Abstract
Tamoxifen, a selective estrogen receptor (ER) modulator (SERM), remains a frontline clinical therapy for patients with ERα-positive breast cancer. However, the relatively rapid development of resistance to this drug in the metastatic setting remains an impediment to a durable response. Although drug resistance likely arises by many different mechanisms, the consensus is that most of the implicated pathways facilitate the outgrowth of a subpopulation of cancer cells that can either recognize tamoxifen as an agonist or bypass the regulatory control of ERα. Notable in this regard is the observation here and in other studies that expression of anterior gradient homology 2 (AGR2), a known proto-oncogene and disulfide isomerase, was induced by both estrogen (17β-estradiol, E2) and 4-hydroxytamoxifen (4OHT) in breast cancer cells. The importance of AGR2 expression is highlighted here by the observation that (a) its knockdown inhibited the growth of both tamoxifen-sensitive and -resistant breast cancer cells, and (b) its increased expression enhanced the growth of ERα-positive tumors in vivo and increased the migratory capacity of breast cancer cells in vitro. Interestingly, as with most ERα-target genes, the expression of AGR2 in all breast cancer cells examined requires the transcription factor FOXA1. However, in tamoxifen-resistant cells, the expression of AGR2 occurs in a constitutive manner, requiring FOXA1, but loses its dependence on ER. Taken together, these data define the importance of AGR2 in breast cancer cell growth and highlights a mechanism where changes in FOXA1 activity obviate the need for ER in the regulation of this gene.
Keywords: AGR2, FOXA1, estrogen receptor, ERα, resistance, tamoxifen, breast cancer
Introduction
Approximately 40,000 women die from breast cancer each year, making it the leading cause of early mortality in women (1). Most breast cancers are estrogen receptor alpha (ERα) positive and recognize estrogens as mitogens. Therefore, not surprisingly, strategies that interfere with ERα action have emerged as frontline therapies for the treatment and prevention of breast cancer. Specifically, ERα can be effectively targeted at the level of (a) receptor activity, using selective estrogen receptor modulators (SERMs), such as tamoxifen, and selective estrogen receptor degraders (SERDs) or (b) ligand availability, using aromatase (cyp19) inhibitors. However, close to 60% of ERα-positive tumors exhibit intrinsic resistance or rapidly acquire resistance to endocrine interventions (especially in the metastatic setting), requiring the subsequent use of largely ineffective cytotoxic therapies (2-4). Unfortunately, it has been difficult to predict de novo resistance to endocrine therapy and/or assess the likelihood of early relapse. Some progress in this regard was made with the development of the HoxB13/IL17RB gene predictor (5), although clinically this test has not been widely used and its utility remains unclear. Thus, there is an unmet medical need to define the fundamental processes underlying endocrine resistance with a view to developing mechanism based diagnostic tests to assess likely drug responses and to identify new approaches to mitigate the impact of resistance.
Although the mechanisms underlying acquired resistance to tamoxifen treatment are multifaceted and diverse, most studies have implicated alterations in kinase signaling pathway activation or epigenetic changes that enable the tamoxifen:ERα complex to activate transcription (4, 6). Additionally, several recent studies have described the identification of proto-oncogenes whose expression and/or activities are positively up-regulated by endocrine manipulation and which alter tamoxifen pharmacology. One such proto-oncogene, the secreted protein AGR2 (anterior gradient homology 2), has been shown to exhibit oncogenic activities in several malignancies including breast, ovarian, pancreatic and prostate cancer (7-11). Further, this protein is overexpressed in primary tumors and in cellular models of breast cancer (12-14). Although ERα+ breast cancers, in general, exhibit a low histological grade, the expression of AGR2 in this tumor subtype is associated with a poorer outcome (8, 12, 15). Furthermore, in primary ERα+ tamoxifen treated breast cancers, AGR2 overexpression is linked to tamoxifen treatment failure (8). Recently it was demonstrated that AGR2 is a direct ERα target gene in MCF7 cells. More importantly, however, it was observed that tamoxifen induced the expression of this gene similar to the ERα agonist estradiol (8). Thus, it is possible that although tamoxifen is an effective antagonist on most ERα-responsive genes, its ability to activate AGR2 expression may limit its therapeutic response (7, 8, 16).
In addition to serving as a marker of resistance to endocrine therapy, it is likely that AGR2 itself contributes in a significant manner to tumor biology. However, there is minimal information as to how AGR2 manifests its oncogenic activities. It has been demonstrated that AGR2 has the structural features of a protein disulfide isomerase although a potential substrate(s) for these activities has yet to be identified. Further, it is not known if the isomerase activity of this protein is required for its oncogenic functions. Given that it is a secreted protein, it is not surprising that some have considered the possibility that AGR2 may be involved in the folding of the extracellular domains of proteins that influence cell growth and survival (17, 18). Although it has also been shown that the secreted AGR2 protein influences gene expression, it is unclear if this is accomplished as a consequence of its ability to (a) modulate the activity of some cell surface protein, (b) bind to and activate a specific “AGR2” receptor or (c) function as a direct modulator of an intracellular signaling pathway. Regardless, considering our interest in defining the molecular pharmacology of tamoxifen, we embarked on a study to evaluate the impact of AGR2 expression on the transition of breast cancer cells from a tamoxifen sensitive to a tamoxifen resistant state and to define the mechanisms by which tamoxifen manifests agonist activity on the AGR2 promoter. It was anticipated that this study would be informative with respect to the specific processes that enable the ERα:tamoxifen complex to activate AGR2 expression and may highlight the mechanisms underlying the activity of this and other SERMs.
Materials and Methods
Cell culture and treatments
MCF7 and its derivative MCF7 TamR (tamoxifen resistant) were maintained in Dulbecco's Modified Eagle Medium and Ham's F12 Nutrient Mixture (DMEM/F12) with 8% fetal bovine serum (FBS; Gemini), L-glutamine (Invitrogen) and non-essential amino acids (NEAA; Invitrogen). Both cell lines were authenticated by short tandem repeat profiling from ATCC and tested for Mycoplasma. MCF7 GPS ER was maintained in DMEM/F12 supplemented with 8%FBS, L-glutamine and NEAA.
For experiments, cells were plated in media lacking phenol red with 8% charcoal stripped FBS (CFS; Gemini). Unless otherwise indicated, cells were plated for 48 hours and then treated for 24 hours with 17β-estradiol (E2; Sigma), 4-hydroxytamoxifen (4OHT; Sigma) or ICI182, 780 (ICI; Tocris) as indicated and RNA or protein harvested.
Plasmids
The pLenti CMV puro Gal4-DBD (control) and pLenti CMV puro AGR2 plasmids were generated by subcloning Gal4-DBD and AGR2 cDNA into pENTR1a (Invitrogen) and recombining into pLenti CMV puro DEST (Invitrogen).
