Abstract
Thioaptamers targeting the dengue-2 virus (DENV-2) envelope protein domain III (EDIII) were developed. EDIII, which contains epitopes for binding neutralizing antibodies, is the putative host-receptor binding domain and is thus an attractive target for development of vaccines, anti-viral therapeutic and diagnostic agents. Thioaptamer DENTA-1 bound to DENV-2 EDIII adjacent to a known neutralizing antibody binding site with a dissociation constant of 154 nM.
Keywords: Thioaptamer, dengue virus, envelope protein, SELEX, binding, anti-viral, diagnostic agent
Introduction
Dengue virus (DENV) belongs to the family Flaviviridae, genus flavivirus. Four different closely related dengue serotypes (DENV types 1–4) cause human disease including non-specific viral syndrome, fatal dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). The increasing global spread of DENV and the lack of an approved vaccine or anti-viral therapeutic has prompted extensive research [1]. The virus enters the host cell through receptor-mediated endocytosis facilitated by the binding of envelope (E) protein to cell surface receptors. The endosome’s low pH triggers an E protein conformational change, leading to virus-membrane fusion [2]. The 395 residue E ectodomain has three domains, namely EDI, EDII and EDIII (residues 295–394), and the EDIII structure has a typical immunoglobulin-like fold [3]. EDIII folds independent of other E subdomains while retaining its structure, which is well conserved among flaviviruses, [4–8] and its antigenicity. At the virion five-fold symmetry axis, EDIII’s exposed loops form a hydrogen bond network possibly involved in host cell receptor binding [2]. DENV2 neutralizing antibody 1A1D-2 locks the EDIII positions on the viral surface, hampering structural rearrangement of E and preventing membrane fusion [9]. Thus, EDIII is an attractive target for developing drugs, vaccines and anti-viral agents. Thioaptamers are backbone-modified aptamers with decreased susceptibility to nucleases [10] and enhanced binding characteristics [11]. Our goal was to develop thioaptamers that prevent infection by blocking the virus entry into the host cell.
Materials and methods
Cloning, expression and purification of DENV-2 EDIII
DENV-2 EDIII was expressed in a bacterial system using C2566 E.coli cells. A clone containing pET15b plasmid (kind gift from Dr. Alan Barrett, UTMB, Galveston) for expressing the DENV-2 EDIII (WT strain 11608, 103 amino acids, M292-K394, 11.9 kDa) was PCR amplified and cloned into the pET22b plasmid for incorporation of a C-terminal 6x his-tag. The ligated product was transfected into C2566 complement cells (New England Biolabs, Inc) using the manufacturer’s protocol. Uniformly 15N, 13C labeled EDIII protein was expressed as described previously with a few changes that are presented here [5–9] that greatly improved the yield. The denatured protein in the supernatant was purified under partially denaturing conditions (2 M guanidine hydrochloride) by size-exclusion chromatography (SEC) using a Superdex G-75 column fitted to an AKTA Purifier FPLC system. Pure fractions (determined by SDS-PAGE) were pooled together. The protein was slowly refolded by dialysis against native buffer (six exchanges) at 4 °C using Snake Skin™ 3 kDa cut-off membranes. Refolded protein was concentrated and/or exchanged into NMR buffer using 3 kDa cut-off Amicon Centriprep concentrators.
Generation of Thioaptamer Library
The initial 68-base ssDNA (non-thioated) random library was generated commercially to contain a 23-base forward primer, a 21-base reverse primer, and a 24-base central random region (Supplementary Fig. 1) providing for 424 (~1014) different sequences, which were converted to thioaptamers by PCR as described [11–15]. Large scale thio-PCR of 2.4 ml volume in 24 tubes containing 100 μl each and a final template concentration of 0.1 nM were used (diversity of about 1011 sequences). 1 μM of each primer (forward primer has biotin at 5′ end) and 100 μM of each dNTP were used with 1x PCR buffer II, 4mM MgCl2 and 0.05 U/μl Taq polymerse. PCR was set up for initial 94 °C denaturation for 5 minutes, followed by 11 cycles of 94 °C denaturation for 1 minute, 60 °C annealing for 2 minutes and 72 °C extension for 3 minutes and a final extension at 72 °C for 10 minutes. The PCR product was used for single strand isolation using streptavidin coated magnetic beads that bind the biotinylated DNA strand using established protocols [13]. The isolated ssDNA was confirmed by PCR with each individual primer and with both primers together. Since it is the reverse strand ssDNA (library strand) that is used as template, only the forward primer makes double stranded product (no product with reverse primer) while both primers amplified the template. The isolated ssDNA constituted the thioaptamer library that was used for selection.
