Abstract
Trypanosoma brucei is a vector borne, lethal protistan parasite of humans and livestock in sub-Saharan Africa. Antigenic Variation of its cell surface coat enables the parasite to evade adaptive immune responses and to live freely in the blood of its mammalian hosts. The coat consists of ten million copies of variant surface glycoprotein (VSG) that is expressed from a single VSG gene, drawn from a large repertoire and located near the telomere at one of fifteen so-called bloodstream expression sites (BESs). Thus, antigenic variation is achieved by switching to the expression of a different VSG gene. A BES is a tandem array of expression site-associated genes and a terminal VSG gene. It is polycistronically transcribed by a multifunctional RNA polymerase I (RNAPI) from a short promoter that is located 45–60 kb upstream of the VSG gene. The mechanism(s) restricting VSG expression to a single BES are not well understood. There is convincing evidence that epigenetic silencing and transcription attenuation play important roles. Furthermore, recent data indicated that there is regulation at the level of transcription initiation and that, surprisingly, the VSG mRNA appears to have a role in restricting VSG expression to a single gene. Here, we review BES expression regulation and propose a model in which telomere-directed, epigenetic BES silencing is opposed by BES promoter-directed, activated RNAPI transcription.
1. Introduction
The tsetse borne, unicellular parasite Trypanosoma brucei, which belongs to the phylogenetic order Kinetoplastida, is the only known organism that has evolved a multifunctional RNA polymerase I (RNAPI) system. This system is used to transcribe ribosomal gene units (RRNA) in the nucleolus, as in all eukaryotes, yet also to transcribe gene units that encode the parasite’s major cell surface antigens (Kooter and Borst, 1984; Günzl et al., 2003). Trypanosomes have a unique mode of protein coding gene expression that allows them to utilize other RNA polymerases than RNAPII for the production of functional mRNA. In their genome, protein coding genes are arranged in long tandem arrays which are polycistronically transcribed. The precursor RNA is processed by spliced leader (SL) trans splicing and polyadenylation, resulting in mature, monocistronic mRNAs (Günzl, 2010; Michaeli, 2011; Preußer et al., 2012). Since in trans splicing the same capped leader sequence, derived from the SL RNA, is spliced onto the 5/ end of each mRNA, this process represents a post-transcriptional mode of capping that is decoupled from RNAPII transcription. Consequently, trypanosomes, in contrast to mammals (Grummt and Skinner, 1985), are able to use RNAPI to effectively and specifically express endogenous gene units that encode their major cell surface antigens (Rudenko et al., 1991; Zomerdijk et al., 1991a). This antigen, in mammalian-infective metacyclic and bloodstream form (BF) trypanosomes, is known as the variant surface glycoprotein (VSG), while the major cell surface antigen in insect-stage procyclic form trypanosomes is procyclin.
T. brucei causes Human and Animal African Trypanosomiasis (also known as Sleeping Sickness and Nagana, respectively) throughout sub-Saharan Africa (Fevre et al., 2006). The parasite lives freely in the bloodstream of its mammalian host, evading the immune system by antigenic variation of its cell surface coat. The coat consists of ten million copies of the same VSG, shielding invariant membrane proteins from immune recognition (Schwede et al., 2011). T. brucei possesses roughly 2500 different VSG genes and pseudogenes (Cross et al., 2014), and periodic switching to the expression of an alternative VSG gene leads to antigenic variation. VSG genes are located in subtelomeric regions of 11 megabase, 5 intermediate-sized and ~100 minichromosomes covering, in total, ~30% of the genome (Ersfeld, 2011; Horn, 2014). However, the active VSG gene is invariably located next to the telomere within an expression site, with the coding region ending ~200–1800 bp upstream of the telomeric repeats. Metacyclic trypanosomes express a single VSG monocistronically from one of five metacyclic expression sites in which the RNAPI promoter is located ~1–4 kb upstream of the coding region (Ginger et al., 2002; Kolev et al., 2012; Cross et al., 2014). Conversely, BFs express the active VSG from one of fifteen polycistronic “bloodstream expression sites” (BESs) which comprise a tandem array of typically 8–9 expression-site associated genes (ESAGs) and a terminal VSG gene (Figure 1) (Hertz-Fowler et al., 2008). ESAGs appear to be important for the successful infection of the mammalian host since they encode a variant heterodimeric transferrin receptor (ESAG6 and ESAG7), whose varying affinity for transferrins of different host species is thought to expand the parasite’s host range (Bitter et al., 1998). These also encode adenylate cyclases (ESAG4) that inhibit the innate immune system upon trypanosome lysis (Salmon et al., 2012).
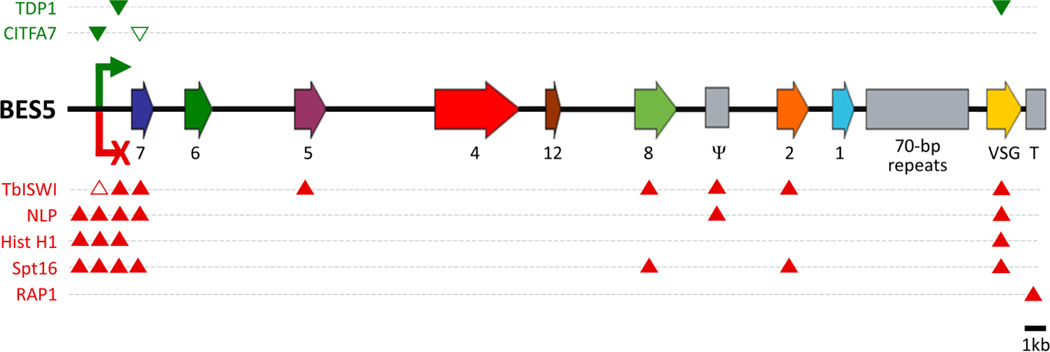
Fig. 1. Schematic outline of BES5 and interacting proteins.