Gene silencing and overexpression
Small interfering RNA (siRNA) were used to transiently silent ERα (Invitrogen), AGR2 (Invitrogen) and FOXA1 (Sigma) with control siRNAs (siLuc; Invitrogen and Mission Control; Sigma). MCF7 and TamR cells were seeded at 20-30 × 104 cells per well on a six-well plate and transfected using Dharmafect IV (Dharmacon) for 48-72 hours, unless otherwise specified.
pLenti CMV puro Gal4-DBD and pLenti CMV puro AGR2 plasmids were cotransfected (Fugene, Roche Applied Science) with the vsvg, gag-pol and rev packaging vectors into 293FT cells. The viral supernatants were filtered and supplemented with 8ug/mL polybrene before infecting MCF7 cells for two serial 24-hour periods. Cells were then selected with 1ug/mL puromycin yielding the MCF7 Gal4 and MCF7 AGR2 cell lines.
RNA preparation and qRT-PCR analyses
Total RNA was prepared using the Aurum Total RNA Mini Kit (BioRad). cDNA was synthesized from 0.5ug total RNA using the BioRad iScript cDNA Synthesis Kit. Quantitative RT-PCR (qRT-PCR) was performed with 2ul 1:20 diluted cDNA, 0.2μmol/L primers and the iQ SYBR Green supermix (BioRad), the results calculated using the 2−ΔΔcT method (31) and data normalized to a 36B4 internal control. Primer sequences are available upon request.
Immunoblotting
Cells were lysed in whole cell extract buffer containing 100mM Tris-HCl pH8.0, 2mM EDTA, 0.02% SDS, 0.5% Nonidet P-40 and 150mM NaCl and 1x protease inhibitors (Sigma). Protein samples were separated on a 10% SDS Page polyacrylamide gel, transferred to nitrocellulose membranes and the following antibodies were used for detection: ERα(D-12, Santa Cruz Biotechnology), AGR2 (Abnova), FOXA1 (Abcam), β-Actin (ACTB; Sigma) and cytokeratin 18 (KRT18; Santa Cruz Biotechnology). Appropriate horseradish peroxidase conjugated secondary antibodies (BioRad) were used and the proteins detected using Western Lightning® Plus ECL chemiluminescence reagents (Perkin-Elmer).
Proliferation Assay
30 × 104 MCF7 and TamR cells were silenced for AGR2 for 48 hours prior to the proliferation assay. 0.2 × 104 siAGR2 cells were plated per well on 96-well plates. One plate of MCF7 siAGR2 and TamR siAGR2 was decanted and frozen on day 1 as a control. The remaining plates were treated identically on day 1 and day 3 with E2 as specified and harvested on day 5. DNA content was analyzed using FluoReporter assay (Invitrogen) per manufacturer instructions.
Migration Assay
Cells were serum starved for 24 hours with DMEM/F12, 0.1% bovine serum albumin and 10mM HEPES. 7.5 × 104 cells in 100uL were plated on BD Biocoat Control Inserts 8.0 micron (BD Biosciences) in duplicate and migrated towards 8% FBS for 16 hours. Migrating cells were stained with 5% crystal violet in 20% methanol and counted. For siRNA migration assay, 40 × 104 cells were silenced for 48 hours and then serum starved for 24 hours prior to plating as previously described. For the spent media experiments, MCF7 and TamR cells were plated as previously described and migrated towards spent media from the MCF7 Gal4 and MCF7 AGR2 cells.
Xenograft Tumor Analyses
All xenograft procedures approved by the Duke University Institute for Animal Care and Use Committee. Ovariectomized estrogenized (0.72 mg/60 days 17β-estradiol (E2) time-released sc pellet; Innovative Research of America) NU/NU mice (˜6 weeks of age) were injected with 5 × 106 MCF7 Gal4 or MCF7 AGR2 into the axial mammary fat pad. Tumors were measured by caliper three times per week until tumor volume ((L2 × W)/2) reached 0.2 cm3. Mice were then randomized for continued treatment with E2 or placebo (E2 withdrawn), with or without tamoxifen co-treatment (injected sc daily with 0.5 mg/mouse dissolved in corn oil).
Bioinformatics Analyses
Forest Plot
To query the prognostic significance of our 4OHT induced genes, we assembled a metaset of breast cancer patients from 25 publicly available datasets, which included 4885 patients. The Affymetrix microarray data (HGU133plus2 and HGU133A) was downloaded from GEO, normalized with fRMA (32) and batch corrected using the COMBAT algorithm within R yielding only the probe sets common between the two platforms. Clinical data was also aggregated from GEO and duplicate patient samples were removed. Each tumor was then classified into tumor subtypes using the PAM50 (33) gene modules in Genefu. Genes identified as being induced by 4OHT were then ranked according to significance and the top 25 were submitted to the R package survcomp (34) to calculate Concordance Index scores for each gene and plotted.
AGR2 Correlation Network
Data from The Cancer Genome Atlas (TCGA-BRCA) (35) was downloaded and compiled locally. The RNA-Seq data comprised of 961 patient samples at the time of download and Spearman correlation was computed for every gene present with respect to AGR2 or ESR1. Clustering analysis of the top correlating genes of AGR2 indicated distinct groups and the package DiffCorr (36) from R (37) was used to derive the differential correlation metric of (AGR2 –ESR1). In addition, mRNA expression from 921 cell lines was downloaded from CCLE (38) and Spearman correlation was similarly obtained. Top AGR2 correlating genes from TCGA data (R>.50) was inputted into Cytoscape (39) with TCGA Spearman values indicated by node color and CCLE Spearman values indicated by node size. The associated table lists the top factors and is ranked by the differential correlation metric.
Statistical Analysis
Migration, qPCR and transfection data are represented as mean ± SEM. Tumor growth data was analyzed by either 2-way ANOVA (non-repeated measures) followed by a Bonferroni's T-test, or by Kaplan-Meier survival analysis.
Results
The ERα-target gene AGR2 is constitutively expressed in models of tamoxifen resistant ERα positive breast cancer
Previously, we identified a set of genes whose expression in MCF7 cells was positively upregulated by 4-hydroxytamoxifen (4OHT) and whose induction by other SERMs reflected the relative agonist activity of the compound tested (19). Analysis of the prognostic significance of these genes in luminal A cancers revealed that the expression of PTGES and AGR2 were among the most significantly associated with poor outcome (Figure 1A). Since previous clinical data suggests that PTGES has no major effect on breast cancer susceptibility or survival among those with invasive breast cancer (20), we chose to focus on AGR2. AGR2 is a proto-oncogene whose expression is shown to be induced by 17β-estradiol (E2) in several cancers of epithelial origin (7, 8, 16). Further, there is considerable interest in using AGR2 as a potential biomarker of drug resistance in breast cancer (17, 18). Given these data, we decided to evaluate the extent to which AGR2 contributes to the pathobiology of ERα-positive breast cancer and influences the response of these cancers to tamoxifen.
