Summary
R-loops, consisting of an RNA-DNA hybrid and displaced single-stranded DNA, are physiological structures that regulate various cellular processes occurring on chromatin. Intriguingly, changes in R-loop dynamics have also been associated with DNA damage accumulation and genome instability, however the mechanisms underlying R-loop induced DNA damage remain unknown. Here we demonstrate in human cells that R-loops induced by the absence of diverse RNA processing factors, including the RNA/DNA helicases Aquarius (AQR) and Senataxin (SETX), or by the inhibition of topoisomerase I, are actively processed into DNA double-strand breaks (DSBs) by the nucleotide excision repair endonucleases XPF and XPG. Surprisingly, DSB formation requires the transcription-coupled nucleotide excision repair (TC-NER) factor Cockayne syndrome group B (CSB), but not the global genome repair protein XPC. These findings reveal an unexpected and potentially deleterious role for TC-NER factors in driving R-loop-induced DNA damage and genome instability.
Keywords: R-loop, RNA processing, genome instability, TC-NER
Introduction
R-loops, structures that contain an RNA-DNA hybrid and displaced single-stranded DNA, can form during transcription when an RNA molecule emerging from the transcription machinery hybridizes with the DNA template. These structures arise naturally in organisms from bacteria to humans, and they have a multitude of roles in the cell (Aguilera and García-Muse, 2012; Skourti-Stathaki and Proudfoot, 2014; Hamperl and Cimprich, 2014). In human cells, R-loops form over switch regions at the immunoglobulin locus to facilitate class switching, a physiological event in which DSBs are initiated through the processing of R-loops (Yu et al., 2003). In addition, R-loops form preferentially at the promoters of genes with a high GC skew to protect these regions from DNA methylation (Ginno et al., 2012). They also form at the termination regions of genes where they promote efficient transcriptional termination (Skourti-Stathaki et al., 2011).
R-loops can form in an unscheduled manner due to defects in RNA processing (Huertas and Aguilera, 2003, Li and Manley, 2005; Paulsen et al., 2009; Stirling et al., 2012; Wahba et al., 2011), and in these situations they are commonly associated with DNA damage. Indeed, R-loops were initially proposed to be the source of the hyper-recombination phenotype in yeast THO/TREX complex mutants, where they form as a result of defects in transcriptional elongation and RNA export (Huertas and Aguilera, 2003). Unscheduled R-loops are also thought to initiate the genomic or epigenomic changes associated with several neurodegenerative diseases, including amyotrophic lateral sclerosis, Fragile X syndrome and Friedreich's ataxia (Chen et al., 2004; Colak et al., 2014; Groh et al., 2014; Loomis et al., 2014), and they can cause genome instability at trinucleotide repeat sequences and common fragile sites, suggesting that they may contribute to cancer (Haeusler et al., 2014; Helmrich et al., 2011).
Cells utilize diverse mechanisms to regulate the formation of R-loops. These structures can be resolved by RNase H, which specifically degrades the RNA moiety in RNA-DNA hybrids (Wahba et al., 2011), or by helicases such as Senataxin, which unwind RNA-DNA hybrids (Mischo et al., 2011; Skourti-Stathaki et al., 2011). R-loop formation is also suppressed by topoisomerase I, which resolves the negative torsional stress behind RNA polymerase II to prevent annealing of the nascent RNA with the DNA template (Tuduri et al., 2009). Other RNA processing factors also preclude R-loop formation, presumably by binding to RNA as it emerges from RNA polymerase (Li et al., 2007). However when these mechanisms fail, R-loops may persist or accumulate, ultimately leading to DNA breaks and genome instability (Huertas and Aguilera, 2003, Li and Manley, 2005; Paulsen et al., 2009; Wahba et al., 2011; Tuduri et al., 2009; Stirling et al., 2012).
How DNA damage arises from an R-loop is an unresolved question. Several studies in bacteria, yeast, and human cells suggest that R-loop-induced DNA damage is associated with defects in replication fork progression (Alzu et al., 2012; Gan et al., 2011; Wellinger et al., 2006; Yuce and West, 2012; Tuduri et al., 2009). Whether it is the R-loop itself or the stalled RNA polymerase resulting from R-loop formation that impairs DNA replication and ultimately causes replication fork collapse and DSB formation is not clear. It has also been proposed that DNA damage may arise from the single-stranded DNA in the R-loop, because this DNA is more susceptible to DNA damaging agents (Lindahl, 1993) and could be targeted by enzymes like activation-induced cytidine deaminase (AID) that act at the immunoglobin locus (Muramatsu et al., 2000). However, AID is not expressed in most cells types, and no other specific factors have been shown to cause DNA damage when R-loops arise in cells. Thus, many questions remain about the mechanisms that underlie the accumulation of DNA damage and genome instability associated with R-loop formation.
Here, we report that R-loops formed in the absence of mRNA processing factors or in the presence of camptothecin, an inhibitor of topoisomerase I, are actively processed by the nucleotide-excision repair (NER) endonucleases XPF and XPG. Moreover, the transcription-coupled nucleotide excision repair (TC-NER) protein CSB is required for this processing suggesting that R-loop processing is coupled to stalled transcription complexes. We also demonstrate that this mechanism is conserved in yeast where it drives genomic instability. These findings reveal a new function for TC-NER factors in R-loop processing, and provide the first molecular insights into the processes underlying R-loop-induced DNA damage.
