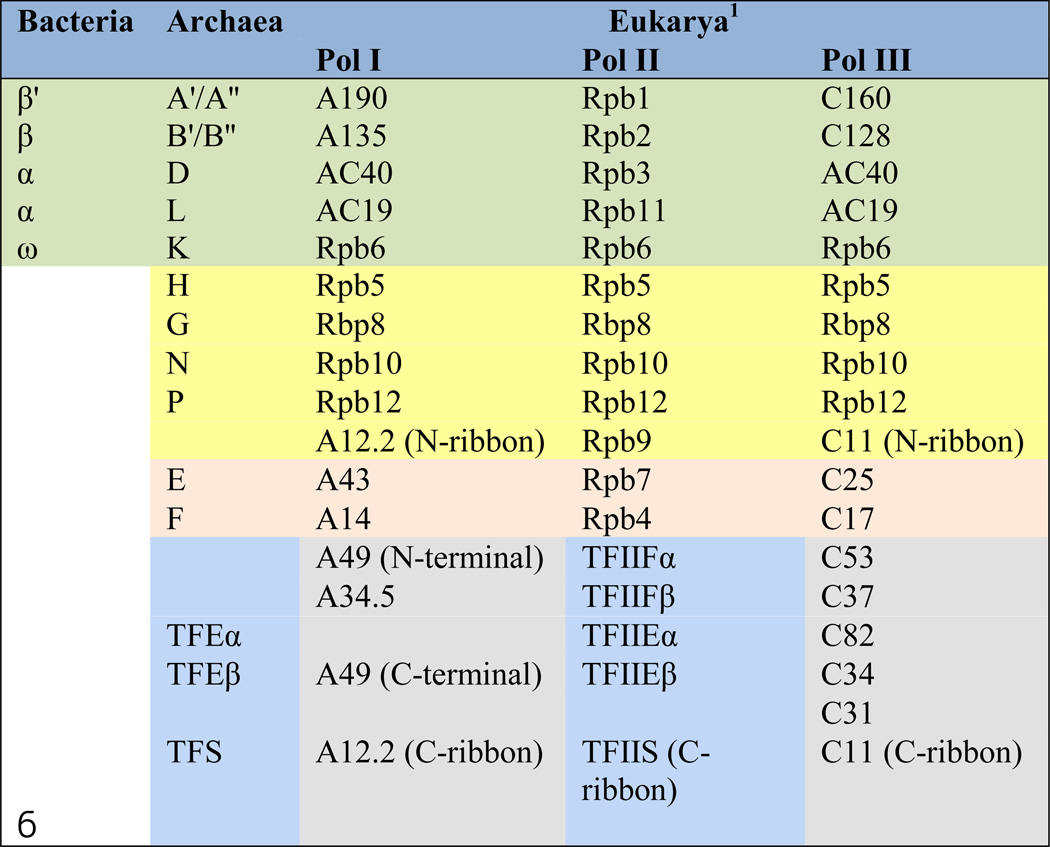 |
Subunit composition and homology of the multisubunit RNA polymerases from the three domains of life are presented. The universally conserved polymerase subunits are shaded in green; the archaeo-eukaryote-specific functional core subunits are in yellow, the polymerase stalk components are in tan. All of the conserved subunits or the Pol I/III subunits (in grey) homologous to Pol II trans-acting factors (in blue) are organized in the same rows.
Eukaryotic RNA polymerase subunits are according to S. cerevisiae nomenclature.
Even though all multisubunit RNA polymerases are highly conserved [reviewed in (Cramer, 2002)], there are important structural and functional differences between the related RNAP complexes (Kuhn et al., 2007; Carter and Drouin, 2009b; Viktorovskaya et al., 2013). Among the three eukaryotic RNA polymerases, Pol I is considered the most divergent (Carter and Drouin, 2009b; Carter and Drouin, 2009a). Ribosome synthesis is closely linked to cell growth; thus, there is intense selective pressure on Pol I. Most reviews on multi-subunit RNA polymerases focus on the similarities between the paralogs (Cramer, 2002; Cramer et al., 2008; Vannini and Cramer, 2012). However, there is a growing body of evidence that identifies key differences between the nuclear RNA polymerases. This review highlights these differences between Pol I and Pol II and focuses on unique features of the Pol I transcription system. We suggest that despite high sequence similarity, Pol I has evolved unique characteristics to suit its specialized cellular role.
The role for Pol I transcription in metabolism
Ribosome biogenesis is a central feature of cell biology. The assembly of the ribosomal particles commences in a special sub-nuclear compartment – the nucleolus, continues in the nucleoplasm and is completed in the cytosol. Cells produce approximately 2000 ribosomes per minute (Warner, 1999), and the rate of ribosome biosynthesis is proportional to the rates of cell growth and proliferation (Waldron and Lacroute, 1975; Kief and Warner, 1981). The energetic investment that cells make in ribosome biosynthesis is greater than the investment in any other process. Transcription of the rDNA by Pol I is the first step in ribosome biogenesis and has been shown to be rate-limiting (Laferte et al., 2006). Despite the cellular commitment to rDNA transcription, there is a hole in our understanding of the unique properties of Pol I transcription and how it is regulated.
Transcription of the rDNA by Pol I is unique in its high rate of initiation, polymerase density, specific organization within the nucleolus and tight connection to ribosome assembly. Transcription by Pol I accounts for more than 60% of total nuclear transcription (Warner, 1999). According to calculations, Pol I transcription initiation must occur every 5 seconds in growing yeast under standard conditions (Reeder and Lang, 1997). To support this high rate of transcription initiation, escape from the promoter must be similarly efficient. To complete the transcript synthesis, Pol I elongates through the 35S rRNA gene at approximately 60 nucleotides per second. Actively transcribed rDNA repeats are densely packed with polymerases, carrying approximately 50 – 60 elongation complexes per 35S rRNA gene (French et al., 2003). In yeast the full size of the pre-rRNA transcript is approximately 6.7 kb, which is considerably longer than the average Pol II or III transcript. Pol I transcription elongation is also functionally coupled to processing of the nascent rRNA transcript (Schneider et al., 2007). Ongoing studies continue to identify factors that affect transcription elongation by Pol I and rRNA biosynthesis [reviewed in (Schneider, 2012)].
Ribosome biosynthesis requires that Pol I exhibit efficient transcription initiation, high processivity, and resistance to pauses or arrests. To meet these demands, Pol I has evolved unique enzymatic properties as well as a set of specialized transcription factors and sophisticated organization of the rDNA within the nucleolus.
RNA polymerase I subunit composition
Pol I consists of 14 subunits (Table 1; Figure 1), seven of which are shared with Pols II and/or III. Rpb5, Rpb6, Rpb8, Rpb10 and Rpb12 are common for all three enzymes, whereas AC40 and AC19 are found only in Pols I and III. The remaining seven subunits (A190, A135, A49, A43, A34.5, A14 and A12.2) are specific for Pol I though they share sequence or structural homology with subunits or trans-acting factors for Pols II and III (Cramer et al., 2008; Vannini and Cramer, 2012).