Figure 1. AGR2 is constitutively expressed in tamoxifen resistant ERα-positive breast cancer.
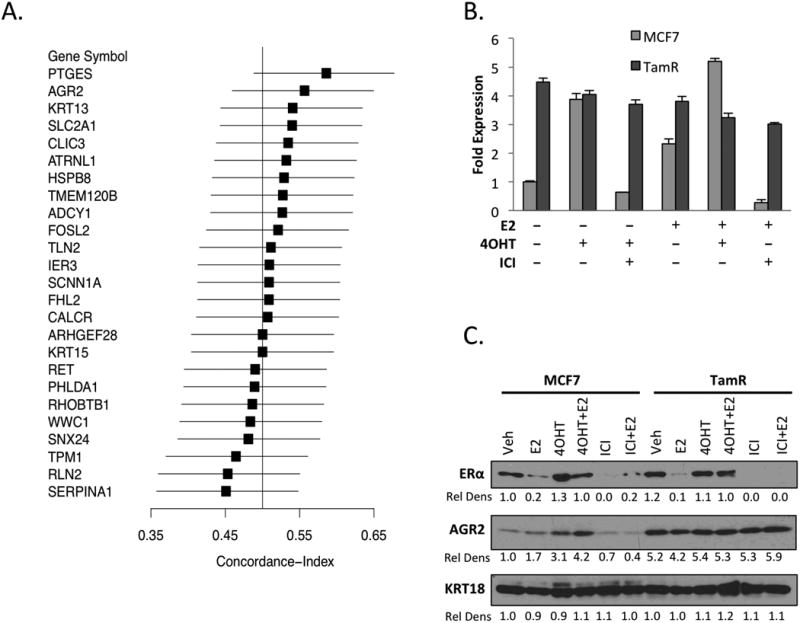
A. AGR2 mRNA expression is associated with a worse prognosis in breast cancer. Forest plot analysis indicating genes associated with prognosis in Luminal A breast cancer.
B. AGR2 expression is constitutively expressed in a cellular model of tamoxifen resistance. MCF7 and TamR cells were seeded in phenol red free media for 48 hours with CFS and then treated with either Vehicle, 1nM E2, 100nM 4OHT alone or in combination with 100nM ICI. AGR2 mRNA expression was analyzed by qRT-PCR, normalized to the expression of the 36B4 housekeeping gene and presented as fold change relative to Vehicle (Veh) treated cells.
C. AGR2 is upregulated by antiestrogens in MCF7 cells, but is constitutively expressed in TamR cells. MCF7 and TamR cells were plated for 48 hours in phenol red free media supplemented with CFS and then treated with 10nM E2, 100nM 4OHT or 100nM ICI for 24 hrs. Whole cell extracts were immunoblotted for ERα, AGR2 and cytokeratin 18 (KRT18) expression and the Relative Density (Rel Dens) of the extracts determined.
We have developed a cellular model of tamoxifen resistance (TamR) from an MCF7 xenograft tumor continually treated with tamoxifen in vivo until the onset of resistance. In contrast to TamR models developed by continuously treating cells with tamoxifen in vitro (21, 22), the cell line derived from the corresponding TamR tumors retains both ERα expression and a robust ERα-dependent transcriptional program. This model has been shown to be very predictive of the human disease and has been used in the development of several endocrine agents for breast cancer ((16, 23); unpublished results). With some exceptions, the ERα dependence of genes was conserved in MCF7 and TamR cells ((16); data not shown). However, several genes showed constitutive high expression in TamR compared to the parental MCF7. Of relevance to this work is the observation that in parental MCF7 cells, the expression of AGR2 was induced following treatment with E2 or 4OHT, but was inhibited by concomitant treatment with the pure antiestrogen ICI 182, 780 (ICI) (Figure 1B). Interestingly, AGR2 levels (both mRNA and protein) were expressed at a high constitutive level in TamR cells and this expression was not influenced by treatment with E2, 4OHT or ICI (Figure 1B, C). Several other ERα target genes such as KRT13 were also expressed in a constitutive manner in the TamR cells, although their expression was inhibited by ICI (data not shown). Further, it was demonstrated that the expression of AGR2 and other tamoxifen responsive genes including KRT13 were induced by tamoxifen treatment when TamR cells were propagated as xenografts (Supplementary Figure 1). As was the case in vitro KRT13, but not AGR2 expression, was inhibited by ICI treatment (Supplementary Figure 1). Thus, while AGR2 is a robust target in ERα-dependent tamoxifen sensitive (treatment naïve) cells, its elevated level of expression in the TamR models seems to no longer require ERα. We next addressed the impact of AGR2 on tumor biology and sought to elucidate the mechanism(s) underlying its expression in TamR cells.
AGR2 is required for proliferation and enhances migration in cellular models of tamoxifen sensitive and tamoxifen resistant breast cancer
Recently, it has been reported that AGR2 is a proto-oncogene that regulates mammary epithelial cell proliferation (24) and that elevated expression of this protein is associated with increased cell proliferation and migration in breast cancer cells (7, 8). However, a role for AGR2 in the pathology of tamoxifen resistant breast cancer has not been defined. Therefore, we performed a comparative analysis of the impact of siRNA-mediated knockdown of AGR2 expression on the growth of MCF7 and TamR cells. The results of this study, performed using two different siRNAs, revealed that AGR2 was required for the growth and survival of both tam sensitive MCF7 and tam resistant TamR cell lines (Figure 2A, Supplementary Figure 2).
Figure 2. Functional activities of AGR2 in tamoxifen sensitive and tamoxifen resistant ERα-positive breast cancer cells.
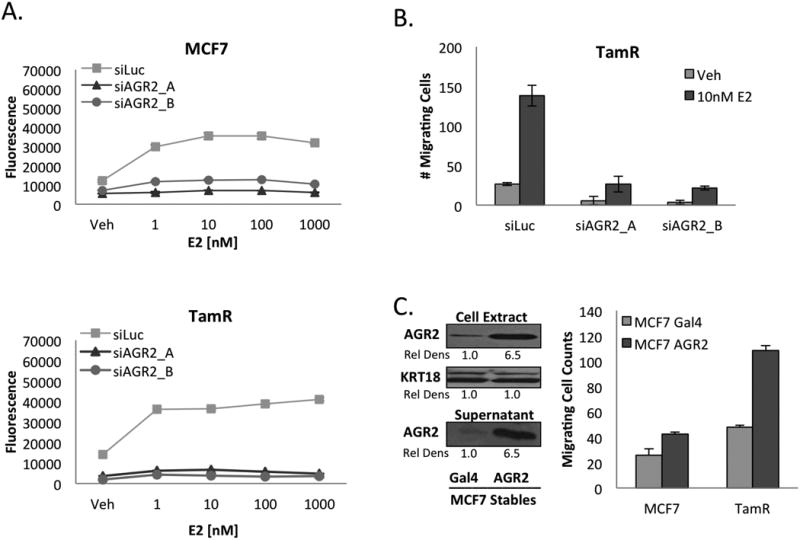
A. AGR2 is required for 17β-estradiol induced proliferation. MCF7 and TamR cells were treated with siAGR2 and plated for 48 hours in phenol red free media supplemented with CFS. 0.2 × 104 Control and siAGR2 cells were then plated on 96-well plates and treated with either Vehicle or 17β-estradiol (E2) as indicated for 5 days.
