Summary
Non-resolving inflammation expands a heterogeneous population of myeloid suppressor cells capable of inhibiting T cell function. This heterogeneity has confounded the functional dissection of individual myeloid subpopulations and presents an obstacle for anti-tumor immunity and immunotherapy. Using genetic manipulation of cell death pathways, we found the monocytic suppressor cell subset, but not the granulocytic subset requires continuous c-FLIP expression to prevent caspase-8-dependent, RIPK3-independent cell death. Development of the granulocyte subset requires MCL-1-mediated control of the intrinsic mitochondrial death pathway. Monocytic suppressors tolerate the absence of MCL-1 provided cytokines increase expression of the MCL-1-related protein A1. Monocytic suppressors mediate T cell suppression, while their granulocytic counterparts lack suppressive function. The loss of the granulocytic subset via conditional MCL-1 deletion did not alter tumor incidence implicating the monocytic compartment as the functionally immunosuppressive subset in vivo. Thus, death pathway modulation defines the development, survival and function of myeloid suppressor cells.
Introduction
Non-resolving inflammation is caused by failure to eliminate a long-lived insulting entity including persisting microbes, implanted medical devices, cholesterol and fat in atherosclerosis and obesity, and self-antigens driving chronic auto-inflammation (Nathan and Ding, 2010). In cancer, non-resolving inflammation is driven by the growing malignancy and is associated with the production of large numbers of mature and immature myeloid cells from the bone marrow (BM). Circulating immature myeloid cells with immunosuppressive functions are collectively called myeloid-derived suppressor cells (MDSCs), and are negatively correlated with cancer outcomes (Gabrilovich et al., 2012; Wu et al., 2014). MDSC expansion is related to a hematopoietic response to inflammation where growth factors such as GM-CSF and G-CSF signal to the BM to transiently increase cellular output. This ‘emergency’ hematopoiesis aids in the destruction and elimination of the insulting entity, and is followed by tissue repair and resolution (Manz and Boettcher, 2014). In non-resolving inflammation the inciting agent remains, and the hematopoietic cycle linked to clearance and resolution becomes dysregulated. MDSCs have attracted attention in cancer biology because they are linked with suppression of lymphocyte activation. The number and activity of cytotoxic CD8+ T cells are correlated with anti-tumor immunity (Gajewski et al., 2013; Galon et al., 2013). Therapies designed to elicit anti-tumor T cell responses must overcome or bypass the local MDSC-mediated immune suppression inside the tumor microenvironment (Motz and Coukos, 2013; Restifo et al., 2012).
The current understanding of MDSC development, lifespan and function has been limited by heterogeneity of the myeloid populations produced from the BM under inflammatory stress (Gabrilovich et al., 2007; Gabrilovich and Nagaraj, 2009; Gabrilovich et al., 2012; Wu et al., 2014). Thus, it remains unclear which type of MDSC to target, and which MDSC sub-population(s) contributes to immunosuppression. MDSCs express combinations of myeloid-associated cell surface markers and have an immature myeloid phenotype, but their hallmark functional characteristic is their ability to suppress T cells. (Gabrilovich et al., 2007; Peranzoni et al., 2010; Schouppe et al., 2013; Talmadge and Gabrilovich, 2013; Youn et al., 2011). MDSCs comprise heterogeneous mixtures of mature and immature granulocytes, monocyte-macrophages, and more primitive cells such as band-form granulocytic precursors (Gabrilovich et al., 2007; Gabrilovich and Nagaraj, 2009; Movahedi et al., 2008; Youn et al., 2011). So far there is no accepted marker system to predict if a MDSC will be suppressive without evaluating its suppressive function using in vitro T cell assays. However, the presence of activated T cells or local inflammatory milieus engenders changes in MDSCs and alters their functional activity (Haverkamp et al., 2011). Thus, functional dissection of MDSCs is a type of ‘Schrödinger’s Cat’ scenario where suppression is monitored using an assay that induces the functional property for which it is testing for (Haverkamp et al., 2011). Because the specific MDSC sub-populations required for T cell suppression remains controversial, efforts to engineer MDSCs has not yet advanced to the point at which a defined cell type is used therapeutically (Highfill et al., 2010; Yin et al., 2010). Similarly, inhibiting the key suppressive subtype(s) of MDSCs to enhance T cell function may be an avenue to improve anti-tumor immunity via interruption of the tumor-induced immunosuppressive milieu (Gajewski et al., 2013; McAllister and Weinberg, 2014; Restifo et al., 2012).
In the mouse, most studies focus on the accumulation of CD11b+Gr-1+ cells in the blood, spleen and local inflammatory site, and human MDSC counterparts have been defined (Talmadge and Gabrilovich, 2013). Murine MDSCs are further subdivided by surface expression of Ly6C and Ly6G. Granulocytic MDSC are Ly6G+, Ly6C+, whereas monocytic MDSC (mixtures of immature monocytes, macrophages and monocyte-derived dendritic cells) are Ly6Glo, Ly6C+. Based on these and other markers including F4/80, CD115 and IL-4Rα, elaborate strategies have been used to separate MDSCs, and both granulocytic (referred to here as PMN-MDSC) and monocytic (Mo-MDSC) MDSCs have demonstrated immunosuppressive properties. However, further examination into the function of these cells has been impeded given MDSC cell surface markers have overlapping expression with other cell types and no transcription factor deficiencies have yet been described to ablate specific MDSC sub-populations (Gabrilovich and Nagaraj, 2009; Movahedi et al., 2008; Priceman et al., 2010; Schouppe et al., 2013; Youn et al., 2011). Thus heterogeneity is an inherent hurdle toward understanding the role of regulatory myeloid cells in chronic inflammation. To overcome this obstacle, we sought to define MDSC sub-populations using properties of intrinsic and extrinsic cell death pathways involved in myeloid lineage development and survival. Herein we demonstrate the hierarchy of cell death pathways required for the development, survival and function of MDSCs, and define monocytic MDSCs as the dominant immunosuppressive subset. Further, we demonstrate that the anti-apoptotic molecule c-FLIP is constitutively required for the development of Mo-MDSCs, and that the induction of the MCL-1-related anti-apoptotic A1 (three closely related A1 isoforms are encoded by Bcl2a1a, Bcl2a1b and Bcl2a1c) by GM-CSF further promotes their survival. In contrast, PMN-MDSC require a different anti-apoptotic molecule MCL-1 for development, and the loss of granulocytes through conditional MCL-1 ablation does not contribute to tumor growth or incidence.