Depiction of BES5 (to scale) as a representative BES according to the published sequence (Hertz-Fowler et al., 2008). The diagram includes ESAGs (labeled 1, 2, 4–8 and 12), a VSG pseudogene (Ψ), 70 bp repeats preceding the terminal VSG gene, and the telomeric repeats (T). Note that some BESs have an additional promoter and an ESAG10 gene ~14 kb upstream of the depicted promoter (not shown). The green arrow and red X represent the promoter when the BES is in the active state and silent state, respectively. Activating factors that are predominantly associated with the active BES are indicated above the diagram in green whereas factors which are implied in BES silencing are listed below the diagram in red. Filled and empty arrowheads indicate positive and negative ChIP results, respectively. Histone H1 and SPT16 associate predominately with silent sites whereas TbISWI and NLP were shown to interact equally with expression sites in both states.
The BES promoter resides 45–60 kb upstream of the telomere (Zomerdijk et al., 1990). It extends only 67 bp upstream of the transcription initiation site and comprises two short sequence elements (Vanhamme et al., 1995; Pham et al., 1996). Both elements are required for efficient binding of the multi-subunit class I transcription factor A (CITFA) which is essential for RNAPI transcription in the trypanosome (Brandenburg et al., 2007). The active BES is transcribed outside the nucleolus (Chaves et al., 1998), apparently in a small compartment termed the expression site body (ESB) (Navarro and Gull, 2001). In BFs the switch to the expression of another VSG occurs by two principal ways: either the active BES is silenced while one of the silent BESs is activated, or a DNA recombination event replaces the VSG gene in the active BES with a VSG gene from the repertoire.
Antigenic variation and mono-allelic VSG expression in T. brucei have been a research focus for decades. Several factors involved in BES silencing have been identified (see below) and BES silencing has been linked to DNA replication/ORC1 (Tiengwe et al., 2012; Benmerzouga et al., 2013), chromosome maintenance (Kim et al., 2013), and association of BESs with the nuclear lamina (DuBois et al., 2012). In addition, cohesin plays a critical role in maintaining the activated state of the BES during the cell cycle (Landeira et al., 2009). Recently, excellent and detailed reviews have addressed antigenic variation in trypanosomes and the biology of BES silencing (Horn and McCulloch, 2010; Rudenko, 2010; Alsford et al., 2012; Glover et al., 2013; Horn, 2014). Here we focus on the most recent findings of factors that appear to be directly involved in BES regulation, and propose a model in which BES-specific telomeric silencing is opposed by a mechanism that activates transcription initiation at the promoter of the active BES.
2. Telomeric Silencing
The active VSG gene, independent of whether it resides in metacyclic or bloodstream expression sites, is invariably located near the telomere, indicating that the telomere has an essential function in regulating VSG expression. Accordingly, repression of RNAPI-mediated transcription by the telomere was directly demonstrated by integrating a plasmid with seeds for de novo telomere formation either at BESs or, internally, at RRNA loci (Glover and Horn, 2006). At the latter, tight repression extended only 2 kb upstream of telomeric repeats whereas, at inactive BESs, repression reached at least 5 kb in these experiments. The more extended repression of silent BESs was consistent with previous findings in which integration of RNAPI promoter-driven reporter cassettes at different positions of a silent BES were repressed, even when placed 14 kb upstream of the telomere (Horn and Cross, 1997). Several lines of evidence suggest that the pronounced silencing of BESs is dependent on the telomere. Depletion of the telomeric protein RAP1 led to de-repression of silent BESs, co-expression of multiple BES-encoded VSG genes, and the formation of additional extranucleolar RNAPI foci (Yang et al., 2009). Furthermore, depletion of the disruptor of telomeric silencing B (DOT1B), which methylates lysine 76 of trypanosome histone H3 (Janzen et al., 2006), similarly led to de-repression of silent BESs (Figueiredo et al., 2008). Direct evidence for repression of a BES from the telomere stems from a recent study in which induced expression of a VSG transgene, inserted into one of the RRNA loci, surprisingly led to a short-term, reversible attenuation of the active BES, indicating that VSG mRNA plays a direct role in the regulation of mono-allelic VSG expression (Batram et al., 2014). Interestingly, a time course experiment showed that this silencing of the active BES spread from the telomere towards the BES promoter in a DOT1B-dependent manner (Batram et al., 2014). Together, these data strongly indicated that BES silencing is directed by the telomere. Furthermore, it is likely that the VSG gene on silent BESs is protected from RNAPI transcription by more than one mechanism because DOT1B knockout cells could still shut down the active VSG gene upon ectopic VSG expression but were unable to attenuate expression of the remainder of the active BES (Batram et al., 2014).
3. BES transcription attenuation
Inactive BESs are completely silent only in regard to their telomere-proximal regions, including the terminal VSG gene. VSG mRNA from inactive BESs is 104 to 105-fold less abundant than that from the active BES (Figueiredo et al., 2008; Yang et al., 2009). Despite this strong difference, several observations have shown that transcription does initiate at silent BESs at a clearly detectable level. The first evidence came from a study in which insertion of a selectable marker gene 1 kb downstream of a “silent” BES promoter led to resistant parasites (Navarro and Cross, 1996). BES sequences are highly similar, especially at the promoter and in the proximal downstream region, differing from each other only by a few single nucleotide polymorphisms. However, the first genes within BESs are ESAG7 and ESAG6 which encode the heteromeric transferrin receptor and harbor short hypervariable regions that distinguish them from each other (Zomerdijk et al., 1991b). Analysis of ESAG6 cDNA sequences, which on BESs are located ~5 kb downstream from the promoter, revealed that 20% of the ESAG6 mRNA in BFs was derived from various silent BESs whereas 80% stemmed from the active BES, demonstrating that, even in the absence of selective pressure, productive transcription did occur in the promoter-proximal domain of inactive BESs (Ansorge et al., 1999). Subsequently, a vast cDNA clone analysis along whole BESs showed that silent BESs contributed much more to the promoter-proximal cDNA pool than to pools of promoter-distant cDNAs, revealing that transcription that initiated at silent BESs was attenuated along the BES (Vanhamme et al., 2000). Recently, this approach was repeated with single cells, confirming that silent BESs are transcribed in their promoter-proximal region and that transcription was attenuated further downstream within a single trypanosome (Kassem et al., 2014). Finally, the demonstration that BES silencing spreads gradually from telomere to promoter and BES reactivation occurs gradually in the opposite direction (Batram et al., 2014) strongly supports the notion that transcription is attenuated at silent BESs.