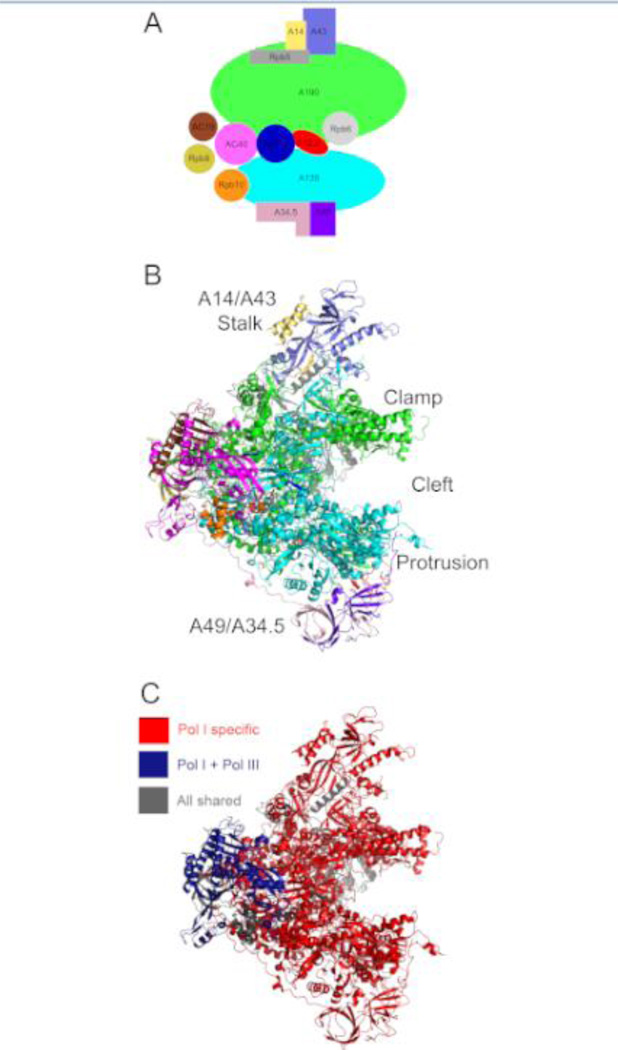
Figure 1. Yeast Pol I complex.
A. Cartoon diagram of the 14 subunit Pol I complex in yeast. B. Crystal structure of yeast RNA polymerase I [4C3H; Fernandez-Tornero et al., 2013). The color code for the 14 subunits is identical to the key in panel A. C. Same as B, except for the color-code: Pol I specific subunits (A190, A135, A49, A43, A34.5, A14, A12.2) are in red; subunits shared between Pols I and III (AC40, AC19) are in blue and subunits common to all three polymerases (Rpb5, Rpb6, Rpb8, Rpb10, Rpb12) are in grey.
Relatively few studies have been devoted to defining the enzymatic properties of Pol I. The overall structure of the polymerase was investigated using EM techniques confirming that the Pol I core is similar to that of Pol II (Klinger et al., 1996; Bischler et al., 1998; Bischler et al., 2002; De Carlo et al., 2003; Kuhn et al., 2007). Most of the distinctions between Pol I and Pol II have been attributed to peripheral surfaces. Thus, for decades there was a general assumption that since multisubunit RNA polymerases are highly conserved, their catalytic features are functionally similar. However, recent structural studies and functional analysis of Pol I uncovered surprising differences between Pols I and II (Engel et al., 2013; Fernandez-Tornero et al., 2013; Viktorovskaya et al., 2013).
The first insights into the functional differences between the most conserved regions of the eukaryotic RNA polymerase paralogs have come from a sequence-based in silico analysis of the two largest subunits from the three eukaryotic RNAPs (Carter and Drouin, 2009b). The variations identified between the polymerases cluster near the active centers and involve regions responsible for the interaction of the polymerase with the RNA-DNA hybrid. These results suggested that RNAPs might differ in their ability to maintain a stable transcription bubble. Consistently, recent analysis using Pol I mutants as well as chimeric Pol II enzymes hosting conserved active center regions of Pol I provided the first experimental evidence of functional divergence in enzymatic properties of Pols I and II (Viktorovskaya et al., 2013). The results of this study suggested that Pols I and II might have evolved different rate-limiting steps during the nucleotide addition cycle. More detailed analysis of the catalytic properties of Pols I, II and III is required to understand molecular mechanisms of the three RNAPs.
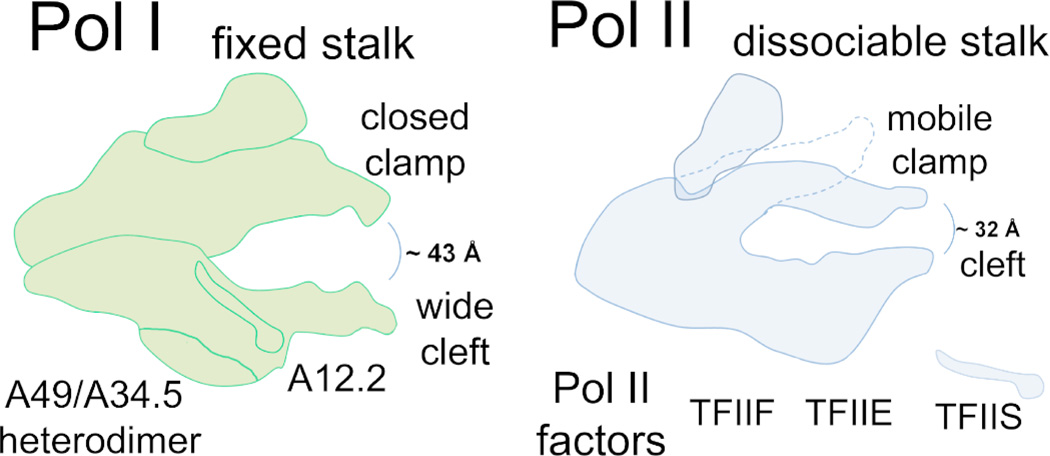
Consistent with the functional studies (Viktorovskaya et al., 2013), recent high resolution structures of the Pol I complex revealed several critical differences in the configuration of the Pol I active center compared to that of Pol II [Figure 2; (Engel et al., 2013; Fernandez-Tornero et al., 2013)]. The active center of the enzyme is located between the two largest Pol I-specific subunits, A190 and A135 which form opposite sides of the central DNA-binding “cleft” (Kuhn et al., 2007). The most unexpected discovery was that the central cleft of Pol I is significantly wider than observed in high resolution structures of other RNA polymerases (Engel et al., 2013; Fernandez-Tornero et al., 2013). An extended loop, unique for the Pol I complex, mimics the DNA backbone in the cleft and might contribute to transcription regulation. Another surprising discovery was that the universally conserved bridge helix domain of the active center was captured in an unwound conformation in the Pol I crystal structure. The bridge helix is predicted to directly affect nucleotide incorporation and polymerase translocation during elongation. Thus, differences in the structure or dynamics of the bridge helices between RNA polymerases would likely result in substantially different catalytic properties. The third major structural feature distinguishing Pol I from the other polymerases was the “closed” confirmation the clamp domain (Figure 2). The clamp is a flexible element which functions in template loading and RNA-DNA hybrid stability. Thus, a divergent position of the clamp might contribute to different properties of Pol I in processivity, elongation complex stability and potentially loading onto the DNA during transcription initiation.