Results
AQR knockdown induces R-loop-dependent DNA damage
To investigate the mechanism of R-loop processing in human cells, we took advantage of the data from a genome-wide siRNA screen we previously carried out to identify factors involved in the maintenance of genome stability; highly enriched amongst the genes that induced DNA damage when knocked down were RNA processing factors. Surprisingly, overexpression of RNase H reversed the DNA damage observed after depletion of many of these RNA processing factors, suggesting that R-loops might be a source of this damage (Paulsen et al., 2009). We were particularly interested in one of these factors, Aquarius (AQR), a protein which is part of a subfamily of proteins possessing a conserved DEAxQ-like domain with putative RNA/DNA helicase activity (Fairman-Williams et al., 2010; Hirose et al., 2006). Interestingly, this subfamily includes Senataxin (SETX), which is thought to promote efficient transcriptional termination by resolving R-loops formed at specific loci (Skourti-Stathaki et al., 2011), and its yeast orthologue, Sen1, which prevents R-loop-mediated genome instability (Alzu et al., 2012; Mischo et al., 2011).
Knockdown of AQR robustly induced the DNA damage response (DDR), as evidenced by the phosphorylation of histone variant H2AX (termed γH2AX), a marker of DNA damage (Figure S1A-C) (Paulsen et al., 2009). We also observed phosphorylation of the transcriptional repressor and DDR target KAP1 (termed P-KAP1) as well as the phosphorylation of CHK1 and RPA-2 (Figure 1A). These findings suggest that AQR knockdown ultimately leads to DSB formation and fork stalling. To test whether knockdown of AQR produced DSBs, or induced DDR signaling by some other mechanism, we performed a neutral comet assay. The significant increase in comet tail moment we observed in AQR-depleted cells provides direct evidence for DSB formation and suggests that AQR knockdown does not simply induce DDR signaling (Figure 1B, C). Importantly, there was no significant difference in cell cycle progression upon AQR knockdown (Figure S1D). After prolonged knockdown, however, AQR-depleted cells accumulate in G2, consistent with the observed DSB formation and checkpoint activation (Figure S1E).
Figure 1. AQR knockdown leads to DSBs formation and R-loop accumulation.
(A) P-KAP1, P-CHK1 and pS33-RPA2 levels in HeLa cells transfected with siLUC and two siAQR for 72 hours. (B) Neutral comet assay in HeLa cells transfected with siLUC or siAQR. Scale bar represents 50 μm. (C) Quantification of comet tail moment for the experiment described in (B). a.u. = arbitrary units. ****p<0.0001. (D) Quantification of RNA-DNA hybrids detected by slot-blot with S9.6 antibody in HeLa cells, with fold enrichment relative to siLUC signal. Errors bars are SEM of three biological replicates. (*p<0.05 by student's t-test). (E) Immunostaining with S9.6 (red) and nucleolin (green) antibodies in HeLa cells transfected with siRNA and fixed after 48 hours. The nucleus (stained with Hoechst) is outlined. Scale bar represents 10 μm. The levels of all panels were adjusted equally in Adobe Photoshop. (F) Quantification of S9.6 signal per nucleus after nucleolar removal for the experiment described in (E), shown as box and whiskers plot. ****p<0.0001. See also Figure S1.
RNase H1 overexpression in AQR-depleted cells decreases γH2AX (Paulsen et al., 2009), and we found that it reduces P-KAP1 as well (Figure S1F). This finding suggests that RNA-DNA hybrids induced by the knockdown of AQR lead to DNA damage. To directly determine whether RNA-DNA hybrids accumulate upon AQR knockdown, we used a monoclonal antibody (S9.6) that specifically detects these hybrids (Boguslawski et al., 1986) to probe genomic DNA extracted from wild-type and AQR-depleted cells. We observed a two-fold enrichment of RNA-DNA hybrids in AQR-depleted cells, which was abolished by pre-treatment of the DNA with RNase H1 (Figure 1D). We also measured the nuclear S9.6 signal using confocal microscopy. Strikingly, high S9.6 signal was present in the nucleolus and mitochondria even before AQR knockdown. Although this is consistent with the known presence of RNA-DNA hybrids in these cellular compartments (Hage et al., 2010; Aguilera and García-Muse, 2012), we also found that the nucleolar S9.6 signal persisted after RNase H1 treatment. This could be due to the presence of RNA species that are resistant to RNase H1, such as more structured RNA-DNA hybrids, or incomplete action of the nuclease in the nucleolus, where RNA-DNA hybrids are abundant. More importantly, in the absence of AQR, we observed an enrichment of nuclear RNA-DNA hybrids (Figure 1E), which we quantified after subtraction of the nucleolar signal (Figure 1F), and this enrichment could be reversed by treatment with RNase H1. Together, these data strongly suggest that the DNA damage observed in the absence of AQR results from the accumulation of R-loops.
