Abstract
Mod5 is the yeast tRNA isopentenyl transferase, an enzyme that is conserved from bacteria to humans. Mod5 is primarily cytoplasmic where it modifies the A37 position of a few tRNAs, and the yeast enzyme has been shown capable of forming heritable, amyloid-like aggregates that confer a selective advantage in the presence of specific antifungal agents. A subpopulation of Mod5 is also found associated with nuclear tRNA genes, where it contributes tRNA-gene mediated (tgm) silencing of local transcription by RNA polymerase II. The tgm-silencing function of Mod5 has been observed in yeast and a Mod5-deletion in yeast can be complemented by the plant and human tRNA isopentenyl transferases, but not the bacterial enzymes, possibly due to the lack of an extended C-terminal domain found in eukaryotes. In light of this additional nuclear role for Mod5 we discuss the proposed role of the human homologue of Mod5, TRIT1, as a tumor suppressor protein.
Mod5 has distinct functions in the nucleus and cytoplasm
Transfer RNA (tRNA) molecules undergo numerous post-transcriptional modifications, with greater than 15% of all the nucleotides of nuclear-encoded tRNAs in S. cerevisiae having covalent modifications (Phizicky and Hopper, 2010). The effects of these modifications include changes in tRNA stability, translation effeciency, and translation fidelity (Urbonavicius et al., 2001; Phizicky and Hopper, 2010). Transfer RNA modifications are prevalent around the anticodon loop, particularly at nucleotide positions A34 and A37. In all domains of life, the A37 nucleotide modification of some tRNAs is conserved and suggests an important evolutionary purpose for these modifications (Phizicky and Hopper, 2010; El Yacoubi et al., 2012). One such modification is the conversion of A37 to N6-(isopentenyl) adenosine (i6A37) by a tRNA isopentenyl transferase. The enzyme responsible for this activity in S. cerevesiae is Mod5, which transfers an isopentenyl group from dimethylallyl pyrophosphate (DMAPP) to N6 of A37 on a small subset of tRNAs (Dihanich et al., 1987).
The MOD5 gene is non-essential, but highly conserved and the tRNA-modifying function of its protein product is conserved in bacteria (Soderberg and Poulter, 2000), yeast (Dihanich et al., 1987; Lamichhane et al., 2011), worm (Lemieux et al., 2001), plant (Golovko et al., 2002), and human (Golovko et al., 2000). There are two highly conserved domains of Mod5 which are responsible for substrate binding (ATP/GTP and DMAPP) (Figure 1) (Soderberg and Poulter, 2000; Zhou and Huang, 2008). Structurally, the bacterial and eukaryotic tRNA isopentenyl transferases are similar in that they both contain a large core domian and insertion domain which interact to form a channel where the anticodon stem loop of the bound tRNA resides (Soderberg and Poulter, 2000; Zhou and Huang, 2008).
Figure 1.

The tRNA isopentenyl transferase is mostly conserved from bacteria to humans, however eukaryotes have evolved an additional C-terminal tail not present in E. coli. The ATP/GTP-domain and DMAPP-binding domain are conserved in all four species. The amyloid, prion-like domain required for Mod5-aggregation (Suzuki et al., 2012) is partially conserved between yeast and humans and suggests potential for human TRIT1 aggregation capabilities. A zinc-finger binding domain in the c-terminal tail is conserved among the eukaryotes and not present in bacteria (Soderberg and Poulter, 2000; Zhou and Huang, 2008).
The bacterial and eukaryotic homologs of this enzyme have been conserved in their tRNA modification activity (Dihanich et al., 1987; Golovko et al., 2000; Soderberg and Poulter, 2000; Golovko et al., 2002), but the eukaryotic versions also have additional capabilities. A major difference between the bacterial and eukaryotic tRNA isopentenyl transferase sequences is that the eukaryotic proteins contain a ~100 residue zinc finger-containing, C-terminus that is absent from the bacterial enzymes (Golovko et al., 2000; Soderberg and Poulter, 2000; Lemieux et al., 2001; Golovko et al., 2002; Zhou and Huang, 2008) (Figure 1). The exact function of this domain is not currently clear, but it has been proposed by Zhou and Huang (2008) to function in protecting the insertion domain from degradation and/or increasing the number of contact points that the protein makes with the tRNA as means to increase the specificity for tRNA-binding (Zhou and Huang, 2008). An increase in discrimination in binding for a tRNA modifying enzyme is in accordance with increased complexity of eukaryotic proteomes, which likely require a greater stringency during translation. Alternatively, the observation that bacterial enzymes do not contain this C-terminus but are still able to effectively modify tRNAs may suggest that the C-terminus has evolved a function for the eukaryotic enzymes in addition to the tRNA-modifying function. Interestingly, the C-terminus contains a bipartite nuclear localization signal (NLS), which directs a pool of Mod5 to the nucleus, specifically the nucleolus (Tolerico et al., 1999). The reason for this nucleolar localization was not originally clear, since it is thought this tRNA modification only occurs in the cytoplasm.
