Abstract
Suppressor tRNAs bear anticodon mutations that allow them to decode premature stop codons in metabolic marker gene mRNAs, that can be used as in vivo reporters of functional tRNA biogenesis. Here, we review key components of a suppressor tRNA system specific to S. pombe and its adaptations for use to study specific steps in tRNA biogenesis. Eukaryotic tRNA biogenesis begins with transcription initiation by RNA polymerase (pol) III. The nascent pre-tRNAs must undergo folding, 5′ and 3′ processing to remove the leader and trailer, nuclear export, and splicing if applicable, while multiple complex chemical modifications occur throughout the process. We review evidence that precursor-tRNA processing begins with transcription termination at the oligo(T) terminator element, which forms a 3′ oligo(U) tract on the nascent RNA, a sequence-specific binding site for the RNA chaperone, La protein. The processing pathway bifurcates depending on a poorly understood property of pol III termination that determines the 3′ oligo(U) length and therefore the affinity for La. We thus review the pol III termination process and the factors involved including advances using gene-specific random mutagenesis by dNTP analogs that identify key residues important for transcription termination in certain pol III subunits. The review ends with a ‘technical approaches’ section that includes a parts lists of suppressor-tRNA alleles, strains and plasmids, and graphic examples of its diverse uses.
Keywords: RNA polymerase III, transcription termination, RPC11, RPC53, RPC37, RPC2
OVERVIEW
Our purpose here is to review a body of published work obtained using a powerful methodological approach in use in ours and others’ laboratories, to investigate tRNA biogenesis in S. pombe. The core approach is tRNA-mediated suppression (TMS) and our objective is to review the mechanistic workings of this intricate system for the purpose of studying tRNA biogenesis. We will first discuss the components of TMS activity including strengths and weaknesses of the approach, and provide examples of strategies of optimization for specific applications. We note however, that descriptions of much of the discussion will benefit from specific perspectives on context, for which we will include a limited number of references to related work in S. cerevisiae, vertebrates and other organisms, but by no means will this represent a general review of tRNA biogenesis. Therefore, for comprehensive review of tRNA biogenesis a number of excellent publications are recommended ((1, 2), also see (3-7)). We will describe various constituents of the S. pombe TMS system including a variety of suppressor-tRNA alleles that sensitize it to different steps in the tRNA processing and maturation pathway, and review data on tRNA 3′ end formation and other aspects of tRNA biogenesis including transcription initiation and termination by RNA polymerase III, and a mutagenesis-based genetic approach to study the trans-acting factors involved.
NONSENSE SUPPRESSION
Suppressor tRNA sequences were isolated as genes that suppress nonsense mutations in protein coding genes (8). These suppressors are derived from normal tRNA genes that undergo an anticodon mutation that enables them to read a stop codon in an mRNA and insert their amino acid. In eukaryotes, suppressor tRNAs in Saccharomyces cerevisiae and Schizosaccharomyces pombe are among the most studied although some have been used in higher eukaryotes as well (9 and refs therein). When a suppressor tRNA is used in combination with a selectable marker carrying a corresponding nonsense mutation as a premature stop codon, effects on the biogenesis of the suppressor tRNA can be monitored by changes in the functional expression of the protein product of the marker gene. This provides opportunity to use the suppressor as a nonessential reporter of tRNA biogenesis; only if it undergoes all steps necessary to produce a mature functional tRNA, will it efficiently activate the selectable marker. In some cases one may direct the suppression assay to be mainly dependent on one or another specific step in the tRNA biogenesis pathway. Eukaryotic suppressor tRNAs have been used to study many aspects of tRNA biogenesis including transcription by RNA polymerase III (pol III), tRNA structure-function relationships, processing, subcellular trafficking, decay and other aspects of tRNA maturation. A large amount of work on tRNA biogenesis has been done using the budding yeast, S. cerevisiae, and many processing and modification steps as well as protein factors and other enzymes involved have been thoroughly reviewed ((1, 2), also see (3-7)).
S. pombe as a model system
This review will focus on advances made using a tRNA mediated suppression (TMS) system in the fission yeast, S. pombe. The fission and budding yeasts diverged more than 1100 million years ago and while neither is closer evolutionarily than the other is to human, researchers have noted that S. pombe appears more similar to human cells in cell cycle control, pol II core promoter structure, transcription by pol III, intron complexity and extent of pre-mRNA splicing, mitochondrial structure, and respiratory function (10-14). The S. pombe TMS system has been useful for examining functionality of human proteins, including La protein and its control by faithful phosphorylation by protein kinase CK2 in the yeast, and its nuclear-cytoplasmic trafficking (15-18). It was also useful in characterizing the RNA binding and chaperone activities of several human La-related proteins (LARPs) (19-21), as well as the tRNA isopentenyl transferase, TRIT1 (22).
The major pol III promoter elements for tRNA genes, known as the A box and B box, are internal, located downstream of the transcription start site (for excellent review see Ref (46)). tRNA gene architecture and the suppressor pathway in S. pombe is presented in Fig. 1. tRNA suppressor genes that were isolated from S. pombe were mostly studied in S. cerevisiae because the genetics, molecular biology and biochemistry of S. pombe were less advanced at the time, and also because tRNA expression in S. pombe appeared restrictively complex (40, 41). S. pombe tRNA genes could be readily expressed in S. cerevisiae whereas apparently similar tRNA genes isolated from S. cerevisiae could not be expressed in S. pombe (40, 41). This conundrum was explained by the discovery that S. pombe requires tRNA genes to have upstream TATA box promoter elements (Fig. 1A), which are absent from the vast majority of S. cerevisiae tRNA genes (42) but present upstream of a majority of tRNA genes in other species including human (43). As noted later in the review, S. pombe and S. cerevisiae also differ in other aspects of transcription, including initiation factor assembly mechanisms and termination, highlighting the possibility that other aspects of tRNA biogenesis may also differ in these yeasts.
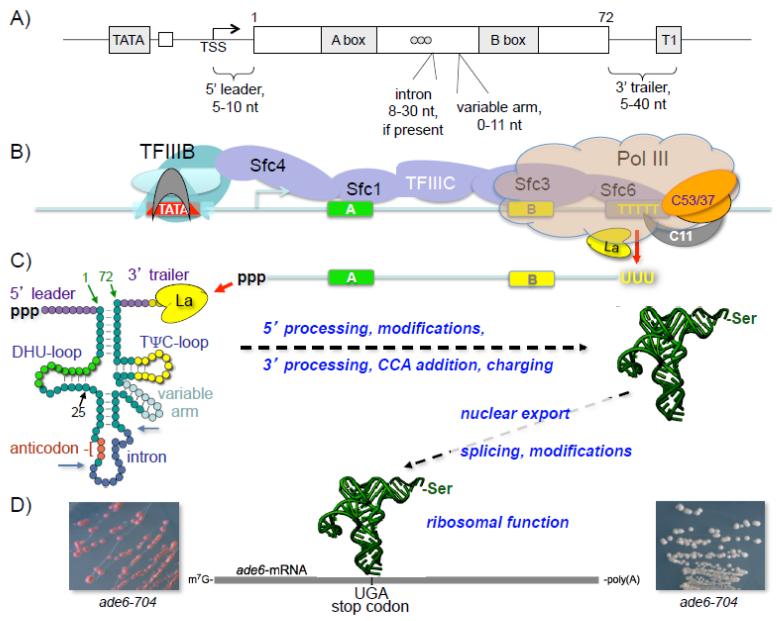
Figure 1. tRNA gene architecture and suppressor pathway in S. pombe.
A) Schematic representation of tRNA gene architecture in S. pombe including variable length regions. The upstream TATA box (at approximate position −30 relative to transcription start site, TSS) promoter element is required for all known pol III transcribed genes in S. pombe. The intragenic A box and B box promoter elements are indicated as is the terminator element, oligo(T) represented by T1. Three adjacent circles between A and B boxes represent the anticodon. Variable loop and intron if present are indicated. B) A S. pombe tRNA gene with promoter elements bound by transcription factors, TFIIIB and TFIIIC is shown, based on studies of S. cerevisiae (131, 185, 186). TFIIIC (purple) is a six subunit complex that binds to A and B boxes and recruits Brf1 (cyan) and Bdp1 (light cyan) to the TATA box-bound TBP (gray). TFIIIB remains bound to DNA after initiation by pol III resulting in a high rate of transcription reinitiation (187). The 17 subunit pol III (light orange) is shown with the three termination related subunits, the C53/37 dimer and C11, highlighted in dark orange and gray respectively (see text). The La protein (yellow) binds in a sequence-specific and length-dependent manner to the 3′ oligo(U) tract of the RNA product that results from transcription termination at the oligo(T) terminator, and is the first protein to associate with newly synthesized nascent pol III transcripts (see text). C) Schematized nascent suppressor pre-tRNAserUCA and its various processing steps; color coding reflect the RNA copies of the A and B box elements. A clover leaf structure of nascent pre-tRNA is shown with the 5′ leader, intron, 3′ trailer and other elements as indicated; dihydrouridine stem loop (DHU-loop), variable arm, and thymidine-pseudouridine cytidine-rich stem loop (TΨC-loop). Arrows indicate intron-exon boundaries. Mature tRNA position 25 is indicated at the base of the DHU-loop (see text) as well as mature tRNA positions 1 and 72. Various steps in the maturation process are indicated on either side of the horizontal dashed arrow. Pre-tRNA splicing occurs in the cytoplasm in yeast (188, 189) and assorted chemical modifications of various nucleotides occur at multiple points during tRNA maturation (reviewed in 2). D) The S. pombe ade6-704 mRNA encoding the AIR carboxylase (see text) bears a premature UGA stop at codon position 215. In the absence of the opal suppressor tRNAserUCA, the ade6-704 mRNA undergoes incomplete translation and cells accumulate a purine biosynthetic pathway intermediate that forms red pigment when grown in limiting adenine media (left panel). In the presence of a functional suppressor tRNAserUCA, the stop codon is decoded and active Ade6 protein is made, the purine pathway intermediates are metabolized, and accumulation of red pigment is prevented, leading to white colonies (right panel).
Mutations in the pol III transcription machinery, responsible for synthesis of tRNAs, 5S rRNA and other small RNAs, were found to be associated with developmental deficiencies in zebra fish and with human disease. Specifically, a genetic screen in zebra fish uncovered a mutation in the second largest pol III subunit (Rpc2) that caused digestive system malformation (23). Introducing this mutation into S. pombe Rpc2 showed that it caused decreased association of Rpc2 and another subunit, Rpc11, arguing that the interaction interface had been conserved from yeast to vertebrates (23). More recently, mutations in the two largest pol III subunits cause hypomyelinating leukodystrophy, and in the Bdp1 subunit of transcription initiation factor, TFIIIB, cause hearing loss (24-26). Thus it is tempting to speculate that the S. pombe TMS system may be useful toward examining the mechanistic basis of these mutations.
Although S. pombe suppressor tRNA genes were used to identify pol III promoter elements as well as to analyze tRNA structure-function, the studies were performed using the S. pombe genes in S. cerevisiae (27, 28). Suppressor tRNAs were also used for genetic screens of fission and budding yeast that identified and characterized genes involved in tRNA biogenesis that include the tRNA isopentenyl transferase, Mod5/sin1+/tit1+, the U34 anticodon modification enzyme, sin3+/Elp3, the pol III repressor, Maf1, and the tRNA nuclear export factor, Los1/XpoT ((29-38), also see (39); for historical perspective and early suppressor analysis in S. pombe see (28)).
tRNA-mediated suppression (TMS) of a reporter allele
The reporter/marker genes carrying premature stop codons that are generally employed for TMS can be classified into two categories: the first includes genes such as for antibiotic resistance or metabolic enzymes that cause specific nutrient prototrophy, and the second includes genes that alter cell color. Examples of the second class include the bacterial LacZ gene which when expressed and allowed to metabolize X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside), cause colonies to be blue (39). Most relevant to this review is the phosphoribosyl-aminoimidazole carboxylase (AIR carboxylase) gene that encodes a key enzyme in adenine biosynthesis (29, 30, 44). AIR carboxylase is encoded by ADE2 in S. cerevisiae and ade6+ in S. pombe. In the presence of limiting amounts of adenine in the growth media, cells lacking AIR carboxylase accumulate a metabolic intermediate which upon oxidation forms red pigment (i.e., in O2 respiring cells, note that anaerobic cells will appear white when ade6-704 is unsuppressed). An ochre nonsense (UAA) allele, ade2-1 in S. cerevisiae and an opal nonsense (UGA) allele, ade6-704 in S. pombe have been used extensively to study suppressor tRNAs in these organisms. The nonsense mutation in the S. pombe ade6-704 allele originally isolated from a Kohli strain was recently mapped to T645A, converting the cysteine codon, UGU at position 215 to a UGA stop codon ((17), Alexander Lorenz, personal communication).
Expression of a functional opal suppressor-tRNA in an ade6-704 strain allows synthesis of the full length AIR carboxylase and thereby prevents accumulation of the red pigment, resulting in white colonies. In addition to fully suppressed and un-suppressed states, different color intensities indicate relative levels of suppressor-tRNA (abundant = white, absence = red, intermediate = shades of pink), although tight correlation may require titration of the specific activity of the suppressor (below).
A consideration of suppressor tRNA use in loss-of-suppression applications should be that the loss may occur at any of a number of known or unknown steps of tRNA biogenesis (see (39)). This approach can lead to the discovery of new genes or pathways involved in tRNA biogenesis ((31), also see (39)). However, to focus on a specific step of interest, the potential complexities of loss-of-function may be circumvented by adopting gain-of-function strategies (below). Other considerations include the expression level of the suppressor and its specific activity, as these can also affect the dynamic range of the assays, toxicity and feasibility of the studies. Various approaches have been used to address some of these issues, including development of two distinct TMS systems, monomeric and dimeric tRNA gene alleles, that vary in cis-elements, suppressor specific activity and requirement for trans-acting factors.