R-loops induced upon AQR knockdown are processed by the NER endonucleases XPF and XPG
R-loops are thought to be open DNA structures with flap extremities, and we speculated that this structure might be recognized and processed by nucleases that act on related structures in the cell. The NER pathway uses two structure-specific flap endonucleases, XPF and XPG, to repair bulky lesions in the genome caused by a variety of DNA damaging agents. This process requires over 30 proteins that collectively recognize distortions caused by the lesion, excise a small lesion-containing oligonucleotide, and fill in the resulting gap by repair synthesis (Fagbemi et al., 2011). Intriguingly, purified XPF and XPG were previously shown to cleave R-loop structures formed at S regions of the immunoglobulin locus in vitro (Tian, 2000). Surprisingly, however, these nucleases were also shown to be dispensable for class switch recombination (CSR), suggesting XPF and XPG have no role in the processing of R-loops formed at S regions (Tian et al., 2004a; 2004b). Indeed, it has been shown that this processing depends upon AID (Muramatsu et al., 2000).
Because R-loops formed upon depletion of AQR induce unprogrammed DNA damage and are likely distinct from those formed at the immunoglobin locus, we wondered whether XPF or XPG might be able to act on R-loops induced by AQR knockdown in cells. To test this hypothesis, we assessed whether DDR signaling is induced upon knockdown of AQR in the absence of XPG. Strikingly, double knockdown of both XPG and AQR dramatically reduced P-KAP1 observed after knockdown of AQR alone (Figure 2A). Importantly, DSB formation was also reduced, as indicated by a neutral comet assay (Figure 2B, C). Next, we investigated the effects of XPG in AQR-depleted cells by using immortalized XPG-deficient fibroblasts from a Xeroderma Pigmentosum (XP) patient, and an isogenic cell line complemented with wild-type XPG. AQR knockdown induced P-KAP1 in the XP patient cell line complemented with XPG (lanes 4 and 6 in Figure 2D). This phosphorylation was dramatically reduced when AQR was down-regulated in the non-complemented XPG-deficient cells (lanes 3 and 5 in Figure 2D). We also tested the role of XPF in this process using an analogous strategy. We found that P-KAP1 was also reduced when AQR was down-regulated in XP patient cells deficient in XPF (Figure 2E). Thus, DSBs resulting from AQR knockdown are dependent upon both XPF and XPG. These findings suggest that XPF and XPG may be able to process R-loops resulting from AQR knockdown in cells.
Figure 2. R-loop-induced DNA damage depends on XPF and XPG.
(A) P-KAP1 level in HeLa cells transfected with siXPG or siLUC 24 hours prior to transfection with siLUC or siAQR. (B) Neutral comet assay in HeLa cells treated as in Figure 2A. Scale bar represents 50 μm. (C) Quantification of comet tail moment for the experiment described in (B). a.u. arbitrary units. ****p<0.0001. (D, E) P-KAP1 level in XPG- and XPF-patient cell lines either complemented or not with the corresponding wild-type proteins, and transfected with siLUC, siAQR#2 or siAQR#3. (F, G) Quantification of percent γH2AX-positive cells in XPG- and XPF-patient cell lines either complemented or not with the wild-type or nuclease-dead proteins, and transfected with indicated siRNA. (SEM, n=3). (H) Immunostaining with S9.6 (red) and nucleolin (green) antibodies in HeLa cells transfected with siXPG or siLUC 24 hours before transfection with siLUC or siAQR. A merge of the two channels is shown, with the nucleus (stained with Hoechst) outlined. Scale bar represents 10 μm. The levels of all panels were adjusted equally in Adobe Photoshop. (I) Quantification of S9.6 immunofluorescence intensity per nucleus for the experiment described in (H), shown as box and whiskers plot. ****p<0.0001.
To further test this idea, we asked whether the endonuclease activities of XPG and XPF are required to generate the DSBs observed after AQR knockdown or whether XPG and XPF simply play a structural role. To do so, we analyzed γH2AX levels by immunofluorescence in immortalized XPG- and XPF-deficient patient fibroblasts complemented with nuclease-dead forms of XPG or XPF proteins, XPG-E791A and XPF-D676A, respectively (Fagbemi et al., 2011). AQR knockdown in XPG- and XPF-deficient fibroblasts complemented with the wild-type form of XPG or XPF showed a high level of γH2AX compared to the control siRNA-transfected cell lines. In contrast, the γH2AX signal was much lower when AQR was knocked down in cells complemented with the nuclease-dead form of XPG or XPF (Figure 2F, G). Thus, the nuclease activities of both XPG and XPF are required to generate the DSBs observed upon AQR depletion.
Our data are consistent with the possibility that DSBs result from the direct processing of R-loops by XPF and XPG. However, another explanation is that decreases in transcription associated with XPF and XPG knockdown (Le May et al., 2010) reduce the levels of R-loops and consequently DNA damage. Alternatively, XPF and XPG might process DNA lesions that arise in the single-stranded DNA of the non-template strand, lesions that would require NER for repair after the RNA-DNA hybrid is resolved. To distinguish between these models, we monitored the fate of RNA-DNA hybrids by confocal microscopy after co-depletion of both XPG and AQR. We observed more RNA-DNA hybrids when both XPG and AQR were knocked down compared to knockdown of AQR alone (Figure 2H, I). If XPG was processing DNA lesions induced by the formation of R-loops through the classical NER pathway, R-loops would not be expected to accumulate. Similarly, decreases in transcription would not lead to R-loop accumulation. This result therefore suggests that XPG can induce DNA damage in cells by directly processing RNA-DNA hybrids.