Results
Genetic manipulation of death pathways skews suppressor populations
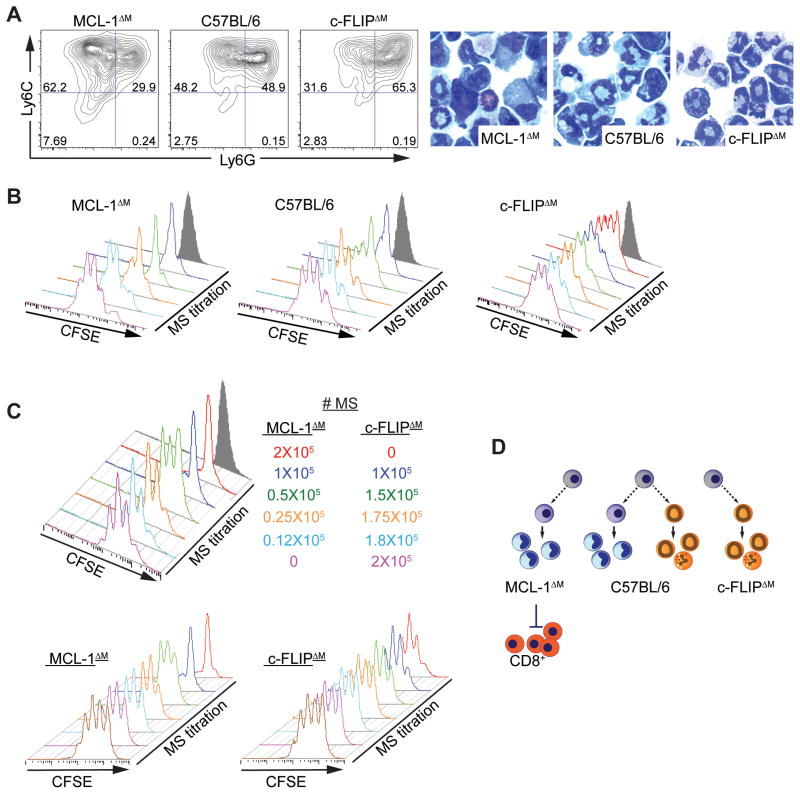
Immature monocytic (Mo) and granulocytic (PMN) cells with can be expanded in vitro from BM myeloid precursors with IL-6 and GM-CSF (Marigo et al., 2010). Like MDSCs isolated from spleens of cancer-bearing mice (Ugel et al., 2012), BM myeloid cells grown in GM-CSF and IL-6 exhibit a continuum of Ly6C and Ly6G expression (Figure 1A). Hereafter we refer to in vitro-generated populations of cells expanded from the BM as ‘myeloid suppressors’ (MS) to avoid contention with specific definitions of in vivo MDSCs accumulating in non-resolving inflammation (Gabrilovich et al., 2007; Talmadge and Gabrilovich, 2013). We harnessed previous observations about the role of extrinsic death receptor mediated and intrinsic mitochondrial anti-apoptotic pathways in regulating the development of myeloid lineages and devised a ‘mirror image’ genetic system to generate MS cultures dominated by either the granulocytic or monocytic populations. To generate MS depleted of granulocytic lineages we used mice bearing a conditional allele of the anti-apoptotic bcl-2 family member Mcl1 crossed to the LysM-Cre deleter (Steimer et al., 2009). These animals (referred to as MCL-1ΔM) have a near complete ablation of mature granulocytes in tissues and blood, but retain normal numbers of monocytes and macrophages (Dzhagalov et al., 2007; Steimer et al., 2009). BM-derived MS cultures from the MCL-1ΔM animals presented an enriched Ly6C+, Ly6G− Mo-MS fraction and reduced PMN-MS fraction relative to controls (Figure 1A, S1A, B). Deletion of MCL-1 in MS cultures was efficient, as immunoblotting showed a near complete absence of MCL-1 protein (Figure S1C). Therefore MCL-1 ablation was a means to skew MS populations such that the proportion of Mo-MS in the cultures was increased relative to control cultures.
Figure 1. Death pathways can be used to manipulate the population structure and activity of MS.
(A–C) BM-generated MS were grown from MCL-1ΔM, C57BL/6, or FLIPΔM mice. (A) Flow cytometry analysis of MS cultures. Numbers indicate the percentage of CD11b+ cells. Cytospins collected from MS cultures. Data are representative of no fewer than 10 independent experiments. (B) Suppressive function of MS was measured using 5 × 105 CFSE-labeled OT-I cells cultured with titrated numbers of MS from each genotype in the presence of SIINFEKL peptide. CFSE dilution in CD8+ cells was evaluated by flow cytometry. Gray shaded histograms show negative control wells cultured without peptide. Plots from 4 independent experiments are shown (n = 2 for each experiment). (C) Representative plots from 2 independent experiments (n = 2 for each experiment) showing CFSE dilution in in CD8+ cells cultured with MCL-1ΔM MS mixed with varying numbers of FLIPΔM MS to a final number of 2 × 105 total MS (top panel) or unmixed control MCL-1ΔM and FLIPΔM MS (bottom panels). (D) Diagram showing how genetics can be used to modulate the composition and suppressive function of MS sub-populations.
To generate a suppressor population skewed toward PMN-MS we used mice lacking c-FLIP (encoded by Cflar) in the late myeloid lineages again using the LysM-Cre deleter (c-FLIPΔM). c-FLIP is an inhibitor of extrinsic death pathways controlled by death receptor (DR) signaling: Caspase-8-mediated apoptosis, and RIPK1 and RIPK3-mediated necroptosis (Green et al., 2011; Newton et al., 2014). c-FLIPΔM mice have hyper-production of granulocytes and a relative depletion of monocyte-derived macrophages (Gordy et al., 2011; Huang et al., 2010) and in our colony, die before weaning. BM cultures from c-FLIPΔM mice had inverse properties from the MCL-1ΔM MS cultures and were enriched for Ly6G+, Ly6C+ PMN-MS (Figure 1A, S1A, B).