The MOD5 gene encodes two in-frame open reading frames (ORFs), each translated from a separate AUG sites at amino acids positions 1 and 12, respectively. Mod5 protein translated from the first AUG site localizes to the mitochondria and cytoplasm (Mod5p-I), whereas Mod5 translated from the second AUG site localizes to the nucleus and cytoplasm (Mod5p-II) (Boguta et al., 1994; Tolerico et al., 1999). The cytoplasmic and mitochondrial Mod5 pools modify distinct tRNA populations: nuclear-encoded tRNAs are modified by the cytoplasmic pool and mitochondrial-encoded tRNAs are modified by the isoform located in the mitochondrial. Interestingly, cytoplasmic Mod5 activity connects sterol biosynthesis and tRNA modification as these two pathways share a common intermediate – dimethyallylpyrophosphate (DMAPP) (Benko et al., 2000) (Figure 2). Thus, decreases in tRNA modification by Mod5 result in increased sterol production via the mevalonate pathway and vice versa (Benko et al., 2000). The human OD5 genecounterpart, TRIT1, also encodes proteins distributed to the cytoplasm and the mitochondria and a mutation of Arg323 to Gln323 in TRIT1 has been shown to cause a defect in tRNA modification resulting in encephalopathy and myoclonic epilepsy due to defects in mitochondrial protein synthesis and respiration (Yarham et al., 2014). Because i6A tRNA modification in yeast occurs in the cytoplasm and mitochondria, it was surprising to learn that a portion of the Mod5 pool is also localized to the nucleus, specifically the nucleolus (12). However, recent studies showed that this nucleolar pool is involved in a novel function - antagonizing local transcription by RNA polymerase II, termed tRNA gene-mediated (tgm) silencing (Boguta et al., 1994; Pratt-Hyatt et al., 2013).
Figure 2.
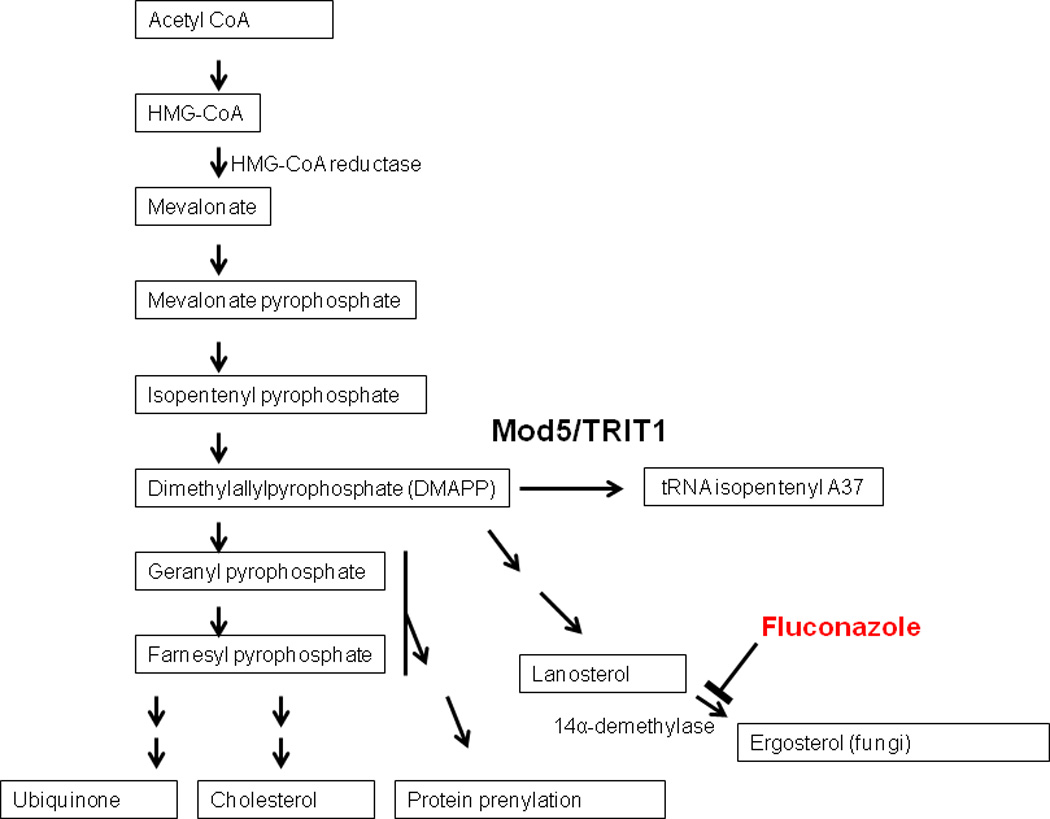
Mod5/TRIT1 compete for DMAPP substrates with essential pathways. Aggregated Mod5 loses its ability to modify tRNAs and thus yeast that contain aggregated-Mod5 draw less DMAPP out the ergosterol pathway and are better able to overcome fluconazole treatment (Benko et al., 2000; Suzuki et al., 2012).