R-loops induced by defects in mRNA processing or camptothecin treatment are processed by XPG
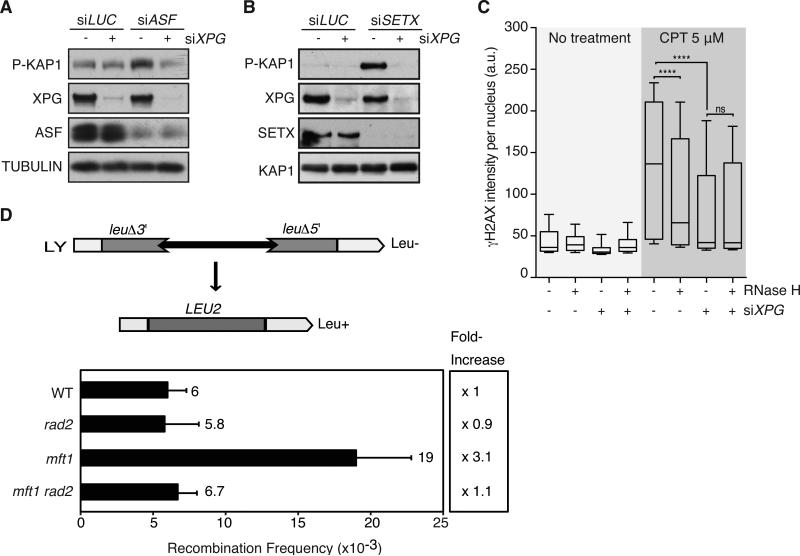
We then asked whether XPG acts exclusively on R-loops induced by AQR knockdown or if our findings could be extended to other factors which perturb R-loop dynamics. First, we tested the relationship between XPG and two mRNA processing factors that have been linked to R-loop formation and strong DDR activation, the splicing factor ASF/SF2 (Li and Manley, 2005) and the AQR-related helicase SETX (Alzu et al., 2012; Skourti-Stathaki et al., 2011). Concurrent knockdown of XPG in ASF- or SETX-depleted cells abrogated the DDR response indicating that XPG processes R-loops induced by depletion of these factors (Figure 3A, B).
Figure 3. The processing of R-loops by the endonucleases XPF and XPG is a general and conserved mechanism.
(A, B). P-KAP1 level in HeLa cells transfected with siXPG or siLUC 24 hours before transfection with siASF or siSETX. (C) γH2AX intensity in HeLa-TetON-RNase H1 cells transfected with siLUC or siXPG for 48 hours and treated for 2 hours with 5μM camptothecin. Doxycycline (500 ng/μl) was added in combination with siRNAs where indicated. a.u. = arbitrary units. (D) Frequencies of recombination in the LY direct-repeat system. Each data represents the median of 3-4 independent experiments. Error bars represent the standard error of the median (SEM, n=3-4). See also Figure S2 and Table S1.
We also tested the effect of XPG knockdown on DNA damage induced by camptothecin (CPT), an inhibitor of topoisomerase I. Previous work showed that overexpression of RNase H1 decreased the induction of γH2AX following CPT treatment in post-mitotic primary neurons and non-cycling HeLa cells (Sordet et al., 2009), indicating that CPT generates R-loops that induce DSBs. To determine if this is also true when cells are cycling, we took advantage of a stable HeLa cell line we generated expressing tetracycline-inducible FLAG-tagged RNase H1. This allowed us to examine the DDR specifically in FLAG-positive cells. We found that overexpression of RNase H1 decreased the induction of γH2AX. We also found that XPG knockdown decreased the induction of γH2AX to a slightly greater extent than RNase H overexpression (Figure 3C). Moreover, the combined effect of XPG knockdown and RNase H1 expression was similar to that of XPG knockdown alone. These data indicate that R-loops formed in response to CPT treatment are processed into DSBs by XPG. Thus, XPG action is not limited to specific R-loops associated with AQR loss. Rather, XPG has a general role in the processing of R-loops into DSBs.
The processing of R-loops by XPG drives genome instability in yeast
Next, we considered whether XPG's influence on the DNA damage associated with R-loop formation is conserved in other species. The THO protein complex composed of the Tho2, Hpr1, Mft1 and Thp2 proteins is involved in transcription and RNA export (Chávez et al., 2000), and previous studies suggest that yeast THO mutants have a hyper-recombination phenotype that is dependent on the formation of R-loops (Huertas and Aguilera, 2003). Thus, we asked whether mutating RAD2, the yeast XPG homolog, would suppress this phenotype. To test this, we measured the frequency of R-loop induced recombination (Prado and Aguilera, 1995, see Figure S2A for the assay description) in a single mutant of the THO complex mft1Δ and in the double mutant rad2Δmft1Δ. The formation of recombinants markedly increased in the single mutant mft1Δcompared to the wild-type strain (Chávez et al., 2000). However, the recombination frequency returned to the wild-type level in the rad2Δmft1Δ double mutant (Figure 3D), indicating that Rad2 promotes genome instability in the absence of mRNA processing factors. We confirmed this result using an alternative recombination assay (Figure S2B). These findings suggest that the molecular mechanisms responsible for the processing of R-loop are broadly conserved.