Selection of Thioaptamers
Magnetic beads with Ni-chelate groups on the surface were purchased from Bio-Rad, Inc. The beads were washed with the protein binding buffer (Buffer B - 50 mM NaH2PO4, 300 mM NaCl and 20 mM imidazole at pH 8.0). The protein binding capacity of the beads was up to 2 mg/ml of bead suspension. The purified DENV-2 EDIII protein with C-terminal his-tag in buffer B (500 μl) at a concentration of 160 μg/ml was added to 50 μl of bead suspension and incubated overnight. The beads with protein were then washed with 200 μl interaction buffer (Buffer I - 50 mM NaH2PO4, 50 mM NaCl, 20 mM imidazole and 0.005% tween 20 at pH 8.0) two times to remove unbound protein, and the protein coated beads were stored in 500 μl of buffer I. The protein concentration on beads was estimated by subtracting the amount of protein eluted in the wash from the amount of protein added to the beads. The protein coated beads were used from this stock for every selection round. 100 μl of the bead suspension was used for the first round of selection and it was reduced to 25 μl in the second through fifth rounds to increase stringency. The thioaptamer library in buffer I (100 μl) was added to protein coated beads and incubated with slow mixing for 30 minutes at room temperature. The unbound thioaptamers were removed and the beads were washed with 200 μl of Buffer I followed by two washes with 400 μl of wash buffer each (Buffer W - 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole and 0.005% tween 20 at pH 8.0). A final wash of 50 μl was made to use as PCR template for the wash fraction. The protein and thioaptamer complexes on the beads were eluted twice using 50 μl elution buffer (Buffer E - 50 mM NaH2PO4, 300 mM NaCl, 300 mM imidazole and 0.005% tween 20 at pH 8.0). The imidazole in the Buffer E released the protein bound to beads as a complex with bound thioaptamers (selected). A PCR was set up using the washes and elutions as template to compare the relative amounts of thioaptamers eluting at each step. The PCR was performed for 20 cycles and the products were checked on 6% PAGE gel. The elution fractions were pooled together and used as a template for the large scale PCR, and ssDNA was isolated to make an enriched thioaptamer library for the next round of selection. This process of selection was conducted five times (Supplementary Fig. 2). The eluted fraction from selection round 5 was PCR amplified then cloned using the TOPO cloning kit, and the plasmids from individual clones were isolated and sequenced. The sequences were aligned using CLUSTALW software [16] to determine the enriched sequences and to identify the conserved regions (see supplementary material for details). Mfold software was used to predict the secondary structures.
Filter binding assay to study DENV-2 EDIII thioaptamer binding
The four most abundant thioaptamers were synthesized with 5′-biotin ends. A fixed thioaptamer concentration (2 nM) in each well was mixed with eleven protein concentrations ranging from 4 μM down to 3.91 nM, or no protein (last column). The 50 μl binding reactions were conducted in duplicate in a 96 well plate at room temperature for 45 minutes using 10 mM Tris buffer (pH 7.4). A 96-well dot-blot apparatus with nitrocellulose and nylon membranes on top and bottom, respectively, was used, and the reaction liquids were driven through the membranes by vacuum [17] and at least three 100 μl buffer washes were performed to remove non-specific binders. The membranes were marked for orientation and placed face down under UV-light to cross link the thioaptamers to the membranes and the protocol for chemiluminescence detection module (Thermo scientific, Inc) was followed. The membranes were developed using streptavidin antibody-HRP conjugation induced chemiluminescence detected using a CCD camera [18]. The membranes were imaged using an Alphaimager (AlphaInnotech, Inc) and the chemiluminescence intensities were analyzed using ImageJ software. These data were used to calculate the percentage of thioaptamers on each membrane, based on the sum of the corresponding dots on both membranes (total intensity) being 100 percent. A plot of percentage of TA bound vs. protein concentration was made to calculate the binding constants using a non-linear regression for single site binding with Hill slope using Graphpad Prism software.