The nuclear function of Mod5 is distinct from its tRNA-isopentenylation activity
Tgm-silencing is a form of transcriptional silencing originally observed in yeast. It was found that RNA polymerase II (pol II) transcription adjacent (within ~500bp) to actively transcribed tRNA genes is antagonized to varied extents, depending on the pol II transcription unit (Hull et al., 1994). This form of silencing occurs outside of the range of steric interference and is independent of the orientation of the tRNA gene (Wang et al., 2005). Clustering of the tRNAs in the nucleolus is essential for tgm-silencing, illustrating the importance of spatial organization of the tRNA genes and chromatin structure (Thompson et al., 2003; Haeusler et al., 2008). Due to Mod5’s involvement in tRNA biogenesis, it was included in a genetic screen for yeast proteins that affect tgm-silencing (Kendall et al., 2000; Wang et al., 2005). Mod5 was shown to be required for tgm-mediated silencing, though not for nucleolar localization of the tRNA genes (14). Chromatin immunoprecipitation experiments demonstrated Mod5 directly associated with tRNA gene transcription complexes, which was reinforced by the result that Mod5 co-immunoprecipitated with the pol III transcription machinery subunits, Brf1, Bdp1, Smc4, Rpc53, Rpc52, and Tfc1. Finally, RNA-immunoprecipitation experiments showed Mod5 bound to pre-tRNAs. Interestingly, Mod5 was found bound to pre-tRNAs whether or not they were substrates for isopentenylation, consistent with the observation that tRNA genes encoding Mod5-substrates and non-substrates both could silence an adjacent polII promoter (Pratt-Hyatt et al., 2013) These data suggest that the substrate specificity for tRNA modification in the cytoplasm is distinct from the nuclear role of Mod5 in silencing. Consistent with this, depletion of DMAPP substrate required for tRNA isopentenylation, does not affect tgm-silencing, suggesting that the tRNA isopentenylation acitivity of Mod5 is not required for silencing (Pratt-Hyatt et al., 2013). Based on the observation that truncation of the pre-tRNA transcript substantially reduces local tgm-silencing (Pratt-Hyatt et al., 2013), it seems likely that Mod5 binding to nascent pre-tRNA via recognition of the pre-tRNA structure is required for full effect of the silencing mechanism.
One model for the role of Mod5 in tgm silencing is that the formation of a Mod5-pre-tRNA complex at the tRNA gene might recruit chromatin remodeling proteins as a mechanism to impose silencing on adjacent promoters. The involvement of nucleosome surfaces and several chromatin modifying and remodeling enzymes in tgm silencing has been documented, though how the tRNA gene communicates physically with the chromatin is not yet understood (Good et al., 2013). Mutations affecting nucleosome covalent modification and remodeling include those in the RSC nucleosome remodeling complex subunits, Rpd3 deacetylase subunits, Hos1 deacetylase, and the Glc7 phosphatase (Good et al., 2013). Taken together with the association of Mod5 with pre-tRNAs and the transcription complexes, we propose that the Mod5-pre-tRNA complex might help recruit one or more of the chromatin-modifying enzymes that exert a repressive effect on nearby promoters through nucleosome rearrangement. In support of this, tandem affinity tagged Mod5 coimmunoprecipitates with multiple chromatin structure remolding complexes (RSC) and histones (Pratt-Hyatt et al., 2013). It is currently not clear how Mod5 is recruited to the tRNA gene initially, and no direct physical link between Mod5 and chromatin-modifying complexes has yet been demonstrated.