TC-NER factors are required for the processing of R-loops in mammalian cells
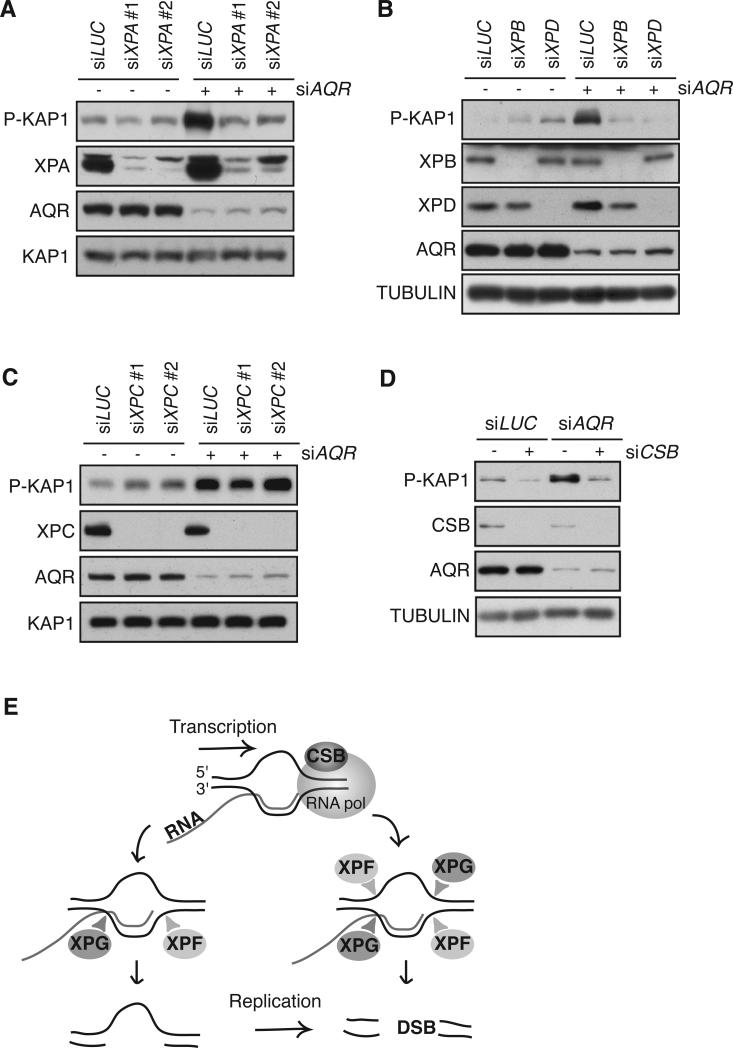
Lastly, we considered whether XPF and XPG act on R-loops through an NER-like pathway or through another mechanism involving their nuclease activities. During canonical NER, XPA plays a critical role in positioning NER factors, including the XPF and XPG nucleases, for incision and repair (Fagbemi et al., 2011). Thus, we asked whether XPA can contribute to R-loop processing. We found that XPA depletion clearly suppressed the DDR activation observed upon knockdown of AQR (Figure 4A). Next, we asked whether the TFIIH complex is required for the processing of R-loops. Two components of TFIIH, XPB and XPD, play an essential role during NER, using their respective DNA-dependent ATPase and helicase activities to open the DNA around the lesion (Fagbemi et al., 2011). We found that concurrent knockdown of either XPB or XPD with AQR dramatically reduced P-KAP1 compared to knockdown of AQR alone (Figure 4B). Similar results were obtained in immortalized XPD-deficient patient fibroblasts, and an isogenic cell line complemented with a wild-type form of XPD; AQR knockdown induced γH2AX in the complemented cell line while the γH2AX signal was reduced in the non-complemented cell line (Figure S3A). These findings indicate that R-loops processing is a concerted action that requires the NER factors, XPA, XPB and XPD. They also indicate that R-loop processing is not due to the unregulated activity of these endonucleases on flap structures, but instead is a result of classical NER-like events.
Figure 4. R-loop processing requires TC-NER factors but not XPC.
(A) P-KAP1 level in HeLa cells transfected with siXPA or siLUC 24 hours before transfection with siLUC or siAQR. (B) P-KAP1 level in HeLa cells transfected with siXPB, siXPD or siLUC 24 hours before transfection with siLUC or siAQR. (C) P-KAP1 level in HeLa cells transfected with siXPC or siLUC 24 hours before transfection with siLUC or siAQR. (D) P-KAP1 level in HeLa cells transfected with siCSB or siLUC 24 hours before transfection with siLUC or siAQR. (E) Model for how an R-loop is processed into a DSB. The stalling of the RNA polymerase allows CSB to recruit the endonucleases XPF and XPG. XPF and XPG generate a gap that can be converted into a DSB through DNA replication and/or XPF and XPG cleave the R-loop on both strands producing a DSB. See also Figure S3.