B. AGR2 affects TamR cell migration. TamR cells were treated with siAGR2 for 48hrs in phenol red free media and then serum starved for 24hrs. 7.5×104 cells were plated and then allowed to migrate towards 10% FBS for 16hrs.
C. Overexpressed AGR2 is secreted and enhances breast cancer cell migration. MCF7 cells overexpressing AGR2 (MCF7 AGR2) or MCF7 Gal4 control cells were plated in phenol red free media with CFS for 24 hours and then changed to phenol red free media containing insulin, transferrin and selenium for 48 hours. Proteins from the supernatant were then precipitated and whole cell extracts and precipitated supernatant immunoblotted for AGR2 expression. For the migration assay, MCF7 and TamR cells were plated for 48 hours in phenol red free media containing CFS before being serum starved for 24 hours. 7.5×104 cells were plated and then allowed to migrate for 16 hours towards spent media harvested from either MCF7 Gal4 or MCF7 AGR2 overexpressing cells.
One of the characteristics of the TamR cells, in comparison to MCF7 cells, is that they have a high migratory potential when assayed in vitro. Given that others have shown that AGR2 influences cell migration (7, 8), we evaluated the effect of AGR2 knockdown on cell migration. As shown in Figure 2B, E2 treatment increased the migration of TamR cells and this effect was ablated upon siRNA mediated knockdown of AGR2 expression. To confirm the role of AGR2 in cell migration, we used a lentiviral-based approach to overexpress AGR2 in MCF7 cells. It has been established that AGR2 is a secreted protein and we were able to demonstrate that the recombinant AGR2 produced in the cell lines we developed was indeed secreted (Figure 2C). Additionally, although treatment with spent media from AGR2 overexpressing cells did not overcome the intracellular proliferative functions of AGR2 when knocked down (Supplementary Figure 3), we demonstrated that the same spent media was sufficient to induce MCF7 cell migration and more importantly, further potentiated the robust migratory behavior of TamR cells (Figure 2C). Taken together, these data indicate that intracellular AGR2 promotes cellular proliferation while secreted AGR2 promotes migration, and that its constitutive expression in models of endocrine resistant cancer is likely to be important in disease pathogenesis.
AGR2 overexpression enhances E2 stimulated growth of ERα-dependent xenografts in vivo
We next undertook a series of studies to assess the impact of AGR2 overexpression on tumor growth and tamoxifen response in MCF7-cell derived xenografts. To this end, MCF7 Gal4 (control) or MCF7 AGR2 cells were injected subcutaneously into the mammary fat pad of ovariectomized athymic nu/nu mice. All mice received estrogen treatment until the tumors reached ∼0.2cm3, and the mice were then randomized into 4 groups: (a) continued estrogen treatment; (b) continued estrogen with co-administration of tamoxifen; (c) estrogen withdrawal; and (d) estrogen withdrawal with tamoxifen administration. This final group was included to determine whether overexpression of AGR2 was sufficient to permit tamoxifen dependent growth of the tumors as observed in the TamR xenografts. One of the most important findings of this study was that the E2 treated tumors overexpressing AGR2 grew significantly faster than their Gal4 expressing counterparts (Figure 3A). It was also observed that when compared to Gal4, AGR2 expression reduced the efficacy of tamoxifen, although the differences between these groups did not reach statistical significance (Figure 3A). The effect of AGR2 on tamoxifen pharmacology was more apparent when the data were presented as time to event (tumors >400mm3)(Figure 3B). Upon E2 withdrawal alone, no changes were observed between the Gal4 and AGR2 expressing tumors (Figure 3C). However, it was observed that upon E2 withdrawal, the efficacy of tamoxifen was reduced in AGR2 expressing tumors (Figure 3C). Taken together, these data indicate that AGR2 impacts the pathobiology and pharmacology of tumors in mice, although its expression alone is not sufficient to confer tamoxifen resistance in the selected models.
Figure 3. AGR2 overexpression induces E2 induced growth in vivo.
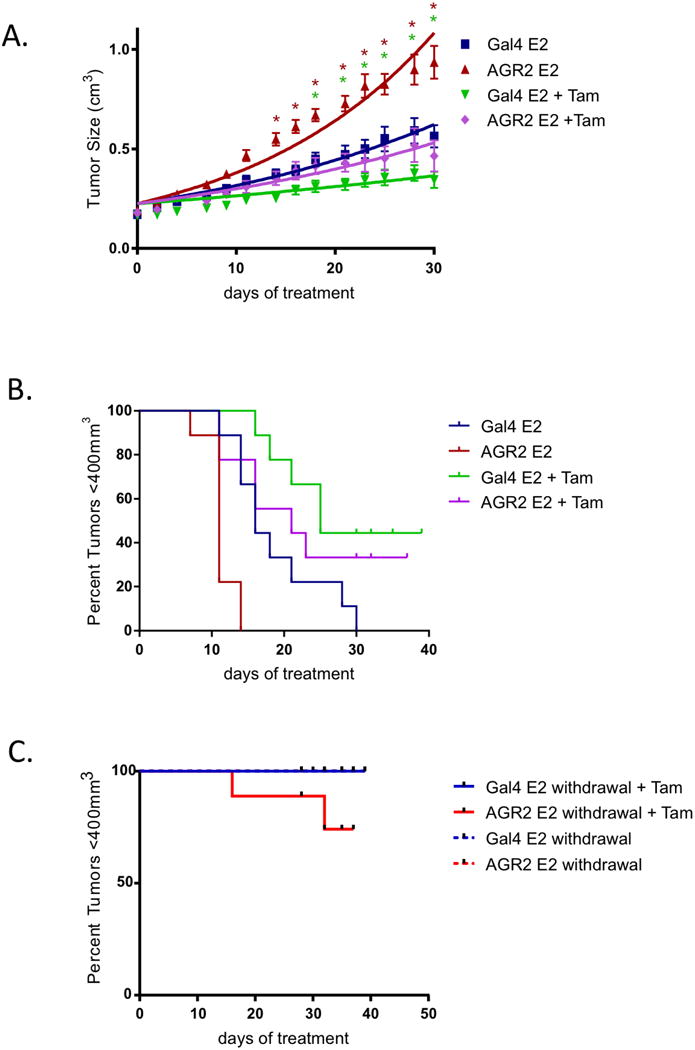
A. Overexpressed AGR2 increases tumor growth in vivo. MCF7 Gal4 and MCF7 AGR2 cells were injected into the axial mammary fat pads of ovariectomiz edestrogenized NU/NU mice. After tumor volume growth to 0.2 cm3, MCF7 Gal4 and MCF7 AGR2 tumors were randomized (10 mice per group) to continue receiving E2 treatment with and without tamoxifen co-treatment (* indicates statistically significant differences with the MCF7 Gal4 E2 group, P<0.05).