Biological implications of Mod5 protein misfolding
Recently, studies by Tanaka and colleagues have demonstrated an interesting tendency of overexpressed yeastMod5-GFP, to misfold into amyloid aggregates that have prion-like heritability that results in resistance to a fungicide, fluconazole (Figure 2). (Suzuki et al., 2012; Suzuki and Tanaka, 2012; Sugiyama and Tanaka, 2014). We have confirmed these results using a chromosomally-integrated, Mod5-GFP in which its expression is driven off the endogenous Mod5 promoter. Following fluconazole-selection, we observe large cytoplasmic Mod-GFP aggregates and diffuse nuclear signal (Figure 3A, right panels), while in untreated cells there is diffuse cytoplasmic and nuclear patterning (Figure 3A, left panels). Fluconazole inhibits an essential enzyme required for ergosterol synthesis in yeast, and at sufficient concentrations is lethal to the cells. The ergosterol pathway competes with other important metabolic processes, including the tRNA-isopentenylation pathway, and these pathways compete for a common precursor, DMAPP. Consistent with its sharing the substrate DMAPP between i6a modification and sterol biosynthesis, Mod5 that is misfolded into amyloid aggregates loses enough of cytoplasmic tRNA-modifying capability to allow more precursors to flow in the ergosterol biosynthesis pathway, overcoming the fluconazole block (Suzuki et al., 2012) (Figure 2). When yeast containing aggregated Mod5 are tested for the nuclear function, however, tgm-silencing is unaffected by the fluconazole selection (Figure 3B). It seems possible that the Mod5 in the nucleus does not misfold into amyloid aggregates, being already engaged in complexes with the tRNA gene transcription complex and/or the pre-tRNA transcripts. Alternatively, tgm-silencing might require lower levels or a separate pool of Mod5 compared to cytoplasmic tRNA modification, and thus incomplete aggregation may leave enough Mod5 present to keep tgm-silencing mechanisms intact.
Figure 3.
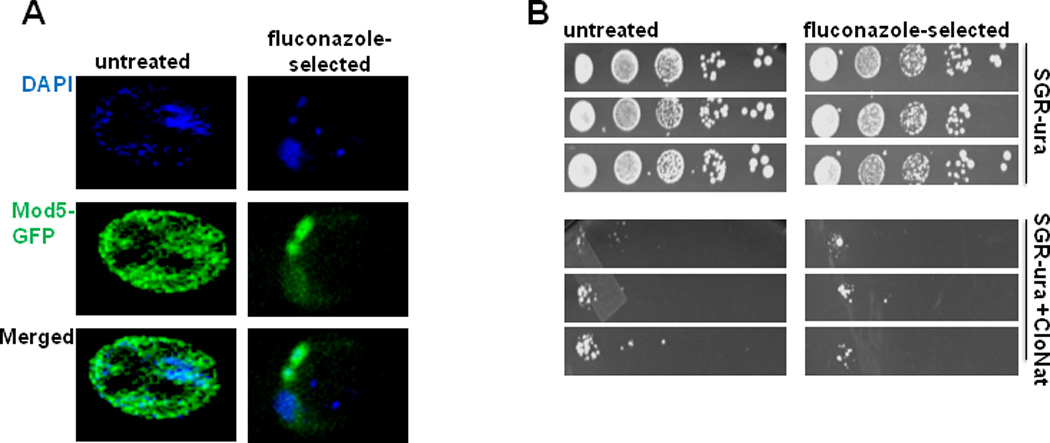
Mod5-GFP aggregates formed under fluconazole selection do not affect nuclear tgm-silencing. (A) Cellular localization of Mod5-GFP (green) in untreated yeast contrasting with cytoplasmic aggregation of Mod5-GFP in yeast following 7 days of fluconazole-selection. (nuclear DAPI staining in blue) (B) Fluconazole-selection does not alleviate nuclear tRNA gene-mediated silencing relative to unselected controls (untreated). Yeast were transformed with a tgm-silencing reporter plasmid consisting of a URA3 selection marker and a tRNATyr (SUP4) gene which silences a nat1 resistance gene under the control of a Gal4 UAS and Gal1 basal promoter(Hull et al., 1994; Pratt-Hyatt et al., 2013).