The endonucleases XPF and XPG, as well as the factors XPA, XPB and XPD, are involved in two forms of NER, global genome repair (GG-NER) and transcription coupled repair (TC-NER) (Svejstrup, 2002). To delineate which of these pathways was responsible for R-loop-induced DNA damage or if both pathways are involved, we examined the effect of depleting factors specific to each form of NER. XPC recognizes helix-distorting lesions during GG-NER and subsequently mobilizes NER proteins (Sugasawa et al., 1998). We found that P-KAP1 did not decrease upon depletion of XPC when AQR was knocked down (Figure 4C), suggesting that R-loop processing is not mediated by the GG-NER pathway. Similar results were observed in immortalized XPC-deficient fibroblasts from an XP patient, and in an isogenic cell line complemented with a wild-type form of XPC; AQR knockdown induced γH2AX and P-KAP1 in both the complemented and non-complemented XP patient cell lines (Figure S3B, C). To probe the role of TC-NER in R-loop processing, we examined the effect of depleting CSB, which recruits downstream NER proteins when a DNA lesion is encountered on the transcribed strand (Fousteri et al., 2006). Surprisingly, we found that knockdown of CSB reduced the high P-KAP1 observed after knockdown of AQR (Figure 4D). The P-KAP1 induced upon knockdown of SETX was strongly reduced upon depletion of CSB as well (Figure S3D). These data suggest that the TC-NER factor CSB is required for the processing of R-loops into DSBs. Since CSB but not XPC is required for the processing of R-loops, we propose that TC-NER factors act in a non-cannonical manner to generate DNA damage when R-loops form during transcription.
Discussion
Our findings reveal a conserved molecular mechanism by which R-loops are actively processed to DSBs, and they indicate that DSBs do not simply result from the collision of a replication fork with an R-loop. Surprisingly, this processing involves several NER factors, including XPA, the TFIIH subunits XPB and XPD, and the endonucleases XPF and XPG. Because we also observe that the TC-NER protein CSB, but not XPC, is required for this processing, we conclude that R-loop processing requires the concerted action of TC-NER factors. The involvement of XPF and XPG in the processing of R-loops induced by depletion of RNA-processing factors or CPT treatment, and not R-loops involved in CSR, indicates there are distinct pathways for processing different types of R-loop structures. We suggest that R-loops associated with paused transcription complexes may be the target of TC-NER factors. This is a new and unexpected role for these factors outside the transcription-coupled repair of DNA damage and suggests that TC-NER factors affect genome stability in diverse, and potentially detrimental, ways.
There are a variety of distinctions between the R-loop structure and the lesion-containing structures processed by XPF and XPG during classical NER which raise interesting questions about precisely how R-loops are cut by TC-NER factors. Because CSB is required, a parsimonious explanation would be that R-loop processing is triggered upon stalling of the RNA polymerase complex by an R-loop; this pause would allow CSB to recruit XPF and XPG for processing, as it does when it stalls at a DNA lesion. However, our observation that NER factors lead to DSB formation is unexpected in the classical NER context, because NER factors typically generate a single-strand DNA gap during excision of a lesion. In the context of R-loops, there are potential substrates for the flap endonucleases on both the transcribed and non-transcribed strand, and at both ends of the R-loop. Thus, processing could lead directly to DSB formation, or to the formation of nicks or gaps which are known to ultimately cause fork collapse and DSBs formation in S phase (Figure 4E). Indeed, diverse studies suggest that DNA replication is required for R-loop-induced genome instability, and the activation of ATR suggested by the phosphorylation of RPA and CHK1 indicates there may be effects of these structures or their processed intermediates in S phase (Alzu et al., 2012; Gan et al., 2011; Tuduri et al., 2009; Wellinger et al., 2006; Yuce and West, 2012). It is also possible that R-loop associated TC-NER factors act directly on forks that collide with R-loop structures or that processing of the R-loop is coordinated with DNA replication. Regardless of the precise structure processed, the requirement for TC-NER factors in generating the DSBs associated with R-loop formation reveals the molecular mechanism of R-loop induced genome instability.
Importantly, R-loops further accumulate in cells depleted for AQR when XPG is also knocked down. This suggests that XPG clears R-loops in the absence of efficient mRNA processing, and it also raises the possibility that NER factors may be a clearance pathway for naturally occurring R-loops. However, whether TC-NER-dependent R-loop processing is beneficial to cells is still unclear. Although TC-NER-dependent processing leads to DNA damage, it is possible that this is preferred to the continued persistence of R-loops, which could pose additional problems for the cells. R-loop processing may be a cost that comes with the ability to rapidly repair lesions during transcription. Given that the effect of XPG on R-loops is observed in both yeast and humans, we speculate that this processing is conserved and has some long-term benefit to the organism.