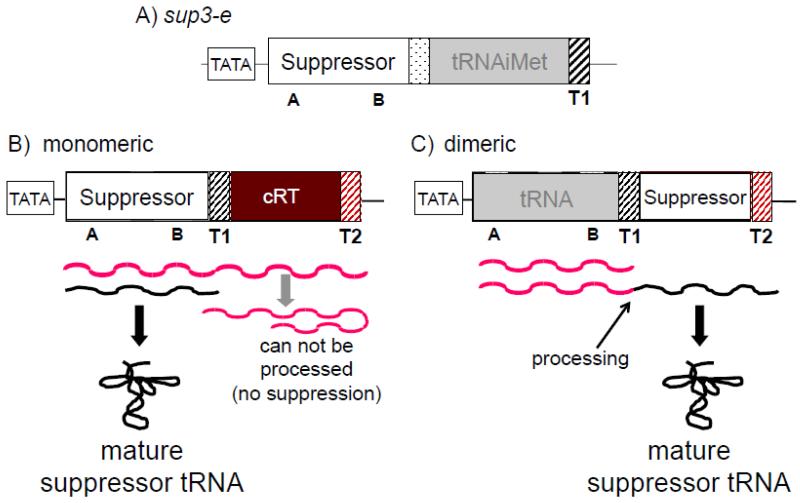
The S. pombe suppressor tRNA gene, sup3-e produces the opal suppressor tRNA with an anticodon UGA→UCA mutation that allows it to read the UGA stop codon in ade6-704 mRNA (28, 45). Sup3-e is one of very few dicistronic pol III-transcribed genes in S. pombe, consisting of a tRNAserUCA sequence followed by a short A+T-rich spacer and tRNAiMet (48). Similar rare dimeric tRNA gene arrangements are also found in S. cerevisiae, human and other species. In some cases, the A and B box promoter arrangement is used to drive pol III-dependent transcription of a downstream snoRNA (small nucleolar) or other small non-coding RNA (46, 49). For sup3-e and other dimeric tRNA genes in S. pombe, only the first tRNA has an upstream TATA promoter element, (42, 50). Thus for sup3-e in S. pombe, transcription of the tRNAiMet is completely dependent on initiation directed by the upstream tRNAserUCA. The nascent dimeric tRNA transcript is posttranscriptionally processed into two tRNAs (51). S. pombe suppressor-tRNAserUCA is aminoacylated with serine and would therefore insert serine at UGA codons (52, 53).
Variable specific activities of suppressor tRNA alleles
Strong opal suppressors are also referred to as efficient, hence the name sup3-e, in part to distinguish them from inefficient suppressors such as sup3-i (an ochre suppressor generated by C -> T mutation in the middle nucleotide of Sup3-e anticidon) (28). Sup3-e is transcribed at high levels comparable to endogenous tRNA genes. Suppression of UGA is expectedly the least toxic of the stop codons since it is used for only ~15% of mRNAs in S. pombe and these are not expressed at high levels, whereas UAA and UAG are used as stops for the remaining 61% and 24%, respectively, including the highest level expressed genes (54). Sup3-e appears to exhibit high specific decoding activity at UGA codons.
Expression of too strong a suppressor-tRNA can be detrimental because it might suppress translation termination of many varied cellular mRNAs. A monomeric allele derived from sup3-e whose only mutations are the G→C at position 35 in the anticodon and a compensatory C→G mutation in the intron position 37:10 that re-establishes base pairing with the anticodon and intron (the G37:10 mutation) to enhance processing (16, 55), causes severe slow growth in S. pombe as compared to alleles with suppressor mutations (unpublished but see Fig. 2I in (56)). Ideally, the amount of suppressor-tRNA mediated suppression activity should be titrated to produce a full range of ade6-704 response but not in excess of this. It is useful to obtain suppressor alleles that vary in specific activity in order to find ones that sensitize the tRNA to processing and maturation factors of interest and minimize toxicity. This was achieved not by repressing transcription but by introducing three mutations that decrease specific activity of the suppressor allele (11). Directed mutagenesis toward this objective was informed by prior extensive analyses of sup3-e by Ian Willis and colleagues in Dieter Söll’s laboratory that produced a catalog of mutations of nearly all nucleotide positions in suppressor-tRNASerUCA (sup3-e) and their effects on transcription, splicing and end-processing of the nascent transcript (see TABLE II in 28).
Analysis of sup3-e mutations included in vitro transcription and RNA processing that helped establish biogenesis pathways for nascent pre-tRNAs in S. cerevisiae ((28) and references therein). This mutation information was later exploited for suppressor alleles for specific applications using S. pombe TMS (Table 1).
Table 1. Suppressor tRNA alleles used in different studies.
Different S. pombe strains used in studies previously published are listed along with the suppressor tRNA allele, number of Ts at the terminator and the purpose of use. References for each published study are given. Mutations shown in the alleles column indicate the nucleotide changes in the tRNA generated by each allele.
| System | Strain | Allele | Number of Ts |
Purpose | Reference |
|---|---|---|---|---|---|
| Monomeric | yYH1 | C40T; T47.3C; C47.6T |
7 | Termination, processing |
(88) |
| ySH9 (sla1Δ) |
Wt | 7 | Termination, processing |
(88) | |
| ySH18 | C47.6T | 7 | La chaperone activity |
(56, 65, 111, 182) |
|
| yYH4B (rrp6Δ) |
C40T; T47.3C; C47.6T |
7 | tRNA maturation | (56) | |
| yYH5a (sla1Δ rrp6Δ) |
C40T; T47.3C; C47.6T |
7 | tRNA maturation | (56) | |
| yNB1 (maf1Δ) |
C40T; T47.3C; C47.6T |
7 | Pol III regulation | in preparation | |
| yNB5 (tit1Δ) |
C40T; T47.3C; C47.6T |
7 | tRNA modification | (183) | |
| yAS110 (sla1Δ) |
C40T; T47.3C; C47.6T |
7 | tRNA maturation | (16) | |
| ySK5 | C40T; T47.3C; C47.6T; G37.10G |
7 | tRNA maturation | (15, 16) | |
| yAMm4T-2 | C40T; T47.3C; C47.6T |
4 | Termination | (88) | |
| yAS56 | No suppressor | NA | RNAse Z | (184) | |
| yAMm5T-1 | C40T; T47.3C; C47.6T |
5 | Termination | (88) | |
| yAM148 | No suppressor, leu1+ |
NA | control | (56) | |
| yAS68 | C40T; T47.3C; C47.6T |
3 | Termination | (64, 129) | |
| yAS99 | No suppressor | NA | control | (64, 129) | |
| yAMm5T-1 | C40T; T47.3C; C47.6T |
5 | Termination | (88) | |
| Dimeric | yKR1 | C37.10G T47.3C |
5 | Termination | (129) |
| yJI1 | C37.10G T47.3C |
6 | Termination | (64) |
The monomeric suppressor tRNA system
The suppressor-tRNASerUCA (sup3-e) was used to create the constructs for the monomeric and dimeric TMS systems (Fig. 2). The sup3-e gene architecture is unusual since most (~97%) of tRNA genes in S. pombe and other model organisms are free-standing tRNA genes of highly typical monomeric structure with dedicated promoters and terminators that accommodate their variable length characteristics (Fig. 1A). Therefore, conversion of sup3-e to a monomeric gene should increase its applicability to normal tRNA biogenesis. The suppressor-tRNA was isolated by deleting the downstream tRNAiMet sequence; in addition, a unique 3′ trailer sequence followed by a dedicated terminator (see Fig. 1 in (44)) and a downstream sequence referred to as complementary read-through (cRT) were introduced to create the pSer allele (Fig. 2B). The suppressor constructs are usually stably integrated at the leu1+ locus for suppression analysis.
Figure 2.
Schematic representation of the S. pombe sup3-e dimeric tRNA gene and the two different construct types derived from it that are used for TMS. In the monomeric TMS system, pol III that fails to terminate at T1 produces a transcript containing the cRT (complementary Read Through) sequence which can fold-back to anneal with the suppressor-tRNA region and thereby inhibit processing and tRNA production. In the dimeric TMS system, transcription read-through beyond T1 is required to generate the suppressor tRNA (A=A box, B=B box, T1=test terminator, T2=second terminator). The dimeric system requires initiation at the upstream tRNA promoter and should therefore select for termination-specific mutants.
Monomeric suppressor tRNA allele-specific applications; the La protein pre-tRNA chaperone
A cartoon of the stages of suppressor tRNA biogenesis and function at ade6-704 is shown in figure 1B-D. An early deterministic step in the processing pathway is whether or not the nascent pre-tRNA productively interacts with the La protein, controlled in large part by its 3′ oligo(U) tail. La is an 3′ oligo(U) length-dependent binding protein that also recognizes the 3′ OH group on newly synthesized RNA; replacement of 3′ OH with phosphate, as results from cleavage by many ribonucleases, decreases affinity for La (57, reviewed in 58). As reviewed previously, pol III-transcribed genes produce nascent transcripts with length heterogeneity in their 3′ oligo(U) tracts (58). Accordingly, tRNA genes with minimal length oligo(T) terminators produce shorter oligo(U) tracts that evade La binding and its consequences to 3′ processing (59, 60). Evidence from S. pombe indicates that nascent pol III transcripts compete for a limited amount of La protein, and increasing 3′ oligo(U) length confers competitive advantage to some pre-tRNAs (61). A review of the relationship of 3′ oligo(U) length and tRNA structural impairments, and the variety of interacting factors involved in pre-tRNA 3′ end processing and decay is available (21). Because La also acts as a RNA chaperone that can assist folding, its positive effects on tRNA maturation are most apparent for pre-tRNAs with some degree of structural impairment (56, 62, 63).
Much of the work that elucidated roles for the La protein in pre-tRNA maturation in S. pombe made use of mutated suppressor tRNA alleles that critically sensitized them to the absence of La. Three point mutations (C40T, T46.3C and C47.6T), each of which alone had no significant effect on suppressor tRNA transcription, processing or suppressor activity in wild type cells (28), were introduced into the monomeric pSer suppressor tRNA allele, creating the MSer allele, tRNASerM (44). When combined, the three mutations debilitated the suppressor tRNA MSer relative to pSer (44). One of these mutations in particular, C47:6U (i.e. the U47:6 allele in ySH18) made it highly dependent on the chaperone function of the La protein (56, 64, 65). An additional mutation in the intron (C37:10G) that restores base pairing with the suppressor anticodon promotes pre-tRNA processing by RNAse P (16, 55). Thus, some mutations decrease processing efficiency of the nascent pre-tRNA that is exacerbated in the absence of La resulting in decreased levels of the mature tRNA. Some mutations may decrease amino acylation, and effects on decoding activity per se in the ribosome are also possible.
Influences of the pre-tRNA 3′ trailer and terminator read-through on processing
The structure-function relationship of the monomeric tRNA suppressor gene reflects interrelated aspects of pol III termination and 3′ end processing. The percentage of transcripts that are terminated at the oligo(T) terminator element at the 3′ end of a tRNA gene is referred to as termination efficiency whereas elongation beyond it is referred to as read-through transcription. The transcript region between the ends of tRNA sequences and their terminators comprise the 3′ trailers, and in natural tRNA genes are of variable length and sequence (Fig. 1A). The A and B box promoter elements together with the terminator element are bound by one or another of the six subunits of the transcription factor TFIIIC complex (Fig. 1B) which can accommodate the inter-element distance variabilities among tRNA genes ((66) and refs therein). Many tRNA genes have non-canonical or degenerate terminators (interrupted T stretches) immediately downstream of the tRNA sequence (67), which if read-through produce extended transcripts that may be correctly processed to mature tRNA, consistent with prior observations and presumably reflecting alternative pathways for tRNA 3′ end processing (68-70).
Secondary structure elements in pre-tRNA are major determinants of recognition by the 3′ processing endonuclease, RNase Z (a.k.a., 3′ tRNase) which removes the trailer (70-75). One or more 3′ exonucleases can mediate pre-tRNA 3′ processing for those not acted on by RNase Z (76-79). Thus, 3′ endo- and 3′ exo- nucleolytic processing comprise two pathways for pre-tRNA 3′ end maturation in eukaryotes, the former of which is in part directed by the oligo(U) 3′ end-binding protein, La (76, 80), and the latter of which can branch into more than one pathway (reviewed in (21).
To serve the purpose of 3′ end pathway-specific studies, a suppressor tRNA gene should be dependent on accurate and efficient termination at its T1 terminator (Fig. 1A) so that read-through does not lead to mature tRNA. Placing only 2Ts at the T1 site of the monomeric gene schematized in Fig. 2B will extend the 3′ trailer length and allow examination of effects of different sequences placed downstream to T1 on in vivo suppression. A similar arrangement is found at a natural S. cerevisiae tRNAPheGAA gene, which has a 37 nt 3′ trailer (81). The construct, pSer-2T-RT37-T2, which contains the tRNAPheGAA 3′ trailer placed between 2T and an 8T terminator indeed produced strong suppression (R.M., unpublished); indicating that read-through of a T1 terminator would also produce suppression. However, certain heterologous sequences placed downstream of an inefficient terminator can interfere with 3′ processing (70). Therefore, to ensure that TMS would be dependent on accurate termination, a synthetic sequence can be placed downstream of T1 to interfere with tRNA processing of read-through transcripts (44). A sequence referred to as cRT, was designed to base pair with the 3′ half of the tRNASerUCA precursor and efficiently block suppression (44) (Fig. 2B). This established a pSer-allele version of the suppressor (a.k.a. tRNASerWT or W (44, 56)). Table 1 lists the suppressor-tRNA alleles.