Mod5’s function in tgm-silencing is conserved in at least the plant and human (TRIT1) homologues as demonstrated by complementation assays in mod5Δ yeast strains (Pratt-Hyatt et al., 2013). This functional conservation suggests an evolutionary importance for the silencing activity. Transfer RNA genes are widely dispersed throughout the linear chromosomes, and it is possible that the far more numerous Short Interspersed Elements (SINEs) might also be affected. Thus, participation of Mod5 in this form of RNA-mediated chromatin regulation might represent a source of genome-wide gene regulation in higher eukaryotes. It is not clear to what extent the eukaryotic-specific C-terminal domain participates in the nuclear function. Attempts at C-terminal deletions of Mod5 (and many other point mutations in the enzyme) appear to destabilize the protein, and expression of a bacterial homolog, MiaA from E. coli, did not complement the silencing function ((Pratt-Hyatt et al., 2013) and unpublished results).
The human Mod5 homologue, TRIT1, is also a proposed tumor suppressor
The human homologue of yeast Mod5, TRIT1, has been proposed as a tumor suppressor protein (Spinola et al., 2005). TRIT1 displays a complex pattern of alternative splicing isoforms in both normal and lung adenocarcinomas tissues, in which only the full length mRNA isoform is able to complement tRNA modification in yeast. Expression of the full length isoform is downregulated 6–14 fold in lung tumor tissue when compared to expression in normal lung tissue (Spinola et al., 2005) and indicated that TRIT1 may have tumor suppressive functions. Overexpression of full length TRIT1 transcripts in lung cancer cell lines reduced colony size formation and migration, and these cells were less capable of forming tumors in a nude-mouse xenograph model when compared to lung cancer cells transfected with a empty vector control plasmid (Spinola et al., 2005).
Several TRIT1 single nucleotide polymorphisms (SNP) (rs11881557, rs2172362, and rs230310) are signficantly associated with increased lymph node metastasis in gastric cancers, tumor classification, and/or tumor site in a Southeast Chinese study population. However, these SNPS do not directly alter TRIT1 protein coding sequence since they reside within noncoding regions, therefore the mechanism of how the TRIT1 SNPS affect cancers is unclear, but may include an effect on TRIT1 transcription, stability, or splicing (Chen et al., 2013). Additionally, a Phe202Leu mutation predicts short survival in Italian lung cancer patients, while the same mutation predicts in a Norwegian population an increased survival (Spinola et al., 2005). Although the effect of this mutation is significant with regards to lung cancer survival, it does not however appear to affect i6A modification of tRNAs. These data suggest that although TRIT1 exhibits tumor suppressive behaviors, the effects appears to be tissue type- and population-dependent. Furthermore, the effects of TRIT1 on tumor behavior may not be limited only to TRIT1’s cytoplasmic function in tRNA modification. We hypothesize that like Mod5 in yeast, TRIT1 may have a role in silencing promoters in human cells, whether near tRNA genes or at more widespread loci. As a preliminary test of this hypothesis, we have examined whether TRIT1, like Mod5 (Tolerico et al., 1999), resides in the nucleus as well as the cytoplasm in human cells. Consistent with a nuclear role, we have found a subpopulation of TRIT1 to be present in the nucleus (Figure 4). Whether nuclear localization of TRIT1 plays a role in transcriptional control or tumor suppression has not yet been tested. To separate cytoplasmic effects of TRIT1 from nuclear effects are technically challenging experiments, but may begin to explain conflicting data in which in one ethnic population a TRIT1 mutation decreases cancer patient survival, while the same mutation in a different ethnic population increases their survival (Spinola et al., 2005).
Figure 4.