Using our matched genetic systems we investigated the suppressive capacity of c-FLIPΔM and MCL-1ΔM MS relative to control MS by monitoring OVA-specific CD8+ T (OT-I) cell proliferation during in vitro T cell-MS co-cultures. c-FLIPΔM MS had negligible suppressive activity while MCL-1ΔM MS were enriched for suppression on a cell-for-cell basis (Figure 1B, S1D). To determine if the lack of inhibitory function of PMN-MS was caused by the loss of monocytic cells in c-FLIPΔM animals, we performed experiments wherein MCL-1ΔM and c-FLIPΔM MS were mixed immediately prior to culture with OT-I cells, at a fixed suppressor cell number per well. T cell proliferation was inhibited only in the presence of high numbers of MCL-1ΔM MS (Figure 1C). These data demonstrate suppression is principally found in the monocytic component of the MS pool. To further evaluate MS-mediated suppression of T cell proliferation, we sorted Mo-MS and PMN-MS from wild-type BM cultures based on CD11b-gated Ly6G and Ly6C expression (Figure S1E). Purified Mo-MS were suppressive, with detectable inhibitory effects on T cell proliferation even when cultured at a 1:20 MS: OT-I cell ratio. However, when OT-I cells were cultured with PMN-MS, proliferation was inhibited, but only at the highest ratios, and lost as the number of PMN-MS was decreased by one dilution step (Figure S1F, G).
One explanation for the differences between Mo-MS and PMN-MS in T cell suppressive activity could have been decreased viability of the PMN-MS. We therefore evaluated the viability of both MS populations at 24 h intervals throughout the duration of the T cell suppression assay. The percentage of live cells was measured using a viability dye (V405) detecting cell surface amine groups, a hallmark of dead and dying cells. Viability was equivalent between Mo-MS and PMN-MS at all time points measured. Therefore, PMN-MS fail to suppress T cell proliferation even though they survive equally as well as the suppressive Mo-MS (Figure S1H). Collectively, our genetic and sorting strategies resolve the long-standing issue of which subset in the MS pool is responsible for suppression of T cell proliferation, identifying monocytic MS as the dominant suppressive population (Figure 1D).
c-FLIP controls the development of monocytic suppressors
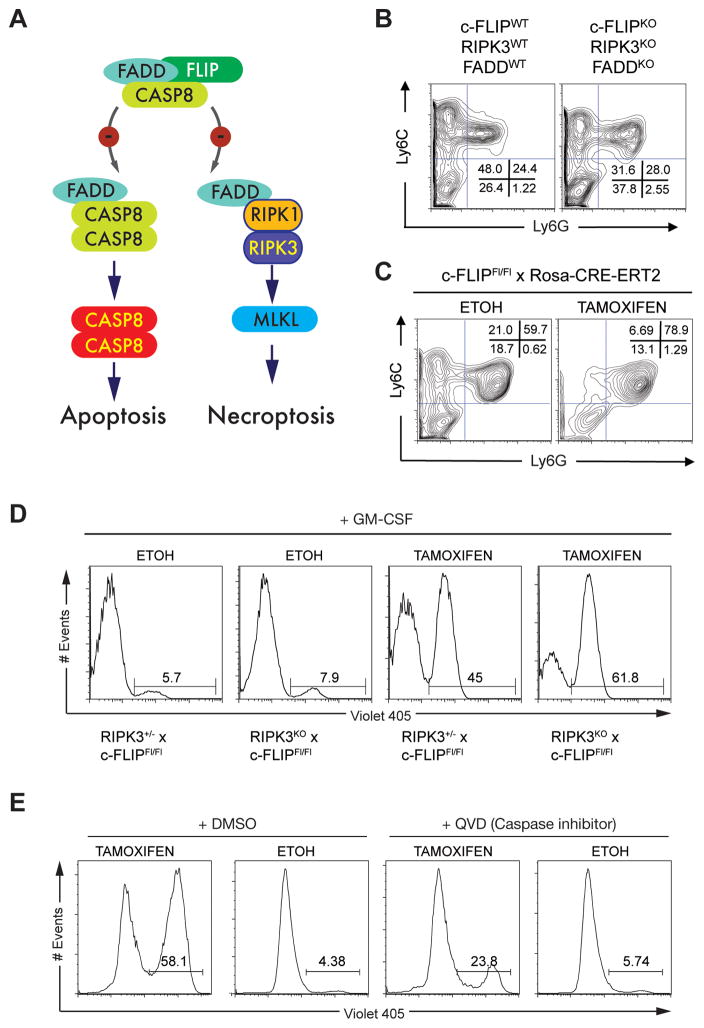
c-FLIP inhibits FADD-caspase-8 apoptotic and RIPK1-RIPK3-mediated necroptotic pathways (Figure 2A) (Green et al., 2011). Therefore if c-FLIP regulates Mo-MS survival as predicted from the c-FLIPΔM results, ablation of FADD and RIPK3 should rescue Mo-MS even in a complete c-FLIP-deficient background. Unlike c-FLIPΔM MS cultures (Figure 1A), BM from mice lacking c-FLIP, FADD and RIPK3 had Mo-MS (Figure 2B). Thus c-FLIP is important for the viability of Mo-MS through development in vitro but dispensable for PMN-MS. We confirmed this conclusion by depleting c-FLIP using tamoxifen-mediated Cre-deletion in Cflarfl/fl; Rosa-CRE-ERT2 BM MS cultures (Figure 2C). Mo-MS were absent from the tamoxifen-treated but not from vehicle control cultures, while PMN-MS were present in all conditions, arguing c-FLIP is dispensable for PMN-MS but continuously required for Mo-MS.
Figure 2. c-FLIP controls Mo-MS viability.
(A) Diagram showing the role of c-FLIP in the inhibition of caspase 8 mediated apoptosis and necroptosis. (B) Flow cytometry analysis of BM-MS generated from FLIPWT, RIPK3WT, FADDWT and c-FLIPKO, RIPK3KO, FADDKO triple deficient mice. Numbers indicate the percentage of live cells (n = 3 independent experiments). (C) Flow cytometry analysis of BM-MS from Cflarfl/fl; Rosa-CreERT2 mice treated with 4-OH tamoxifen at d3 of culture. Ethanol at the same final concentration as the 4-OH tamoxifen cultures served as the control. Numbers indicate the percentage of live cells (n = 3 independent experiments). (D) Cflar was exogenously deleted by 4-OH tamoxifen on d5 of culture using BM-MS from Cflarfl/fl; Rosa-CreERT2 mice on a Ripk3+/− or Ripk3−/− background. Control cells received ethanol. On d6 Mo-MS were sorted and cultured with GM-CSF (50 ng/mL) for 24h, after which viability was assessed using V405 staining. (E) Cflar was exogenously deleted by 4-OH tamoxifen in the presence or absence of QVD (20 μM) on d5 of culture using BM-MS from Cflarfl/fl; Rosa-CreERT2 mice. Control cells received ethanol and DMSO. On d6 Mo-MS were sorted and cultured with GM-CSF (50 ng/mL) in the presence or absence of QVD (20 μM) for 24h. Viability was assessed using V405 staining.