B. Survival curves of MCF7 Gal4 and MCF7 AGR2 tumors, which were randomized (10 mice per group) to continue receiving E2 treatment with and without tamoxifen co-treatment (Gal4 E2 vs AGR2 E2, p=0.0009; Gal4 E2 vs Gal4 E2+Tam, p=0.0156; AGR2 E2 vs AGR2 E2+Tam, p=0.0009; AGR2 E2+Tam vs Gal4 E2+Tam, p=0.367).
C. Survival curves of MCF7 Gal4 and MCF7 AGR2 tumors, which were randomized (10 mice per group) and had E2 treatment withdrawn, or had E2 treatment withdrawn but with continued tamoxifen treatment (p=0.2044).
AGR2 expression in tamoxifen resistant breast cancer cells loses its dependence on ERα
As described above, we determined that while ICI reduces basal and E2-induced AGR2 expression in the MCF7 cells, the expression of AGR2 in the TamR model in vitro and in vivo (Figure 1, Supplementary Figure 1) remained consistently elevated despite treatment with ERα agonists or antagonists. These observations led us to question the extent to which ERα is required for AGR2 expression in the TamR model and how this compares to the parental tamoxifen sensitive MCF7 cells. Using two different siRNAs, it was demonstrated that AGR2 mRNA expression in MCF7 cells was completely dependent on ERα expression (Figure 4A), whereas a knockdown of ERα expression in TamR cells had a minimal effect on AGR2 expression (Figure 4B). The loss of ERα dependence on AGR2 expression in the TamR line was also apparent when examined at the protein level, where it was demonstrated that robust knockdown of ERα expression did not significantly influence AGR2 expression (Figure 4C, Supplementary Figure 4). Therefore, while ERα regulates the expression of AGR2 in the tamoxifen sensitive MCF7 cells, the expression of AGR2 is both elevated and loses its dependence on ERα expression in our validated model of tamoxifen resistance.
Figure 4. AGR2 expression in tamoxifen resistant breast cancer cells loses its dependence on ERα.
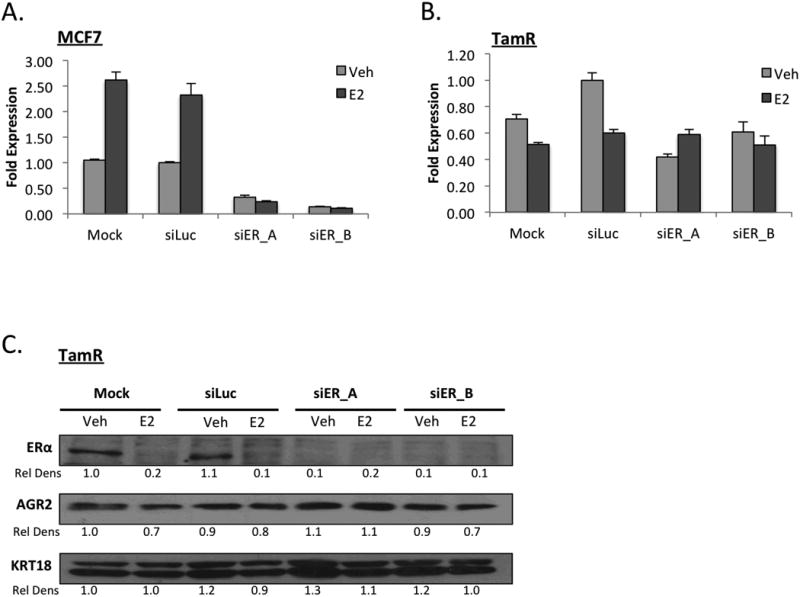
A. AGR2 expression is regulated by ERα in MCF7 cells. MCF7 cells were treated with siERα and plated for 48 hours in phenol red free media supplemented with CFS and then treated with 1nM E2 for 24hrs. AGR2 expression was analyzed by qRT-PCR, normalized to the expression of the 36B4 housekeeping gene and presented as fold change relative to siLuc-Vehicle treated cells.
B. ERα regulation of AGR2 is diminished in TamR cells. TamR cells were treated with siERα and plated for 48 hours in phenol red free media supplemented with CFS and then treated with 1nM E2 for 24hrs. AGR2 expression was analyzed by qRT-PCR.
C. ERα regulation of AGR2 is diminished in TamR cells. TamR cells were treated with siERα for 48hrs and then treated with E2 for 24 hrs. The resulting whole cell extracts were immunoblotted for ERα, AGR2 and cytokeratin 18 (KRT18) expression.
FOXA1 is required for AGR2 expression in both tamoxifen sensitive and resistant cells
The observation that AGR2 expression did not require ERα in tam resistant TamR cells, but that ERα expression requires AGR2 in both tam sensitive MCF7 and tam resistant TamR cells (Supplementary Figure 5) raised the question as to the identity of the transcription factor(s) required for the expression of AGR2 in TamR cells. To resolve this issue, we performed a comparative analysis of the Cancer Cell Encyclopedia (CCLE) and The Cancer Genome Atlas (TCGA) Breast Cancer datasets to identify genes that are highly co-expressed with AGR2 in ERα-positive breast tumors. In this manner, we determined (at the mRNA level) that the expression of tetraspanin-13 (TSPAN13) and forkhead box transcription factor 1 (FOXA1) was most closely correlated with AGR2 mRNA expression (Figure 5A). Interestingly, the genes for AGR2 and TSPAN13 map to the same locus on chromosome 7. We therefore elected to focus our studies on FOXA1 as (a) it has been shown to function as a pioneer factor for ERα on many genes, (b) its expression has been associated with tamoxifen resistance in published studies (25, 26), (c) we and others (27, 28) have demonstrated that the expression of an AGR2 promoter-luciferase reporter could be induced in heterologous cells by expressing FOXA1 (data not shown) and (d) that the interaction of FOXA1 with this promoter could be demonstrated by ChIP (Supplementary Figure 6). For these studies, two independent siRNAs were used to knockdown FOXA1 expression in TamR cells. Using this approach, quantitative ablation of FOXA1 mRNA expression could be accomplished and this dramatically reduced AGR2 expression in both MCF7 and TamR cells at the level of mRNA (Figure 5B) and protein (Figure 5C). Interestingly, confounding the interpretation of these data was the observation, somewhat expectedly, that ERα expression was completely dependent on FOXA1 expression in both cell lines. Thus, it was unclear if FOXA1 was having a direct role on AGR2 expression or if it participated in an indirect manner by regulating ERα expression; an issue of particular importance in understanding AGR2 regulation in TamR cells. This mechanistic question was addressed by expressing GFP-tagged ERα under the control of a heterologous CMV promoter (GPS ER) in MCF7 cells. Knockdown of FOXA1 expression decreased the expression of endogenous ERα mRNA and protein in the MCF7 cell line, whereas the expression of the exogenous GFP-ERα cells was not drastically effected (Figure 5D, 5E, Supplementary Figure 7). Of note, however, was the observation that even in the background of stable ERα overexpression, siRNA mediated knockdown of FOXA1 abrogated AGR2 expression. It is important to note that FOXA1 expression is equivalent in both MCF7 and TamR cells suggesting that in the latter cells, the increased activity of a factor (or process) diminishes the need for ERα. Taken together, these data suggest that the expression of the AGR2 proto-oncogene in both MCF7 and TamR cells requires FOXA1 but that the requirement for ERαin the regulation of this gene is lost as cells progress to a state of tamoxifen resistance.