R-loops have been found to cover a substantial portion of the genome and to play fundamental roles in various cellular processes (Chan et al., 2014; Ginno et al., 2012; Ginno et al., 2013; Skourti-Stathaki and Proudfoot, 2014) suggesting that not all R-loops induce genome instability. Thus, our work raises the fascinating question of how certain types of R-loops are specifically protected from the deleterious effects of the TC-NER machinery and what drives DSB formation and genome instability at other R-loop sites. In the absence of splicing factors or in the presence of the topoisomerase inhibitor CPT, we and others have shown that R-loops accumulate (Alzu et al., 2012; Li and Manley, 2005; Skourti-Stathaki et al., 2011; Sordet et al., 2009). Thus, one possibility is that these unscheduled R-loops saturate the clearance pathways that normally act to resolve these structures or prevent their formation, thereby allowing TC-NER factors to cleave R-loops aberrantly. Another possibility is that unscheduled R-loops exist in different chromosomal contexts which may affect their processing. Consistent with this idea, R-loops arise at new genomic loci in yeast mutants of RNaseH or Senataxin (Chan et al., 2014). Lastly, the dynamics of unscheduled R-loops may differ from those of regulatory R-loops, allowing the latter to escape the deleterious effect of TC-NER factors. Indeed, the dynamic formation and resolution of R-loops is needed for efficient transcriptional termination (Skourti-Stathaki et al., 2011).
In summary, we demonstrate that the processing of unscheduled R-loops by the TC-NER pathway poses a threat to genome stability. R-loops have been observed at some common fragile sites (Helmrich et al., 2011) as well as the proto-oncogene MYC (Duquette et al., 2005), and based on our results, we propose that TC-NER-dependent R-loop processing contributes to genome instability and cancer progression by stimulating recombination at R-loop sites. Recently, the accumulation of R-loops has also been implicated in the silencing of critical genes that are associated with neurodegenerative diseases and which contain repeated DNA sequences (Colak et al., 2014; Groh et al., 2014; Haeusler et al., 2014; Loomis et al., 2014). Thus, the formation of R-loops may be detrimental in different ways; R-loop processing by TC-NER factors might promote genome rearrangements leading to cancer, while R-loop stabilization may be more relevant to neurodegenerative diseases. Lastly, because TC-NER factors may also play a role in the clearance of R-loops, it is tempting to speculate that some phenotypes observed in XP/CS patients result from a defect in R-loop processing. The mechanistic insights provided by this work may ultimately point the way to strategies for the modulation of R-loop formation and processing that could be used for the treatment of these and others human diseases.
EXPERIMENTAL PROCEDURES
Cell culture, western blotting, and immunofluorescence were performed using standard methods. Neutral comet assay were performed as described previously (Tuduri et al., 2009). For slot blot total genomic DNA was blotted on Nylon membrane and probed with either S9.6 antibody or denatured and probed with the single strand DNA antibody. The yeast LY recombination assays were performed as previously described (Luna et al., 2005). Detailed information regarding methodology and any associated references are available in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Julie Rageul (Stony Brook University), Frederic Chedin (University of California Davis), and Xialu Li (National Institute of Biological Sciences, Beijing) for providing helpful reagents and advice. We would also like to thank Yea-Lih Lin (Institute of Human Genetics, France) for technical advice and members of the Cimprich lab for thoughtful discussion. This work was supported by the Spanish Ministry of Economy and Competitiveness (grants BFU2010-16372 and CSD2007-00015), the European Union (FEDER) to A.A., and an NIH grant (GM100489) to K.A.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
K.A.C. and J.S. designed the study and analyzed the data. J.S., C.T.S., and M.L.G.R. performed the experiments and analyzed the data. R.D.P. generated the preliminary data motivating the study. A.A. analyzed the data and edited the manuscript. J.S. and K.A.C wrote the manuscript.
SUPPLEMENTAL INFORMATION
Supplemental Information including Supplemental Experimental Procedures and three figures can be found with this article online at
The authors declare no conflicts of interest.
REFERENCES
- Aguilera A, García-Muse T. R Loops: From Transcription Byproducts to Threats to Genome Stability. Mol. Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, Saponaro M, Brambati A, Cocito A, Foiani M, et al. Senataxin Associates with Replication Forks to Protect Fork Integrity across RNA-Polymerase-II-Transcribed Genes. Cell. 2012;151:835–846. doi: 10.1016/j.cell.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods. 1986;89:123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- Chan YA, Aristizabal MJ, Lu PYT, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P. Genome-Wide Profiling of Yeast DNA:RNA Hybrid Prone Sites with DRIP-Chip. PLoS Genet. 2014;10:e1004288. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez S, Beilharz T, Rondón AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. The EMBO Journal. 2000;19:5824–5834. doi: 10.1093/emboj/19.21.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-Z, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am J Hum Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak D, Zaninovic N, Cohen MS, Rosenwaks Z, Yang WY, Gerhardt J, Disney MD, Jaffrey SR. Promoter-Bound Trinucleotide Repeat mRNA Drives Epigenetic Silencing in Fragile X Syndrome. Science. 2014;343:1002–1005. doi: 10.1126/science.1245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette ML, Pham P, Goodman MF, Maizels N. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 2005;24:5791–5798. doi: 10.1038/sj.onc.1208746. [DOI] [PubMed] [Google Scholar]