Microscale Thermophoresis
DENTA-1 was synthesized with Cy5 fluorophore at the 5′ end and purified by HPLC. Cy5 labeled DENTA-1 (50 nM) was used to observe for fluorescence emission in a thin standard glass capillary tube (obtained from Nanotemper Gmbh) using 6–8 μl of sample. The binding reactions with 15 concentrations of serially two-fold diluted protein were set up in tubes starting from 20 μM protein down to 1.22 nM protein and one with no protein as a control. The samples were loaded into the center of the capillaries and plugged with wax at the ends. The experiments were performed at room temperature with 40% LASER intensity that increases the temperature up to +6 °C at the center of the capillary for 30 seconds. The data were fitted to equation 1 using the Nanotemper software to obtain the binding constant for DENTA-1, where BL is the bound complex, B0 is total protein, L0 is total ligand and Kd is the equilibrium dissociation constant. Knowing BL, B0 and L0, the value of Kd was calculated [19].
| equation 1 |
NMR 2-D HSQC to determine binding site
The DENV-2 EDIII protein (50 μM) was exchanged into NMR buffer (20 mM Tris D11, 50 mM NaCl, 1 mM NaN3, 10 % (v/v) D2O at pH 6) to make the NMR sample. The 600 μL NMR sample was transferred to an 8″ NMR tube (Wilmad 541-PP) for data collection. Experiments were acquired on a 600 MHz Bruker Advance instrument (25 °C) equipped with a triple resonance cold probe and field gradients (Rice University). The 13C and 15N dimensions were referenced indirectly using frequency ratios. 2D 1H, 15N-HSQC NMR experiments were performed with DENV-2 EDIII protein alone and in complex with 25 μM DENTA-1.
Results
Expression, purification and characterization of DENV-2 EDIII
Previously published methods for purifying affinity-tagged flavivirus EDIII proteins using refolding before purification caused protein aggregation and low yields (data not shown). However, purity greater than 95% was achieved in a single step without affinity tags by performing the size exclusion chromatography under partially denaturing conditions. Protein aggregation is very minimal while refolding after purification, enabling efficient recovery of purified protein yielding 35 mg/L of LB culture and 25 mg/L of minimal media culture. UV-Vis spectroscopy was used to quantify the protein at OD280 and also the OD260/OD280 ratio was calculated to confirm the absence of nucleic acids. The purified and refolded form of isotopically labeled protein was verified by 2D 1H, 15N–HSQC NMR experiment to confirm the proper folding by observing characteristic amide proton chemical shifts that are sharp and well separated in the 1H and 15N dimensions (Supplementary Fig. 3).
Thioaptamer selection
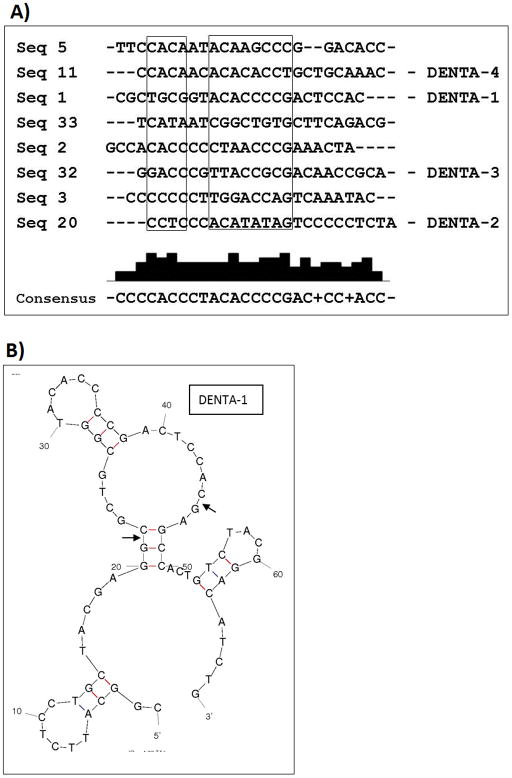