Figure 5. FOXA1 drives AGR2 expression in both tamoxifen sensitive and tamoxifen resistant estrogen receptor positive breast cancer.
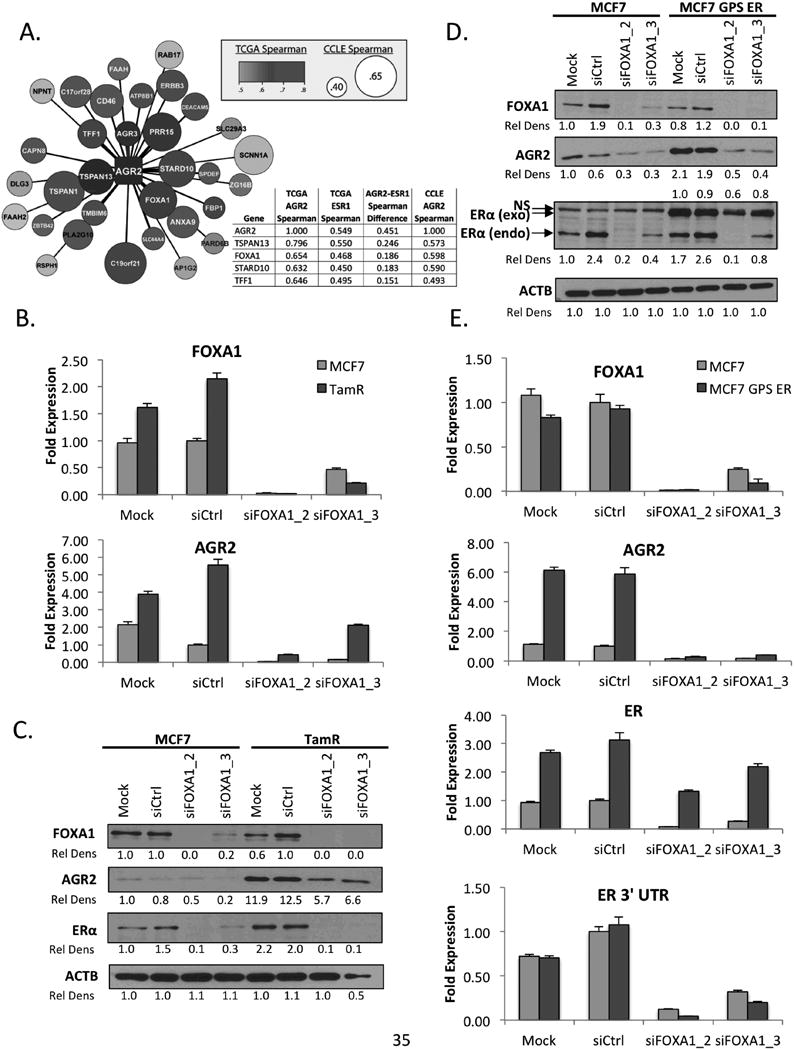
A. Definition of the AGR2 expression network identifies FOXA1 as a regulator of AGR2 expression. A comparative analysis of CCLE and TCGA was performed to predict genes whose expression is highly associated with AGR2. From the genes identified, FOXA1 was one of the top represented genes of the list. CCLE correlation is represented by size with larger circles predicting higher co-expression and TCGA correlation is represented based on color, Red>Purple>Blue>White.
B. FOXA1 regulates AGR2 mRNA expression. MCF7 and TamR cells were treated with siFOXA1 and plated for 4 days in phenol red free media supplemented with CFS. FOXA1 and AGR2 expression was analyzed by qRT-PCR, normalized to the expression of the 36B4 housekeeping gene and presented as fold change relative to siCtrl treated cells.
C. FOXA1 regulates AGR2 expression in MCF7 and TamR cells. MCF7 and TamR cells were treated with siFOXA1 for 4 days in phenol red free media with CFS. The resulting whole cell extracts were immunoblotted for FOXA1, AGR2, Erα and β-Actin (ACTB) expression.
D. FOXA1 regulates AGR2 expression when ERα expression is induced. MCF7 and MCF7 GPS ER cells were treated with siFOXA1 for 4 days in phenol red free media with CFS. The resulting whole cell extracts were immunoblotted for FOXA1, AGR2, ERα and β-Actin (ACTB) expression.
E. FOXA1 regulates AGR2 mRNA expression with induced ER expression. MCF7 and MCF7 GPS ER cells were treated with siFOXA1 and plated for 4 days in phenol red free media supplemented with CFS. FOXA1, ERα 3′ UTR, ERα and AGR2 expression was analyzed by qRT-PCR, normalized to the expression of the 36B4 housekeeping gene and presented as fold change relative to siCtrl treated cells.
Discussion
Recently, there has been renewed interest in identifying specific genes that are causally linked to the development of resistance to endocrine therapy in breast cancer and the identification of pathways downstream of ERα that may be amenable to targeted therapeutic intervention in this disease. It is within this context that we undertook a study to probe the role of the proto-oncogene AGR2 in tamoxifen pharmacology in breast cancer. In general, ERα+ breast tumors are associated with a more favorable prognosis, considered to be less aggressive, and are likely to respond to SERMs such as tamoxifen. However, ERα+ AGR2 overexpressing breast tumors treated with tamoxifen are significantly more aggressive and exhibit a poor prognosis (8, 12, 15). Consequently, ERα+ AGR2 overexpressing tumors are considered to be intrinsically resistant to tamoxifen and/or are poised to develop resistance to endocrine therapy. It was of significance, therefore, that we observed that heterologous expression of AGR2 increased the growth of MCF7-cell derived xenografts and altered their response to tamoxifen. These findings suggest a causal role for AGR2 in the processes of pathological importance in breast cancer and highlight its likely role in the development of tamoxifen resistance.