Because c-FLIP is required to inhibit both necroptosis and Caspase-8-mediated apoptosis, we next tested if one or both pathways were involved in the viability of Mo-MS. We generated Cflarfl/fl; Rosa-CRE-ERT2 mice on a RIPK3-deficient background. Following tamoxifen-mediated Cre-deletion, Mo-MS died, even in GM-CSF, arguing viability of Mo-MS is controlled by c-FLIP inhibiting caspase-8-mediated apoptosis rather than RIPK1-RIPK3 necroptosis (Figure 2D). Concurrent treatment of MS cultures with the pan-caspase inhibitor QVD at the time of tamoxifen mediated Cflar ablation reduced cell death (Figure 2E). Therefore the viability of Mo-MS is controlled by c-FLIP inhibition of caspase-8 mediated apoptosis rather than RIPK1-RIPK3 necroptosis.
Suppressors are dependent on the MCL-1 – A1 axis
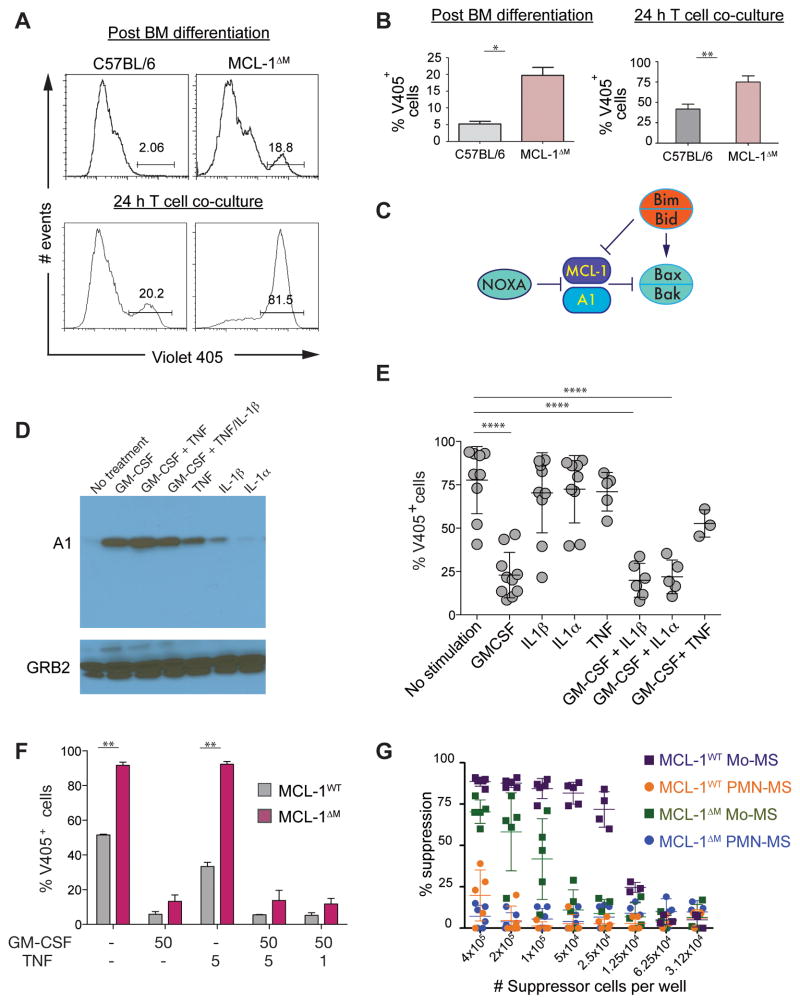
Genetic experiments have shown nearly all cells in the body have an absolute requirement for MCL-1 (Opferman et al., 2005; Opferman et al., 2003; Wang et al., 2013; Weber et al., 2010). Monocytic cells are an exception, as they seem to develop normally in the absence of MCL-1. However, MCL-1ΔM Mo-MS were more sensitive to death compared to control Mo-MS within BM MS and T cell co-cultures (Figure 3A, B). Therefore, we hypothesized expression of another anti-apoptotic protein was being induced to compensate for the loss of MCL-1. We performed gene expression studies in human and mouse MDSCs and observed high mRNA expression of the MCL-1-related factor A1 (Figure S2A–D). Like MCL-1, A1 is inhibited by the BH3-only protein NOXA and couples to the BAX-BAK mitochondrial intrinsic death pathway (Figure 3C). A1 gene expression is regulated by NF-κB, and the short half-life of A1 protein is controlled post-translationally (Lee et al., 1999; Wang et al., 1999). We therefore reasoned induction of A1 might sustain Mo-MS viability in the absence of MCL-1 and investigated what factors promoted A1 expression. GM-CSF was a strong inducer of A1 protein relative to TNF, IL-1α or IL-1β (Figure 3D) and rescued the viability of MCL-1ΔM Mo-MS (Figure 3E). Therefore, GM-CSF is a key factor sustaining A1 expression and regulates the viability of Mo-MDSCs in the absence of MCL-1.
Figure 3. A1 and MCL-1 are needed to maintain the survival of Mo-MS.
(A, B) BM-MS were grown from C57BL/6 or MCL-1ΔM mice. The percentage of cell death was measured by V405 staining of control or MCL-1ΔM Mo-MS at d6 of BM-MS culture (top) or after 24 h T cell suppression assay co-culture (bottom). (B) Quantification of V405+ cells from the experiments representative in (A). Data expressed as the mean ± the s.d. and are representative of two independent experiments. Statistical analysis with unpaired t tests was performed; *p ≤ .05, **p≤ .005. (C) Diagram showing the role of A1 and MCL-1 in inhibiting the mitochondrial death pathway. (D) Lysates from C57BL/6 MS cultures were subjected to immunoblotting for A1 following 24 h stimulation with the cytokines shown, all at 50 ng/mL. GRB2 (~ 26 kDa) was used as the loading control. (E) The percentage of non-viable V405+ Mo-MS after stimulation with cytokines (50 ng/mL). Data expressed as the mean ± s.d. and are representative of no less than 3 independent experiments (n = 2 samples for each experiment). Statistical analysis with unpaired t tests was performed; ****p≤ 0.0001. (F) The percentage of non-viable V405+ Mo-MS was evaluated by V405 staining after 24 h stimulation with the indicated cytokines (ng/mL) (n = 3 independent experiments). Quantification of data, expressed as the mean ± s.d. from one independent experiment. Statistical analysis with unpaired t tests was performed; **p ≤ 0.005. (G) Suppressive function of MS was measured using 5 × 105 CFSE-labeled OT-I cells co-cultured with titrated MS in the presence of SIINFEKL peptide. CFSE dilution was evaluated by flow cytometry after 72 h. Data are compiled from 3 independent experiments, and are presented as mean ± s.d.