Sequence control elements that direct termination by Pol III
Evidence had suggested that the oligo(T) tract length required for termination varied among species, and that sequence context surrounding the T tract could influence termination efficiency (82-84). Monomeric TMS showed that S. pombe requires 5Ts to efficiently terminate pol III transcription (44). Parallel biochemical analyses indicated that S. pombe pol III can terminate at 5Ts, one nucleotide shorter than S. cerevisiae and more similar to human pol III (44). Vertebrate pol III can terminate with high efficiency at 4Ts but only in the context of particular flanking sequences (44, 60, 84, 85). A 5T tract is an efficient terminator for human pol III even in what would be considered poor context for 4Ts (85). 6T is an efficient terminator in S. cerevisiae but 5T may be sufficient in certain contexts (86). While 5T is efficient in S. pombe, potential effects of context on a 4T tract have not been reported. However, genome sequence data suggest that the related fission yeast, S. Japonicus may require only 4Ts at a significant number of its tRNA gene terminators (87).
Although the species-specific differences in terminator T-length are subtle, the data suggest that they are relevant not only to termination but also to downstream function via oligo(U) length-dependent binding by La and the pathway used for pre-tRNA processing (88). Species-specific terminator length is inversely correlated with the sensitivity of the corresponding pol III to α-amanitin (44). Alpha-amanitin binds between the bridge helix and trigger loop motifs of the largest polymerase subunit, at the base of the funnel region, and inhibits the elongation mechanism (89, 90). Thus, the correlation may be meaningful because a major function of the pol III terminator is to slow elongation.
RNA 3′ oligo(U) length can be modulated by the activity of the pol III subunit, C11, during transcript termination (88). RNA 3′ oligo(U) length can be linked to posttranscriptional processing and requirement for chaperone activity of the 3′ oligo(U)-binding protein, La, and functional tRNA maturation (56, 88). In a later section we review data on pol III subunits involved in termination.
The majority of pre-tRNAs including their 5′ leaders, 3′ trailers and oligo(U) termini are less than 100 nt in length. Pol III has a very high rate of transcript production in vivo, with fast elongation, high density of pol III molecules per gene, and fast reinitiation. The yeast 5S rRNA gene of 132 bp has an observed in vivo occupancy of up to 3 pol III molecules, an elongation rate of 60-75 nt/second, and reinitiation interval of 1.2 seconds (91). This is highly consistent with independent calculations of 2-4 tRNAs/gene/second or successive pol III terminations every 0.25-0.3 seconds (92). High efficiency transcription reinitiation by pol III is likely facilitated by a simple and unrestricted action at its terminator (93).
As will be reviewed in detail below, evidence indicates that transcription termination by pol III may facilitate efficient reinitiation. We will therefore briefly consider aspects of transcription initiation by pol III that are relevant to the S. pombe TMS system.
Species-specific transcription initiation factor recruitment to tRNA genes
As noted above, unlike in S. cerevisiae, pol III-transcribed genes in S. pombe require an upstream TATA element for transcription initiation (42). Evidence that plant tRNA genes rely on upstream TATA elements for pol III transcription ((47, 94), and refs therein) were supported by genome-wide analysis (42, 86). A majority of human tRNA genes engage TBP at upstream TATA promoters (43). Earlier attempts to identify TATA upstream of human tRNA genes revealed only very minor enrichment in a 625 tRNA gene set (95), although it is now recognized that a large number of pseudogenes occupied that set. The new study uncovered 386 human tRNA genes with an upstream TATA that was cross-linked to TBP, presumably reflecting a functional tRNA gene set (43).
A mechanistic basis for the difference is that the S. pombe TFIIIB subunit Brf1 does not form a stable association with TATA-binding protein (TBP) in the absence of DNA, while S. cerevisiae Brf1 does (96). The S. pombe TFIIIC complex also revealed homology of Sfc6/TFC6 to human TFIIICβ and a stable association with spBrf1 but not spTBP (96, 97). Thus, while TFIIIB subunits share sequence homology in the two yeasts, they differ in interaction mechanisms. Similarly, features specific to plant TBP and Brf1 rather than tRNA gene upstream architecture per se, are responsible for TATA box dependence (47). The physiologic ramifications of these differences in TBP recruitment mechanisms remain to be uncovered.
Co-transcriptional pre-tRNA processing is unlikely
Given the timescale of 1-2 seconds from initiation to termination of pre-tRNA synthesis, one can consider the potential for cotranscriptional folding. While some RNAs have been observed to undergo functional co-transcriptional folding, their lengths and the timescales are significantly longer than for tRNA in part due to transcriptional pausing which can assist proper folding (98-100). Correct folding can be context dependent (98) suggesting that co-transcribed flanking sequences can affect folding of an embedded RNA. A role for RNase P in coordinating pol III transcription with early steps in tRNA processing has been proposed (101), similar to pre-mRNA processing (102). Earlier work in yeast noted an interaction between RNAse P and the pol III transcription factor TFIIIB (103).
Simple stem loops can form in a fraction of a second after synthesis (reviewed in (99)). Therefore, one can ask for tRNAs, to what degree of extension would pol III have to proceed before a functionally-recognized element would emerge from the polymerase? Beginning at the 5′ end, the first tRNA functional element synthesized during elongation is the tRNA dihydrouridine stem-loop (DHUSL) (Fig. 1C). According to mature tRNA numbering, the 3′ end of the DHUSL is at position 25. However, pre-tRNAs have 5′ leaders of 5-9 nts, an additional ~5 nt in the pol III RNA exit channel,plus the RNA-DNA hybrid in the active center, ~9 nt. Thus, pol III would have to synthesize a minimum of 42 nt of RNA, about half of the tRNA gene length, before the DHUSL could exist outside of the polymerase. Although other simple structures might compete with formation of the DHUSL depending on the sequence of the tRNA gene at hand, these would reflect an offline folding pathway and would have to be melted to allow formation of the DHUSL. According to this scenario, the rate limiting step for formation of the DHUSL, a functional element recognized by processing machinery, would appear to be limited by the pol III elongation rate rather than folding rate per se.
Major determinants of substrate recognition by RNase P are the DHUSL and the double stranded tRNA acceptor stem (104-106). For acceptor stem formation, the 3′ end region must be synthesized, emerge from pol III and be available to base pair with the 5′ end region. Based on the architectures of most yeast tRNA genes (Fig. 1A) and steric hindrance by pol III, transcription termination may be a prerequisite of acceptor stem formation. Thus it would seem that formation of a functional structure that could undergo what is usually the first step of processing, 5′ end maturation by RNase P (76), would be limited by the pol III elongation rate and synthesis of the 3′ terminal part of the tRNA.
Despite the theoretical possibility that a nascent elongating transcript may form a tRNA secondary structure by the time pol III arrives at the terminator, an experimental observation described above in another context, argues against this. If pre-tRNA secondary structure formed by the time pol III passed the T1 terminator, and RNase Z was readily active, the cRT region of the suppressor tRNA should not be able to block its processing (Fig. 2B). Yet, the cRT region does indeed block processing of both the mutated and non-mutated versions of the monomeric suppressor tRNA alleles (11).
A transcriptional role for RNase P should not imply co-transcriptional processing because this proposed function has also been reported to apply to many noncoding RNA genes synthesized by pol II and pol I, whose transcripts are not its processing substrates (107, 108). The collective considerations noted above argue that co-transcriptional folding to functionally processable forms is unlikely a significant aspect of tRNA biogenesis.
RNA chaperone activity of La protein
Some structural RNAs can form off-pathway misfolding intermediates that become kinetically trapped in nonproductive forms (109, 110). Studies in yeast indicate a difficult folding process for some pre-tRNAs that can be assisted by the chaperone function of La (56, 62, 65, 111). In this capacity, La appears to be one of several tRNA maturation factors that interact with tRNAs during biogenesis, that can assist structurally impaired pre-tRNAs in achieving productive structure (112, 113). Such pre-tRNAs that are deficient for productive interactions and not efficiently processed, succumb to degradation by nuclear surveillance and/or other decay pathways ((56, 113, 114), detailed in (21, 58)). RNA chaperone and other maturation-associated activities have also been demonstrated for the La-related proteins (LARPs), some of whose targets are small nuclear RNAs and others are cytoplasmic mRNAs (21, 115-117).
Pre-tRNA processing begins after transcription termination by pol III
Studies in S. cerevisiae were the first to indicate that events at the 3′ oligo(U) terminus of a nascent pre-tRNA direct the order of 5′ processing by RNase P (76), confirmed in S. pombe (118-120). More specifically, for the great majority of pre-tRNAs, the eukaryote-ubiquitous La protein binds the oligo(U)-3′ terminus of nascent pre-tRNA and directs RNase P to cleave the 5′ leader before the 3′ trailer is removed (76, 119)(121). For these tRNAs, consistent with biochemical findings that indicate that La binding inhibits 3′ processing by RNase Z (122), the pre-tRNA 3′ end determines processing order in presence of La and when La is absent, 3′ end processing occurs first (76). That the 3′ end is a major determinant of transcript fate is supported by the compelling finding that La proteins bearing mutations that activate its premature nuclear export, carry their pre-tRNA ligands with their intact 5′ leaders and 3′ trailers, to the cytoplasm, bypassing processing by RNases P and Z (18, 120).
The available evidence indicate that all links between transcription and processing begin with formation of the pre-tRNA 3′ oligo(U) end, which includes pol III transcription termination (88). The cumulative observations strongly support the idea that La is the first protein to interact with newly synthesized nascent pre-tRNAs, and other pol III transcripts (Fig. 1B-C), consistent with its association with pol III-active genes in chromatin in yeast and human cell nuclei (91, 123).
Genetic screens for trans-acting factors involved in pol III termination
An advance in pol III terminator recognition came from Ben Hall and colleagues who introduced an oligo(dT) tract into the intron of an S. cerevisiae ochre suppressor-tRNA as part of a genome-wide, spontaneous mutation, loss-of-suppression screen (124). This identified Ret1, the second largest pol III subunit (Rpc2, a.k.a., C2) (125). Detailed information on the structure and function of Ret1/C2 in termination will be reviewed in a later section.
A modified approach led to a dimeric TMS construct for use in a pol III subunit-specific screen in S. pombe (126). An key distinction between the sup3-e dimeric gene and the dimeric suppressor constructed for this purpose is the relative position of the suppressor tRNA in the construct. In sup3-e the suppressor is the 5′ proximal tRNA, followed by tRNAiMet (44) whereas it is the second tRNA in the dimeric construct used to study termination in S. pombe (Fig. 2A, C).
The dimeric suppressor designed for TMS use in S. pombe is very similar to the arrangement used by Willis and colleagues to study pol III initiation (127, 128). Because the A and B box promoter sequences are also important for function of the tRNA product (Fig. 1B-C), the dimeric arrangement allows promoter mutagenesis without affecting the downstream suppressor tRNA. A similar approach in S. pombe may afford further examination of TATA-dependent pol III initiation (42, 50).
The dimeric TMS system
The monomeric suppressor with its cRT processing inhibitory element is not ideal for assessing terminator read-through because the expected outcome, decreased suppression might occur due to loss of function of any number of steps. The dimeric TMS system can produce positive gain of suppression when pol III fails to terminate at the test terminator T1 and instead reads-through (Fig. 2C). Here, the upstream tRNA is a wild-type tRNAser gene that provides the promoter for initiation, a 3′ trailer and test terminator, T1, and downstream of this is the suppressor tRNAserUCA followed by a default terminator, T2, comprised of 21 T residues (64). This arrangement dictates that the suppressor tRNA should be produced only if pol III initiates properly but fails to terminate at T1. Thus, any mutants obtained should be generally competent for transcription initiation but specifically deficient for termination (129).
It should be noted that unlike the mutated Mser alleles used as monomeric genes, the suppressor in the dimeric system exhibits significantly higher specific activity for TMS (44), albeit dependent on strain background (see 56). This results not only from lack of the MSer mutations, but in addition a gain-of-function mutation in the intron (C37:10G) that restores base pairing between anticodon and intron that increases processing efficiency and tRNA maturation in S. pombe (55, 64, 119). There are two predictable consequences of this. First, the dimeric suppressor should not be as dependent on La protein as is the MSer allele. Second, the higher specific activity relative to the MSer allele suggests that full TMS (white colonies) should be observed for as little as 20% or more read-through of the T1 terminator, and lower levels of read-through should produce partial suppression (pink). This is consistent with that observed by northern blotting which can reveal relative quantitative differences in read-through (64, 129).
The dimeric TMS has been used in two contexts, first to analyze terminator strength by transforming S. pombe with dimeric constructs that differ at the T1 terminator, to assess how well they block pol III from reading-through. This approach corroborated results from the monomeric system; while ~50% termination efficiency occurs at 4Ts and much less at 3Ts, T tracts of 5 or more produce ≥90% termination efficiency (44, 64).
The second context was to use strains with an integrated dimeric suppressor allele to isolate by screening of gene-specific randomly mutagenized pol III subunits, mutants that fail to terminate efficiently at T1 and instead read-through. By this approach a large number of mutants in three pol III subunits, C11, C37 (64, 129) as well as C1 (K.R. and R.M., manuscript in preparation) were readily isolated (below). For two other proteins, C53, the heteromeric binding partner of C37 (130), and Sfc6, a TFIIIC subunit whose S. cerevisiae homolog, Tfc6, localizes to the terminator region of tRNA genes (131), yielded no mutants (K.R. and R.M., manuscript in preparation).