The partitioning efficiency in each selection round is vital for success [20]. We used magnetic nickel chelate agarose beads (Qiagen, Inc) for selection over five iterations. Pooled eluted fractions from round 1 were brought up to 500 mM imidazole for negative selection against beads without target protein. The protein-thioaptamer complexes remained in solution (due to the presence of imidazole) and the thioaptamers that bound to the beads (if any) were removed. The supernatant containing protein-thioaptamer complexes was then dialyzed to remove imidazole and was used as template for the thio- PCR to prepare the round 2 library. Relative thioaptamer concentrations of each wash and elution were assessed by PCR. The partitioning efficiency was improved by performing two washes. The volume of wash used was two to three folds higher than binding volume and about 10 fold higher than elution volume to reduce non-specific interactions while selecting for thioaptamers that bind strongly with low off rates [21]. The amount of thioaptamers eluted with the protein as a bound complex was always found to be more than the amount released in the same volume of the wash showing positive selection. The 6% PAGE gel shows the relative amount of thioaptamers eluted at each selection step (Supplementary Fig. 4). This method enabled us to monitor the efficiency of partitioning between target-bound and unbound thioaptamer molecules at each selection round. TOPO cloning and sequencing was performed after the fifth round of selection, and 38 sequences were obtained. One sequence occurred 4 times and three sequences occurred 3 times. Five others occurred twice. These eight sequences were aligned using ClustalW (Fig. 1A). The sequence 1 that occurred 4 times was named as DENTA-1 and the three sequences 20, 32 and 11 that occurred thrice were named DENTA-2, -3, and -4, respectively. The complete sequences of the four selected DENTAs are shown in Table 1. Mfold software was used to predict the secondary structures for thioaptamer sequences that occurred in higher frequency. The structure of DENTA1 is shown in Fig. 1B.
Figure 1.
A) CLUSTALW sequence alignment is shown - sequences 1, 20, 32 and 11 are DENTA-1, 2, 3 and 4, respectively. The 24 base random regions were used in the alignment. The conserved region is boxed and the consensus sequence is also shown at the bottom. B) The Mfold software predicted structure for DENTA-1 is shown. Conserved region is identified in the loop. Arrows denote the start and end of the 24 base random region (bases 21 – 45).
Table 1.
TOPO sequencing revealed the enriched dengue thioaptamer sequences. The 24-base random region is in the middle. The thiol-modification is at the 5′ end of adenine (A) present in the random region and the 3′ end primer region. The 5′ end primer region does not have the thiol-modification since it originated from the primers used in the PCR reaction which were not thiol-modified.
| Name | Sequence (5′ - 3′) |
|---|---|
| DENTA-1 | CGGCATTCTCCTGCTACGAGG-CGCTGCGGTACACCCCGACTCCAC – GAGCCACTGTCTACGGACATCTG |
| DENTA-2 | CGGCATTCTCCTGCTACGAGG-CCTCCCACATATAGTCCCCCTCTA – GAGCCACTGTCTACGGACATCTG |
| DENTA-3 | CGGCATTCTCCTGCTACGAGG-GGACCCGTTACCGCGACAACCGCA – GAGCCACTGTCTACGGACATCTG |
| DENTA-4 | CGGCATTCTCCTGCTACGAGG-CCACAACACACACCTGCTGCAAAC-GAGCCACTGTCTACGGACATCTG |
Characterization of thioaptamer binding to DENV-2 EDIII protein
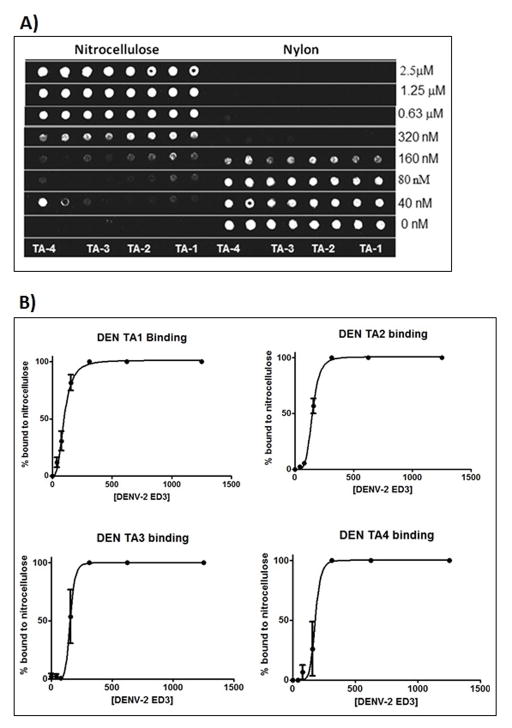