The absence of MCL-1 primes Mo-MS for death
Our data raised the possibility that the viability of MCL-1ΔM Mo-MS might be compromised when A1 inducing factors were limiting. Therefore we determined if sustained A1 expression was required to maintain the suppressive function of Mo-MS in the absence of MCL-1. When we sorted MCL-1ΔM or control C57BL/6 Mo-MS from BM cultures we found MCL-1ΔM Mo-MS were more sensitive to death in the absence of cytokines compared to control Mo-MS (Figure 3F, S2E). Viability was rescued only in the presence of GM-CSF, presumably due to induction of A1 (Figure 3D, S2E, F). When MCL-1ΔM Mo-MS were used in T cell suppression assays we noted a striking reduction in their ability to block CD8+ proliferation compared to control Mo-MS (Figure 3G) or the unsorted MCL-1ΔM Mo-MS cultures (Figure 1). The mechanism involved rapid loss of viability of MCL-1ΔM Mo-MS during T cell suppression assay (Figure 3A) causing an apparent decrease in suppressive function. Therefore, MCL-1ΔM Mo-MS require continuous exposure to cytokines to sustain A1 expression. These data also provide an explanation for the survival of monocyte-lineage cells in the MCL-1ΔM mice, where high circulating amounts of GM-CSF are likely enforcing A1 expression (Steimer et al., 2009).
In control Mo-MS cells ‘constitutive’ MCL-1 expression should be ‘dominant’ to the effects of cytokine regulated A1. In other words, the absence of A1 alone should not affect Mo-MS viability. To test this we used a Vav-regulated shRNA system to simultaneously deplete all three murine A1 isoforms in mouse MDSC cultures (Ottina et al., 2012). As expected, A1-deficient Mo-MS retained their suppressive effects arguing A1 is required for sustaining the viability of MO-MDSCs but not for their suppressive function (Figure S2G–H).
Requirement for c-FLIP in Mo-MS cannot be bypassed
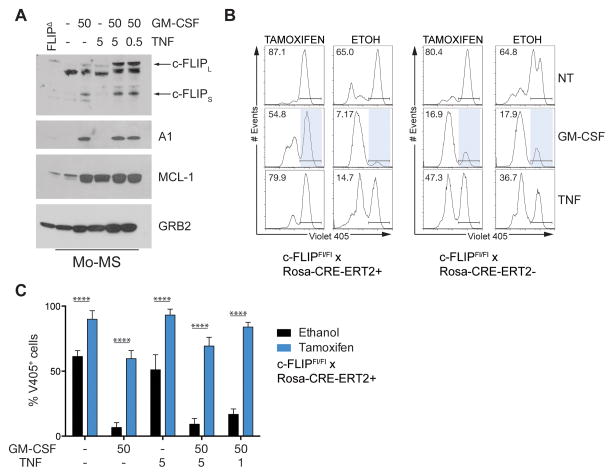
A prediction from the experiments described above is Mo-MS survive because they express sufficient c-FLIP and require either MCL-1 or exogenous stimulation (GM-CSF) to induce A1. What is the factor inducing c-FLIP? At multiple stages during development, DR signaling induces c-FLIP as a regulatory mechanism to suppress Caspase-8 and RIPK3 death. A candidate DR ligand regulating Mo-MS survival is TNF (Hu et al., 2013; Sade-Feldman et al., 2013; Zhao et al., 2012). We therefore asked if TNF induced the expression of c-FLIP in Mo-MS. TNF and GM-CSF-treated Mo-MDSCs had increased expression of c-FLIP and maintained A1 amounts similar to GM-CSF-treated cells (Figure 4A). TNF + GM-CSF treated PMN-MDSCs expressed c-FLIP and MCL-1, and did not express A1 (Figure S3).
Figure 4. Exogenous regulation of A1 partially rescues c-FLIP loss.
(A) BM-MS were grown from C57BL/6 mice. Mo-MS cell lysates were subjected to immunoblotting for the indicated targets following 24 h stimulation with the cytokines shown (ng/mL). GRB2 (~ 26 kDa) was used as the loading control. Protein lysates from FLIPΔM MS served as a specificity control for c-FLIP expression. (B–C) BM-MS cultures from Cflarfl/fl Rosa-Cre-ERT2+ or Cflarfl/fl Rosa-Cre-ERT2- mice were treated with tamoxifen or ethanol as a control on D5 of BM culture. (B) On d6 Mo-MS were sorted and cultured with cytokines for 24 h: GM-CSF (50 ng/mL), TNF (5 ng/mL). Viability was measured by V405 staining. Representative histograms are gated on Ly6C+ cells. (C) Mo-MS were sorted and cultured with the indicated cytokines for 24h. The percentage of non-viable V405+ cells in Mo-MS cultures. Data are expressed as the mean ± s.d. from one independent experiment and are representative of 2 independent experiments (2 mice per group). Statistical analysis with unpaired t tests was performed; ****p≤ 0.0001.
Given TNF + GM-CSF treatment induced both A1 and c-FLIP in Mo-MS, we next sought to determine if A1 in the absence of c-FLIP could maintain Mo-MDSC viability in response to GM-CSF. To test this, we again used tamoxifen-mediated Cre-deletion of c-FLIP in MS cultures using Cflarfl/fl; Rosa-CRE-ERT2 mice (Figure 4B, C). In the absence of c-FLIP the viability of Mo-MS could not be restored by GM-CSF (Figure 4B, C). Thus, in Mo-MS, A1 induction by GM-CSF to inhibit the intrinsic death pathway cannot bypass the absolute requirement for c-FLIP to inhibit the extrinsic Caspase-8 death pathway. Together these data demonstrate c-FLIP is the dominant survival factor for Mo-MS development. However, development of Mo-MS also requires sufficient expression of either MCL-1 or A1 to ensure their survival (Figure 4).