Two stages of termination; terminator recognition followed by pausing and release
The three major RNA polymerases in yeast and animal cells, I, II & III, are ancestrally related, share 5 common subunits and homology in several others, and use similar mechanisms for transcription initiation (132, 133). However, the cis-acting termination signals that trigger the first step of their termination processes are quite divergent (134). The two major steps are recognition of a termination signal followed by pausing and destabilization of an otherwise stable elongation complex (EC) with release of the 3′ RNA from the polymerase active center (135). For pol II the two steps are clearly separated in space and time, and most pol II ‘termination’ studies focus on the first (134). However, the extent to which the three pols may share mechanisms for the later step, destabilization of the EC and transcript release, is largely unknown.
Termination of pre-mRNA transcription by pol II involves multiple accessory factors and a termination signal that can be one hundred or so base pairs upstream of the site at which release occurs (134). By contrast, the pol III termination signal is a simple T tract on the non-template strand, oligo(dA) in the template; pol III recognizes this, pauses and releases its transcript at the same site (135). This is a unique feature of pol III as other RNA polymerase ternary complexes do not spontaneously disassemble at oligo(T) tracts (135).
Termination by other RNA polymerases requires a terminator-proximal RNA hairpin or other RNA duplex structure in addition to T-rich tract, and in some cases use a mechanism that requires accessory factors (134, 136, 137). The initial evidence that pol III was sufficient to terminate transcription at an oligo(T) tract without the need for RNA structure and in the absence of accessory factors (84, 138, 139) was confirmed by studies of bona fide pol III that more directly distinguish pausing and transcript release (140). It was recently reported that a terminator-proximal RNA hairpin structure was required for termination by pol III (141). However, it was subsequently shown that this requirement was peculiar to an unusual, ill-defined form of polymerase used in those experiments, whereas efficient termination by bona-fide pol III does not require terminator-proximal RNA structure in vitro or in vivo (140).
Pol III protein motifs involved in termination, elongation and RNA 3′ cleavage
Extensive mutagenesis identified three regions in the Rpc2 pol III subunit, (C2/Ret1) that are important for either increased or decreased termination at oligo(T) (142-145). The mutations altered elongation by pol III, in accord with a ‘kinetic coupling’ model in which termination efficiency is inversely related to elongation rate. To put it another way, the longer the polymerase spends dwelling on a terminator, the more likely it will terminate and vice versa, if other conditions of EC instability are met (146, 147).
The three regions identified in C2 that alter elongation and termination are the lobe, fork loops and anchor, recently reviewed in the context of overall polymerase structure and inter-subunit interactions (135). Studies of elongation by bacterial RNA polymerase and pol II had identified the elongation factors, Gre A & B and TFIIS, respectively, which function by an intriguing mechanism, stimulation of polymerase-associated intrinsic transcript 3′ cleavage activity (148-151). While GreB/A and TFIIS are ancillary factors that transiently associate with their polymerase, the intrinsic cleavage activity of pol III is more integral (152). This intrinsic 3′ transcript cleavage activity was found to be greatly altered in many of the pol III termination/elongation mutants (142-145). However, as reviewed below these probably reflect effects of the C2 mutations on the cleavage activity of another subunit, C11, and possibly C37.
An advance in identifying additional pol III subunits involved in termination came with discovery of the integral subunit C11, a two domain protein of which its N-terminal is homologous to Rpb9 and its C-terminal is homologous to the transcript cleavage domain of TFIIS (88, 153). Heterocomplementation of C11-deficient S. cerevisiae with S. pombe C11 produces a novel form of purified pol III referred to as pol IIIΔ that lacks intrinsic transcript cleavage activity and reads through certain oligo(T) terminators (153). However, pol IIIΔ also lacks the C53/37 heterodimer in addition to C11 (154).
Pol IIIΔ exhibits more defeciency on some terminators than others (153, 154). Later data showed that termination by pol IIIΔ is oligo(T) length-dependent (130). The oligo(T) length required for efficient termination by pol IIIΔ is ≥9Ts, which is significantly longer than required by intact pol III from S. cerevisiae and the great majority of pol III terminators found in vivo. The inherent ability of pol IIIΔ to efficiently terminate at 9Ts would appear to reflect a core mechanism of termination, i.e., autonomous disassembly, possibly induced by acute sensitivity to thermal instability of oligo(rU:dA) hybrid (130, 155). For efficient termination on 6T tracts, C53/C37 & C11 augment the core mechanism via an auxiliary mechanism (130).
The C53/37 subcomplex appears to associate with the same surface of the C2 lobe domain identified as important for termination in Hall’s laboratory; moreover, this general region appears juxtaposed to the docking site for the N-terminal domain of C11 (156-159). This is consistent with mutation of a segment of the analogous C2 lobe of zebra fish pol III as well as S. pombe pol III that causes loss of C11 (160). A major biochemical activity of C53/37 is to decrease the elongation rate of pol III, consistent with the kinetic coupling model of termination and the C2 lobe mutations (154). Mutations in Rpb9-homologous N-terminal domain of C11 also cause terminator read-through further supporting the lobe region’s involvement in association of C53/37 and kinetic coupling (64). Presumably from the peripheral lobe location of the N-terminal domain of C11 extends its TFIIS-homologous C-terminal ribbon into the pol III active center during intrinsic transcript cleavage (88, 153) (Fig. 1B).
Other pol III subunits involved in elongation, termination and RNA 3′ cleavage
To identify additional motifs important for termination, specific subunits were subjected to random mutagenesis and dimeric TMS screening in S. pombe (64, 88, 129). Use of dNTP analogs produces a wide spectrum of mutations (129, 161) that when coupled with gain-of-function TMS can reveal specific motif(s) in the target subunits (64, 88, 129).
When applied to the monomeric TMS system, phenotypic gain-of-function mutations that cluster in the C-terminal domain of C11 were found to impair intrinsic 3′-5′ transcript cleavage (88). Inhibition of cleavage was associated with extended 3′ oligo(U) tracts on the pre-tRNA transcripts which increased their affinity for La protein, directing them to an efficient processing pathway, resulting in gain of suppression function (reviewed in 21, 58, 88). Notably however, despite expectations otherwise, the C11 C-terminal mutants did not cause terminator read-through by multiple criteria (64, 88). By contrast and as noted above, mutations in the N-terminal C11 domain anchored at the pol III lobe near C53/37, do indeed cause oligo(T) read-through (64), similar to mutations in C37 itself (below).
Similar TMS screening of C37 identified a cluster of termination mutations in an acidic region near its C-terminus (129). This very same region had been physically cross-linked to the fork loop 2 region of C2 in the active center of a S. cerevisiae pol III elongation complex (159). This suggests that this C37 region might affect termination by altering the elongation-related dynamics of fork loop 2 (129). We wish to emphasize here that a major set of Ret1/C2 mutants localized to the fork loop 2 region of C2 (142). Mutation of pol II fork loop 2 increases pausing (162). Thus, it would appear that this C-terminal acidic region of C37 physically interacts with the active center and contributes to pausing by pol III and promotes termination at oligo(T).
Conserved involvement of Rpb2 in termination by pol II
A mutagenesis gain-of-function screening approach was taken for S. cerevisiae pol II; its termination signal, the poly(A) addition site, was placed in an intron of a LacZ reporter and used to screen mutagenized Rpb2 (163). Remarkably, some of the mutations uncovered are analogous to the Rpc2 lobe of pol III, in positions that interact with TFIIF subunits which share sequence homology with the RpC2 lobe-interacting domains of the pol III termination subunits, C53/37 (129, 163). This suggests that termination by pols II and III exhibit similarities despite divergence in the cis-signals and mechanisms that initiate the process. While the pol III-peripheral domains of C53/37 and C11 dock on the C2 lobe region, there is functional genetic and physical evidence that indicate that others parts of these subunits reach into and alter the active center during elongation and termination (88, 126, 129, 159, 164). The extent if any to which pol II associated factors may access the active center during termination is presently unknown.
A model of transcription termination by pol III
From genetic screening using TMS in S. pombe and S. cerevisiae as well as biochemical studies of purified pol III, structural and mechanistic models for termination can be derived. A most prominent feature is the link between elongation rate and termination efficiency. Initial deductions based on mechanistic studies of Ret1/C2 have been strengthened by results obtained with C53/C37 (142, 143, 154). Additional data suggest that C11 may also contribute to pausing on the terminator (130, 145). Thus, the sensitivity of pol III to elongation control as the principal determinant of termination is paramount. In this regard it is noteworthy that elongation control by pol II is largely reserved for critical gene regulatory mechanisms and these are in large part controlled by TFIIF and TFIIS (165, 166) Likewise bacterial RNAP uses GreB/A and other elongation-related proteins as transient factors (151, 167, 168). By contrast, elongation control by pol III would appear to principally serve termination, mediated by integral subunits.
Any model of pol III termination should consider the fact that pol IIIΔ terminates autonomously at ≥8 Ts, and that an auxiliary mechanism contributed by C53/37 and C11 augments this at shorter T tracts such as found in vivo (130). However, recent studies indicate that reduced elongation rate is just one of two components of the pol III termination mechanism. C53/37 itself surely slows elongation while C11 confers additional activities. The N-terminal domain of C11 contributes to the pausing and oligo(T) read-through component via C2 lobe-associated interaction with C53/C7 (126). It should be noted that a rare set of mutations in C2 that did not conform to the kinetic coupling model, as they reduced elongation rate and termination, and vice versa, suggesting that some of these mutations may affect the core mechanism itself (143).
The second component of the auxiliary mechanism of termination requires an anti-arrest activity mediated by a cleavage-competent C11 C-terminal domain (130). The model posits that while slowing elongation is necessary, this increases the potential for terminator arrest, i.e., pol III inactivation without dissociation (130, 143). Thus, the second component of the auxiliary mechanism circumvents this deadly outcome of reduced elongation at oligo(T) but requires cleavage-competent C11 to efficiently prevent terminator arrest (130). Furthermore, there is a downstream consequence of the 3′ length heterogeneous pol III termination mechanism including C11mutant-induced 3′ length extension. Heterogeneity of 3′ oligo(U) length directs nascent pre-tRNA products via their binding or not to La protein, a feature that has apparently been functionally assimilated into alternate pre-tRNA processing pathways (21, 88).
The cumulative results lead to the idea that the pol III active center is a complex and busy place during termination with regions of four subunits that appear to be in proximity; fork loop 2 of C2/Ret1, a C-terminal acidic region of C37, transcript cleavage domain of C11 and specific motifs of C1, that alter pausing, read-through and RNA cleavage (88, 129, 143, 145) (K.R. & R.M., manuscript in preparation). However, while these regions appear to contribute to elongation control, the subunits/motifs involved in more direct destabilization of the EC with transcript release, remain to be resolved before a better understanding of termination by pol III can be achieved.
METHODS AND TECHNICAL APPROACHES
Suppressor constructs
Both the monomeric and dimeric system suppressor-tRNA construct types were created in the pJK148 plasmid backbone, which carries a selectable leu1+ marker that integrates into the S. pombe chromosome (169) (Fig. 4). Upon linearization of the pJK148-suppT plasmid with the restriction enzyme, Nde1, and transformation, it undergoes efficient homologous recombination at the leu1-32 locus, converting it to leu1+, providing leucine prototrophy to the transformants (169). This transformation approach can be used as an assay to compare different suppressor-tRNA alleles bearing mutations in cis-sequences to determine their TMS efficiencies, or to isolate stable integrants of the suppressor-tRNA allele that can later be used as strains to examine the effects of trans-acting factors, e.g., pol III subunits, and/or processing factors such as C11, La, and Rrp6.
Figure 4. Plasmid maps for the suppressor-tRNA genes used in the monomeric (A) and dimeric (B) systems.
The ‘x’ shown in plasmid names inside the circular maps indicate the number of T residues in terminator T1. The T2 for the monomeric system is 8Ts and for the dimeric system it is 21Ts. In both systems the plasmid backbone is pJK148. Prior to integration, the plasmid is pre-digested with NdeI, indicated as N within the leu1+ gene, which directs it for homologous recombination at the leu1-32 locus; it is therefore necessary that cells with the leu1-32 allele, which carries a single nucleotide mutation that renders it inactive, rather than a leu1-deletion allele, be used. In the monomeric system, the test terminator T1 can be replaced using its flanking restriction sites BglII and SacI (represented as Bg and S). The Sap1 site downstream to the construct in dimeric system is not a unique site (indicated by asterisk) as SapI also cuts downstream of T2. The cRT region represents sequence complementary to the suppressor tRNA, that can fold back to form a duplex with the suppressor-tRNA sequence that perturbs proper tRNA folding and inhibits correct processing. A and B boxes (represented as A and B in boxes) are the internal promoter elements. Only the upstream tRNA has a TATA box to ensure transcription is initiated only from the upstream tRNA sequence. Several different alleles of suppressor tRNA that have various combinations of the 4 mutations shown in A and varying strengths have been used in monomeric system (see Table 1). The dimeric system suppressor-tRNA allele produces a suppressor-tRNA with high specific activity (see text).