Filter binding assays are well characterized for studying protein-nucleic acid interactions [18]. The results of filter binding assays show the binding of thioaptamers to the target protein in a concentration dependent manner on the nitrocellulose membrane. The thioaptamers that do not bind the nitrocellulose membrane are trapped by the nylon membrane beneath the nitrocellulose membrane. All the thioaptamer molecules were bound at the highest concentration of protein used and the amount of bound thioaptamers decreased with decreasing protein concentrations. The percentage of unbound thioaptamers (relative to total amount of thioaptamers) trapped in the nylon membrane increases from higher to lower protein concentration (Fig. 2A). The spot intensities corresponding to nitrocellulose and nylon membranes were used to calculate the percentage of total thioaptamers per well that is distributed between the nitrocellulose and nylon membranes. The plot of percentage of thioaptamers bound with increasing protein concentration was made and the data were fitted to a single site binding equation with Hill slope to obtain the Kd. The binding of sequences DENTA-1 through DENTA-4 with DENV-2 EDIII were characterized in duplicate and the data were fit using GraphPad Prism (Fig. 2B), the results of which are shown in Table 2. Microscale thermophoresis results for DENV-2 EDIII and DENTA-1 binding show increasing complex formation with increasing protein concentrations, as observed from changes in thermophoresis values. The fluorescence intensity was measured to observe the Cy-5 labeled DENTA-1 molecules (by itself and in complex with DENV-2 EDIII) diffusing out of the heating center when the laser is turned on (300 seconds) and when the laser is off, they diffuse back to the center (Fig. 3A). The rate of diffusion was measured for all of the 16 samples at least three times. The differences in the diffusion rates of DENTA-1 alone and with increasing concentrations of DENV-2 EDIII protein were measured. The thermophoresis values were calculated and the data were fit to equation 1 to provide a calculated Kd of 154 ± 40 nM for the DENTA-1 and DENV-2 EDIII binding interaction (Fig. 3B).
Figure 2.
A) Filter binding assay showing the chemiluminescence intensities on nitrocellulose and nylon membranes. The biotin-labeled DENTAs were used for development of chemilunescence using the streptavidin-HRP conjugate. B) The percentage of thioaptamers bound to nitrocellulose membrane are plotted against the protein concentration (nM) to fit the single site binding equation with Hill slope to determine the binding constants for DENTAs binding to DENV-2 EDIII.
Table 2.
Binding constants for the four selected thioaptamers calculated from the filter binding assays are tabulated showing the fitting parameters and the equations used in the fit. Y is the percentage chemiluminescence intensity on the nitrocellulose membrane and X is the protein concentration in nM.
| Single site binding model with hill slope (h)a | Four parameter logistic fit modelb | |||||
|---|---|---|---|---|---|---|
| Sequence | Kd (nM) | h | R2 | EC50 (nM) | h | R2 |
| DENTA-1 | 99 ± 5 | 2.9 | 0.98 | 98 ± 11 | 3.1 | 0.98 |
| DENTA-2 | 148 ± 3 | 4.8 | 0.99 | 147 ± 8 | 4.9 | 0.99 |
| DENTA-3 | 153 ± 18 | 7.7 | 0.96 | 153 ± 21 | 7.7 | 0.93 |
| DENTA-4 | 178 ± 34 | 7.9 | 0.96 | 177 ± 65 | 8.5 | 0.95 |
Y=Bmax*Xh/(KDh + Xh)
Y=100/(1+10(LogEC50−X)*h)
Figure 3.