Figure 2. Major structural differences between Pol I and Pol II.
Cartoon diagram of Pol I (green) and Pol II (blue) complexes is shown highlighting several structural divergences between the paralogs, such as the width of the DNA cleft, conformation of the clamp and interactions of the core with the stalk. Pol I specific subunits A12.2, A49 and A34.5 as well as Pol II trans-acting factors TFIIS, TFIIE and TFIIF that share homology with these Pol I subunits (as in Table 1) are indicated.
Taken together, these computational, biochemical and structural findings support the model that Pol I has evolved unique features. Logically, these features likely render the enzyme more efficient for its unique cellular role. Given the critical relationship between ribosome biogenesis and cell proliferation, these findings demonstrate the need for more detailed characterization of Pol I function and its regulation.
The A12.2 core subunit and Pol I’s cleavage activity
Unlike Pol II or prokaryotic RNAPs, Pol I has a “composite active center”. In addition to the two catalytic subunits, the active center of Pol I is also occupied by a domain of the A12.2 subunit (Figure 2). This unique positioning of A12.2 is supported by both crystallographic and cross-link mass-spectrometry studies (Ruan et al., 2011; Jennebach et al., 2012; Engel et al., 2013; Fernandez-Tornero et al., 2013).
A12.2 has two functional domains: the N-terminus of A12.2 is homologous to Pol II’s Rpb9 subunit whereas the C-terminal ribbon domain of A12.2 shows sequence similarity to the Pol II transcription factor TFIIS [Table 1; (Van Mullem et al., 2002)]. The N-terminal zinc binding domain of A12.2 anchors the subunit to the Pol I complex and plays a role in Pol I complex integrity (Nogi et al., 1993; Van Mullem et al., 2002). Consistent with its homology to Rpb9, the N-terminal domain of A12.2 occupies a location within the Pol I complex similar to the known position of Rpb9 in Pol II (Kuhn et al., 2007).
The C-terminal domain of A12.2 is homologous to the part of the transcription factor TFIIS that stimulates RNA cleavage activity of Pol II. In agreement with sequence homology, in vitro studies confirmed that the C-terminus of A12.2 is required for RNA hydrolysis by Pol I (Kuhn et al., 2007). Cross-link mass spectrometry and structural analysis of the Pol I complex further suggested that the C-terminal zinc ribbon of A12.2 reaches into the active site and transiently interacts with the polymerase nucleotide triphosphate entry pore (Ruan et al., 2011; Jennebach et al., 2012). Such positioning of the A12.2 in the active center potentially explains two of the known, distinguishing characteristics of Pol I: insensitivity to α-amanitin and strong nascent RNA hydrolysis activity.
The stable association of A12.2 with Pol I and its positioning in the active center explains why the intrinsic RNA cleavage activity of Pol I is strong, in contrast to the weak cleavage activity of Pol II. RNA cleavage activity of Pol II has been shown to be important for reactivation of an arrested complex and for fidelity (Koyama et al., 2007; Cheung and Cramer, 2011). It is likely that A12.2 contributes directly to pause/arrest avoidance and proofreading by Pol I as well; however, this prediction has not yet been tested.
A12.2 is the only non-essential component of the active center of the enzyme. The absence of A12.2 results in a slow growth and heat-sensitive phenotype (Nogi et al., 1993). The C-terminal zinc ribbon domain of the A12.2 subunit is necessary for stimulation of RNA cleavage activity (Kuhn et al., 2007); however, deletion of this domain did not result in significant growth defects (Van Mullem et al., 2002). These apparently paradoxical observations suggest that additional overlapping features of Pol I or associated factors contribute to stimulation of the RNA hydrolysis activity or to avoidance or clearance of transcriptional arrests in vivo. Alternatively, A12.2 may not influence pause/arrest properties of the enzyme. Perhaps, a primary role for A12.2 is in transcription termination, since the rpa12Δ mutant was shown to result in read-through beyond the transcription termination site on rDNA (Prescott et al., 2004; Braglia et al., 2011; Ruan et al., 2011). Structural and biochemical data suggest that A12.2 is a “built-in” cleavage factor, but the molecular details of its function and its roles in rRNA synthesis remain unknown.
The peripheral subunits of Pol I
The A49, A43, A34.5, and A14 are peripheral subunits of Pol I that are thought to play roles in transcription initiation and elongation. The A43 and A14 subunits form the stalk of the polymerase, involved in transcription initiation, whereas the A49 and A34.5 sub-complex is thought to primarily function as an intrinsic transcription elongation factor [Figure 1; (Kuhn et al., 2007)].
The A14/43 sub-complex
The three eukaryotic RNA polymerases as well as archaeal RNA polymerases share a homologous structure that protrudes from the core enzyme named the stalk [Table 1; Figure 1; reviewed in (Grohmann and Werner, 2011)]. Pol I’s stalk is formed by the A14/A43 heterodimer and is structurally similar to its counterparts in other polymerases (Todone et al., 2001; Bischler et al., 2002; Hu et al., 2002; Peyroche et al., 2002; Meka et al., 2003; Kuhn et al., 2007; Geiger et al., 2008). However, unlike Pol II and archaeal RNA polymerase, Pol I’s stalk is tightly anchored to the polymerase core by large interaction surfaces [Figure 2; (Fernandez-Tornero et al., 2013)]. Whereas Pol II’s stalk is dissociable in vitro and in vivo (Mosley et al., 2013), Pol I’s stalk seems to be a permanent part of the enzyme. Such strong association of the stalk with Pol I core may contribute to efficient transcription initiation rates. Alternatively, perhaps the dissociable stalk of Pol II may serve a greater role in regulation of protein-coding genes.
Early studies revealed that the A43 subunit is essential for cell viability, whereas A14 is dispensable in vivo (Smid et al., 1995; Thuriaux et al., 1995). Either the lack of A14 or a C-terminal truncation of A43 results in temperature – sensitivity. Purification of Pol I from these strains yields transcriptionally inactive enzymes deprived of A14, A43 and ABC23 subunits, suggesting that A14 and A43 contribute to Pol I structural integrity (Smid et al., 1995; Peyroche et al., 2002). When ABC23 is added back to these polymerase preparations, the enzyme lacking A14/A43 is active in non-specific but not in promoter-dependent transcription in vitro (Lanzendorfer et al., 1997; Peyroche et al., 2000; Peyroche et al., 2002). Functional studies demonstrated that the A14/A43 sub-complex is crucial for recruitment of Pol I to the rDNA promoter via direct association with the transcription initiation factor Rrn3 (Peyroche et al., 2000). Interaction of Pol I and Rrn3 via the A43 subunit is essential for the formation of a complex competent for transcription initiation, and this association is a key target for regulation of rRNA synthesis.
The A34.5/A49 sub-complex
The A34.5 and A49 subunits form a stable sub-complex within Pol I that shares similarity to Pol III’s C37/53 heterodimer but does not have a counterpart among Pol II’s subunits [Table 1; (Kuhn et al., 2007)]. Instead, the A34.5/A49 heterodimer structurally resembles the Pol II transcription factors TFIIF and TFIIE [Figure 2; (Geiger et al., 2010)]. X-ray crystallographic studies suggest that the dimerization module of the A34.5/A49 sub-complex is similar to TFIIF whereas the C-terminal domain of A49 may be related to domains of TFIIE. The location of the A34.5/A49 heterodimer on the polymerase (Figure 1) mirrors the location of respective regions of TFIIF and TFIIE on Pol II as shown by cross-link mass spectrometry (Jennebach et al., 2012). The positioning of the A34.5/A49 sub-complex in proximity to the cleft and the clamp also agrees with the earliest EM data (Bischler et al., 2002).