4. Regulation of BES transcription Initiation
Although the promoters of inactive BESs are not “silent”, there is now convincing evidence that there is substantial regulation at BES promoters. Consistently, promoter-proximal RNA levels were found to be much higher from the active versus silent BESs. Thus, when the neomycin phosphotransferase gene (NEO) was inserted 1 kb downstream of the promoter of an inactive BES, it conferred parasite resistance to a low concentration of the drug G418 (1 µg/ml) while the same gene, when inserted at the identical position of an active BES, boosted resistance at least 100-fold (Navarro and Cross, 1996). The finding that in BFs 80% of ESAG6 mRNA stemmed from the active BES suggested that there is at least a 50-fold stronger ESAG6 expression from the active BES than from the average silent BES. When Yang et al. (2009) introduced a luciferase gene immediately downstream of the active or a silent BES promoter, the active BES produced 1500–4000-fold more light units than the silent BES.
More direct evidence for BES regulation at the level of transcription initiation came from the analysis of CITFA. CITFA consists of seven subunits, CITFA1–7, which are conserved only among kinetoplastid organisms, and the dynein light chain DYNLL1 (also known as LC8). Silencing of CITFA1, CITFA2 and CITFA7 was lethal to BFs grown in culture and strongly and specifically reduced the abundance of rRNA and VSG mRNA (Brandenburg et al., 2007; Nguyen et al., 2012; Park et al., 2014). Accordingly, depletion of CITFA2 from extract virtually abolished RNAPI transcription in vitro, as assayed by ~100 bp-long primer extension products, and the purified CITFA complex produced a specific gel shift with the BES promoter (Brandenburg et al., 2007). Moreover, a ChIP-seq analysis indicated that within a BES, CITFA7 occupancy was restricted to the promoter region (Nguyen et al., 2014). Together, these findings identified CITFA as a basal and general transcription initiation factor for RNAPI transcription in trypanosomes.
Interestingly, marking the active BES and a silent BES ~500 bp downstream of the transcription initiation site (Figueiredo et al., 2008) revealed that CITFA2 and CITFA7 predominantly occupied the promoter of the active BES relative to that of the marked silent BES, a phenotype that was maintained after consecutive in situ switches between the two marked sites (Nguyen et al., 2014). In accordance with CITFA’s role as an RNAPI transcription initiation factor, higher CITFA occupancy at the active versus the silent BES promoter correlated with a ~70-fold higher abundance of promoter-proximal, unspliced RNA and a ~17-fold higher occupancy of the RNAPI-specific subunit RPB6z at the marker gene (Nguyen et al., 2014). Finally, CITFA7 silencing led to a strong reduction of RNAPI occupancy and of promoter-proximal RNA levels, which directly demonstrated that CITFA binding to the promoter is required for high transcription rates in vivo (Nguyen et al., 2014). These data unequivocally showed that mono-allelic BES expression entails a mechanism that functions at the BES promoter, apparently limiting access of CITFA to silent BES promoters and/or ensuring maximal promoter occupancy of CITFA at the active BES.
It should be noted that this mechanism is not an “all or nothing”-mechanism because, in these experiments, the marked silent BES promoter was consistently occupied by CITFA above the level of negative control experiments. This finding is in accordance with promoter-proximal transcription occurring at silent BESs (see above) and it likely explains why hypersensitive DNase I sites in the promoter region, indicative of a bound transcription factor, were not restricted to the active BES but were also detected at a silent BES (Navarro and Cross, 1998). It appears that trypanosomes cannot completely shut down transcription initiation from silent BESs. Alternatively, low level transcription of the promoter-proximal part of silent BESs might serve a biological function. For instance, co-expression of different forms of the heteromeric transferrin receptor, e.g. ESAG6 and ESAG7, could ensure initial survival in different mammalian hosts.
5. Factors involved in BES transcription regulation
There is strong evidence that inactive BESs are silenced epigenetically. Thus, while silent BESs have a nucleosomal structure, the active BES is largely depleted of nucleosomes (Figueiredo and Cross, 2010; Stanne and Rudenko, 2010). Direct evidence that nucleosomes are important for BES promoter silencing stems from depleting histone H3, which rapidly led to a ~11-fold de-repression of a GFP gene introduced downstream of the promoter of a silent BES (Alsford and Horn, 2012). In addition, CAF-1b, a replication-dependent histone chaperone, and the replication-independent chaperone ASF1A, were shown to be important for the inheritance and maintenance of the silenced state of BESs (Alsford and Horn, 2012). Interestingly, silencing the gene of either chaperone led to apparent nucleosome depletion and a de-repression of the promoter-proximal BES region. However, it did not affect expression of the corresponding VSG gene suggesting that nucleosomal structure is particularly important for the regulation of BES promoter activity.
Several chromatin remodeling and modifying proteins have been implicated in BES repression so far. The first epigenetic factor found to play a role in BES regulation was the chromatin remodeler TbISWI (Hughes et al., 2007). Depletion of this factor increased the mRNA abundance of a reporter gene inserted promoter-proximally into a silent BES up to 60-fold, whereas only a fivefold increase of the corresponding silent VSG mRNA was observed. TbISWI was found to occupy the entire length of both silent and active BESs, but was not enriched at BES promoters (Stanne et al., 2011). Although the specific function of TbISWI remains to be determined, these results suggest that TbISWI controls RNAPI transcription elongation rather than initiation.
Similar de-repression of a promoter-proximal reporter gene was observed when the histone deacetylase DAC3 (Wang et al., 2010), the linker histone H1 (Povelones et al., 2012; Pena et al., 2014), or the nucleoplasmin-like protein NLP (Narayanan et al., 2011) was depleted. The function of DAC3 appears to be promoter-specific since expression of the VSG gene in the marked BES was unaffected at both the mRNA and the protein level (Wang et al., 2010). However, direct association of DAC3 with BESs has not been demonstrated yet and it remains a possibility that DAC3’s control of BES silencing is indirect.