Death signaling and immunosuppressive function of MDSCs in cancer
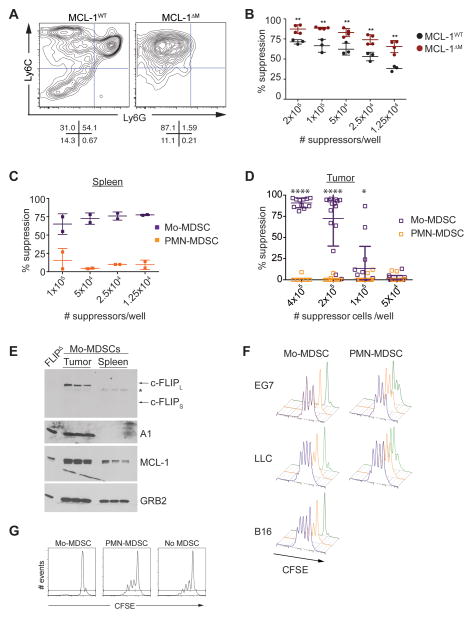
We next sought to determine if the death pathways controlling in vitro-generated MS similarly regulated the survival of MDSCs in malignancy. MDSCs accumulate in spleen, blood and local site of tissue damage as inflammation increases (Gabrilovich and Nagaraj, 2009; Gabrilovich et al., 2012; Ugel et al., 2012). Several studies have demonstrated MDSCs have different functional properties based on the tissue site they are recovered from, and it is now clear different signals control MDSC development or expansion and acquisition of immunosuppressive function. However, an unexplored mechanism to account for the role that different organ sites play in MDSC function is the acquisition of survival signaling. We therefore tested how the anatomical site influences the suppressive function and expression of death signaling inhibitors in MDSCs during tumor growth. Bulk populations of splenic MDSC from control or MCL-1ΔM mice bearing EG7 tumors were enriched for Mo-MDSC and had stronger suppressive activity relative to controls (Figure 5A, B). When the suppressive activity of MDSC subsets isolated from the spleens (Figure 5C) or tumors (Figure 5D) of EG7 tumor bearing C57BL/6 mice was evaluated we found suppressive activity was principally found in the Mo-MDSC fraction (Figure 5C–D). Expression of death inhibitors in Mo-MDSC isolated from spleens and tumors was evaluated by immunoblotting. MCL-1 was expressed in both spleen and tumor resident MDSCs, while c-FLIP and A1 expression were predominantly found in Mo-MDSCs isolated from the tumor site (Figure 5E). Therefore, Mo-MDSC expressed the highest amount of anti-death molecules at the tumor site, and constitutive expression of c-FLIP and A1 distinguishes suppressive tumor resident Mo-MDSC from their splenic counterparts (Figure 5C–E).
Figure 5. Suppressive function and enhanced expression of anti-apoptotics distinguishes Mo-MDSC at the tumor site.
(A) MDSC populations in control and MCL-1ΔM mice bearing EG7 tumors. Contour plots show spleen CD11b+-gated MDSCs. Data are representative of 3 independent experiments (n= 2 mice per group). (B) The suppressive activity of spleen MDSC populations isolated from EG7 tumor-bearing mice. Data are representative of 3 independent experiments. Percent suppression is calculated as described in the methods. Statistical analysis with unpaired t tests was performed; **p ≤ .01. (C–D) MDSCs were isolated from the spleen (C) or tumor (D) of EG7 tumor bearing C57BL/6 mice. Suppressive activity of Mo and PMN-MDSCs was evaluated by CFSE dilution in CD8+ OT-I T cells in the presence of SIINFEKL and titrated MDSCs. (D) Statistical analysis with unpaired t tests was performed; *p <0.01, ****p<0.0001. (E) Protein expression in freshly isolated spleen and tumor Mo-MDSCs from EG7-bearing mice. * indicates a band reactive to anti-c-FLIP antibodies not present in the FLIPΔM negative control that may represent an alternative isoform or processed c-FLIP product. (F) Suppressive function of tumor resident MDSCs using 5 × 105 CFSE-labeled OT-I cells with 4 × 105 (green), 2 × 105 (orange), 1 × 105 (purple) MDSCs with SIINFKEL peptide (gated on CD8+ cells). Cascade plots from 2 experiments are shown (n = 5–8 for each experiment). (G) The ability of tumor resident Mo and PMN-MDSCs to inhibit polyclonal T cell proliferation was evaluated by monitoring CFSE dilution. Representative data from 3 independent experiments are shown (gated on CD8+ cells).
Our data using the EG7 model demonstrates Mo-MDSC are the principle immunosuppressive MDSC population during tumor growth. However, MDSC expansion and population structure can be affected by the source of the primary tumor (Gabrilovich et al., 2012). Therefore, we next asked if Mo-MDSC were the dominant immunosuppressive population of MDSCs in other transplantable tumor models. Mice bearing Lewis lung carcinoma (LLC) or B16 melanoma (B16) tumors had expanded populations of Mo- and PMN-MDSC in the spleen and tumor (Figure S4A, B). Mo-MDSCs isolated from EG7, LLC and B16 tumors strongly inhibited T cell proliferation whereas PMN-MDSCs isolated from EG7 or LLC tumors failed to inhibit T cell proliferation (Figure 5D, F, S4C). Evaluation of PMN-MDSC from B16 tumors was not possible given the low number of PMN-MDSCs infiltrating these tumors (Figure S4B). Further, tumor resident Mo-MDSC but not PMN-MDSC inhibited polyclonal T cell activation in vitro (Figure 5G). Taken together these data identify Mo-MDSC as the principle immunosuppressive population of MDSCs during cancer.