Both A34.5 and A49 are non-essential: deletions of either of the genes encoding these two polypeptides as well as the double mutant are viable but defective in rRNA production (Liljelund et al., 1992; Gadal et al., 1997). The lack of A34.5 subunit does not generate noticeable growth defects or conditional lethality; however, it manifests a moderate caffeine-sensitivity. In the absence of A34.5, there is a slight decrease in A49 stoichiometry within the polymerase, suggesting that A34.5 stabilizes association of the heterodimer with the Pol I core (Gadal et al., 1997; Beckouet et al., 2008).
In contrast to A34.5, deletion of the A49 subunit dramatically impairs cell growth and leads to inviability at lower temperatures (Liljelund et al., 1992). Functional studies of the rpa49Δ strain suggest roles in Pol I transcription initiation and elongation (Beckouet et al., 2008; Albert et al., 2011; Beckouet et al., 2011). Structural insight and biochemical analysis of the A34.5/A49 heterodimer suggest that it primarily acts as a “built-in” transcription elongation and Pol I processivity factor (Huet et al., 1975; Gadal et al., 1997; Kuhn et al., 2007; Geiger et al., 2010); however, roles in promoter opening and transcription initiation were proposed as well (Geiger et al., 2010; Jennebach et al., 2012). The proposed functions of A34.5/A49 in Pol I transcription initiation and elongation are in agreement with structural similarity to TFIIF and TFIIE.
Results of recent structural and cross-link mass-spectrometry studies raise an interesting question (Jennebach et al., 2012; Engel et al., 2013; Fernandez-Tornero et al., 2013): to what extent is the peripheral A34.5/A49 sub-complex involved in Pol I catalytic activities? It has been shown that A49 interacts with A12.2 and contributes to its stimulation of RNA hydrolysis activity (Kuhn et al., 2007; Engel et al., 2013). Furthermore, these “peripheral subunits” might directly affect catalysis since the structural studies found evidence that domains of A34.5/A49 may be mobile on the surface of Pol I with regions potentially reaching the funnel and even the active center of the enzyme (Jennebach et al., 2012). Many interesting questions surrounding the roles for A34.5/A49 in rDNA transcription remain unanswered.
Altogether, structural data suggest that overall architecture of the Pol I complex is evolutionarily related to the Pol II complex in association with TFIIS, TFIIF and TFIIE [Figure 2; (Jennebach et al., 2012; Vannini and Cramer, 2012)]. The Pol I-specific subunits A12.2, A34.5 and A49 have likely evolved from Pol II transcription factors, becoming permanent components of the polymerase (Carter and Drouin, 2010). Functionally, however, much remains to be learned about the detailed contributions of individual subunits to Pol I activity.
Pol I-specific transcription factors
Transcription of the rDNA requires a specific set of transcription factors (Table 2). Unlike Pol II, efficient transcription initiation by yeast Pol I is achieved with the aid of four, relatively simple factors: Rrn3, core factor, TATA-binding protein and upstream activating factor (UAF). Mammalian Pol I requires a similar set of transcription factors, with the exception of UAF, which is identified only in fungi. A number of recent reviews have described each of these factors and their similarity to Pol II and/or III factors in detail (Schneider, 2012; Vannini and Cramer, 2012). However, the one transcription initiation factor that apparently lacks a functional analogue in transcription by Pols II or III is Rrn3.
Table 2.
Yeast RNA polymerase I General Transcription Factors.
| RNA Polymerase | I General Factors | RNA Polymerase II homologs |
References |
|---|---|---|---|
| Rrn3 | monomer | - | Yamamoto et al., 1996 |
| TATA-binding protein (TBP) | monomer | Shared factor | Cormack and Struhl, 1992 |
| Core Factor (CF) | Rrn6, Rrn7, Rrn11 | Related to TFIIB | Keys et al., 1994; Lalo et al., 1996; Lin et al., 1996; Knutson and Hahn, 2011 |
| Upstream Activation Factor (UAF) | Histones H3 and H4, Rrn5, Rrn9, Rrn10 and Uaf30 | Distantly related to SWI/SNF | Keener et al., 1997; Keys et al., 1996; Siddiqi et al., 2001b Liu et al., 2002 |
Four basal transcription factors are required for efficient rRNA synthesis. Those factors are listed with subunit composition, homologous Pol II transcription factors (where applicable) and references to characterization of each factor.
Rrn3 associates with Pol I directly, via the A43 subunit, and it renders the polymerase competent for transcription initiation. Rrn3 forms known contacts with components of the Pol I pre-initiation complex and is thought to aid recruitment of the polymerase to the promoter via these interactions. In this regard, Rrn3 may serve a role similar to sigma factors in bacterial transcription (Grummt, 2003), but different than any described roles for other nuclear transcription factors. Additional support for this analogy is provided by the recent observation that mammalian Rrn3 directly binds promoter DNA, in addition to its known protein:protein interactions (Stepanchick et al., 2013).
The ribosomal DNA locus
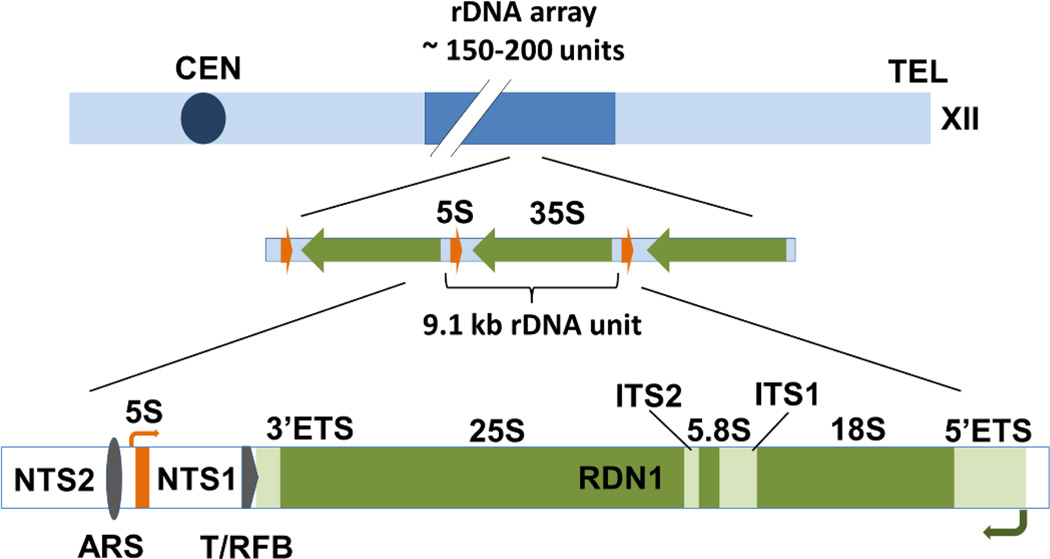
The rDNA loci of eukaryotes are organized in head-to-tail tandem arrays. In yeast the 35S and 5S pre-rRNA genes are located beside one another in a 9,081-bp unit that is repeated approximately 150–200 times to form one large array (Figure 3). This array is located on chromosome XII and is estimated to be approximately 1.4 Mb long, accounting for about 10% of the genome (Schweizer et al., 1969; Kobayashi et al., 1998). The organization of rDNA in most other eukaryotes is similar to that in yeast except that the 5S genes (transcribed by Pol III) are found in distinct arrays apart from the Pol I-transcribed rDNA repeats. Additionally, in higher eukaryotes, the rDNA repeats are located in more than one locus.