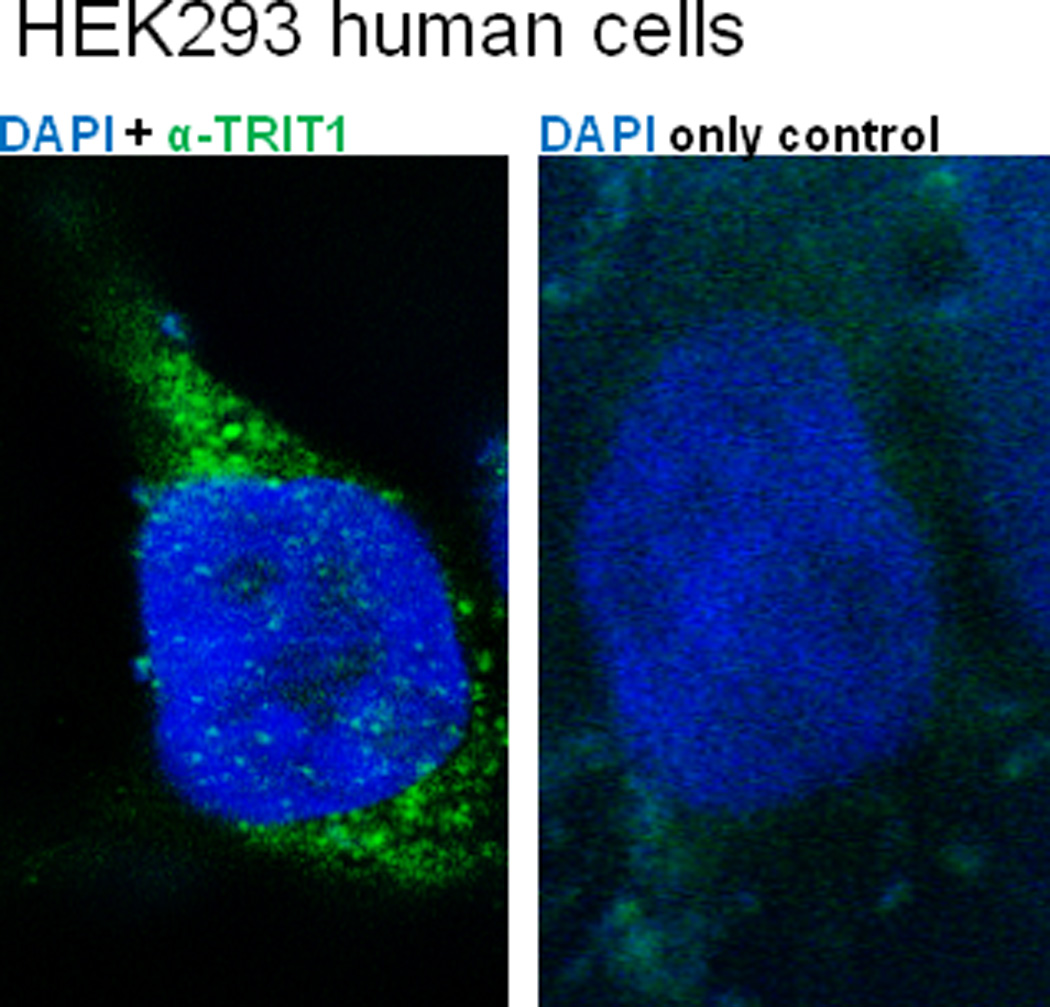
TRIT1 is present in the cytoplasm and nucleus in human cells. Representative 0.25 μM slices in human cells are shown taken through the nuclei and cytoplasm from a 3D-image that was generated using AutoQuant 3D-deconvolution software. Human HEK293A cells with nuclear DAPI staining (blue). (Left panel) Cell probed with commercial TRIT1 primary antibody, with specificity verified by western blot (not shown). Signal was detected with fluorescein-conjugated secondary antibody (green). (Right panel) Cell stained with only fluorescein-conjugated secondary antibody (green) as a control. TRIT1 is primarily cytoplasmic, but there are consistently small amounts of punctate signal in optical slices through the nucleus.
Nuclear tgm silencing is one of several forms of chromatin-mediated negative regulation in yeast, and might have specific selective advantages. For example, Ty retrotransposons are yeast retroviral elements that transpose by RNA being reverse transcribed into cDNA and then reinserted back into the genome (Curcio and Morse, 1996). The result of Ty integration into the genome is thought to have provided a source of genetic variation and fitness throughout evolution. Ty retrotransposons are most highly enriched around tRNAs in yeast (Sandmeyer et al., 1988; Hani and Feldmann, 1998) and the relationship has also been found in other simple eukaryotes (Hofmann et al., 1991; Szafranski et al., 1999). Therefore, a selective advantage for tgm-silencing might be to control this relationship, either through inhibiting Ty transcription when not specifically induced or through controlling recombinatorial deletion of the Ty elements (Pratt-Hyatt et al., 2006). In a chronic ageing model of yeast, mutations which decrease Ty retrotransposition events also decease measures of genomic instability including loss of heterozygosity and chromosome loss events (Curcio and Morse, 1996).
In multicellular organisms such as humans, a controlled form of localized silencing might have a large number of diverse uses. Control of retroviral elements is one, in that it might reduce transpositions that can cause genomic instability; a known hallmark of cancers (Hanahan and Weinberg, 2011). Alternatively, the Mod5 homologs in nuclei of eukaryotes with larger genomes might well contribute to a broader-based regulation of pol II transcription and chromatin effects. It is interesting that selected SINE element transcripts can have broad negative regulatory effects on RNA polymerase II transcription (Espinoza et al., 2004; Goodrich and Kugel, 2006; Goodrich and Kugel, 2009; Yakovchuk et al., 2009). Although there is evidence that the small, structured SINE RNAs can interact directly with RNA polymerase II to inhibit transcription (Espinoza et al., 2004; Mariner et al., 2008; Yakovchuk et al., 2009), the involvement of chromatin structure in vivo has not been tested. Lastly, the presence of RNA polymerase III transcription complexes, including those on SINE elements, might contribute subtle effects to chromatin events wherever they occur. In the case of vertebrates, it can be estimated that there is a potential RNA polymerase III promoter approximately every few thousand base pairs, on average, including in promoter regions and introns.