The role of histone H1 in BES promoter repression has been more deeply investigated. Histone H1 is important for chromatin architecture and generally functions in chromatin condensation and transcription repression (Happel and Doenecke, 2009). Accordingly, co-silencing of the T. brucei H1 multigene family opened up chromatin globally with the strongest effect on silent BES promoters (Pena et al., 2014). Metabolic labeling of nascent RNA then showed that histone H1 depletion resulted in an approximately six-fold higher promoter-proximal transcription rate at a silent BES, indicating that relaxation of the nucleosome structure in the promoter region led to an increase of the transcription initiation rate at the silent BES (Pena et al., 2014).
NLP is a ubiquitous nuclear protein and, accordingly, was found to be associated with all genomic loci analyzed, including the active and silent BESs (Narayanan et al., 2011). Despite this apparent general association with genomic DNA, NLP seems to be particularly important for BES promoter regulation. Depletion of NLP de-repressed a silent BES 45–65-fold, as measured by fluorescence derived from a promoter-proximal GFP gene. Moreover, NLP silencing also reduced promoter-proximal gene expression from the active BES about threefold (Narayanan et al., 2011). While it was speculated that NLP may have a dual function in BES silencing and in promoting processive transcription at the active BES (Narayanan et al., 2011), it is equally possible that loss of NLP enabled competition between silent and the active BES for the RNAPI transcription machinery. However, the specific function of NLP in BES regulation remains to be determined.
SPT16 is a subunit of the trypanosome FACT (“facilitates chromatin transcription”) complex (Patrick et al., 2008) and appears to have a direct role in BES promoter silencing because it was found highly enriched at a silent BES promoter (Denninger et al., 2010). Accordingly, SPT16 silencing increased promoter-proximal GFP expression from a silent BES up to 25-fold, yet de-repression did not extend to the VSG genes of silent BESs. However, the de-repression effect was strongly correlated with an arrest in the G2/early M cell cycle phase, raising the possibility that SPT16 does not generally facilitate BES repression in the bloodstream trypanosome. Moreover, SPT16 depletion strongly reduced VSG expression from the active BES, suggesting a separate BES-related function of SPT16 in facilitating processive RNAPI transcription. Overall, the specific function of FACT in the multifunctional RNAPI system remains unclear. While SPT16 has been co-purified with RNAPII of the related organism Leishmania major (Martinez-Calvillo et al., 2007), its association with T. brucei RNAPI remains to be shown.
The epigenetic factors discussed so far, including RAP1 and DOT1B (see section 2, Telomeric silencing, above), function in BES silencing. The only such factor found to be important for efficient transcription of the active BES is the high mobility group protein TDP1, which belongs to a family of architectural chromatin proteins (Narayanan and Rudenko, 2013). Interestingly, TDP1 exhibited an inverse occupancy pattern to the core histone H3 at RNAPI-transcribed loci and was up to fivefold more abundant at the active BES promoter relative to a silent BES promoter. Accordingly, TDP1 depletion decreased the abundance of pre-rRNA and VSG mRNA from the active BES. In addition, TDP1, a nuclear protein, exhibited predominant localization to the nucleolus and the ESB, and its DNA association was found throughout the active BES and RRNA gene units. Thus, it appears that TDP1 facilitates high rates of processive RNAPI transcription required for trypanosome survival (Narayanan and Rudenko, 2013).
6. A model of BES regulation
It is difficult to integrate the data from BES de-repression studies because, for most factors, specific functions in BES silencing have not been determined yet. Nevertheless, recent data strongly indicated that BES regulation occurs at both ends of expression sites. RAP1 and DOT1B depletion studies have clearly shown that BESs are silenced by a telomere-directed mechanism. Moreover, the demonstration that BES silencing spreads from the telomere towards the promoter (Batram et al., 2014) strongly supports a telomere-directed BES silencing mechanism. However, it is unlikely that this is the only mechanism regulating mono-allelic BES expression. If this was the case one would expect full activation of promoter-proximal transcription once telomeric silencing retreats beyond the promoter region, which should result in a leveling of the transcription rate between active and silent BESs (given the extremely high expression level of the active BES, it is unlikely that a trypanosome can support full activation of all fifteen BESs). However, in all cases reported, de-repression of silent BESs is, at best, moderate with the promoter-proximal expression level remaining manifold below that of the active BES. Furthermore, RAP1 silencing did not strongly affect promoter-proximal transcription of de-repressed BESs and had only a minor influence on the high expression level of the active BES (Yang et al., 2009). Similarly, DOT1B silencing did not affect expression of the active BES at all (Figueiredo et al., 2008). These results strongly argue for the presence of a separate mechanism involved in BES regulation.
Transcription attenuation has been proposed to be that mechanism and, as discussed, there is clear evidence that it does occur on silent BESs (Vanhamme et al., 2000; Kassem et al., 2014). Moreover, some data suggested that transcription attenuation is caused by inefficient transcript processing (Vanhamme et al., 2000) rather than by repressive chromatin. However, recent data do not support this scenario. The finding that RAP1 silencing caused gradual BES de-repression with the greatest effect on telomere-proximal genes, strongly indicated that transcription elongation on silent BESs is “antagonized” by telomere-directed spreading of repressive chromatin (Yang et al., 2009). Furthermore, upon removal of the apparent transcription elongation barrier, e.g. pronounced telomeric silencing, by RAP1 (Yang et al., 2009) or DOT1B (Figueiredo et al., 2008) depletion, promoter-proximal expression remained magnitudes below that of the active site, making it unlikely that transcription attenuation accounts for the strong difference in promoter-proximal transcription observed between active and silent BESs. Hence, transcription attenuation appears to be a consequence of epigenetic silencing rather than a regulatory mechanism, and seems to be in place to prevent the low level of transcription that does initiate at silent BESs from reaching the distally located VSG gene.
Based on the RAP1 silencing results on BES de-repression, Yang et al. (2009) suggested that there has to be a mechanism functioning on the BES promoter that could explain the striking difference in promoter-proximal expression levels between the active and de-repressed/silent BESs. The strongest support for this idea stems from the demonstration that CITFA, which is absolutely required for RNAPI transcription, is predominantly associated with the active BES promoter, versus a silent site, strongly indicating that there is a mechanism in place that allows CITFA to preferentially interact with the active BES. In addition, the fact that depletion of several epigenetic factors increased promoter-proximal transcription with no or very little effect on the downstream VSG gene further supports the notion of a promoter-dependent regulatory mechanism.