Figure 3. Organization of the ribosomal DNA locus (RDN1) in yeast.
Cartoon diagram of the rDNA array with position of the locus relative to the centromere (CEN) and telomere (TEL) of chromosome XII. Head-to-tail tandem arrangement of 9.1 kb repeat, carrying alternating copies of 35S and 5S rDNA is illustrated. The expanded view on bottom shows the positions of 25S, 5.8S, 18S and 5S rRNA regions within a single repeat, as well as the positions of an autonomously replicating sequence (ARS), the 35S terminator and the replication fork barrier (T/RFB) site, the external and internal transcribed (ETS and ITS) and non-transcribed (NTS) sequences.
In wild type yeast, about half of the rDNA repeats is not transcribed and this characteristic is conserved in eukaryotes. EM analysis of rDNA chromatin spreads (Miller spreads) can visualize actively transcribed rDNA units and inactive repeats [observed as gaps between active repeats (French et al., 2003)]. Although inactive rDNA repeats are not essential for regulation of Pol I transcription (French et al., 2003), the presence of the inactive repeats is thought to function in genome stability (Ide et al., 2010).
Concerted evolution of the rDNA and Pol I transcription machinery
One outstanding characteristic of the rDNA is the disparate rates of evolution in different regions of the repeat: from the highly conserved coding region to species-specific non-coding or non-transcribed spacers. Unlike Pol II and Pol III, which initiate transcription from multiple types of promoters, Pol I utilizes a single promoter within each rDNA unit repeated hundreds of times per genome. Due to concerted evolution of the rDNA repeats the promoter sequences are homogenized within any single genome but are highly divergent between taxa (Ganley and Kobayashi, 2007). The molecular coevolution hypothesis predicts that the rate of evolution of promoter-interacting factors correlates with the rate of changes within their target promoter sequences (Arnheim, 1983; Dover and Flavell, 1984). Based on this model, transcription factors that interact with the fast evolving rDNA promoters as well as the regions of Pol I that bind to these factors are expected to have higher rates of evolution compared to Pol II and Pol III transcription machineries. Indeed, a recent study using in silico approaches supports this molecular coevolution hypothesis (Carter and Drouin, 2009a).
The size of the rDNA array
Due to its highly repetitive nature, the total size of the rDNA is not stable; changes in the number of repeats are governed by either homologous recombination or DNA replication (Kobayashi et al., 1998; Houseley and Tollervey, 2011). Recombination between repeats is minimized in cells carrying a deletion of the gene that encodes the replication fork block protein Fob1. Deletion of FOB1 has been exploited to generate a collection of yeast strains with fixed rDNA copies of various numbers (Kobayashi et al., 1998). Among these mutants, there are strains with dramatically reduced rDNA copy number (containing ~25 and ~42 repeats per haploid genomes compared to ~150–200 copies in WT cells). Even when the rDNA copy number is decreased more than four-fold compared to WT, cell growth rate and rRNA synthesis rate are not perturbed (Kobayashi et al., 1998; French et al., 2003). EM analysis of rDNA from strains with reduced copy number revealed an increase in the percentage of actively transcribed repeats (approaching 100%) and increased RNA polymerase density per gene. In fact, in reduced copy strains, there was a polymerase located every 41 nucleotides on average, which is near the upper limit for polymerase packing (French et al., 2003). These data demonstrate that rDNA copy number influences transcription initiation rate at the rDNA, furthermore, the potential to have variation in template copy number between cells is a feature unique to transcription of the rDNA.
Consequences of high RNA polymerase density
Unlike genes transcribed by Pols II or III, the rDNA carries 50–60 densely packed RNA polymerases per transcribed repeat (French et al., 2003). Such polymerase clustering might have interesting consequences on rRNA synthesis. Gadal and colleagues proposed that high Pol I density might help efficient transcription elongation (Albert et al., 2011). Molecular modeling, EM analysis and co-immunoprecipitation assays suggested that the A43 subunit of the leading polymerase contacts the A49 subunit of the trailing elongation complex, resulting in a concerted “push” forward (Beckouet et al., 2008; Albert et al., 2011). Genetic support for this model was drawn from studies using the rpa49Δ mutation in the high- and low-rDNA copy number backgrounds. These data demonstrated that defects caused by the absence of the A49 subunit are partially suppressed by the high polymerase density in the reduced rDNA copy number strains.
If the trailing polymerase “pushes” the leading elongation complex, then selective pressure on the translocation step of the nucleotide addition cycle would be reduced in Pol I compared to paralogous enzymes that typically encounter fewer polymerases per gene. Another consequence of high polymerase density would be increased sensitivity to intrinsic pausing. If a polymerase on the rDNA pauses, there will be a nearly instantaneous pileup of polymerases behind that enzyme (much like cars on a crowded highway). To avoid these “traffic jams”, it is reasonable to hypothesize that Pol I might have evolved catalytic properties that render the enzyme less sensitive to pausing. Genetic data suggest that Pol I may indeed have evolved such properties (Viktorovskaya et al., 2013); however, detailed studies devoted to describing the mechanisms of Pol I catalysis and its comparison to other RNA polymerases are required to directly test these hypotheses.
Functional coupling of rRNA processing to transcription elongation by Pol I
Ribosome biogenesis starts with rRNA synthesis by Pol I in the nucleolus. The Pol I –derived 35S pre-rRNA is processed via digestion, folding and modifications. Co- and post-transcriptional association of the rRNA molecules with ribosomal and non-ribosomal proteins results in formation of the small and large ribosome subunit precursors. The precursors of ribosomal subunits are then exported from the nucleus into the cytoplasm where they undergo final steps of maturation to form functional 40S and 60S particles. The process of ribosome assembly is complex, organized and controlled [reviewed in (Fatica and Tollervey, 2002)].
For many years it was believed that yeast rRNA maturation occurs post-transcriptionally; however, recent studies showed that early steps of rRNA processing and ribosome subunit folding begin on the nascent pre-rRNA transcript (Osheim et al., 2004; Kos and Tollervey, 2010). To test whether rRNA processing efficiency was influenced directly by Pol I transcription elongation rate, mutations in Pol I that reduce the enzyme’s transcription elongation rate were isolated, and rRNA processing was measured in those cells. Indeed, defects in transcription elongation resulted in defects in multiple steps in pre-rRNA processing (Schneider et al., 2007). Together, all of these data demonstrate that transcription by Pol I and rRNA processing are carefully orchestrated events, and the factors that organize these activities remain poorly understood.