A method for determining in vivo pol III termination efficiency
An advantage of this system is that both the monomeric and dimeric versions can be subjected to some degree of relative quantitative analyses. One can transform a leu1-32 ade6-704 strain with or without other mutations of interest (e.g., sla1-Δ, rrp6-Δ) with suppressor-tRNA allele constructs, e.g., with varying length terminators or other mutations, and score the percentage of colonies with suppression relative to negative and positive control constructs (see bar graphs, Figure 2 in reference 44, and Figure 2C in reference 64). Negative controls can be the pJK148 vector lacking a suppressor-tRNA gene (as was done to create the yeast strain yAM148, Table 1) and/or a suppressor-tRNA gene construct with only 2 or 3 Ts at the test terminator, T1 (as in yeast strain yAS68, Table 1) (44, 64). Tabulations of the percentage of colonies that show suppression provide quantitative estimates of the relative termination and/or read-through (44, 56) (see cautionary technical notes below). The quantitation of termination as assessed in vivo was shown to match fairly well with the in vitro data (44).
Read-through transcripts from the monomeric suppressor-tRNA system can be detected using probes to the cRT region (44, 64, 129). As noted above, these read-through transcripts accumulate at detectable levels in cells but are not processed to mature tRNAs. Therefore, northern blotting of read-through transcripts can reveal relative levels of termination read-through among different mutants. This is done with comparison to the suppressor-tRNA gene construct with only 3 Ts at the T1 test terminator and can provide an additional degree of relative quantitation of termination read-through (64, 129).
Nascent pre-tRNA levels reflect relative transcription rates
As noted above the integrated suppressor tRNA-MSer allele, e.g., in yYH1, must produce a significant amount of mature functional tRNA in order to produce suppression, comparable in transcription output to other tRNA genes (44). Transcription termination is kinetically coupled to slow elongation or pausing, and efficient termination also enhances the availability of pol III for subsequent rounds of transcription reinitiation (135). Pol III transcription initiation complexes are very stable, supporting a large number of reinitiations by pol III. Thus, the vast majority of the tRNA produced in yeast results from reinitiation by pol III (93, 130). Accordingly, mutations that affect termination may affect other stages of transcription as well. Therefore, understanding how termination-altering mutations may affect overall transcription rate may provide mechanistic insight into the other stages.
We wish to emphasize that in order to obtain gain of suppression in the dimeric system, initiation, and reinitiation must be efficient. This suggests that the termination read-through mutants obtained using the dimeric system should be specific for termination defects and that mutations that would also impair initiation and/or reinitiation, may not be readily obtained by this approach (129).
Pre-tRNAs have a short half-life largely because they undergo efficient processing that includes nucleolytic removal of the 5′ leader, 3′ trailer and introns if present (173). Mature tRNAs, once formed, have a very long half life, hence their levels do not reflect relative levels of transcription. Because nascent pre-tRNAs are rapidly converted to processed forms, they represent species that can be used for assessment of transcription levels (174). In general, northern analysis using probes that are specific to the 5′ leader, the 3′ trailer or more preferably, the intron if present, can be used to differentiate the most nascent pre-tRNA species if they can be electrophoretically separated or otherwise distinguished (e.g., by RT-PCR) from the processed forms. Specifically, the most nascent of the pre-tRNA species contain both a 5′ leader, an intact 3′ trailer and an intron if present, and they are used in several laboratories to compare relative differences in transcription rates under different conditions or in different strains (129, 174-178). A caveat here is that any mutations that affect the processing of nascent pre-tRNAs either productively or toward decay can change the distribution of the nascent pre-tRNA species and thereby affect the accuracy of the comparison (173, 176). Accordingly, this method is most useful when the mutations being examined do not differentially affect processing and/or decay of the nascent pre-tRNAs, or if the samples/strains compared do not differentially vary in processing-sensitive tRNA mutations.
RNA 3′ end formation and assessment of nascent oligo(U) length
Not all nascent pre-tRNAs are correctly processed to mature tRNAs with the same efficiency. Evidence suggests that some are subjected more than others to nuclear surveillance-mediated 3′ exonucleolytic digestion by the exosome with subsequent decay (78, 176, 179). This decay pathway is especially relevant to pre-tRNAs that contain structural impairments or imperfections (reviewed in 173). RNA polymerase III terminates synthesis of its nascent RNA products within the oligo(T) terminator, producing transcripts that end with oligo(U)-3′OH. It had been known for many years that the 3′ oligo(U) tract on nascent pol III transcripts was heterogeneous in length even when produced from a single tRNA gene or other gene but the mechanisms involved and potential relevance to physiology, RNA processing and phenotype remained unknown (reviewed in 180). Using the monomeric system in a study of the pol III subunit and RNA 3′ cleavage factor, C11, it was shown that mutations that affect the intrinsic RNA 3′ cleavage activity of pol III also affect posttranscriptional 3′ end processing and maturation of the nascent pre-tRNA (88). C11 mutations that decrease its pol III-intrinsic 3′ RNA cleavage activity lead to an increased length of the oligo(U) tract present at the 3′ end of nascent pre-tRNAs. The pre-tRNA chaperone La protein specifically binds to pre-tRNAs in a 3′ oligo(U) length-dependent manner. Therefore, increased 3′ oligo(U) length leads to higher affinity binding of La protein to the pre-tRNAs and thereby protects them from 3′ exonucleolytic digestion and enhances their folding and maturation (56, 88, 111, reviewed in 173).
Natural heterogeneity of 3′ oligo(U) length exists in pre-tRNAs in wild type S. pombe cells (88). La protein is present in limiting amounts for TMS in S. pombe cells and requires four or more Us at the 3′ end for efficient binding (e.g., see Figure 4D) (88). A significant fraction of the nascent pre-tRNAs are produced with three or fewer Us and are not efficiently engaged by La protein while a significant fraction contain four or more Us and do follow the La-dependent pathway of pre-tRNA maturation (88, reviewed in 173). A potential ramification is illustrated by the idea that the ability for any particular pre-tRNA to access the La-dependent pathway may be more critical for its maturation than is La for other pre-tRNAs. As alluded to above regarding COVE analysis, this is likely dependent on their relative structural characteristics, determined by sequence variance inherent in different tRNAs (173).
Methodology was developed to analyze the 3′ oligo(U) length of nascent pre-tRNAs. This involves ligation of an adaptor to the 3′-OH end of RNAs followed by reverse transcription (RT) with a primer that is complementary to the adaptor but carries an extra dA residue at its 3′ end, enriching specifically for nascent RNAs that end with a 3′ U-OH. This is followed by PCR using a gene-specific sense primer, preferably in an intron or 5′ leader (88, 129). The PCR products are inserted into a TA or similar cloning vector (Invitrogen) followed by sequencing of plasmids from 40-50 colonies to assess 3′ oligo(U) length distribution (88, 129).
Random mutagenesis for targeted genetic screening
The TMS systems have been more recently useful when combined with PCR-mediated random mutagenesis of genes that encode pol III subunits (64, 88, 129). Mutagenesis that utilizes the dNTP analogs, 8-oxo-2′-deoxyguanosine-5′-triphosphate (8-oxo dGTP) and 2′-deoxy-P-nucleoside-5-triphosphate (dPTP) produce significantly more broad spectrum mutations than other PCR-mediated methods (181). In the presence of the normal dNTPs these analogs are randomly incorporated into both strands of DNA during the first two cycles of the PCR reaction and are then fixed as mutations in subsequent cycles; the mutations they induce will be distributed and propagated in subsequent PCR cycles. Analysis of several pol III subunit libraries prepared using this method show that every nucleotide is substituted by every other nucleotide, albeit at different rates, and the mutations are distributed throughout the amplicon without significant clustering (129). With careful control one can vary the concentrations of the dNTP analogs and obtain different libraries of the same gene with different overall mutation rates (129). By this approach one can sample a very large number of mutations in an objective manner. Though we have successfully mutagenized genes of length up to 1.5 kb, when analyzing the largest subunits of pol III, which are encoded by large genes spanning ≥3 kbp, we prefer to make three different libraries that separately cover the N terminal, C terminal and the middle region of the gene. This is because sequencing the mutants obtained is more easily manageable and because it also optimizes the yield of single site mutations, which are usually the most informative. When data from these three regions are combined, we are able to get valuable information about the whole subunit (K.R. and R.M., in preparation).
Cautionary technical notes
The pJK148-suppressor-leu1+ construct undergoes homologous recombination at the leu1-32 locus which naturally does not contain a tRNA gene. Therefore it should not be surprising that a variable number (10-15%) of leu1+ transformants remain unsuppressed after transformation to the leu1+ state by constructs that contain strong suppressor-tRNA alleles (see Figure 2A in 44). This is because the transformants did not contain the suppressor-tRNA region of the construct, presumably deleted upon integration of the leu1+ part of the construct (R.M., unpublished observations). Moreover, the leu1+ locus is a hotspot for recombination (169). Indeed, up to 10% or so of transformants carry two or more copies of the leu1+-suppressor-tRNA (R.M., unpublished observations), consistent with the previous observation of approximately 20% of transformants that carry reiterations of the pJK148 plasmid at the leu1 locus (169). These features account for some of the notable heterogeneity in raw transformation suppression data including variable fractions of the transformants with partial (pink) versus full (white) suppression (see Figure 2 in 44, and Figure 2C in 64). Specifically, while most of the MSer-7T transformants are pink a small fraction are lighter shade of pink and fewer still are white; upon further analysis it was found that the light pink and white ones carried 2-3 copies of the pJK148-MSer-tRNAsequence while the darker pink transformants carried one copy and this was approximately correlated with the levels of the mature suppressor-tRNA (unpublished). Also as a result of the hotspot nature of the leu1 locus another caution is that the tandem copy number of leu1+-suppressor-tRNA locus can vary over time in a single strain in response to unintentional selective pressure (J.R. Iben, K.R & R.M., unpublished observations). It is therefore important to freeze strain stocks early after their establishment and to return to them regularly rather than continually passage a strain (K.R & R.M., unpublished observations).
Highlights.
We describe the workings of a suppressor tRNA mediated reporter system in S. pombe.
We review results obtained with the system toward understanding tRNA biogenesis.
We review results obtained on transcription and termination by RNA polymerase III.
We review results on tRNA 3′ end processing and subcellular trafficking.
We describe technical approaches employed and a parts list of reagents.
Figure 3.
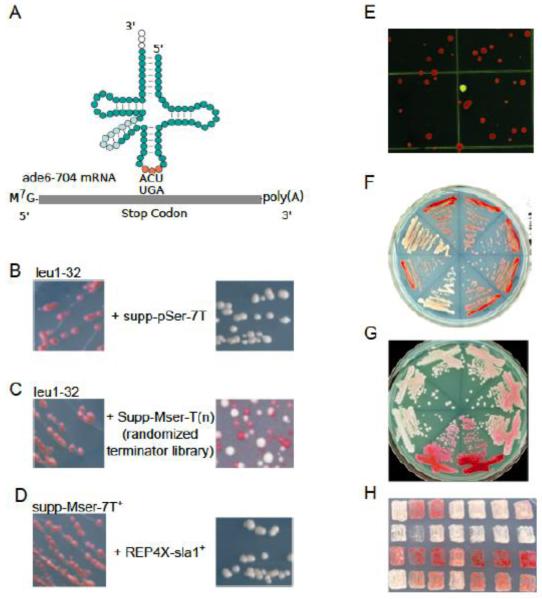
tRNA mediated suppression systems and representative assays in S. pombe. A) The S. pombe ade6-704 allele carries a premature UGA stop at codon position 215 (17). In the absence of the opal suppressor tRNAserUCA, the ade6-704 mRNA undergoes premature translation termination while in the presence of a functional suppressor-tRNA, active Ade6 protein is made and its substrate is further metabolized, preventing accumulation of red pigment. B) An S. pombe leu1-32 strain that carries the ade6-704 allele is shown before and after transformation (left and right panels respectively) with a pJK148 plasmid carrying the pSer-7T allele of the suppressor-tRNA and a leu1+ selection marker, grown on plates lacking leucine and with limiting adenine. C) Similar to B, except that the tRNA allele was constructed with a randomized terminator library, which generates colonies with hues that vary from red to white depending on the strength of the terminator. Colonies were isolated and purified, and plasmids were extracted and sequenced (not shown). Each colony differs by the sequence of the tRNA gene terminator and its immediate flanking sequences. D) A strain deleted of the La sla1 gene (sla1-Δ, yAS110, Table I) and carrying a stably-integrated Mser-7T suppressor allele (left panel) was transformed with a multicopy S. pombe expression plasmid (pRex-Sla1) to over express the tRNA chaperone, La protein (right panel). E) An example of the raw output of a genetic screening for pol III mutants with decreased terminator recognition. A strain that carries the dimeric suppressor tRNA (yKR1, Table I) was transformed with a randomly mutagenized library of the pol III subunit C37 cloned in the pRep4X expression vector. The white colony was picked, purified, the plasmid was isolated and retransformed into the parent strain, yKR1, which produced uniformly white colonies (not shown); sequencing of the plasmid insert identified the mutated residue (129). F) A plate showing several streaks of a monomeric suppressor tRNA-containing (MSer-7T) strain (yYH1) after transformation with plasmids encoding different isolated mutants of the pol III subunit C11. G) A plate showing several streaks of a monomeric suppressor tRNA-containing strain (ySH9) after transformation with plasmids encoding different mutants of human La. H) Patch assay after transformations of yYH1 with plasmids encoding various different trans-acting mutants that show loss or gain of suppression; patch in upper right corner shows the basal suppression level of yYH1 after transformation with an empty vector. (A=A box, i=intron, B=B box, T1=test terminator, T2=second terminator).
ACKNOWLEDGEMENTS
We thank former and current lab members, M. Bayfield, R. Intine, Y. Huang, A. Sakulich, S. Koduru, S. Hasson, A. Mozlin, and J. Mazeika for their contributions toward developing TMS. This work was supported by the Intramural Research Program on Genomics of Differentiation in the Eunice Kennedy Shriver National Institute of Child Health and Human Development, at the National Institutes of Health.