Highlights.
Mod5 and its dual roles in cytoplasmic tRNA modification and nuclear tRNA-gene mediated silencing
Mod5-aggregation, a result of fungicide-selection, does not affect nuclear function
The human homologue, TRIT1, resides both in the cytoplasm and nucleus in human cells
We propose dual cytoplasmic and nuclear functions of TRIT1 and a potential link to human cancers
Acknowledgments
This work was supported by NIH R01 GM063142 to DRE and GM27930 to AKH. DFR is a fellow of the University of Michigan Undergraduate Research Opportunities Program and the Chemistry Department Summer Undergraduate Program. We thank May Tsoi for her technical assistance.
Abbreviations
- tRNA
transfer RNA
- TRIT1
tRNA isopentenyl transferase 1
- i6A37
N6-(isopentenyl) adenosine
- DMAPP
dimethylallyl pyrophosphate
- NLS
nuclear localization signal
- ORF
open reading frames
- tgm
tRNA gene-mediated
- pol II
RNA polymerase II
- pol III
RNA polymerase III
- RSC
chromatin structure remolding complexes
- SINES
Short Interspersed Elements
- SNP
single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Benko AL, Vaduva G, Martin NC, Hopper AK. Competition between a sterol biosynthetic enzyme and tRNA modification in addition to changes in the protein synthesis machinery causes altered nonsense suppression. Proc Natl Acad Sci U S A. 2000;97:61–66. doi: 10.1073/pnas.97.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguta M, Hunter LA, Shen WC, Gillman EC, Martin NC, Hopper AK. Subcellular locations of MOD5 proteins: mapping of sequences sufficient for targeting to mitochondria and demonstration that mitochondrial and nuclear isoforms commingle in the cytosol. Mol Cell Biol. 1994;14:2298–2306. doi: 10.1128/mcb.14.4.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zheng Z, Tang J, Lin X, Wang X, Lin J. Association of polymorphisms and haplotype in the region of TRIT1, MYCL1 and MFSD2A with the risk and clinicopathological features of gastric cancer in a southeast Chinese population. Carcinogenesis. 2013;34:1018–1024. doi: 10.1093/carcin/bgt010. [DOI] [PubMed] [Google Scholar]
- Curcio MJ, Morse RH. Tying together integration and chromatin. Trends Genet. 1996;12:436–438. doi: 10.1016/0168-9525(96)30107-8. [DOI] [PubMed] [Google Scholar]
- Dihanich ME, Najarian D, Clark R, Gillman EC, Martin NC, Hopper AK. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:177–184. doi: 10.1128/mcb.7.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Bailly M, de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- Golovko A, Hjalm G, Sitbon F, Nicander B. Cloning of a human tRNA isopentenyl transferase. Gene. 2000;258:85–93. doi: 10.1016/s0378-1119(00)00421-2. [DOI] [PubMed] [Google Scholar]
- Golovko A, Sitbon F, Tillberg E, Nicander B. Identification of a tRNA isopentenyltransferase gene from Arabidopsis thaliana. Plant Mol Biol. 2002;49:161–169. doi: 10.1023/a:1014958816241. [DOI] [PubMed] [Google Scholar]
- Good PD, Kendall A, Ignatz-Hoover J, Miller EL, Pai DA, Rivera SR, Carrick B, Engelke DR. Silencing near tRNA genes is nucleosome-mediated and distinct from boundary element function. Gene. 2013;526:7–15. doi: 10.1016/j.gene.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7:612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Kugel JF. From bacteria to humans, chromatin to elongation, and activation to repression: The expanding roles of noncoding RNAs in regulating transcription. Crit Rev Biochem Mol Biol. 2009;44:3–15. doi: 10.1080/10409230802593995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hani J, Feldmann H. tRNA genes and retroelements in the yeast genome. Nucleic Acids Res. 1998;26:689–696. doi: 10.1093/nar/26.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J, Schumann G, Borschet G, Gosseringer R, Bach M, Bertling WM, Marschalek R, Dingermann T. Transfer RNA genes from Dictyostelium discoideum are frequently associated with repetitive elements and contain consensus boxes in their 5' and 3'-flanking regions. J Mol Biol. 1991;222:537–552. doi: 10.1016/0022-2836(91)90495-r. [DOI] [PubMed] [Google Scholar]