The relationship between rRNA processing and Pol I transcription elongation is unique in its specific cellular localization and in the complexity of the assembly process occurring on the nascent transcript; however, modulation of transcription elongation by Pol II or bacterial RNAP is also known to influence the assembly of nascent RNP complexes [reviewed in (Perales and Bentley, 2009)]. The rate of transcription elongation, especially pausing, is proposed to affect the folding pathway of the nascent transcript and co-transcriptional recruitment of the trans-acting factors to RNA [reviewed in (Pan and Sosnick, 2006)]. Thus, coupling transcription to metabolism of the RNA product appears to be a general theme among RNA polymerases.
Chromatin organization of the rDNA
DNA-dependent polymerases have to elongate through chromatin; thus, the unique epigenetic state of the rDNA might have contributed to evolution of Pol I properties. Each of the rRNA genes contains an identical subset of cis-acting elements and can be transcribed by Pol I. However, only approximately half of the rDNA units is transcribed in growing yeast cells. Inactive rDNA repeats have nucleosomes packed in a “beads-on-a-string” structure with a density of approximately 35–40 nucleosomes per gene as estimated by EM (Dammann et al., 1993; Merz et al., 2008; Johnson et al., 2013). This “closed” heterochromatin structure is predicted to inhibit Pol I transcription (Rickards et al., 2007; Birch et al., 2009).
There is more controversy concerning the chromatin structure of the actively transcribed units. As mentioned above, active repeats are densely packed with Pol I transcription elongation complexes (French et al., 2003). Using psoralen-crosslinking or chromatin endogenous cleavage (ChEC) assays, it has been shown that actively transcribed repeats are devoid of nucleosomes in the coding region (Dammann et al., 1993; Merz et al., 2008). EM analysis further confirms that visible portions of the coding region free from polymerases appear “smooth” and nucleosome-free (Johnson et al., 2013). Instead, it has been proposed that such “open” chromatin is organized in a specialized structure containing Hmo1 (Merz et al., 2008). Hmo1 is a non-essential protein that associates with the rDNA promoter and coding regions facilitating transcription by Pol I (Gadal et al., 2002; Hall et al., 2006; Kasahara et al., 2007).
On the other hand, ChIP assays using the low-copy-number strains containing only active rDNA repeats suggested that histones remain associated with the rDNA (Jones et al., 2007). For that reason, the “open” rRNA genes were proposed to have “unphased” or dynamic nucleosome structures. Though it is clear that canonical nucleosomes are absent on the active rDNA (Johnson et al., 2013), histones might still be present forming alternative structures or sub-nucleosomal particles in the “open” chromatin [as suggested in (Jones et al., 2007; Albert et al., 2012; Chen et al., 2012)]. Consistent with this model, multiple chromatin remodeling factors or histone chaperones are suggested to affect Pol I transcription in yeast: Rpd3, Spt16/FACT, SWI/SNF complex, Asf1/Rtt109, Spt6, Chd1, Isw1 and Isw2 (Oakes et al., 2006; Jones et al., 2007; Beckouet et al., 2011; Chen et al., 2012; Johnson et al., 2013; Zhang et al., 2013). Though the roles of these factors are not exclusive to the rDNA, their effects were shown to be rDNA-specific in several cases (Oakes et al., 2006; Chen et al., 2012; Johnson et al., 2013).
Potential benefits of characterization of Pol I transcription
Studies from many labs using multiple experimental approaches have shown that Pol I and related factors have evolved specialized properties to suit their unique cellular role. Understanding the selective pressures that resulted in the evolution of three nuclear transcription machineries is a topic of fundamental biological significance. However, there are also practical reasons to pursue intensive characterization of all three nuclear polymerases, including Pol I. Selective inhibition of individual RNA polymerases appears to have important clinical benefits (Drygin et al., 2010).
The connection between ribosome biosynthesis and cell growth rate is well known in all living cells. Recently, however, a number of labs have begun to exploit this connection, with the goal of developing selective inhibitors of Pol I for use in anti-cancer chemotherapy. In the previous three years, multiple inhibitors of Pol I transcription have been isolated (Drygin et al., 2011; Andrews et al., 2013; Peltonen et al., 2014; Rothblum et al., 2014). Preclinical animal studies demonstrate therapeutic efficacy of Pol I inhibition with limiting toxicity (Bywater et al., 2012; Peltonen et al., 2014), and one Pol I-specific inhibitor, CX-5461, is currently being tested in phase I clinical trials. The future for therapeutic regulation of transcription by Pol I appears bright. To support this progress, continued investigation of transcription by Pol I and related factors is imperative.
This review describes critical differences between RNA polymerases I and II. Biochemical and structural data demonstrate significant divergence in enzymatic properties between these paralogs, despite high conservation. Key differences in target genes, catalytic features and related transcription factors are highlighted with a focus on the biologically distinct roles for the polymerases. Potential practical applications of defining and targeting unique characteristics of RNA polymerase I are described.
Acknowledgements
We thank members of the Pol I research community for their ongoing efforts in characterizing this key cellular activity. We also apologize to the many investigators whose research was not described herein. For comparison of structure and biochemical functions, most of this review focused on the yeast system, but this focus was not intended to detract from the excellent ongoing studies in higher eukaryotes. This work was supported by NIH grant #GM084946 to D.A.S.