Abbreviations
- pol III
RNA polymerase III
- TMS
tRNA-mediated suppression
- pre-tRNA
precursor-tRNA
- TFIII
transcription factor for pol III
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hopper AK, Phizicky EM. tRNA transfers to the limelight. Genes Dev. 2003;17:162–80. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- 2.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–60. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshihisa T. tRNA, new aspects in intracellular dynamics. Cell Mol Life Sci. 2006;63:1813–8. doi: 10.1007/s00018-006-6092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelke DR, Hopper AK. Modified View of tRNA: Stability amid Sequence Diversity. Mol Cell. 2006;21:144–5. doi: 10.1016/j.molcel.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Hopper AK, Pai DA, Engelke DR. Cellular dynamics of tRNAs and their genes. FEBS Lett. 2010;584:310–7. doi: 10.1016/j.febslet.2009.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wichtowska D, Turowski TW, Boguta M. An interplay between transcription, processing, and degradation determines tRNA levels in yeast. Wiley interdisciplinary reviews. 2013;4:709–22. doi: 10.1002/wrna.1190. [DOI] [PubMed] [Google Scholar]
- 7.Hopper AK. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics. 2013;194:43–67. doi: 10.1534/genetics.112.147470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capecchi MR, Hughes SH, Wahl GM. Yeast super-suppressors are altered tRNAs capable of translating a nonsense codon in vitro. Cell. 1975;6:269–77. doi: 10.1016/0092-8674(75)90178-6. [DOI] [PubMed] [Google Scholar]
- 9.Koukuntla R, Ramsey WJ, Young WB, Link CJ. U6 promoter-enhanced GlnUAG suppressor tRNA has higher suppression efficacy and can be stably expressed in 293 cells. J Gene Med. 2013;15:93–101. doi: 10.1002/jgm.2696. [DOI] [PubMed] [Google Scholar]
- 10.Schafer B. Genetic conservation versus variability in mitochondria: the architecture of the mitochondrial genome in the petite-negative yeast Schizosaccharomyces pombe. Curr Genet. 2003;43:311–26. doi: 10.1007/s00294-003-0404-5. [DOI] [PubMed] [Google Scholar]
- 11.Hamada M, Sakulich AL, Koduru SB, Maraia R. Transcription termination by RNA polymerase III in fission yeast: A genetic and biochemical model system. J Biol Chem. 2000;275:29076–81. doi: 10.1074/jbc.M003980200. [DOI] [PubMed] [Google Scholar]
- 12.Forsburg SL. The best yeast? Trends in genetics: TIG. 1999;15:340–4. doi: 10.1016/s0168-9525(99)01798-9. [DOI] [PubMed] [Google Scholar]
- 13.Moreno S, Klar A, Nurse P. In: Methods in Enzymology: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink GR, editors. Vol. 194. Academic Press; San Diego: 1991. pp. 795–823. [Google Scholar]
- 14.Hayles J, Nurse P. Genetics of the fission yeast Schizosaccharomyces pombe. Annu Rev Genet. 1992;26:373–402. doi: 10.1146/annurev.ge.26.120192.002105. [DOI] [PubMed] [Google Scholar]
- 15.Intine RV, Dundr M, Misteli T, Maraia RJ. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Molecular cell. 2002;9:1113–23. doi: 10.1016/s1097-2765(02)00533-6. [DOI] [PubMed] [Google Scholar]
- 16.Intine RV, Sakulich AL, Koduru SB, Huang Y, Pierstorff E, Goodier JL, Maraia RJ. Control of transfer RNA maturation by phosphorylation of the human La antigen on serine 366. Molecular cell. 2000;6:339–48. doi: 10.1016/s1097-2765(00)00034-4. [DOI] [PubMed] [Google Scholar]
- 17.Park JM, Intine RV, Maraia RJ. Mouse and human La proteins differ in kinase substrate activity and activation mechanism for tRNA processing. Gene expression. 2007;14:71–81. doi: 10.3727/105221607783417619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayfield MA, Kaiser TE, Intine RV, Maraia RJ. Conservation of a masked nuclear export activity of La proteins and its effects on tRNA maturation. Mol. Cell Biol. 2007;27:3303–12. doi: 10.1128/MCB.00026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He N, Jahchan NS, Hong E, Li Qiang, Bayfield MA, Maraia RJ, Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Molecular cell. 2008;29:588–99. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain RH, Zawawi M, Bayfield MA. Conservation of RNA chaperone activity of the human La-related proteins 4, 6 and 7. Nucleic Acids Res. 2013;41:8715–25. doi: 10.1093/nar/gkt649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs) Biochim Biophys Acta. 2010;1799:365–78. doi: 10.1016/j.bbagrm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yarham John W., Lamichhane Tek N., Pyle Angela, Mattijssen Sandy, Baruffini Enrico, Bruni Francesco, Taylor RW. Defective i6A37 Modification of Mitochondrial and Cytosolic tRNAs Results from Pathogenic Mutations in TRIT1 and its Substrate tRNA. PLoS Genet. 2014 doi: 10.1371/journal.pgen.1004424. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yee NS, Gong W, Huang Y, Lorent K, Dolan AC, Maraia RJ, Pack M. Mutation of RNA polymerase III subunit rpc2/polr3b leads to deficiency of the RNA cleavage subunit, Rpc11/Polr3k, and disrupts zebrafish digestive system development. PLoS Biol. 2007;5:2484–92. doi: 10.1371/journal.pbio.0050312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernard G, Chouery E, Putorti ML, Tetreault M, Takanohashi A, Carosso G, Brais B. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. American journal of human genetics. 2011;89:415–23. doi: 10.1016/j.ajhg.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitsu H, Osaka H, Sasaki M, Takanashi J, Hamada K, Yamashita A, Matsumoto N. Mutations in POLR3A and POLR3B encoding RNA Polymerase III subunits cause an autosomal-recessive hypomyelinating leukoencephalopathy. American journal of human genetics. 2011;89:644–51. doi: 10.1016/j.ajhg.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girotto G, Abdulhadi K, Buniello A, Vozzi D, Licastro D, d’Eustacchio A, Gasparini P. Linkage Study and Exome Sequencing Identify a BDP1 Mutation Associated with Hereditary Hearing Loss. PloS one. 2013;8:e80323. doi: 10.1371/journal.pone.0080323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nichols M, Willis I, Soll D. Yeast suppressor mutations and transfer RNA processing. Methods in enzymology. 1990;181:377–94. doi: 10.1016/0076-6879(90)81137-j. [DOI] [PubMed] [Google Scholar]
- 28.Kohli J, Munz P, Soll D. In: Molecular Biology of the Fission Yeast. Nazim A, Young P, Johnson BF, editors. Academic Press; San Diego: 1989. pp. 75–96. [Google Scholar]
- 29.Hopper AK, Schultz LD, Shapiro RA. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell. 1980;19:741–51. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- 30.Laten H, Gorman J, Bock RM. Isopentenyladenosine deficient tRNA from an antisuppressor mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 1978;5:4329–42. doi: 10.1093/nar/5.11.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hopper AK. Genetic methods for study of trans-acting genes involved in processing of precursors to yeast cytoplasmic transfer RNAs. Methods in enzymology. 1990;181:400–21. doi: 10.1016/0076-6879(90)81139-l. [DOI] [PubMed] [Google Scholar]
- 32.Boguta M. Maf1, a general negative regulator of RNA polymerase III in yeast. Biochimica et biophysica acta. 2013;1829:376–84. doi: 10.1016/j.bbagrm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Boguta M, Czerska K, Zoladek T. Mutation in a new gene MAF1 affects tRNA suppressor efficiency in Saccharomyces cerevisiae. Gene. 1997;185:291–6. doi: 10.1016/s0378-1119(96)00669-5. [DOI] [PubMed] [Google Scholar]
- 34.Boguta M, Hunter LA, Shen WC, Gillman EC, Martin NC, Hopper AK. Subcellular locations of MOD5 proteins: mapping of sequences sufficient for targeting to mitochondria and demonstration that mitochondrial and nuclear isoforms commingle in the cytosol. Mol Cell Biol. 1994;14:2298–306. doi: 10.1128/mcb.14.4.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murawski M, Szczesniak B, Zoladek T, Hopper AK, Martin NC, Boguta M. maf1 mutation alters the subcellular localization of the Mod5 protein in yeast. Acta Biochim Pol. 1994;41:441–8. [PubMed] [Google Scholar]
- 36.Willis I, Hottinger H, Pearson D, Chisholm V, Leupold U, Soll D. Mutations affecting excision of the intron from a eukaryotic dimeric tRNA precursor. EMBO J. 1984;3:1573–80. doi: 10.1002/j.1460-2075.1984.tb02013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moir RD, Lee J, Haeusler RA, Desai N, Engelke DR, Willis IM. Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc Natl Acad Sci U S A. 2006;103:15044–9. doi: 10.1073/pnas.0607129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamichhane TN, Blewett NH, Cherkasova VA, Crawford AK, Iben JR, Farabaugh PJ, Maraia RJ. Lack of tRNA modification isopentenyl-A37 alters mRNA decoding and causes metabolic deficiencies in fission yeast. Mol Cell Biol. 2013;33:2918–29. doi: 10.1128/MCB.00278-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Zyl WH, Wills N, Broach JR. A general screen for mutant of Saccharomyces cerevisiae deficient in tRNA biosynthesis. Genetics. 1989;123:55–68. doi: 10.1093/genetics/123.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krupp G, Thurianx P, Willis I, Gamulin V, Soll D. First identification of an amber nonsense mutation in Schizosaccharomyces pombe: major differences in the efficiency of homologous versus heterologous yeast suppressor tRNA genes. Molecular & general genetics: MGG. 1985;201:82–7. doi: 10.1007/BF00397990. [DOI] [PubMed] [Google Scholar]
- 41.Willis I, Nichols M, Chisholm V, Soll D, Heyer WD, Szankasi P, Kohli J. Functional complementation between mutations in a yeast suppressor tRNA gene reveals potential for evolution of tRNA sequences. Proc Natl Acad Sci U S A. 1986;83:7860–4. doi: 10.1073/pnas.83.20.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamada M, Huang Y, Lowe TM, Maraia RJ. Widespread use of TATA elements in the core promoters for RNA polymerases III, II, and I in fission yeast. Mol Cell Biol. 2001;21:6870–81. doi: 10.1128/MCB.21.20.6870-6881.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venters BJ, Pugh BF. Genomic organization of human transcription initiation complexes. Nature. 2013;502:53–8. doi: 10.1038/nature12535. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Hamada M, Sakulich AL, Koduru SB, Maraia RJ. Transcription termination by RNA polymerase III in fission yeast. A genetic and biochemically tractable model system. The Journal of biological chemistry. 2000;275:29076–81. doi: 10.1074/jbc.M003980200. [DOI] [PubMed] [Google Scholar]
- 45.Hottinger H, Pearson D, Yamao F, Gamulin V, Cooper T, Soll D. Nonsense suppression in Schizosaccharomyces pombe: the S. pombe Sup3-e tRNASerUGA gene is active in S. cerevisiae. Molecular & general genetics: MGG. 1982;188:219–24. doi: 10.1007/BF00332678. [DOI] [PubMed] [Google Scholar]
- 46.Orioli A, Pascali C, Pagano A, Teichmann M, Dieci G. RNA polymerase III transcription control elements: Themes and variations. Gene. 2012;493:185–94. doi: 10.1016/j.gene.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 47.Dieci G, Yukawa Y, Alzapiedi M, Guffanti E, Ferrari R, Sugiura M, Ottonello S. Distinct modes of TATA box utilization by the RNA polymerase III transcription machineries from budding yeast and higher plants. Gene. 2006;379:12–25. doi: 10.1016/j.gene.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 48.Mao J, Schmidt O, Soll D. Dimeric transfer RNA precursors in S. pombe. Cell. 1980;21:509–16. doi: 10.1016/0092-8674(80)90488-2. [DOI] [PubMed] [Google Scholar]
- 49.Ikegami K, Lieb JD. Integral nuclear pore proteins bind to Pol III-transcribed genes and are required for Pol III transcript processing in C. elegans. Mol Cell. 2013;51:840–9. doi: 10.1016/j.molcel.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Y, McGillicuddy E, Weindel M, Dong S, Maraia R. The fission yeast TFIIB-related factor limits RNA polymerase III to a TATA-dependent pathway of TBP recruitment. Nucleic Acids Res. 2003;31:2108–16. doi: 10.1093/nar/gkg301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt O, Mao J, Ogden R, Beckmann J, Sakano H, Abelson J, Soll D. Dimeric tRNA precursors in yeast. Nature. 1980;287:750–2. doi: 10.1038/287750a0. [DOI] [PubMed] [Google Scholar]
- 52.Kohli J, Kwong T, Altruda F, Soll D, Wahl G. Characterization of a UGA-suppressing serine tRNA from Schizosaccharomyces pombe with the help of a new in vitro assay system for eukaryotic suppressor tRNAs. The Journal of biological chemistry. 1979;254:1546–51. [PubMed] [Google Scholar]
- 53.Rafalski A, Kohli J, Agris P, Soll D. The nucleotide sequence of a UGA suppressor serine tRNA from Schizosaccharomyces pombe. Nucleic Acids Res. 1979;6:2683–95. doi: 10.1093/nar/6.8.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forsburg SL. Codon use table for Schizosaccharomyces pombe. Yeast. 1994;10:1045–7. doi: 10.1002/yea.320100806. [DOI] [PubMed] [Google Scholar]
- 55.Willis I, Frendewey D, Nichols M, Hottinger-Werlen A, Schaack J, Soll D. A single base change in the intron of a serine tRNA affects the rate of RNase P cleavage in vitro and suppressor activity in vivo in Saccharomyces cerevisiae. The Journal of biological chemistry. 1986;261:5878–85. [PubMed] [Google Scholar]
- 56.Huang Y, Bayfield MA, Intine RV, Maraia RJ. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat Struct Mol Biol. 2006;13:611–8. doi: 10.1038/nsmb1110. [DOI] [PubMed] [Google Scholar]