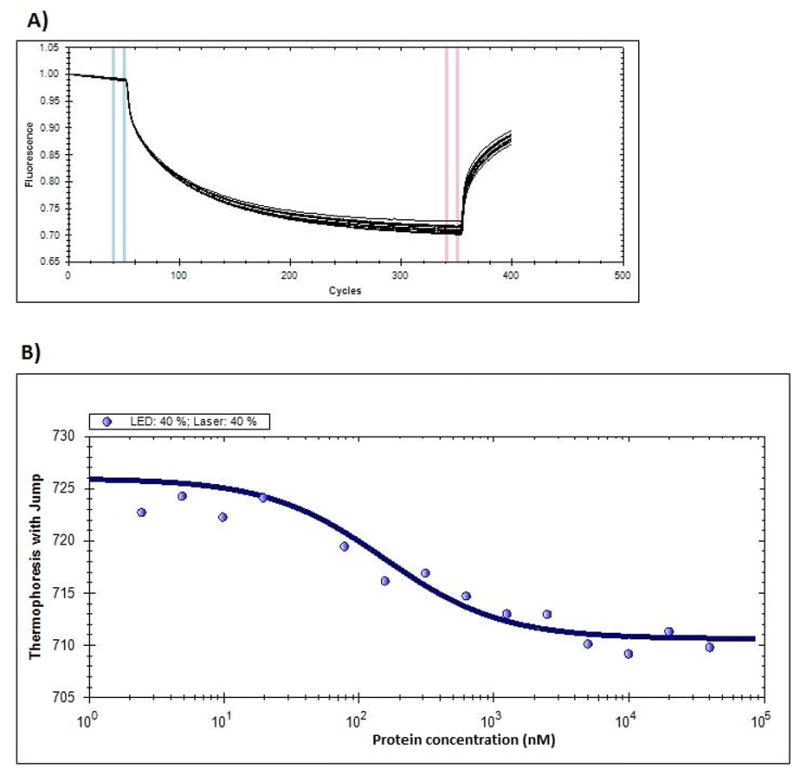
Microscale thermophoresis results – A) Fluorescence change upon switching on and off of LASER at 40% intensity is shown. B) The thermophoresis values were plotted against protein concentration in nM and fitted to the binding equation 1 to obtain the Kd for DENTA-1 and DENV-2 EDIII interaction.
Binding site mapping using 2-D HSQC NMR experiment
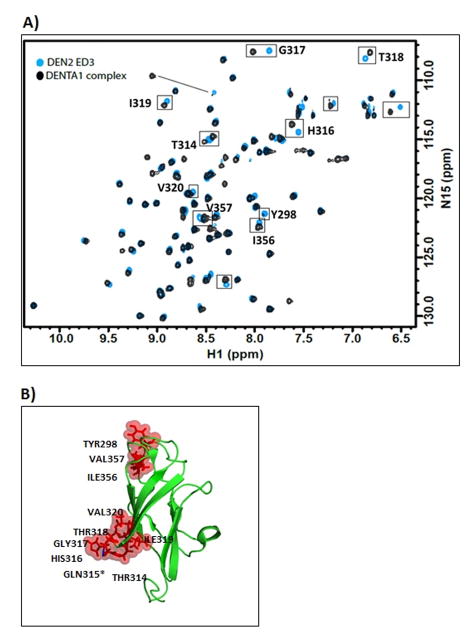
The 2-D 1H,15N-HSQC NMR spectra show chemical shift changes upon addition of DENTA-1, indicating the interaction between DENTA-1 and DENV-2 EDIII (Fig. 4A). The residues that show maximum chemical shift changes constitute the probable DENTA-1 binding site. The amide proton chemical shift for residues H316, G317, T318 and I319 show significant changes, while Y298 and V320 are line broadened. Residue T314 shows modest change and Q315 is unassigned. Spatially close residues I356 and V357 in the FG loop and Y298 in the N-terminus loop show chemical shift changes upon complex formation. Residues showing shifts were mapped onto the loop that follows the β1-strand and the β2-strand in DENV-2 EDIII [25] (Fig. 4B). Contiguous largely perturbed residues signify the predominant binding site. Smaller changes (FG loop) may indicate a weaker second binding site or slight structural changes due to binding elsewhere.
Figure 4.
A) NMR 1H, 15N – 2D – HSQC overlay of spectra from DENV-2 EDIII protein by itself and upon addition of DENTA1 is shown. The cyan peaks represent the amide protons from protein alone and the black peaks represent the amide proton shifts from protein in complex with DENTA-1. B) The residues that showed significant chemical shift changes (>0.5 ppm) upon addition of DENTA-1 were mapped onto the DENV-2 EDIII NMR structure [25] to show the binding region.