Outcome of a genetic model of cancer devoid of PMN-MDSCs
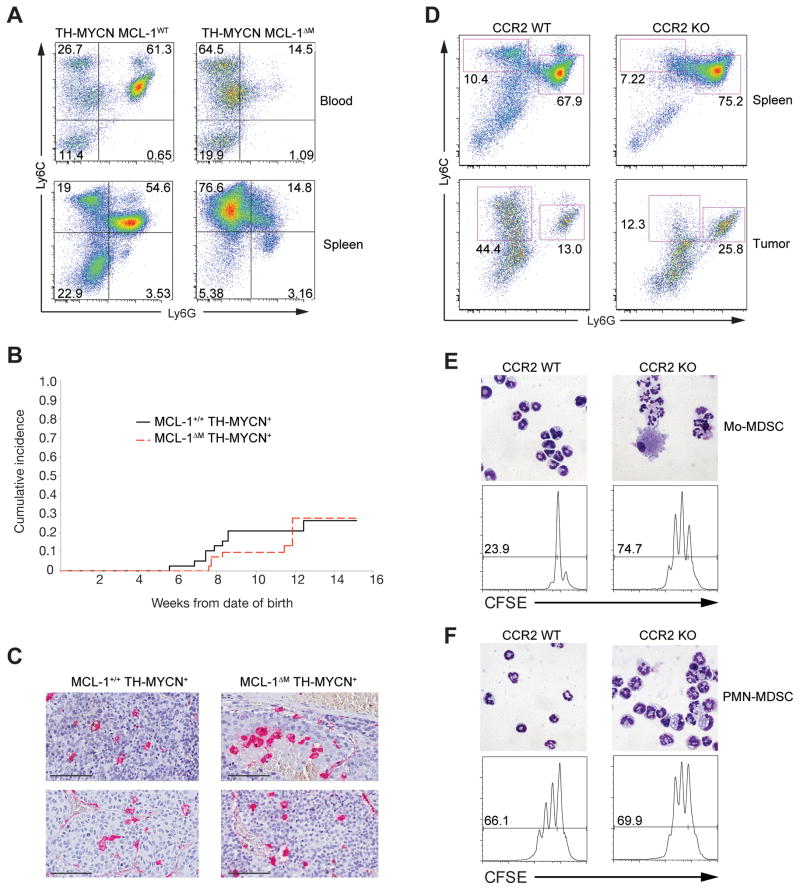
We reasoned that ablation of the non-suppressive PMN-MDSC subset using the MCL-1ΔM background would have minimal affects on tumor growth. To test this, we used the TH-MYCN genetic model of neuroblastoma where elevated numbers of MDSCs accumulate in the blood and spleen (Weiss et al., 1997). We created MCL-1ΔM; TH-MYCN mice and screened for tumors in the kidney-adrenal region by ultrasound imaging (Teitz et al., 2011). Neuroblastoma+ mice on the MCL-1ΔM background had lowered numbers of PMN-MDSCs, and a relative enrichment of Mo-MDSCs in both the blood and spleen (Figure 6A). The absence of PMN-MDSCs and other granulocytes on the MCL-1ΔM background did not affect tumor incidence, pathological appearance (Figure 6B), and tumors had intra-tumoral macrophage numbers equivalent to TH-MYCN+ mice of the same genetic background (Figure 6C). The growth rate of tumors was not significant between genotypes. As noted above MCL-1ΔM mice have high circulating amounts of GM-CSF, which by virtue of A1 regulation, may account for the apparent normal numbers of monocytes and their progeny (Steimer et al., 2009). To further examine the functional role of PMN-MDSCs we used Ccr2−/− mice. In these animals the recruitment of monocytes to inflammatory sites is severely impaired (Ginhoux and Jung, 2014). MDSCs isolated from spleens and tumors of CCR2-deficient mice were enriched for PMN-MDSC (Figure 6D–F). While Ccr2−/− mice had a small population of Ly6G- Ly6C+ cells, they were predominately granulocytic and were unable to inhibit T cell proliferation (Figure 6E–F). Only Mo-MDSC isolated from WT mice contained a functionally suppressive population of Mo-MDSC (Figure 6E–F). Collectively, these data demonstrate that Mo-MDSCs are the main immunosuppressive MDSC population within the tumor microenvironment.
Figure 6. PMN-MDSCs do not influence tumor incidence in vivo and are not immunosuppresive.
(A) Representative flow plots of MDSC populations in neuroblastoma+ MCL-1ΔM mice. Contour plots from blood (top) or spleen (bottom) gated on CD11b+ cells. (B) Cumulative tumor incidence measured by ultrasound imaging in MCL-1ΔM (n = 43) or control mice (n = 38) (tumor incidence is ~20–30 % in TH-MYCN+ mice). All mice were generated from Mcl1fl/+; LysM-Cre+; TH-MCYN+ intercrossed housed in the same rack. All mice were screened beginning at 7–8 weeks after birth and screened for 7 consecutive weeks. Tumor incidence between the strains shows no differences (p = 0.1139, Wilcoxon rank sum test). (C) Macrophage density inside neuroblastomas. Paraffin sections were stained with anti-Mac2 (red) to visualize macrophages. (D–F) EG7 tumors were implanted in Ccr2−/− or WT control mice. (D) Phenotypic analysis of MDSC populations in the spleen (top) and tumor (bottom). Plots are gated on CD11b+ cells. (E–F). Mo and PMN-MDSCs were isolated from tumor tissue as shown in (D). Cytospins of sorted MDSC fractions (top panels) and CFSE dilution in CD8+ OT-I cells (bottom panels) cultured in the presence or absence of 2 × 105 Mo and PMN-MDSCs
Discussion
Our data establish the underlying hierarchy of survival signaling required for myeloid suppressors: Mo-MS require either MCL-1 or A1 and have an absolute requirement for c-FLIP to inhibit caspase-8 mediated apoptosis. By contrast, PMN-MS require MCL-1, and probably A1 (Ottina et al., 2012), but are completely independent of death regulation by c-FLIP.
Our results argue viability of MS influences suppression assays. For example, TNF was linked with increased suppressive capacity of MDSCs (Sade-Feldman et al., 2013; Zhao et al., 2012). In our hands TNF induces c-FLIP expression so long as the suppression of the intrinsic death pathway is simultaneously engaged by MCL-1 or A1, providing a mechanistic explanation for suppression: we predict sufficient c-FLIP expression will prolong the viability of Mo-MS or Mo-MDSCs in culture, allowing a longer ‘window’ of suppression within T cell co-cultures. The same concept can be extended from our in vivo findings tracking c-FLIP, A1 and MCL-1 expression in tumor-bearing mice where both c-FLIP and A1 are increased at the tumor site, likely extending their viability in the tumor microenvironment where necrosis, hypoxia and cell debris are present. Therefore, our data have implications in cancer if the objective is to reduce the number and activity of myeloid suppressors: either c-FLIP or A1/MCL-1 must be targeted to weaken the most suppressive cells.