- 57.Stefano JE. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–54. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- 58.Maraia RJ, Lamichhane TN. 3′ processing of eukaryotic precursor tRNAs. WIRES RNA. 2011;2:362–75. doi: 10.1002/wrna.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maraia RJ, Kenan DJ, Keene JD. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol. Cell. Biol. 1994;14:2147–58. doi: 10.1128/mcb.14.3.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maraia RJ, Chang DY, Wolffe AP, Vorce RL, Hsu K. The RNA polymerase III terminator used by a B1-Alu element can modulate 3′ processing of the intermediate RNA product. Mol Cell Biol. 1992;12:1500–6. doi: 10.1128/mcb.12.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang Y, Intine RV, Mozlin A, Hasson S, Maraia RJ. Mutations in the RNA Polymerase III Subunit Rpc11p That Decrease RNA 3′ Cleavage Activity Increase 3′-Terminal Oligo(U) Length and La-Dependent tRNA Processing. Mol Cell Biol. 2005;25:621–36. doi: 10.1128/MCB.25.2.621-636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chakshusmathi G, Kim SD, Rubinson DA, Wolin SL. A La protein requirement for efficient pre-tRNA folding. EMBO J. 2003;22:6562–72. doi: 10.1093/emboj/cdg625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bayfield MA, Maraia RJ. Precursor-product discrimination by La protein during tRNA metabolism. Nat Struct & Mol Biol. 2009;16:430–7. doi: 10.1038/nsmb.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iben JR, Mazeika JK, Hasson S, Rijal K, Arimbasseri AG, Russo AN, Maraia RJ. Point mutations in the Rpb9-homologous domain of Rpc11 that impair transcription termination by RNA polymerase III. Nucleic Acids Res. 2011;39:6100–13. doi: 10.1093/nar/gkr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Naeeni AR, Conte MR, Bayfield MA. RNA chaperone activity of human La protein is mediated by variant RNA recognition motif. The Journal of biological chemistry. 2012;287:5472–82. doi: 10.1074/jbc.M111.276071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nagarajavel V, Iben JR, Howard BH, Maraia RJ, Clark DJ. Global ‘bootprinting’ reveals the elastic architecture of the yeast TFIIIB-TFIIIC transcription complex in vivo. Nucleic Acids Res. 2013;41:8135–43. doi: 10.1093/nar/gkt611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Orioli A, Pascali C, Quartararo J, Diebel KW, Praz V, Romascano D, Dieci G. Widespread occurrence of non-canonical transcription termination by human RNA polymerase III. Nucleic Acids Res. 2011;39:5499–512. doi: 10.1093/nar/gkr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adeniyi-Jones S, Romeo PH, Zasloff M. Generation of long read-through transcripts in vivo and in vitro by deletion of 3′ termination and processing sequences in the human tRNAimet gene. Nucleic Acids Res. 1984;12:1101–15. doi: 10.1093/nar/12.2.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vnencak-Jones CL, Wahab SZ, Zehner ZE, Holmes WM. A human tRNA(iMet) gene produces multiple transcripts. Molecular and cellular biology. 1987;7:4134–8. doi: 10.1128/mcb.7.11.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Furter R, Snaith M, Gillespie DE, Hall BD. Endonucleolytic cleavage of a long 3′-trailer sequence in a nuclear yeast suppressor tRNA. Biochemistry. 1992;31:10817–24. doi: 10.1021/bi00159a024. [DOI] [PubMed] [Google Scholar]
- 71.Castano JG, Tobian JA, Zasloff M. Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3′ terminus of human pre-tRNA-Met(i) (3′ pre-tRNase) The Journal of biological chemistry. 1985;260:9002–8. [PubMed] [Google Scholar]
- 72.Levinger L, Greene V, Birk A, Bourne R, Kolla S, Whyte S. RNase P and 3′-tRNase processing matrices in the analysis of Drosophila transfer RNA D/T loop tertiary contacts. Nucleic Acids Symp Ser. 1995;33:82–4. [PubMed] [Google Scholar]
- 73.Levinger L, Vasisht V, Greene V, Bourne R, Birk A, Kolla S. Sequence and structure requirements for Drosophila tRNA 5′- and 3′-end processing. The Journal of biological chemistry. 1995;270:18903–9. doi: 10.1074/jbc.270.32.18903. [DOI] [PubMed] [Google Scholar]
- 74.Nashimoto M, Tamura M, Kaspar RL. Minimum requirements for substrates of mammalian tRNA 3′ processing endoribonuclease. Biochemistry. 1999;38:12089–96. doi: 10.1021/bi9911942. [DOI] [PubMed] [Google Scholar]
- 75.Zhao Z, Su W, Yuan S, Huang Y. Functional conservation of tRNase ZL among Saccharomyces cerevisiae, Schizosaccharomyces pombe and humans. The Biochemical journal. 2009;422:483–92. doi: 10.1042/BJ20090743. [DOI] [PubMed] [Google Scholar]
- 76.Yoo CJ, Wolin SL. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]
- 77.Copela LA, Fernandez CF, Sherrer RL, Wolin SL. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA. 2008;14:1214–27. doi: 10.1261/rna.1050408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ozanick SG, Wang X, Costanzo M, Brost RL, Boone C, Anderson JT. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:298–308. doi: 10.1093/nar/gkn925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Skowronek E, Grzechnik P, Spath B, Marchfelder A, Kufel J. tRNA 3′ processing in yeast involves tRNase Z, Rex1, and Rrp6. RNA. 2014;20:115–30. doi: 10.1261/rna.041467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wolin SL, Cedervall T. The La protein. Annual review of biochemistry. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 81.Valenzuela P, Venegas A, Weinberg F, Bishop R, Rutter WJ. Structure of yeast phenylalanine-tRNA genes: an intervening DNA segment within the region coding for the tRNA. Proc Natl Acad Sci U S A. 1978;75:190–4. doi: 10.1073/pnas.75.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allison DS, Hall BD. Effects of alterations in the 3′ flanking sequences on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. #133. 1985;4:2657–64. doi: 10.1002/j.1460-2075.1985.tb03984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mazabraud A, Scherly D, Muller F, Rungger D, Clarkson SG. Structure and transcription termination of a lysine tRNA gene from Xenopus laevis. J Mol Biol. 1987;195:835–45. doi: 10.1016/0022-2836(87)90488-8. [DOI] [PubMed] [Google Scholar]
- 84.Bogenhagen DF, Brown DD. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–70. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- 85.Goodier JL, Maraia RJ. Terminator-specific recycling of a B1-Alu transcription complex by RNA polymerase III is mediated by the RNA terminus-binding protein La. The Journal of biological chemistry. 1998;273:26110–6. doi: 10.1074/jbc.273.40.26110. [DOI] [PubMed] [Google Scholar]
- 86.Braglia P, Percudani R, Dieci G. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J Biol Chem. 2005;280:19551–62. doi: 10.1074/jbc.M412238200. [DOI] [PubMed] [Google Scholar]
- 87.Iben JR, Maraia RJ. Yeast tRNAomics: tRNA gene copy number variation and codon use provide bioinformatics evidence of a new wobble pair in a eukaryote. RNA. 2012;18:1358–72. doi: 10.1261/rna.032151.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang Y, Intine RV, Mozlin A, Hasson S, Maraia RJ. Mutations in the RNA polymerase III subunit Rpc11p that decrease RNA 3′ cleavage activity increase 3′-terminal oligo(U) length and La-dependent tRNA processing. Mol Cell Biol. 2005;25:621–36. doi: 10.1128/MCB.25.2.621-636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bushnell DA, Cramer P, Kornberg RD. Structural basis of transcription: alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc Natl Acad Sci U S A. 2002;99:1218–22. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30:547–56. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.French SL, Osheim YN, Schneider DA, Sikes ML, Fernandez CF, Copela LA, Beyer AL. Visual analysis of the yeast 5S rRNA gene transcriptome: regulation and role of La protein. Mol Cell Biol. 2008;28:4576–87. doi: 10.1128/MCB.00127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moir RD, Willis IM. Regulation of pol III transcription by nutrient and stress signaling pathways. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbagrm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arimbasseri AG, Rijal K, Maraia RJ. Comparative overview of RNA polymerase II and III transcription cycles, with focus on RNA polymerase III termination and reinitiation. Transcription. 2013;4 doi: 10.4161/trns.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yukawa Y, Sugita M, Choisne N, Small I, Sugiura M. The TATA motif, the CAA motif and the poly(T) transcription termination motif are all important for transcription re-initiation on plant tRNA genes. The Plant journal: for cell and molecular biology. 2000;22:439–47. doi: 10.1046/j.1365-313x.2000.00752.x. [DOI] [PubMed] [Google Scholar]
- 95.Hamada M, Huang Y, Lowe TM, Maraia RJ. Widespread Use of TATA Elements in the Core Promoters for RNA Polymerases III, II, and I in Fission Yeast. Mol. Cell. Biol. 2001;21:6870–81. doi: 10.1128/MCB.21.20.6870-6881.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang Y, Hamada M, Maraia RJ. RNA polymerase III from the fission yeast, Schizosaccharomyces pombe. Methods in enzymology. 2003;370:165–73. doi: 10.1016/S0076-6879(03)70014-7. [DOI] [PubMed] [Google Scholar]
- 97.Huang Y, Hamada M, Maraia RJ. Isolation and Cloning of Four Subunits of a Fission Yeast TFIIIC Complex That Includes an Ortholog of the Human Regulatory Protein TFIIICbeta. J. Biol. Chem. 2000;275:31480–7. doi: 10.1074/jbc.M004635200. [DOI] [PubMed] [Google Scholar]
- 98.Koduvayur SP, Woodson SA. Intracellular folding of the Tetrahymena group I intron depends on exon sequence and promoter choice. RNA. 2004;10:1526–32. doi: 10.1261/rna.7880404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–75. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 100.Watson PY, Fedor MJ. Determination of intracellular RNA folding rates using self-cleaving RNAs. Methods Enzymol. 2009;468:259–86. doi: 10.1016/S0076-6879(09)68013-7. [DOI] [PubMed] [Google Scholar]
- 101.Reiner R, Ben-Asouli Y, Krilovetzky I, Jarrous N. A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes Dev. 2006;20:1621–35. doi: 10.1101/gad.386706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14:153–65. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- 103.Ishiguro A, Kassavetis GA, Geiduschek EP. Essential roles of Bdp1, a subunit of RNA polymerase III initiation factor TFIIIB, in transcription and tRNA processing. Mol Cell Biol. 2002;22:3264–75. doi: 10.1128/MCB.22.10.3264-3275.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Carrara G, Calandra P, Fruscoloni P, Doria M, Tocchini-Valentini GP. Site selection by Xenopus laevis RNAase P. Cell. 1989;58:37–45. doi: 10.1016/0092-8674(89)90400-5. [DOI] [PubMed] [Google Scholar]
- 105.Lee Y, Kindelberger DW, Lee JY, McClennen S, Chamberlain J, Engelke DR. Nuclear pre-tRNA terminal structure and RNase P recognition. RNA. 1997;3:175–85. [PMC free article] [PubMed] [Google Scholar]
- 106.Xiao S, Scott F, Fierke CA, Engelke DR. Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Annual review of biochemistry. 2002;71:165–89. doi: 10.1146/annurev.biochem.71.110601.135352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jarrous N, Reiner R. Human RNase P: a tRNA-processing enzyme and transcription factor. Nucleic Acids Res. 2007;35:3519–24. doi: 10.1093/nar/gkm071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reiner R, Krasnov-Yoeli N, Dehtiar Y, Jarrous N. Function and assembly of a chromatin-associated RNase P that is required for efficient transcription by RNA polymerase I. PloS one. 2008;3:e4072. doi: 10.1371/journal.pone.0004072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wan Y, Suh H, Russell R, Herschlag D. Multiple unfolding events during native folding of the Tetrahymena group I ribozyme. J Mol Biol. 2010;400:1067–77. doi: 10.1016/j.jmb.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Woodson SA. Taming free energy landscapes with RNA chaperones. RNA biology. 2010;7:677–86. doi: 10.4161/rna.7.6.13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bayfield MA, Maraia RJ. Precursor-product discrimination by La protein during tRNA metabolism. Nat Struct Mol Biol. 2009;16:430–7. doi: 10.1038/nsmb.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Hinnebusch AG. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–62. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Copela LA, Chakshusmathi G, Sherrer RL, Wolin SL. The La protein functions redundantly with tRNA modification enzymes to ensure tRNA structural stability. RNA. 2006;12:644–54. doi: 10.1261/rna.2307206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–40. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hussain RH, Zawawi M, Bayfield MA. Conservation of RNA chaperone activity of the human La-related proteins 4, 6 and 7. Nucl Acids Res. 2013 doi: 10.1093/nar/gkt649. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Naeeni AR, Conte MR, Bayfield MA. RNA chaperone activity of the human La protein is mediated by a variant RNA recognition motif. J Biol Chem. 2012;287:5472–82. doi: 10.1074/jbc.M111.276071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stone MD, Mihalusova M, O’Connor C,M, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–61. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Van Horn DJ, Yoo CJ, Xue D, Shi H, Wolin SL. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA. 1997;3:1434–43. [PMC free article] [PubMed] [Google Scholar]
- 119.Intine RVA, Sakulich AL, Koduru SB, Huang Y, Pierstorrf E, Goodier JL, Maraia RJ. Control of transfer RNA maturation by phosphorylation of the human La antigen on serine 366. Mol Cell. 2000;6:339–48. doi: 10.1016/s1097-2765(00)00034-4. [DOI] [PubMed] [Google Scholar]
- 120.Intine RV, Dundr M, Misteli T, Maraia RJ. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Mol Cell. 2002;9:1113–23. doi: 10.1016/s1097-2765(02)00533-6. [DOI] [PubMed] [Google Scholar]
- 121.Kufel J, Tollervey D. 3′-processing of yeast tRNATrp precedes 5′-processing. RNA. 2003;9:202–8. doi: 10.1261/rna.2145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nashimoto M, Nashimoto C, Tamura M, Kaspar RL, Ochi K. The inhibitory effect of the autoantigen La on in vitro 3′ processing of mammalian precursor tRNAs. J Mol Biol. 2001;312:975–84. doi: 10.1006/jmbi.2001.5026. [DOI] [PubMed] [Google Scholar]