Discussion
Thioaptamers were selected that bind to DENV-2 EDIII using the SELEX method [22–24]. The partitioning efficiency of the selection rounds were monitored by PCR amplification and PAGE. DNA sequencing of the final selected thioaptamer pool identified the sequences that were repeated most often and it was determined that they are the strongest binders. Filter binding assays demonstrated that the selected thioaptamers bound tightly to DENV-2 EDIII (Table 2). DENTA-1 binding to DENV-2 EDIII was measured using microscale thermophoresis (Kd=154±40 nM) where the binding was measured in solution. EMSA could not be used to measure binding constants since the protein-bound DENTA-1 could not be observed (Supplementary Fig. 6). This may be due to the association and dissociation kinetics of binding during the run leading to diffusion of the protein bound thioaptamers. A 2D HSQC NMR experiment indicated which protein residues were perturbed showing they were directly or indirectly involved in the binding interaction. It is interesting to note that the residues F305-A312 constitute the neutralizing epitope for the monoclonal anti-body 1A1D-2 that binds the EDIII in DENV-2 to neutralize the virus. The cryo-EM and crystal structure of the fab part of antibody 1A1D-2 bound to DENV-2 EDIII showed that the binding occurs on the β1-strand which is not surface exposed in the static structure of mature virus. This also suggests that the EDIII is dynamic on the viral surface and the parts that are buried in the static structure could in fact be exposed to the solvent, allowing antibody binding to this region. The binding of antibody 1A1D-2 locks the virus in a form that is different from the mature form and incapacitates the virus by blocking the conformational change in E that is required for membrane fusion [10]. Applying a similar principle, and by comparing the binding sites for the monoclonal antibody 1A1D-2 and DENTA-1 (Supplementary Fig. 7); it is possible that DENTA-1 may bind the DENV-2 EDIII and neutralize the virus in a similar fashion. Further characterization is needed to evaluate the neutralization efficiency of DENTA-1. Also, these binding molecules can be tethered using molecular scaffolds like dendrimers and can be designed to bind and perhaps lock the EDIII positions at the five-fold or three-fold symmetry axes on the surface of DENV-2. This will also increase the avidity of the DENTAs for better neutralization of the virus. This approach is unique for anti-viral drug discovery and can be useful in designing potent inhibitors particularly against enveloped viruses that inherently possess surface symmetry that can be exploited.
Supplementary Material
Highlights.
Thioaptamers were developed using SELEX targeting DENV-2 EDIII protein
Selected thioaptamers were synthesized and used in filter binding assay to determine binding constants
Site of binding for DENTA-1 was deduced by 2D NMR HSQC and mapped onto DENV-2 EDIII structure
Acknowledgments
We thank Drs. Anoma Somasunderam and Varatharasa Thiviyanathan for useful discussions on thioaptamer selection, Xin Li for help with protein expression and purification, Dr. Rodrigo Maillard for the initial protein expression and purification protocol (pre-publication), and Loren Stagg for help with the microscale thermophoresis instrument in the Ladbury lab at the UT MD Anderson Cancer Center.
Funding Sources:
Welch Foundation (AU-1296), NCI (CA151668 and U55), NIAID (HHSN272200800048C and AI054827), NHLBI (HHSN268201000037C), NIGMS (RC2GM092599ARRA), NIH (T90 DK070109 and 5 R90 DK071505).