Our interest in AGR2 originated from the observation that it was an estrogen responsive gene that was overexpressed in breast cancer. However, it has since been found to be expressed in many cancer types including pancreatic, ovarian and esophageal carcinomas (10, 11, 29), where it plays critical roles in cell migration, growth factor secretion and proliferation. Likewise, in breast cancer, we and others have shown that AGR2 expression promotes cell survival, proliferation and metastasis (7, 8). Of particular importance was the finding that elevated expression of AGR2 in the index biopsy of primary tumors is associated with early progression in patients treated with tamoxifen (8), a result that has led to the suggestion that this protein may contribute to tamoxifen resistance. These findings heightened interest in using AGR2 as a potential biomarker of resistance to endocrine therapy in ERα+ breast cancers. However, until this current study, the mechanisms by which AGR2 influences the development of tamoxifen resistance had yet to be explored. In this study, it was shown that AGR2 was absolutely required for the growth of both tamoxifen sensitive and tamoxifen resistant breast cancer cells in vitro. Further, it was demonstrated that heterologous overexpression of AGR2 enhanced the estrogen dependent growth of MCF-7 derived tumor xenografts and that the resultant tumors were less sensitive to the antiestrogenic actions of tamoxifen. These findings also highlight the importance of the observation that AGR2 expression was elevated in a validated model of tamoxifen resistant breast cancer and that its expression loses its dependence on ERα.
Previously, we determined that the induction of AGR2 expression by tamoxifen in MCF-7 cells is a direct, ERα dependent, transcriptional response. However, we are as yet unable to reconcile these findings with those of others in the same cell line, which indicates that the induction of AGR2 gene expression in response to tamoxifen occurs as a result of the non-genomic activity of ERα on signaling molecules in an AKT dependent manner (30). This encouraged us to undertake a comparative bioinformatics analysis to define proteins and/or processes that may be involved in AGR2 expression in this context. The results of this analysis informed a series of genetic studies that indicated that FOXA1 was required for AGR2 expression in the assayed breast cancer cell types. However, the observation that FOXA1 expression levels did not change as cells progressed from a tamoxifen sensitive to a tamoxifen resistant state suggested that increased FOXA1 activity was a key contributor to the resistance phenotype. Identification of the proteins and/or processes that impact FOXA1 activity is thus a major focus of our efforts in this area.
In both MCF7 and TamR cellular models, we found that knockdown of AGR2 expression resulted in a quantitative downregulation of ERα gene transcription and a loss of the receptor protein. A similar conclusion was arrived at by others studying ERα expression in T47D and ZR751 breast cancer cell models (7). However, the mechanisms by which this occurs are elusive and resolution of this issue awaits a more comprehensive analysis of the biochemical activities and potential targets of AGR2. Regardless, it appears that ERα can regulate AGR2 expression and that this in turnsup regulates ERα expression. Interestingly, although AGR2 expression in tamoxifen resistant cells no longer requires ERα, the sustained expression of ERα still requires AGR2. Thus, the ERα/AGR2 step of the normal ERα/AGR2/ERα regulatory loop is dysregulated in tamoxifen resistant cancer as a consequence, we believe, of an alteration in FOXA1 activity.
This study highlights the importance of the FOXA1/ERα/AGR2 signaling axis in luminal breast cancers and its likely utility as a therapeutic target for the treatment of breast cancer. The importance of FOXA1 in breast cancer physiology has been established, although in and of itself is not likely to be a druggable target. On the other hand, since it is a secreted protein, AGR2 may be a viable target using a neutralizing antibody approach. Such an intervention may have utility as an alternative to, or in conjunction with SERMs or SERDs, as a treatment for ERα-positive cancers. It also remains to be determined how, with no change in expression, FOXA1 is able to sustain the expression of AGR2 in a manner that no longer requires ERα. Indeed, it is inferred from our work that the pathways that impinge on FOXA1 to facilitate its ERα independent activity are also likely to be useful targets for the treatment of breast cancer. Taken together, this study shows that the FOXA1/ERα/AGR2 signaling axis is a viable option for therapeutic intervention and that the specific targeting of the proto-oncogene AGR2 is likely to provide an option for the future therapeutic treatment of breast cancer.
Supplementary Material
Implications: These findings reveal the transcriptional interplay between FOXA1 and ERα in controlling AGR2 during the transition from therapy-sensitive to -resistant breast cancer and implicate AGR2 as a relevant therapeutic target.
Acknowledgments
We thank Christina A Chao and Nicole J Carver for assisting with the in vivo work as well as members of the McDonnell laboratory for their insightful discussions.
This work was supported by a National Institutes of Health Grant R37DK048807 (DPM), a research grant from Pfizer Pharmaceuticals, Inc. (DPM) and a DOD Breast Cancer Postdoctoral Fellowship W81XWH11-1-0601 (TMW).
Footnotes
Disclosure of Potential Conflicts of Interest: The authors disclose no potential conflicts of interest.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1.American Cancer Society. Facts and Figures. 2013 Available from: http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-036845.pdf.