- Hull MW, Erickson J, Johnston M, Engelke DR. tRNA genes as transcriptional repressor elements. Mol Cell Biol. 1994;14:1266–1277. doi: 10.1128/mcb.14.2.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall A, Hull MW, Bertrand E, Good PD, Singer RH, Engelke DR. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc Natl Acad Sci U S A. 2000;97:13108–13113. doi: 10.1073/pnas.240454997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane TN, Blewett NH, Maraia RJ. Plasticity and diversity of tRNA anticodon determinants of substrate recognition by eukaryotic A37 isopentenyltransferases. RNA. 2011;17:1846–1857. doi: 10.1261/rna.2628611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux J, Lakowski B, Webb A, Meng Y, Ubach A, Bussiere F, Barnes T, Hekimi S. Regulation of physiological rates in Caenorhabditis elegans by a tRNA-modifying enzyme in the mitochondria. Genetics. 2001;159:147–157. doi: 10.1093/genetics/159.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt-Hyatt M, Pai DA, Haeusler RA, Wozniak GG, Good PD, Miller EL, McLeod IX, Yates JR, 3rd, Hopper AK, Engelke DR. Mod5 protein binds to tRNA gene complexes and affects local transcriptional silencing. Proc Natl Acad Sci U S A. 2013;110:E3081–E3089. doi: 10.1073/pnas.1219946110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt-Hyatt MJ, Kapadia KM, Wilson TE, Engelke DR. Increased recombination between active tRNA genes. DNA Cell Biol. 2006;25:359–364. doi: 10.1089/dna.2006.25.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer SB, Bilanchone VW, Clark DJ, Morcos P, Carle GF, Brodeur GM. Sigma elements are position-specific for many different yeast tRNA genes. Nucleic Acids Res. 1988;16:1499–1515. doi: 10.1093/nar/16.4.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg T, Poulter CD. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: essential elements for recognition of tRNA substrates within the anticodon stem-loop. Biochemistry. 2000;39:6546–6553. doi: 10.1021/bi992775u. [DOI] [PubMed] [Google Scholar]
- Spinola M, Galvan A, Pignatiello C, Conti B, Pastorino U, Nicander B, Paroni R, Dragani TA. Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer. Oncogene. 2005;24:5502–5509. doi: 10.1038/sj.onc.1208687. [DOI] [PubMed] [Google Scholar]
- Sugiyama S, Tanaka M. Self-propagating amyloid as a critical regulator for diverse cellular functions. J Biochem. 2014;155:345–351. doi: 10.1093/jb/mvu026. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336:355–359. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Tanaka M. Active conversion to the prion state as a molecular switch for cellular adaptation to environmental stress. Bioessays. 2012;35:12–16. doi: 10.1002/bies.201200121. [DOI] [PubMed] [Google Scholar]
- Szafranski K, Glockner G, Dingermann T, Dannat K, Noegel AA, Eichinger L, Rosenthal A, Winckler T. Non-LTR retrotransposons with unique integration preferences downstream of Dictyostelium discoideum tRNA genes. Mol Gen Genet. 1999;262:772–780. doi: 10.1007/s004380051140. [DOI] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolerico LH, Benko AL, Aris JP, Stanford DR, Martin NC, Hopper AK. Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics. 1999;151:57–75. doi: 10.1093/genetics/151.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Haeusler RA, Good PD, Thompson M, Nagar S, Engelke DR. Silencing near tRNA genes requires nucleolar localization. J Biol Chem. 2005;280:8637–8639. doi: 10.1074/jbc.C500017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc Natl Acad Sci U S A. 2009;106:5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarham JW, Lamichhane TN, Pyle A, Mattijssen S, Baruffini E, Bruni F, Donnini C, Vassilev A, He L, Blakely EL, Griffin H, Santibanez-Koref M, Bindoff LA, Ferrero I, Chinnery PF, McFarland R, Maraia RJ, Taylor RW. Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA. PLoS Genet. 2014;10:e1004424. doi: 10.1371/journal.pgen.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Huang RH. Crystallographic snapshots of eukaryotic dimethylallyltransferase acting on tRNA: insight into tRNA recognition and reaction mechanism. Proc Natl Acad Sci U S A. 2008;105:16142–16147. doi: 10.1073/pnas.0805680105. [DOI] [PMC free article] [PubMed] [Google Scholar]