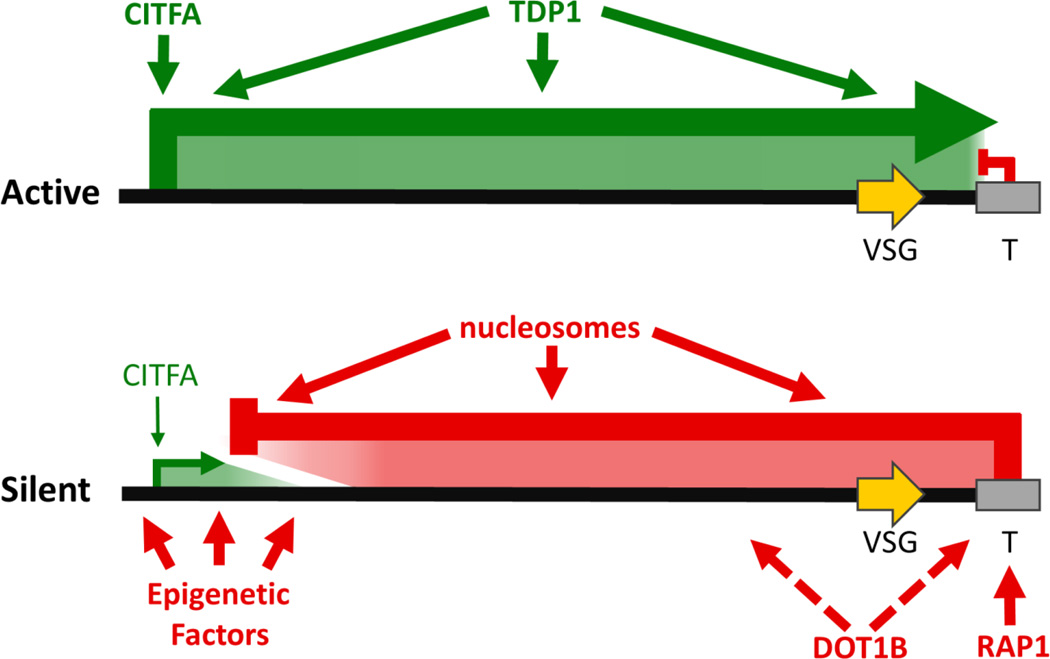
Taking these data into account, we propose a model in which BESs are regulated by two opposing forces, namely telomere-directed epigenetic silencing acting on silent BESs and activated transcription initiation at the active BES (Figure 2). In this model, the active BES promoter has unrestricted access to CITFA and RNAPI, allowing it to achieve the high transcription rate necessary for productive VSG expression. In addition, productive RNAPI transcription at the active BES is ensured by the presence of TDP1. At the same time, telomeric silencing at the active BES is impaired or pushed back so far that RNAPI transcription can extend productively past the VSG gene. In contrast, in this model, silent BES promoters are unable to recruit CITFA and RNAPI in sufficient amounts to allow for high transcription rates. In addition, a telomere-directed repressive epigenetic gradient spreading from the telomere into the BES causes transcription attenuation to prevent the low level of transcription, initiating at inactive BESs, from reaching the VSG gene.
Fig. 2. Model of BES regulation in T. brucei.
In the model of BES regulation, two opposing forces antagonize each other. The active BES is characterized by high transcription initiation rates and the lack of telomere-dependent epigenetic silencing, allowing unrestricted transcription elongation past the terminal VSG gene (green arrow). High processive transcription rates are facilitated by CITFA and TDP1. In silent BESs, low level RNAPI transcription initiation is opposed by BES-specific telomeric silencing that spreads towards the BES promoter causing transcription attenuation. This silencing depends on a nucleosomal structure, DOT1B and RAP1. RAP1 was shown to bind to telomeric repeats but the association of DOT1B with BESs remains to be determined (dotted line). Epigenetic factors may work together to build up a repressive chromatin structure at the promoter of silent BESs, preventing efficient binding of CITFA to its cognate DNA sequence elements. Alternatively, CITFA sequestration may limit the availability of the initiation factor for inactive BESs (not shown).
How are BES promoters differentially regulated? An obvious mechanism that could prevent CITFA from interacting with silent BES promoters and from recruiting RNAPI is a repressive chromatin structure at the promoter. The epigenetic factors whose depletion led to promoter-proximal BES de-repression, e.g. DAC3, histone H1 and NLP, may be important to build up a repressive chromatin structure at the promoter. If this is correct, depletion of these factors should lead to higher CITFA and RNAPI occupancies at de-repressed promoters. An alternative idea for the low promoter activity at “silent BESs” has been put forward in studies of CITFA. CITFA was found to be concentrated in both the nucleolus and the ESB (Nguyen et al., 2014), and it retained this localization even when its promoter-binding capability was impaired by depletion of the essential subunit CITFA1 (Park et al., 2014). It was therefore suggested that sequestration of CITFA into the nucleolus and the ESB could restrict maximal RNAPI transcription to these compartments. This idea is in line with a previous study in which BFs were forced to co-express two BESs simultaneously by antibiotic selection. The two marked BESs were consistently detected in close spatial proximity (Chaves et al., 1999), as if they were competing for an essential expression factor. Furthermore, it may explain why de-repressed BES promoters remain much less active than the promoter from the active BES.
Finally, this model of two opposing forces is supported by the monitoring of the shutdown/reactivation of the active BES upon ectopic expression of VSG mRNA (Batram et al., 2014). In these experiments the active BES was gradually inactivated from the telomere towards the promoter, most likely by an active, telomere-directed process of repressive chromatin spreading, whereas the reactivation of the same BES occurred in the reverse direction, possibly by removal of nucleosomes by the transcription machinery. An important question emanating from this study is how does the ectopically expressed VSG mRNA cross-talk to the active BES? Although this question is beyond the scope of this article, it is tempting to speculate that the VSG mRNA sequestered an important factor for RNAPI transcription, allowing repressive chromatin to spread onto, and silence, the active BES.