- 2.Arpino G, De Angelis C, Giuliano M, Giordano A, Falato C, De Laurentiis M, et al. Molecular mechanism and clinical implications of endocrine therapy resistance in breast cancer. Oncology. 2009;77(Suppl 1):23–37. doi: 10.1159/000258493. [DOI] [PubMed] [Google Scholar]
- 3.Berstein LM, Zheng H, Yue W, Wang JP, Lykkesfeldt AE, Naftolin F, et al. New approaches to the understanding of tamoxifen action and resistance. Endocrine-related cancer. 2003;10:267–77. doi: 10.1677/erc.0.0100267. [DOI] [PubMed] [Google Scholar]
- 4.Hurvitz SA, Pietras RJ. Rational management of endocrine resistance in breast cancer: a comprehensive review of estrogen receptor biology, treatment options, and future directions. Cancer. 2008;113:2385–97. doi: 10.1002/cncr.23875. [DOI] [PubMed] [Google Scholar]
- 5.Ma XJ, Hilsenbeck SG, Wang W, Ding L, Sgroi DC, Bender RA, et al. The HOXB13:IL17BR expression index is a prognostic factor in early-stage breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:4611–9. doi: 10.1200/JCO.2006.06.6944. [DOI] [PubMed] [Google Scholar]
- 6.McDonnell DP, Wardell SE. The molecular mechanisms underlying the pharmacological actions of ER modulators: implications for new drug discovery in breast cancer. Current opinion in pharmacology. 2010;10:620–8. doi: 10.1016/j.coph.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanderlaag KE, Hudak S, Bald L, Fayadat-Dilman L, Sathe M, Grein J, et al. Anterior gradient-2 plays a critical role in breast cancer cell growth and survival by modulating cyclin D1, estrogen receptor-alpha and survivin. Breast cancer research : BCR. 2010;12:R32. doi: 10.1186/bcr2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hrstka R, Nenutil R, Fourtouna A, Maslon MM, Naughton C, Langdon S, et al. The pro-metastatic protein anterior gradient-2 predicts poor prognosis in tamoxifen-treated breast cancers. Oncogene. 2010;29:4838–47. doi: 10.1038/onc.2010.228. [DOI] [PubMed] [Google Scholar]
- 9.Hu Z, Gu Y, Han B, Zhang J, Li Z, Tian K, et al. Knockdown of AGR2 induces cellular senescence in prostate cancer cells. Carcinogenesis. 2012;33:1178–86. doi: 10.1093/carcin/bgs141. [DOI] [PubMed] [Google Scholar]
- 10.Ramachandran V, Arumugam T, Wang H, Logsdon CD. Anterior gradient 2 is expressed and secreted during the development of pancreatic cancer and promotes cancer cell survival. Cancer research. 2008;68:7811–8. doi: 10.1158/0008-5472.CAN-08-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Hao Y, Lowe AW. The adenocarcinoma-associated antigen, AGR2, promotes tumor growth, cell migration, and cellular transformation. Cancer research. 2008;68:492–7. doi: 10.1158/0008-5472.CAN-07-2930. [DOI] [PubMed] [Google Scholar]
- 12.Innes HE, Liu D, Barraclough R, Davies MP, O'Neill PA, Platt-Higgins A, et al. Significance of the metastasis-inducing protein AGR2 for outcome in hormonally treated breast cancer patients. British journal of cancer. 2006;94:1057–65. doi: 10.1038/sj.bjc.6603065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritzsche FR, Dahl E, Pahl S, Burkhardt M, Luo J, Mayordomo E, et al. Prognostic relevance of AGR2 expression in breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:1728–34. doi: 10.1158/1078-0432.CCR-05-2057. [DOI] [PubMed] [Google Scholar]
- 14.Wu ZS, Wu Q, Ding XD, Wang HQ, Shen YX, Fang SY. [Expression of a novel metastasis-inducing protein human anterior gradient-2 (AGR2) in breast cancer and its clinical and prognostic significance] Zhonghua bing li xue za zhi Chinese journal of pathology. 2008;37:109–13. [PubMed] [Google Scholar]
- 15.Barraclough DL, Platt-Higgins A, de Silva Rudland S, Barraclough R, Winstanley J, West CR, et al. The metastasis-associated anterior gradient 2 protein is correlated with poor survival of breast cancer patients. The American journal of pathology. 2009;175:1848–57. doi: 10.2353/ajpath.2009.090246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wardell SE, Nelson ER, Chao CA, McDonnell DP. Bazedoxifene exhibits antiestrogenic activity in animal models of tamoxifen-resistant breast cancer: implications for treatment of advanced disease. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:2420–31. doi: 10.1158/1078-0432.CCR-12-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hengel SM, Murray E, Langdon S, Hayward L, O'Donoghue J, Panchaud A, et al. Data-independent proteomic screen identifies novel tamoxifen agonist that mediates drug resistance. Journal of proteome research. 2011;10:4567–78. doi: 10.1021/pr2004117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salmans ML, Zhao F, Andersen B. The estrogen-regulated anterior gradient 2 (AGR2) protein in breast cancer: a potential drug target and biomarker. Breast cancer research : BCR. 2013;15:204. doi: 10.1186/bcr3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wardell SE, Kazmin D, McDonnell DP. Research resource: Transcriptional profiling in a cellular model of breast cancer reveals functional and mechanistic differences between clinically relevant SERM and between SERM/estrogen complexes. Mol Endocrinol. 2012;26:1235–48. doi: 10.1210/me.2012-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abraham JE, Harrington P, Driver KE, Tyrer J, Easton DF, Dunning AM, et al. Common polymorphisms in the prostaglandin pathway genes and their association with breast cancer susceptibility and survival. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:2181–91. doi: 10.1158/1078-0432.CCR-08-0716. [DOI] [PubMed] [Google Scholar]
- 21.Hurtado A, Holmes KA, Geistlinger TR, Hutcheson IR, Nicholson RI, Brown M, et al. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;456:663–6. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knowlden JM, Hutcheson IR, Jones HE, Madden T, Gee JM, Harper ME, et al. Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology. 2003;144:1032–44. doi: 10.1210/en.2002-220620. [DOI] [PubMed] [Google Scholar]
- 23.Connor CE, Norris JD, Broadwater G, Willson TM, Gottardis MM, Dewhirst MW, et al. Circumventing tamoxifen resistance in breast cancers using antiestrogens that induce unique conformational changes in the estrogen receptor. Cancer research. 2001;61:2917–22. [PubMed] [Google Scholar]
- 24.Verma S, Salmans ML, Geyfman M, Wang H, Yu Z, Lu Z, et al. The estrogen-responsive Agr2 gene regulates mammary epithelial proliferation and facilitates lobuloalveolar development. Developmental biology. 2012;369:249–60. doi: 10.1016/j.ydbio.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nature genetics. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–93. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Ali TZ, Zhou H, D'Souza DR, Lu Y, Jaffe J, et al. ErbB3 binding protein 1 represses metastasis-promoting gene anterior gradient protein 2 in prostate cancer. Cancer research. 2010;70:240–8. doi: 10.1158/0008-5472.CAN-09-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng W, Rosenstiel P, Huse K, Sina C, Valentonyte R, Mah N, et al. Evaluation of AGR2 and AGR3 as candidate genes for inflammatory bowel disease. Genes and immunity. 2006;7:11–8. doi: 10.1038/sj.gene.6364263. [DOI] [PubMed] [Google Scholar]
- 29.Park K, Chung YJ, So H, Kim K, Park J, Oh M, et al. AGR2, a mucinous ovarian cancer marker, promotes cell proliferation and migration. Experimental & molecular medicine. 2011;43:91–100. doi: 10.3858/emm.2011.43.2.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hrstka R, Murray E, Brychtova V, Fabian P, Hupp TR, Vojtesek B. Identification of an AKT-dependent signalling pathway that mediates tamoxifen-dependent induction of the pro-metastatic protein anterior gradient-2. Cancer letters. 2013;333:187–93. doi: 10.1016/j.canlet.2013.01.034. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–53. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroder MS, Culhane AC, Quackenbush J, Haibe-Kains B. survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics. 2011;27:3206–8. doi: 10.1093/bioinformatics/btr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research N. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nature genetics. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukushima A. DiffCorr: an R package to analyze and visualize differential correlations in biological networks. Gene. 2013;518:209–14. doi: 10.1016/j.gene.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 37.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. [Google Scholar]
- 38.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DuSell CD, Umetani M, Shaul PW, Mangelsdorf DJ, McDonnell DP. 27-hydroxycholesterol is an endogenous selective estrogen receptor modulator. Mol Endocrinol. 2008;22:65–77. doi: 10.1210/me.2007-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