- 123.Fairley JA, Kantidakis T, Kenneth NS, Intine RV, Maraia RJ, White RJ. Human La is Found at RNA Polymerase III-Transcribed Genes In Vivo. Proc Nat Acad Sci, USA. 2005;102:18350–5. doi: 10.1073/pnas.0506415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.James P, Hall BD. ret1-1, a yeast mutant affecting transcription termination by RNA polymerase III. Genetics. 1990;125:293–303. doi: 10.1093/genetics/125.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.James P, Whelen S, Hall BD. The RET1 gene of yeast encodes the second-largest subunit of RNA polymerase III. J. Biol. Chem. 1991;266:5616–24. [PubMed] [Google Scholar]
- 126.Iben JR, Mazeika JK, Hasson S, Rijal K, Arimbasseri AG, Russo AN, Maraia RJ. Point mutations in the Rpb9-homologous domain of Rpc11 that impair transcription termination by RNA polymerase III. Nucleic Acids Res. 2011;39:6100–13. doi: 10.1093/nar/gkr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Willis I, Schmidt P, Soll D. A selection for mutants of the RNA polymerase III transcription apparatus: PCF1 stimulates transcription of tRNA and 5S RNA genes. EMBO J. 1989;8:4281–8. doi: 10.1002/j.1460-2075.1989.tb08614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lopez-De-Leon A, Librizzi M, Puglia K, Willis IM. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:211–20. doi: 10.1016/0092-8674(92)90350-l. [DOI] [PubMed] [Google Scholar]
- 129.Rijal K, Maraia RJ. RNA polymerase III mutants in TFIIFα-like C37 cause terminator readthrough with no decrease in transcription output. Nucleic acids research. 2013;41:139–55. doi: 10.1093/nar/gks985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Arimbasseri AG, Maraia RJ. Distinguishing core and holoenzyme mechanisms of transcription termination by RNA polymerase III. Mol Cell Biol. 2013;33:1571–81. doi: 10.1128/MCB.01733-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bartholomew B, Kassavetis GA, Braun BR, Geiduschek EP. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990;9:2197–205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Carter R, Drouin G. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol Biol Evol. 2009;27:1035–43. doi: 10.1093/molbev/msp316. [DOI] [PubMed] [Google Scholar]
- 133.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Molecular cell. 2012;45:439–46. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 134.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–69. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Arimbasseri AG, Rijal K, Maraia RJ. Transcription termination by the eukaryotic RNA polymerase III. Biochimica et biophysica acta. 2013;1829:318–30. doi: 10.1016/j.bbagrm.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Peters JM, Vangeloff AD, Landick R. Bacterial Transcription Terminators: The RNA 3′-end Chronicles. J Mol Biol. 2011;412:793–813. doi: 10.1016/j.jmb.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Arimbasseri AG, Rijal K, Maraia RJ. Transcription termination by the eukaryotic RNA polymerase III. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbagrm.2012.10.006. S1874-9399(12)00177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cozzarelli NR, Gerrard SP, Schlissel M, Brown DD, Bogenhagen DF. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983;34:829–35. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- 139.Wang X, Folk WR. Termination of transcription by RNA polymerase III from wheat germ. J Biol Chem. 1994;269:4993–5004. [PubMed] [Google Scholar]
- 140.Arimbasseri AG, Kassavetis GA, Maraia RJ. Comment on “Mechanism of eukaryotic RNA polymerase III transcription termination”. Science. 2014;345:524. doi: 10.1126/science.1253783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nielsen S, Yuzenkova Y, Zenkin N. Mechanism of eukaryotic RNA polymerase III transcription termination. Science. 2013;340:1577–80. doi: 10.1126/science.1237934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shaaban SA, Krupp BM, Hall BD. Termination-altering mutations in the second-largest subunit of yeast RNA polymerase III. Mol. Cell. Biol. 1995;15:1467–1478. doi: 10.1128/mcb.15.3.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shaaban SA, Bobkova EV, Chudzik DM, Hall BD. In vitro analysis of elongation and termination by mutant RNA polymerases with altered termination behavior. Mol Cell Biol. 1996;16:6468–76. doi: 10.1128/mcb.16.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bobkova EV, Hall BD. Substrate specificity of the RNase activity of yeast RNA polymerase III. The Journal of biological chemistry. 1997;272:22832–9. doi: 10.1074/jbc.272.36.22832. [DOI] [PubMed] [Google Scholar]
- 145.Bobkova EV, Habib N, Alexander G, Hall BD. Mutational analysis of the hydrolytic activity of yeast RNA polymerase III. The Journal of biological chemistry. 1999;274:21342–8. doi: 10.1074/jbc.274.30.21342. [DOI] [PubMed] [Google Scholar]
- 146.Jin DJ, Burgess RR, Richardson JP, Gross CA. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci U S A. 1992;89:1453–7. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.von Hippel PH, Yager TD. The elongation-termination decision in transcription. Science. 1992;255:809–12. doi: 10.1126/science.1536005. [DOI] [PubMed] [Google Scholar]
- 148.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–66. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 149.Jeon C, Yoon H, Agarwal K. The transcription factor TFIIS zinc ribbon dipeptide Asp-Glu is critical for stimulation of elongation and RNA cleavage by RNA polymerase II. Proc Natl Acad Sci U S A. 1994;91:9106–10. doi: 10.1073/pnas.91.19.9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Johnson TL, Chamberlin MJ. Complexes of yeast RNA polymerase II and RNA are substrates for TFIIS-induced RNA cleavage. Cell. 1994;77:217–24. doi: 10.1016/0092-8674(94)90314-x. [DOI] [PubMed] [Google Scholar]
- 151.Conaway RC, Kong SE, Conaway JW. TFIIS and GreB: two like-minded transcription elongation factors with sticky fingers. Cell. 2003;114:272–4. doi: 10.1016/s0092-8674(03)00607-x. [DOI] [PubMed] [Google Scholar]
- 152.Whitehall SK, Bardeleben C, Kassavetis GA. Hydrolytic cleavage of nascent RNA in RNA polymerase III ternary transcription complexes. J Biol Chem. 1994;269:2299–306. [PubMed] [Google Scholar]
- 153.Chedin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12:3857–71. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006;25:118–28. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Martin FH, Tinoco I., Jr. DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 1980;8:2295–9. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fernandez-Tornero C, Bottcher B, Riva M, Carles C, Steuerwald U, Ruigrok RW, Schoehn G. Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Molecular cell. 2007;25:813–23. doi: 10.1016/j.molcel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 157.Fernandez-Tornero C, Bottcher B, Rashid UJ, Steuerwald U, Florchinger B, Devos DP, Muller CW. Conformational flexibility of RNA polymerase III during transcriptional elongation. EMBO J. 2010 doi: 10.1038/emboj.2010.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 159.Wu CC, Lin YC, Chen HT. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol Cell Biol. 2011;31:2715–28. doi: 10.1128/MCB.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yee NS, Gong W, Huang Y, Lorent K, Dolan AC, Maraia RJ, Pack M. Mutation of RNA Pol III subunit rpc2/polr3b Leads to Deficiency of Subunit Rpc11 and disrupts zebrafish digestive development. PLoS Biol. 2007;5:e312. doi: 10.1371/journal.pbio.0050312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zaccolo M, Williams DM, Brown DM, Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J Mol Biol. 1996;255:589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 162.Kireeva ML, Domecq C, Coulombe B, Burton ZF, Kashlev M. Interaction of RNA polymerase II fork loop 2 with downstream non-template DNA regulates transcription elongation. The Journal of biological chemistry. 2011;286:30898–910. doi: 10.1074/jbc.M111.260844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kubicek CE, Chisholm RD, Takayama S, Hawley DK. RNA polymerase II mutations conferring defects in poly(A) site cleavage and termination in Saccharomyces cerevisiae. G3 (Bethesda, Md.) 2013;3:167–80. doi: 10.1534/g3.112.004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kassavetis GA, Prakash P, Shim E. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. The Journal of biological chemistry. 2010;285:2695–706. doi: 10.1074/jbc.M109.074013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–43. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schweikhard V, Meng C, Murakami K, Kaplan CD, Kornberg RD, Block SM. Transcription factors TFIIF and TFIIS promote transcript elongation by RNA polymerase II by synergistic and independent mechanisms. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1405181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kolb KE, Hein PP, Landick R. Antisense oligonucleotide-stimulated transcriptional pausing reveals RNA exit channel specificity of RNA polymerase and mechanistic contributions of NusA and RfaH. J Biol Chem. 2014;289:1151–63. doi: 10.1074/jbc.M113.521393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;22:6322–34. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Keeney JB, Boeke JD. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–56. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Preiser P, Vasisht V, Birk A, Levinger L. Poly(U)-binding protein inhibits Drosophila pre-5 S RNA 3′-exonuclease digestion. The Journal of biological chemistry. 1993;268:11553–7. [PubMed] [Google Scholar]
- 171.Maraia RJ, Kenan DJ, Keene JD. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol Cell Biol. 1994;14:2147–58. doi: 10.1128/mcb.14.3.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lin-Marq N, Clarkson SG. Efficient synthesis, termination and release of RNA polymerase III transcripts in Xenopus extracts depleted of La protein. #837. 1998;17:2033–41. doi: 10.1093/emboj/17.7.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Maraia RJ, Lamichhane TN. 3′ processing of eukaryotic precursor tRNAs. Wiley interdisciplinary reviews. RNA. 2011;2:362–75. doi: 10.1002/wrna.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cormack BP, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–96. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 175.Johnson SS, Zhang C, Fromm J, Willis IM, Johnson DL. Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Molecular cell. 2007;26:367–79. doi: 10.1016/j.molcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 176.Karkusiewicz I, Turowski TW, Graczyk D, Towpik J, Dhungel N, Hopper AK, Boguta M. Maf1 protein, repressor of RNA polymerase III, indirectly affects tRNA processing. The Journal of biological chemistry. 2011;286:39478–88. doi: 10.1074/jbc.M111.253310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Reina JH, Azzouz TN, Hernandez N. Maf1, a new player in the regulation of human RNA polymerase III transcription. PloS one. 2006;1:e134. doi: 10.1371/journal.pone.0000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Molecular cell. 2002;10:1489–94. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 179.McPheeters DS, Cremona N, Sunder S, Chen HM, Averbeck N, Leatherwood J, Wise JA. A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat Struct Mol Biol. 2009;16:255–64. doi: 10.1038/nsmb.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Maraia RJ, Intine RV. La protein and its associated small nuclear and nucleolar precursor RNAs. Gene expression. 2002;10:41–57. [PMC free article] [PubMed] [Google Scholar]
- 181.Zaccolo M, Williams DM, Brown DM, Gherardi E. An Approach to Random Mutagenesis of DNA Using Mixtures of Triphosphate Derivatives of Nucleoside Analogues. J Mol Biol. 1996;255:589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 182.Bayfield MA, Kaiser TE, Intine RV, Maraia RJ. Conservation of a masked nuclear export activity of La proteins and its effects on tRNA maturation. Mol Cell Biol. 2007;27:3303–12. doi: 10.1128/MCB.00026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lamichhane TN, Blewett NH, Maraia RJ. Plasticity and diversity of tRNA anticodon determinants of substrate recognition by eukaryotic A37 isopentenyltransferases. RNA. 2011;17:1846–57. doi: 10.1261/rna.2628611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gan X, Yang J, Li J, Yu H, Dai H, Liu J, Huang Y. The fission yeast Schizosaccharomyces pombe has two distinct tRNase Z(L)s encoded by two different genes and differentially targeted to the nucleus and mitochondria. The Biochemical journal. 2011;435:103–11. doi: 10.1042/BJ20101619. [DOI] [PubMed] [Google Scholar]
- 185.Bartholomew B, Kassavetis GA, Geiduschek EP. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol Cell Biol. 1991;11:5181–9. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Geiduschek EP, Kassavetis GA. In: Transcriptional Regulation. McKnight S, Yamamoto K, editors. Cold Spring Harbor Laboratory Press; New York: 1992. pp. 247–80. [Google Scholar]
- 187.Kassavetis GA, Braun BR, Nguyen LH, Geiduschek EP. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–45. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 188.Yoshihisa T, Ohshima C, Yunoki-Esaki K, Endo T. Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae. Genes Cells. 2007;12:285–97. doi: 10.1111/j.1365-2443.2007.01056.x. [DOI] [PubMed] [Google Scholar]
- 189.Yoshihisa T, Yunoki-Esaki K, Tanaka N, Endo T. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell. 2003;14:3266–79. doi: 10.1091/mbc.E02-11-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]