- Fagbemi AF, Orelli B, Schärer OD. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair. 2011;10:722–729. doi: 10.1016/j.dnarep.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther U-P, Jankowsky E. SF1 and SF2 helicases: family matters. Current Opinion in Structural Biology. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LHF. Cockayne Syndrome A and B Proteins Differentially Regulate Recruitment of Chromatin Remodeling and Repair Factors to Stalled RNA Polymerase II In Vivo. Mol. Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes & Development. 2011;25:2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. R-Loop Formation Is a Distinctive Characteristic of Unmethylated Human CpG Island Promoters. Mol. Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lim YW, Lott PL, Korf I, Chédin F. GC skew at the 52 and 32 ends of human genes links R-loop formation to epigenetic regulation and transcriptional termination. Genome Research. 2013;23:1590–1600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh M, Lufino MMP, Wade-Martins R, Gromak N. R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome. PLoS Genet. 2014;10:e1004318. doi: 10.1371/journal.pgen.1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EAJ, Shaw PG, Kim M-S, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage, El A, French SL, Beyer AL, Tollervey D( Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes & Development. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, Cimprich KA. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair. 2014;19:84–94. doi: 10.1016/j.dnarep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Tora L. Collisions between Replication and Transcription Complexes Cause Common Fragile Site Instability at the Longest Human Genes. Mol. Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Hirose T, Ideue T, Nagai M, Hagiwara M, Shu M-D, Steitz JA. A Spliceosomal Intron Binding Protein, IBP160, Links Position-Dependent Assembly of Intron-Encoded Box C/D snoRNP to Pre-mRNA Splicing. Mol. Cell. 2006;23:673–684. doi: 10.1016/j.molcel.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Le May N, Mota-Fernandes D, VElez-Cruz R, Iltis I, Biard D, Egly J-M. NER Factors Are Recruited to Active Promoters and Facilitate Chromatin Modification for Transcription in the Absence of Exogenous Genotoxic Attack. Mol. Cell. 2010;38:54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL. Inactivation of the SR Protein Splicing Factor ASF/SF2 Results in Genomic Instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Li X, Niu T, Manley JL. The RNA binding protein RNSP1 alleviates ASF/SF2 depletion-induced genomic instability. RNA. 2007;13:2108–2115. doi: 10.1261/rna.734407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Loomis EW, Sanz LA, Chédin F, Hagerman PJ. Transcription-Associated R-Loop Formation across the Human FMR1 CGG-Repeat Region. PLoS Genet. 2014;10:e1004294. doi: 10.1371/journal.pgen.1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna R, Jimeno S, Marín M, Huertas P, García-Rubio M, Aguilera A. Interdependence between Transcription and mRNP Processing and Export, and Its Impact on Genetic Stability. Mol. Cell. 2005;18:711–722. doi: 10.1016/j.molcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Mischo HE, Gómez-González B, Grzechnik P, Rondón AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. Yeast Sen1 Helicase Protects the Genome from Transcription-Associated Instability. Mol Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee M-C, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. A Genome-wide siRNA Screen Reveals Diverse Cellular Processes and Pathways that Mediate Genome Stability. Mol. Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Aguilera A. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes & Development. 2014;28:1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sitesto Promote Xrn2-Dependent Termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordet O, Redon CE, Guirouilh-Barbat JEE, Smith S, Solier SEP, Douarre CEL, Conti C, Nakamura AJ, Das BB, Nicolas E, et al. Ataxia telangiectasia mutated activated by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Reports. 2009;10:887–893. doi: 10.1038/embor.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, Kobor MS, Hieter P. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes & Development. 2012;26:163–175. doi: 10.1101/gad.179721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ. Transcription mechanisms of transcription-coupled repair. Nat. Rev. Mol. Cell Biol. 2002;3:21–29. doi: 10.1038/nrm703. Svejstrup, J.Q. [DOI] [PubMed] [Google Scholar]
- Tian M. Transcription-induced Cleavage of Immunoglobulin Switch Regions by Nucleotide Excision Repair Nucleases in Vitro. Journal of Biological Chemistry. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- Tian M, Jones DA, Smith M, Shinkura R, Alt FW. Deficiency in the Nuclease Activity of Xeroderma Pigmentosum G in Mice Leads to Hypersensitivity to UV Irradiation. Molecular and Cellular Biology. 2004a;24:2237–2242. doi: 10.1128/MCB.24.6.2237-2242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Shinkura R, Shinkura N, Alt FW. Growth Retardation, Early Death, and DNA Repair Defects in Mice Deficient for the Nucleotide Excision Repair Enzyme XPF. Molecular and Cellular Biology. 2004b;24:1200–1205. doi: 10.1128/MCB.24.3.1200-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuduri S, Crabbé L, Conti C, Tourrière H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell. Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and Multiple RNA Biogenesis Factors Cooperate to Prevent RNA:DNA Hybridsfrom Generating Genome Instability. Mol. Cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RE, Prado F, Aguilera A. Replication Fork Progression Is Impaired by Transcription in Hyperrecombinant Yeast Cells Lacking a Functional THO Complex. Molecular and Cellular Biology. 2006;26:3327–3334. doi: 10.1128/MCB.26.8.3327-3334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Chédin F, Hsieh C-L, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- Yuce O, West SC. Senataxin, Defective in the Neurodegenerative Disorder Ataxia with Oculomotor Apraxia 2, Lies at the Interface of Transcription and the DNA Damage Response. Molecular and Cellular Biology. 2012;33:406–417. doi: 10.1128/MCB.01195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.