7. Conclusion
Mono-allelic VSG expression in T. brucei differs from other allelic exclusion systems, such as var gene expression in Plasmodium falciparum (Kirkman and Deitsch, 2012; Guizetti and Scherf, 2013) or olfactory receptor expression in mammals (Magklara and Lomvardas, 2013) by the fact that the active VSG gene must be transcribed at an extremely high rate to enable rapidly proliferating trypanosomes to completely cover themselves with VSG. The careful measurement of RNA abundances and half lives indicated that the active VSG gene is transcribed at a 50-fold higher rate than a β tubulin gene (Ehlers et al., 1987). At the same time, T. brucei must ensure that VSG genes on other BESs are not expressed. The parasite achieves this balancing act apparently by restricting full RNAPI transcription initiation to the active BES and by shielding VSG genes on silent BESs by a telomere-dependent silencing mechanism that causes attenuation of RNAPI transcription.
Highlights.
VSG expression from telomeric “bloodstream expression sites” (BESs) is mono-allelic
BES transcription is mediated by RNA polymerase I outside the nucleolus
BESs appear to be regulated by telomere-dependent silencing and by modulation of promoter activity
Acknowledgments
This work was supported by grant R01 AI059377 to A.G. from the National Institute of Allergy and Infectious Diseases (NIAID) of the U.S. National Institutes of Health (NIH).
Abbreviations
- BES
bloodstream expression site
- BF
bloodstream form [trypanosome]
- CITFA
class I transcription factor A
- DOT1B
disruptor of telomeric silencing
- ESAG
expression-site associated gene
- ESB
expression site body
- FACT
“facilitates chromatin transcription” factor
- NEO
neomycin phosphotransferase
- NLP
nucleoplasmin-like protein
- ORC1
Origin Recognition Complex subunit 1
- RNAP
RNA polymerase
- RRNA
ribosomal RNA gene unit
- SL
spliced leader
- VSG
variant surface glycoprotein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alsford S, duBois K, Horn D, Field MC. Epigenetic mechanisms, nuclear architecture and the control of gene expression in trypanosomes. Expert. Rev. Mol. Med. 2012;14:e13. doi: 10.1017/erm.2012.7. [DOI] [PubMed] [Google Scholar]
- Alsford S, Horn D. Cell-cycle-regulated control of VSG expression site silencing by histones and histone chaperones ASF1A and CAF-1b in Trypanosoma brucei. Nucleic Acids Res. 2012;40:10150–10160. doi: 10.1093/nar/gks813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge I, Steverding D, Melville S, Hartmann C, Clayton C. Transcription of 'inactive' expression sites in African trypanosomes leads to expression of multiple transferrin receptor RNAs in bloodstream forms. Mol. Biochem. Parasitol. 1999;101:81–94. doi: 10.1016/s0166-6851(99)00060-2. [DOI] [PubMed] [Google Scholar]
- Batram C, Jones NG, Janzen CJ, Markert SM, Engstler M. Expression site attenuation mechanistically links antigenic variation and development in Trypanosoma brucei. Elife. 2014;3:e02324. doi: 10.7554/eLife.02324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerzouga I, Concepcion-Acevedo J, Kim HS, Vandoros AV, Cross GA, Klingbeil MM, Li B. Trypanosoma brucei Orc1 is essential for nuclear DNA replication and affects both VSG silencing and VSG switching. Mol Microbiol. 2013;87:196–210. doi: 10.1111/mmi.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter W, Gerrits H, Kieft R, Borst P. The role of transferrin-receptor variation in the host range of Trypanosoma brucei. Nature. 1998;391:499–502. doi: 10.1038/35166. [DOI] [PubMed] [Google Scholar]
- Brandenburg J, Schimanski B, Nogoceke E, Nguyen TN, Padovan JC, Chait BT, Cross GA, Günzl A. Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex. EMBO J. 2007;26:4856–4866. doi: 10.1038/sj.emboj.7601905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Rudenko G, Dirks MA, Cross M, Borst P. Control of variant surface glycoprotein gene-expression sites in Trypanosoma brucei. EMBO J. 1999;18:4846–4855. doi: 10.1093/emboj/18.17.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Zomerdijk J, Dirks MA, Dirks RW, Raap AK, Borst P. Subnuclear localization of the active variant surface glycoprotein gene expression site in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 1998;95:12328–12333. doi: 10.1073/pnas.95.21.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross GA, Kim HS, Wickstead B. Capturing the variant surface glycoprotein repertoire (the VSGnome) of Trypanosoma brucei Lister 427. Mol. Biochem. Parasitol. 2014;195:59–73. doi: 10.1016/j.molbiopara.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Denninger V, Fullbrook A, Bessat M, Ersfeld K, Rudenko G. The FACT subunit TbSpt16 is involved in cell cycle specific control of VSG expression sites in Trypanosoma brucei. Mol. Microbiol. 2010;78:459–474. doi: 10.1111/j.1365-2958.2010.07350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBois KN, Alsford S, Holden JM, Buisson J, Swiderski M, Bart JM, Ratushny AV, Wan Y, Bastin P, Barry JD, Navarro M, Horn D, Aitchison JD, Rout MP, Field MC. NUP-1 Is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biol. 2012;10:e1001287. doi: 10.1371/journal.pbio.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers B, Czichos J, Overath P. RNA turnover in Trypanosoma brucei. Mol. Cell. Biol. 1987;7:1242–1249. doi: 10.1128/mcb.7.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersfeld K. Nuclear architecture, genome and chromatin organisation in Trypanosoma brucei. Res. Microbiol. 2011;162:626–636. doi: 10.1016/j.resmic.2011.01.014. [DOI] [PubMed] [Google Scholar]
- Fevre EM, Picozzi K, Jannin J, Welburn SC, Maudlin I. Human African trypanosomiasis: Epidemiology and control. Adv. Parasitol. 2006;61:167–221. doi: 10.1016/S0065-308X(05)61005-6. [DOI] [PubMed] [Google Scholar]