ABREVIATIONS
- DENTA-1
dengue thioaptamer 1
- DENV-2
dengue virus type-2
- EDIII
envelope protein domain III
- ssDNA
single stranded DNA
Footnotes
Disclosure:
The authors declare the following competing financial interest(s): D.G.G. and the University of Texas Health Science Center at Houston have research-related financial interests in AptaMed Inc. and AM Biotechnologies LLC (Houston, TX).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, et al. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8:S7–16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, et al. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhardwaj S, Holbrook M, Shope RE, Barrett AD, Watowich SJ. Biophysical characterization and vector-specific antagonist activity of domain III of the tick-borne flavivirus envelope protein. J Virol. 2001;75:4002–4007. doi: 10.1128/JVI.75.8.4002-4007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volk DE, Lee YC, Li X, Thiviyanathan V, Gromowski GD, Li L, et al. Solution structure of the envelope protein domain III of dengue-4 virus. Virology. 2007;364:147–154. doi: 10.1016/j.virol.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volk DE, Anderson KM, Gandham SH, May FJ, Li L, Beasley DW, et al. NMR assignments of the sylvatic dengue 1 virus envelope protein domain III. Biomol NMR Assign. 2008;2:155–157. doi: 10.1007/s12104-008-9109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volk DE, May FJ, Gandham SH, Anderson A, Von Lindern JJ, Beasley DW, et al. Structure of yellow fever virus envelope protein domain III. Virology. 2009;394:12–18. doi: 10.1016/j.virol.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volk DE, Gandham SH, May FJ, Anderson A, Barrett AD, Gorenstein DG. NMR assignments of the yellow fever virus envelope protein domain III. Biomol NMR Assign. 2007;1:49–50. doi: 10.1007/s12104-007-9000-9. [DOI] [PubMed] [Google Scholar]
- 10.Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol. 2008;15:312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Wang H, Beasley DW, Volk DE, Zhao X, Luxon BA, et al. Selection of thioaptamers for diagnostics and therapeutics. Annal New York Acad Sci. 2006;1082:116–119. doi: 10.1196/annals.1348.065. [DOI] [PubMed] [Google Scholar]
- 12.Bunka DH, Platonova O, Stockley PG. Development of aptamer therapeutics. Curr Opin Pharm. 2010;10:557–562. doi: 10.1016/j.coph.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Somasunderam A, Thiviyanathan V, Tanaka T, Li X, Neerathilingam M, Lokesh GL, et al. Combinatorial selection of DNA thioaptamers targeted to the HA binding domain of human CD44. Biochemistry. 2010;49:9106–9112. doi: 10.1021/bi1009503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King DJ, Ventura DA, Brasier AR, Gorenstein DG. Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry. 1998;37:16489–16493. doi: 10.1021/bi981780f. [DOI] [PubMed] [Google Scholar]
- 15.Kang J, Lee MS, Copland JA, Luxon BA, Gorenstein DG. Combinatorial selection of a single stranded DNA thioaptamer targeting TGF-beta1 protein. Bioorg Med Chem Lett. 2008;18:1835–1839. doi: 10.1016/j.bmcl.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 17.Woodbury CP, Jr, von Hippel PH. On the determination of deoxyribonucleic acid-protein interaction parameters using the nitrocellulose filter-binding assay. Biochemistry. 1983;22:4730–4737. doi: 10.1021/bi00289a018. [DOI] [PubMed] [Google Scholar]
- 18.Kang J, Lee MS, Gorenstein DG. Quantitative analysis of chemiluminescence signals using a cooled charge-coupled device camera. Anal Biochem. 2005;345:66–71. doi: 10.1016/j.ab.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Comm. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 20.Irvine D, Tuerk C, Gold SELEXION L. Systematic evolution of ligands by exponential enrichment with integrated optimization by non-linear analysis. J Mol Biol. 1991;222(3):739–61. doi: 10.1016/0022-2836(91)90509-5. [DOI] [PubMed] [Google Scholar]
- 21.Lou X, Qian J, Xiao Y, Viel L, Gerdon AE, Lagally ET, et al. Micromagnetic selection of aptamers in microfluidic channels. P N A S. 2009;106:2989–2994. doi: 10.1073/pnas.0813135106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 23.Gopinath SC. Methods developed for SELEX. Anal Bioanal Chem. 2007;387:171–182. doi: 10.1007/s00216-006-0826-2. [DOI] [PubMed] [Google Scholar]
- 24.Hamula C, Guthrie J, Zhang H, Li X-F, Le XC. Selection and analytical applications of aptamers. Trends in Anal Chem. 2006;25(7):681–691. [Google Scholar]
- 25.Huang KC, et al. Solution structure and neutralizing antibody binding studies of domain III of the dengue-2 virus envelope protein. Proteins. 2008;70(3):1116–9. doi: 10.1002/prot.21806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.