Abbreviations
- Pol
RNA polymerase
- rDNA
ribosomal DNA
- rRNA
ribosomal RNA
- CF
core factor
- UAF
upstream activating factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Albert B, Leger-Silvestre I, Normand C, Ostermaier MK, Perez-Fernandez J, Panov KI, Zomerdijk JC, Schultz P, Gadal O. RNA polymerase I-specific subunits promote polymerase clustering to enhance the rRNA gene transcription cycle. J Cell Biol. 2011;192:277–293. doi: 10.1083/jcb.201006040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert B, Perez-Fernandez J, Leger-Silvestre I, Gadal O. Regulation of ribosomal RNA production by RNA polymerase I: does elongation come first? Genet Res Int. 2012;2012:276–948. doi: 10.1155/2012/276948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews WJ, Panova T, Normand C, Gadal O, Tikhonova IG, Panov KI. Old drug, new target: ellipticines selectively inhibit RNA polymerase I transcription. J Biol Chem. 2013;288:4567–4582. doi: 10.1074/jbc.M112.411611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheim N. Concerted evolution of multigene families. In: Koehn R, Nei M, editors. Evolution of Genes and Proteins. Sinauer, Sunderland, MA: 1983. pp. 38–61. [Google Scholar]
- Beckouet F, Labarre-Mariotte S, Albert B, Imazawa Y, Werner M, Gadal O, Nogi Y, Thuriaux P. Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription. Mol Cell Biol. 2008;28:1596–1605. doi: 10.1128/MCB.01464-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckouet F, Mariotte-Labarre S, Peyroche G, Nogi Y, Thuriaux P. Rpa43 and its partners in the yeast RNA polymerase I transcription complex. FEBS Lett. 2011;585:3355–3359. doi: 10.1016/j.febslet.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Birch JL, Tan BC, Panov KI, Panova TB, Andersen JS, Owen-Hughes TA, Russell J, Lee SC, Zomerdijk JC. FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 2009;28:854–865. doi: 10.1038/emboj.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischler N, Balavoine F, Milkereit P, Tschochner H, Mioskowski C, Schultz P. Specific interaction and two-dimensional crystallization of histidine tagged yeast RNA polymerase I on nickel-chelating lipids. Biophys J. 1998;74:1522–1532. doi: 10.1016/S0006-3495(98)77864-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischler N, Brino L, Carles C, Riva M, Tschochner H, Mallouh V, Schultz P. Localization of the yeast RNA polymerase I-specific subunits. EMBO J. 2002;21:4136–4144. doi: 10.1093/emboj/cdf392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braglia P, Kawauchi J, Proudfoot NJ. Co-transcriptional RNA cleavage provides a failsafe termination mechanism for yeast RNA polymerase I. Nucleic Acids Res. 2011;39:1439–1448. doi: 10.1093/nar/gkq894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, Huser N, Proffitt C, Bliesath J, Haddach M, Schwaebe MK, Ryckman DM, Rice WG, Schmitt C, Lowe SW, Johnstone RW, Pearson RB, McArthur GA, Hannan RD. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell. 2012;22:51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter R, Drouin G. The evolutionary rates of eukaryotic RNA polymerases and of their transcription factors are affected by the level of concerted evolution of the genes they transcribe. Mol Biol Evol. 2009a;26:2515–2520. doi: 10.1093/molbev/msp164. [DOI] [PubMed] [Google Scholar]
- Carter R, Drouin G. Structural differentiation of the three eukaryotic RNA polymerases. Genomics. 2009b;94:388–396. doi: 10.1016/j.ygeno.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Carter R, Drouin G. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol Biol Evol. 2010;27:1035–1043. doi: 10.1093/molbev/msp316. [DOI] [PubMed] [Google Scholar]
- Chen H, Fan M, Pfeffer LM, Laribee RN. The histone H3 lysine 56 acetylation pathway is regulated by target of rapamycin (TOR) signaling and functions directly in ribosomal RNA biogenesis. Nucleic Acids Res. 2012;40:6534–6546. doi: 10.1093/nar/gks345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- Cramer P. Multisubunit RNA polymerases. Curr Opin Struct Biol. 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, Jawhari A, Jennebach S, Kamenski T, Kettenberger H, Kuhn CD, Lehmann E, Leike K, Sydow JF, Vannini A. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carlo S, Carles C, Riva M, Schultz P. Cryo-negative staining reveals conformational flexibility within yeast RNA polymerase I. J Mol Biol. 2003;329:891–902. doi: 10.1016/s0022-2836(03)00510-2. [DOI] [PubMed] [Google Scholar]
- Dover GA, Flavell RB. Molecular coevolution: DNA divergence and the maintenance of function. Cell. 1984;38:622–623. doi: 10.1016/0092-8674(84)90255-1. [DOI] [PubMed] [Google Scholar]
- Drygin D, Lin A, Bliesath J, Ho CB, O'Brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A, Streiner N, Quin JE, Sanij E, Bywater MJ, Hannan RD, Ryckman D, Anderes K, Rice WG. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011;71:1418–1430. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. RNA polymerase I structure and transcription regulation. Nature. 2013;502:650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- Fernandez-Tornero C, Moreno-Morcillo M, Rashid UJ, Taylor NM, Ruiz FM, Gruene T, Legrand P, Steuerwald U, Muller CW. Crystal structure of the 14-subunit RNA polymerase I. Nature. 2013;502:644–649. doi: 10.1038/nature12636. [DOI] [PubMed] [Google Scholar]
- French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Labarre S, Boschiero C, Thuriaux P. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 2002;21:5498–5507. doi: 10.1093/emboj/cdf539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Mariotte-Labarre S, Chedin S, Quemeneur E, Carles C, Sentenac A, Thuriaux P. A34.5, a nonessential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machine. Mol Cell Biol. 1997;17:1787–1795. doi: 10.1128/mcb.17.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley AR, Kobayashi T. Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 2007;17:184–191. doi: 10.1101/gr.5457707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger SR, Kuhn CD, Leidig C, Renkawitz J, Cramer P. Crystallization of RNA polymerase I subcomplex A14/A43 by iterative prediction, probing and removal of flexible regions. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:413–418. doi: 10.1107/S174430910800972X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger SR, Lorenzen K, Schreieck A, Hanecker P, Kostrewa D, Heck AJ, Cramer P. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol Cell. 2010;39:583–594. doi: 10.1016/j.molcel.2010.07.028. [DOI] [PubMed] [Google Scholar]
- Grohmann D, Werner F. Cycling through transcription with the RNA polymerase F/E (RPB4/7) complex: structure, function and evolution of archaeal RNA polymerase. Res Microbiol. 2011;162:10–18. doi: 10.1016/j.resmic.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- Hall DB, Wade JT, Struhl K. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:3672–3679. doi: 10.1128/MCB.26.9.3672-3679.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. Repeat expansion in the budding yeast ribosomal DNA can occur independently of the canonical homologous recombination machinery. Nucleic Acids Res. 2011;39:8778–8791. doi: 10.1093/nar/gkr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Wu S, Sun Y, Yuan CC, Kobayashi R, Myers MP, Hernandez N. Characterization of human RNA polymerase III identifies orthologues for Saccharomyces cerevisiae RNA polymerase III subunits. Mol Cell Biol. 2002;22:8044–8055. doi: 10.1128/MCB.22.22.8044-8055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J, Buhler JM, Sentenac A, Fromageot P. Dissociation of two polypeptide chains from yeast RNA polymerase A. Proc Natl Acad Sci U S A. 1975;72:3034–3038. doi: 10.1073/pnas.72.8.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide S, Miyazaki T, Maki H, Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010;327:693–696. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- Jennebach S, Herzog F, Aebersold R, Cramer P. Crosslinking-MS analysis reveals RNA polymerase I domain architecture and basis of rRNA cleavage. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, French SL, Osheim YN, Li M, Hall L, Beyer AL, Smith JS. Rpd3- and spt16-mediated nucleosome assembly and transcriptional regulation on yeast ribosomal DNA genes. Mol Cell Biol. 2013;33:2748–2759. doi: 10.1128/MCB.00112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HS, Kawauchi J, Braglia P, Alen CM, Kent NA, Proudfoot NJ. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat Struct Mol Biol. 2007;14:123–130. doi: 10.1038/nsmb1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara K, Ohtsuki K, Ki S, Aoyama K, Takahashi H, Kobayashi T, Shirahige K, Kokubo T. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:6686–6705. doi: 10.1128/MCB.00876-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kief DR, Warner JR. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger C, Huet J, Song D, Petersen G, Riva M, Bautz EK, Sentenac A, Oudet P, Schultz P. Localization of yeast RNA polymerase I core subunits by immunoelectron microscopy. EMBO J. 1996;15:4643–4653. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama H, Ito T, Nakanishi T, Sekimizu K. Stimulation of RNA polymerase II transcript cleavage activity contributes to maintain transcriptional fidelity in yeast. Genes Cells. 2007;12:547–559. doi: 10.1111/j.1365-2443.2007.01072.x. [DOI] [PubMed] [Google Scholar]
- Kuhn CD, Geiger SR, Baumli S, Gartmann M, Gerber J, Jennebach S, Mielke T, Tschochner H, Beckmann R, Cramer P. Functional architecture of RNA polymerase I. Cell. 2007;131:1260–1272. doi: 10.1016/j.cell.2007.10.051. [DOI] [PubMed] [Google Scholar]
- Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzendorfer M, Smid A, Klinger C, Schultz P, Sentenac A, Carles C, Riva M. A shared subunit belongs to the eukaryotic core RNA polymerase. Genes Dev. 1997;11:1037–1047. doi: 10.1101/gad.11.8.1037. [DOI] [PubMed] [Google Scholar]
- Liljelund P, Mariotte S, Buhler JM, Sentenac A. Characterization and mutagenesis of the gene encoding the A49 subunit of RNA polymerase A in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992;89:9302–9305. doi: 10.1073/pnas.89.19.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meka H, Daoust G, Arnvig KB, Werner F, Brick P, Onesti S. Structural and functional homology between the RNAP(I) subunits A14/A43 and the archaeal RNAP subunits E/F. Nucleic Acids Res. 2003;31:4391–4400. doi: 10.1093/nar/gkg652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz K, Hondele M, Goetze H, Gmelch K, Stoeckl U, Griesenbeck J. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 2008;22:1190–1204. doi: 10.1101/gad.466908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley AL, Hunter GO, Sardiu ME, Smolle M, Workman JL, Florens L, Washburn MP. Quantitative proteomics demonstrates that the RNA polymerase II subunits Rpb4 and Rpb7 dissociate during transcriptional elongation. Mol Cell Proteomics. 2013;12:1530–1538. doi: 10.1074/mcp.M112.024034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y, Yano R, Dodd J, Carles C, Nomura M. Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol Cell Biol. 1993;13:114–122. doi: 10.1128/mcb.13.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes ML, Siddiqi I, French SL, Vu L, Sato M, Aris JP, Beyer AL, Nomura M. Role of histone deacetylase Rpd3 in regulating rRNA gene transcription and nucleolar structure in yeast. Mol Cell Biol. 2006;26:3889–3901. doi: 10.1128/MCB.26.10.3889-3901.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- Peltonen K, Colis L, Liu H, Trivedi R, Moubarek MS, Moore HM, Bai B, Rudek MA, Bieberich CJ, Laiho M. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell. 2014;25:77–90. doi: 10.1016/j.ccr.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D. "Cotranscriptionality": the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche G, Levillain E, Siaut M, Callebaut I, Schultz P, Sentenac A, Riva M, Carles C. The A14-A43 heterodimer subunit in yeast RNA pol I and their relationship to Rpb4-Rpb7 pol II subunits. Proc Natl Acad Sci U S A. 2002;99:14670–14675. doi: 10.1073/pnas.232580799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 2000;19:5473–5482. doi: 10.1093/emboj/19.20.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott EM, Osheim YN, Jones HS, Alen CM, Roan JG, Reeder RH, Beyer AL, Proudfoot NJ. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc Natl Acad Sci U S A. 2004;101:6068–6073. doi: 10.1073/pnas.0401393101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder RH, Lang WH. Terminating transcription in eukaryotes: lessons learned from RNA polymerase I. Trends Biochem Sci. 1997;22:473–477. doi: 10.1016/s0968-0004(97)01133-x. [DOI] [PubMed] [Google Scholar]
- Rickards B, Flint SJ, Cole MD, LeRoy G. Nucleolin is required for RNA polymerase I transcription in vivo. Mol Cell Biol. 2007;27:937–948. doi: 10.1128/MCB.01584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothblum K, Hu Q, Penrod Y, Rothblum LI. Selective Inhibition of rDNA Transcription by a Small-Molecule Peptide that Targets the Interface Between RNA Polymerase I and Rrn3. Mol Cancer Res. 2014 doi: 10.1158/1541-7786.MCR-14-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan W, Lehmann E, Thomm M, Kostrewa D, Cramer P. Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J Biol Chem. 2011;286:18701–18707. doi: 10.1074/jbc.M111.222273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA. RNA polymerase I activity is regulated at multiple steps in the transcription cycle: recent insights into factors that influence transcription elongation. Gene. 2012;493:176–184. doi: 10.1016/j.gene.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, Michel A, Sikes ML, Vu L, Dodd JA, Salgia S, Osheim YN, Beyer AL, Nomura M. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol Cell. 2007;26:217–229. doi: 10.1016/j.molcel.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer E, MacKechnie C, Halvorson HO. The redundancy of ribosomal and transfer RNA genes in Saccharomyces cerevisiae. J Mol Biol. 1969;40:261–277. doi: 10.1016/0022-2836(69)90474-4. [DOI] [PubMed] [Google Scholar]
- Smid A, Riva M, Bouet F, Sentenac A, Carles C. The association of three subunits with yeast RNA polymerase is stabilized by A14. J Biol Chem. 1995;270:13534–13540. doi: 10.1074/jbc.270.22.13534. [DOI] [PubMed] [Google Scholar]
- Stepanchick A, Zhi H, Cavanaugh AH, Rothblum K, Schneider DA, Rothblum LI. DNA binding by the ribosomal DNA transcription factor rrn3 is essential for ribosomal DNA transcription. J Biol Chem. 2013;288:9135–9144. doi: 10.1074/jbc.M112.444265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuriaux P, Mariotte S, Buhler JM, Sentenac A, Vu L, Lee BS, Nomura M. Gene RPA43 in Saccharomyces cerevisiae encodes an essential subunit of RNA polymerase I. J Biol Chem. 1995;270:24252–24257. doi: 10.1074/jbc.270.41.24252. [DOI] [PubMed] [Google Scholar]
- Todone F, Brick P, Werner F, Weinzierl RO, Onesti S. Structure of an archaeal homolog of the eukaryotic RNA polymerase II RPB4/RPB7 complex. Mol Cell. 2001;8:1137–1143. doi: 10.1016/s1097-2765(01)00379-3. [DOI] [PubMed] [Google Scholar]
- Van Mullem V, Landrieux E, Vandenhaute J, Thuriaux P. Rpa12p, a conserved RNA polymerase I subunit with two functional domains. Mol Microbiol. 2002;43:1105–1113. doi: 10.1046/j.1365-2958.2002.02824.x. [DOI] [PubMed] [Google Scholar]
- Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- Viktorovskaya OV, Engel KL, French SL, Cui P, Vandeventer PJ, Pavlovic E, Beyer AL, Kaplan CD, Schneider DA. Divergent contributions of conserved active site residues to transcription by eukaryotic RNA polymerases I and II. Cell Reports. 2013;4:974–984. doi: 10.1016/j.celrep.2013.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron C, Lacroute F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol. 1975;122:855–865. doi: 10.1128/jb.122.3.855-865.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Anderson SJ, French SL, Sikes ML, Viktorovskaya OV, Huband J, Holcomb K, Hartman JLt, Beyer AL, Schneider DA. The SWI/SNF chromatin remodeling complex influences transcription by RNA polymerase I in Saccharomyces cerevisiae. PLoS One. 2013;8:e56793. doi: 10.1371/journal.pone.0056793. [DOI] [PMC free article] [PubMed] [Google Scholar]