- Figueiredo LM, Cross GA. Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot. Cell. 2010;9:148–154. doi: 10.1128/EC.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo LM, Janzen CJ, Cross GA. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 2008;6:e161. doi: 10.1371/journal.pbio.0060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginger ML, Blundell PA, Lewis AM, Browitt A, Günzl A, Barry JD. Ex vivo and in vitro identification of a consensus promoter for VSG genes expressed by metacyclic-stage trypanosomes in the tsetse fly. Eukaryot. Cell. 2002;1:1000–1009. doi: 10.1128/EC.1.6.1000-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover L, Horn D. Repression of polymerase I-mediated gene expression at Trypanosoma brucei telomeres. EMBO Rep. 2006;7:93–99. doi: 10.1038/sj.embor.7400575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover L, Hutchinson S, Alsford S, McCulloch R, Field MC, Horn D. Antigenic variation in African trypanosomes: the importance of chromosomal and nuclear context in VSG expression control. Cell. Microbiol. 2013;15:1984–1993. doi: 10.1111/cmi.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I, Skinner JA. Efficient transcription of a protein-coding gene from the RNA polymerase I promoter in transfected cells. Proc. Natl. Acad. Sci. USA. 1985;82:722–726. doi: 10.1073/pnas.82.3.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizetti J, Scherf A. Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell. Microbiol. 2013;15:718–726. doi: 10.1111/cmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günzl A. The pre-mRNA splicing machinery of trypanosomes: complex or simplified? Eukaryot. Cell. 2010;9:1159–1170. doi: 10.1128/EC.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günzl A, Bruderer T, Laufer G, Schimanski B, Tu LC, Chung HM, Lee PT, Lee MG. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell. 2003;2:542–551. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Hertz-Fowler C, Figueiredo LM, Quail MA, Becker M, Jackson A, Bason N, Brooks K, Churcher C, Fahkro S, Goodhead I, Heath P, Kartvelishvili M, Mungall K, Harris D, Hauser H, Sanders M, Saunders D, Seeger K, Sharp S, Taylor JE, Walker D, White B, Young R, Cross GA, Rudenko G, Barry JD, Louis EJ, Berriman M. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One. 2008;3:e3527. doi: 10.1371/journal.pone.0003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D. Antigenic variation in African trypanosomes. Mol Biochem Parasitol. 2014;195:123–129. doi: 10.1016/j.molbiopara.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D, Cross GA. Position-dependent and promoter-specific regulation of gene expression in Trypanosoma brucei. EMBO J. 1997;16:7422–7431. doi: 10.1093/emboj/16.24.7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D, McCulloch R. Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Curr. Opin. Microbiol. 2010;13:700–705. doi: 10.1016/j.mib.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K, Wand M, Foulston L, Young R, Harley K, Terry S, Ersfeld K, Rudenko G. A novel ISWI is involved in VSG expression site downregulation in African trypanosomes. EMBO J. 2007;26:2400–2410. doi: 10.1038/sj.emboj.7601678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen CJ, Hake SB, Lowell JE, Cross GA. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol. Cell. 2006;23:497–507. doi: 10.1016/j.molcel.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Kassem A, Pays E, Vanhamme L. Transcription is initiated on silent variant surface glycoprotein expression sites despite monoallelic expression in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 2014;111:8943–8948. doi: 10.1073/pnas.1404873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Park SH, Günzl A, Cross GA. MCM-BP is required for repression of life-cycle specific genes transcribed by RNA polymerase I in the mammalian infectious form of Trypanosoma brucei. PLoS One. 2013;8:e57001. doi: 10.1371/journal.pone.0057001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman LA, Deitsch KW. Antigenic variation and the generation of diversity in malaria parasites. Curr. Opin. Microbiol. 2012;15:456–462. doi: 10.1016/j.mib.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Ramey-Butler K, Cross GA, Ullu E, Tschudi C. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 2012;338:1352–1353. doi: 10.1126/science.1229641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooter JM, Borst P. Alpha-amanitin-insensitive transcription of variant surface glycoprotein genes provides further evidence for discontinuous transcription in trypanosomes. Nucleic Acids Res. 1984;12:9457–9472. doi: 10.1093/nar/12.24.9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Bart JM, Van Tyne D, Navarro M. Cohesin regulates VSG monoallelic expression in trypanosomes. J. Cell. Biol. 2009;186:243–254. doi: 10.1083/jcb.200902119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, Lomvardas S. Stochastic gene expression in mammals: lessons from olfaction. Trends Cell Biol. 2013 doi: 10.1016/j.tcb.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Calvillo S, Saxena A, Green A, Leland A, Myler PJ. Characterization of the RNA polymerase II and III complexes in Leishmania major. Int. J. Parasitol. 2007;37:491–502. doi: 10.1016/j.ijpara.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol. 2011;6:459–474. doi: 10.2217/fmb.11.20. [DOI] [PubMed] [Google Scholar]
- Narayanan MS, Kushwaha M, Ersfeld K, Fullbrook A, Stanne TM, Rudenko G. NLP is a novel transcription regulator involved in VSG expression site control in Trypanosoma brucei. Nucleic Acids Res. 2011;39:2018–2031. doi: 10.1093/nar/gkq950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan MS, Rudenko G. TDP1 is an HMG chromatin protein facilitating RNA polymerase I transcription in African trypanosomes. Nucleic Acids Res. 2013;41:2981–2992. doi: 10.1093/nar/gks1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro M, Cross GA. DNA rearrangements associated with multiple consecutive directed antigenic switches in Trypanosoma brucei. Mol. Cell. Biol. 1996;16:3615–3625. doi: 10.1128/mcb.16.7.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro M, Cross GA. In situ analysis of a variant surface glycoprotein expression-site promoter region in Trypanosoma brucei. Mol. Biochem. Parasitol. 1998;94:53–66. doi: 10.1016/s0166-6851(98)00049-8. [DOI] [PubMed] [Google Scholar]
- Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- Nguyen TN, Müller LS, Park SH, Siegel TN, Günzl A. Promoter occupancy of the basal class I transcription factor A differs strongly between active and silent VSG expression sites in Trypanosoma brucei. Nucleic Acids Res. 2014;42:3164–3176. doi: 10.1093/nar/gkt1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TN, Nguyen BN, Lee JH, Panigrahi AK, Günzl A. Characterization of a novel class I transcription factor A (CITFA) subunit that is indispensable for transcription by the multifunctional RNA polymerase I of Trypanosoma brucei. Eukaryot. Cell. 2012;11:1573–1581. doi: 10.1128/EC.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Nguyen BN, Kirkham JK, Nguyen TN, Günzl A. A new strategy of RNA interference that targets heterologous sequences reveals CITFA1 as an essential component of class I transcription factor A in Trypanosoma brucei. Eukaryot. Cell. 2014;13:785–795. doi: 10.1128/EC.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick KL, Luz PM, Ruan JP, Shi H, Ullu E, Tschudi C. Genomic rearrangements and transcriptional analysis of the spliced leader-associated retrotransposon in RNA interference-deficient Trypanosoma brucei. Mol. Microbiol. 2008;67:435–447. doi: 10.1111/j.1365-2958.2007.06057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena AC, Pimentel MR, Manso H, Vaz-Drago R, Pinto-Neves D, Aresta-Branco F, Rijo-Ferreira F, Guegan F, Pedro Coelho L, Carmo-Fonseca M, Barbosa-Morais NL, Figueiredo LM. Trypanosoma brucei histone H1 inhibits RNA polymerase I transcription and is important for parasite fitness in vivo. Mol. Microbiol. 2014;93:645–663. doi: 10.1111/mmi.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VP, Qi CC, Gottesdiener KM. A detailed mutational analysis of the VSG gene expression site promoter. Mol. Biochem. Parasitol. 1996;75:241–254. doi: 10.1016/0166-6851(95)02513-8. [DOI] [PubMed] [Google Scholar]
- Povelones ML, Gluenz E, Dembek M, Gull K, Rudenko G. Histone H1 plays a role in heterochromatin formation and VSG expression site silencing in Trypanosoma brucei. PLoS Pathog. 2012;8:e1003010. doi: 10.1371/journal.ppat.1003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preußer C, Jae N, Günzl A, Bindereif A. Pre-mRNA splicing in Trypanosoma brucei: factors, Mechanisms, and Regulation. In: Bindereif A, editor. RNA Metabolism in Trypanosomes. Springer Press; 2012. pp. 49–76. [Google Scholar]
- Rudenko G. Epigenetics and transcriptional control in African trypanosomes. Essays Biochem. 2010;48:201–219. doi: 10.1042/bse0480201. [DOI] [PubMed] [Google Scholar]
- Rudenko G, Chung HM, Pham VP, Van der Ploeg LH. RNA polymerase I can mediate expression of CAT and neo protein- coding genes in Trypanosoma brucei. EMBO J. 1991;10:3387–3397. doi: 10.1002/j.1460-2075.1991.tb04903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon D, Vanwalleghem G, Morias Y, Denoeud J, Krumbholz C, Lhomme F, Bachmaier S, Kador M, Gossmann J, Dias FB, De Muylder G, Uzureau P, Magez S, Moser M, De Baetselier P, Van Den Abbeele J, Beschin A, Boshart M, Pays E. Adenylate cyclases of Trypanosoma brucei inhibit the innate immune response of the host. Science. 2012;337:463–466. doi: 10.1126/science.1222753. [DOI] [PubMed] [Google Scholar]
- Schwede A, Jones N, Engstler M, Carrington M. The VSG C-terminal domain is inaccessible to antibodies on live trypanosomes. Mol. Biochem. Parasitol. 2011;175:201–204. doi: 10.1016/j.molbiopara.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanne TM, Kushwaha M, Wand M, Taylor JE, Rudenko G. TbISWI regulates multiple polymerase I (Pol I)-transcribed loci and is present at Pol II transcription boundaries in Trypanosoma brucei. Eukaryot. Cell. 2011;10:964–976. doi: 10.1128/EC.05048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanne TM, Rudenko G. Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes. Eukaryot. Cell. 2010;9:136–147. doi: 10.1128/EC.00281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiengwe C, Marcello L, Farr H, Dickens N, Kelly S, Swiderski M, Vaughan D, Gull K, Barry JD, Bell SD, McCulloch R. Genome-wide analysis reveals extensive functional interaction between DNA replication initiation and transcription in the genome of Trypanosoma brucei. Cell Rep. 2012;2:185–197. doi: 10.1016/j.celrep.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme L, Pays A, Tebabi P, Alexandre S, Pays E. Specific binding of proteins to the noncoding strand of a crucial element of the variant surface glycoprotein, procyclin, and ribosomal promoters of Trypanosoma brucei. Mol. Cell. Biol. 1995;15:5598–5606. doi: 10.1128/mcb.15.10.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme L, Poelvoorde P, Pays A, Tebabi P, Xong HV, Pays E. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol. Microbiol. 2000;36:328–340. doi: 10.1046/j.1365-2958.2000.01844.x. [DOI] [PubMed] [Google Scholar]
- Wang QP, Kawahara T, Horn D. Histone deacetylases play distinct roles in telomeric VSG expression site silencing in African trypanosomes. Mol. Microbiol. 2010;77:1237–1245. doi: 10.1111/j.1365-2958.2010.07284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009;137:99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomerdijk JC, Kieft R, Borst P. Efficient production of functional mRNA mediated by RNA polymerase I in Trypanosoma brucei. Nature. 1991a;353:772–775. doi: 10.1038/353772a0. [DOI] [PubMed] [Google Scholar]
- Zomerdijk JC, Kieft R, Duyndam M, Shiels PG, Borst P. Antigenic variation in Trypanosoma brucei: a telomeric expression site for variant-specific surface glycoprotein genes with novel features. Nucleic Acids Res. 1991b;19:1359–1368. doi: 10.1093/nar/19.7.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomerdijk JC, Ouellette M, ten Asbroek AL, Kieft R, Bommer AM, Clayton CE, Borst P. The promoter for a variant surface glycoprotein gene expression site in Trypanosoma brucei. EMBO J. 1990;9:2791–2801. doi: 10.1002/j.1460-2075.1990.tb07467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]