The role of c-FLIP expression in the monocytic compartment requires further evaluation because it remains unclear when and how c-FLIP is induced to counteract DR-death inducing signaling in the BM. One model is tonic signaling from TNFRs in the BM induces sufficient c-FLIP to protect from the effects of FAS and allow maturation of monocytic cells. However, Mo-MDSCs in the inflammatory site are associated with increased c-FLIP expression, resulting in increasing resistance to DR signaling from excessive TNF or possibly TRAIL, and results in enhanced viability (Condamine et al., 2014). However, as the lifespan of monocytic cells in inflammatory sites is limited, and monocytic cells continuously re-seed unresolved inflammatory sites, Mo-MDSCs are likely subject to a multitude of signals that exogenously regulate c-FLIP and A1, limiting or extending their lifespan accordingly.
Our experiments provide guidance on controversial aspects of MDSC subsets and their immunosuppressive activity. Monocytic lineage cells are the dominant immunosuppressive population of MDSCs. Furthermore, these data provide rational for using our genetic platform approach to revisit unresolved aspects of MDSC function. For example, the role of the PMN-MDSCs, which are produced in some tumor models in excess of monocytic MDSCs, are poorly understood (Youn et al., 2012). Part of this confusion may arise from the heterogeneity of PMN-MDSC studied. In cancer patients, sorted PMN-MDSCs as defined by CD115+, DRlow cells commonly contained monocytic cells (Vasquez-Dunddel et al., 2013). In our hands, PMN-MS and PMN-MDSCs have no suppressive capacity toward T cells and this data is supported by showing loss of granulocytes did not influence the outcome of neuroblastoma development. We note other studies have documented T cell suppressive effects of PMN-MDSCs (Movahedi et al., 2008; Schouppe et al., 2013; Youn et al., 2012), along with anti-tumor effects caused by depleting granulocytic cells with anti-Gr-1 or anti-Bv8 antibodies (Kowanetz et al., 2010; Srivastava et al., 2012). However, our genetic models offer the advantage of separating the Mo- and PMN- MDSC lineages in a more thorough manner than previously possible, and when coupled with antibody depletion studies, a more precise picture of the contribution of myeloid subtypes to cancer can be gained. In addition, issues of antibody specificity towards a particular MDSC can be established with greater certainty, given many surface molecules of myeloid cells, like Gr-1, are expressed on other cells of the immune system. This point is further illustrated by our data showing Ly6G- Ly6C+ granulocytes contaminate Mo-MDSC sorting gates due to overlapping surface marker expression. PMN-MDSC may also have functions beyond T cell suppression (Kowanetz et al., 2010; Ryan et al., 2013). One alternative explanation is PMN-MDSCs are produced as a consequence of deregulated hematopoiesis and are ‘bystanders’. Collectively, the role of PMN-MDSCs in cancer and other forms of non-resolving inflammation remains unclear but based on our findings can be elucidated using the genetic platforms we have established.
The segregated requirement for the activity of c-FLIP in Mo-MS and Mo-MDSCs but their granulocytic counterparts was unexpected because both cell types originate from the common myeloid progenitor (CMP). Our data suggest the requirement for c-FLIP is acquired at some point after the CMP because granulocytic cells are independent of c-FLIP. Therefore, a ‘branch point’ in myeloid lineage development exists where c-FLIP expression becomes important for one branch but not the other. This concept can be extended to interrogate the timing and strength of signal from DR receptors during myeloid development, since signaling from FAS, TRAIL or TNF receptors in the absence or low amounts of c-FLIP would induce death (Perlman et al., 1999). Therefore, one or more signals must induce c-FLIP in monocytic precursors to ensure survival, while further enforcement of c-FLIP expression, as we found in the tumor versus spleen Mo-MDSCs, would increase viability.
An additional implication of the requirement for c-FLIP in Mo-MDSCs and their monocytic precursors comes from in vivo use of chemotherapy drugs. Several chemotherapy agents cause a selective depletion of monocytic cells, including gemcitabine and trabectadin (Germano et al., 2013; Ugel et al., 2012). As the latter was linked with TRAIL-mediated death, it suggests that chemotherapy drugs might affect c-FLIP expression in monocytic cells directly or indirectly. When cells with low c-FLIP encounter a DR signal, their viability is compromised. Therefore, one way to weaken Mo-MDSCs would be to exogenously adjust the ability of cells to express c-FLIP to protective amounts, while exposing them to DR signaling. Our data provide new insights that will help direct development of novel treatments for non-resolving inflammation wherein the survival of immunosuppressive MDSCs can be modulated to eliminate them during tumor growth or enhance their survival for the treatment of autoimmune diseases.
Experimental Procedures
Mice
C57BL/6, Ccr2−/− and OT-I mice were obtained from The Jackson Laboratory (Bar Harbor, ME), Mcllfl/fl and Cflarfl/fl mice were bred to the LysM-Cre deleter (B6.129P2-Lyz2tm1(cre)Ifo/J) and the specific alleles used here were reported (Huang et al., 2010; Steimer et al., 2009). TH-MYCN mice were screened for tumors by weekly ultrasound imaging (Teitz et al., 2011; Weiss et al., 1997). Deletion of c-FLIP or MCL-1 was confirmed for all mice by flow cytometry of the blood (for CD11b and Ly6C/Ly6C) or by immunoblot. RIPK3 and FADD-deficient mice and their compound crosses have been described (Dillon et al., 2012; Weinlich et al., 2013). Mice bearing shRNAs against all A1 isoforms were published previously (Ottina et al., 2012). All mice in this study were used according to protocols approved by the Institutional Animal Care and Use Committee at St. Jude Children’s Research Hospital.
Flow cytometry
Single cell suspensions were blocked with normal mouse serum (1:10) for 5 minutes at room temperature and were then stained with antibodies at 1:100 dilutions. Pacific blue anti-mouse CD11b, APC anti-mouse Ly6C, APC anti-mouse CD8, and PE anti-mouse Ly6G were purchased from BioLegend. Viability was assessed using a LIVE/DEAD fixable dead cell stain kit (Invitrogen). V405 staining was performed according to the manufacturer’s protocol and analyzed on APC+ Mo-MDSC populations. Flow analysis was performed on a FACS Canto II and data were analyzed using FlowJo software. Sorting was performed using an iCyte Reflection system.
OT-I T cell suppression assays
Lymph node cells from OT-I transgenic mice were labeled with 5 μM CFSE, washed and added to all wells at 5 × 105 cells per well. SIINFEKL peptide (1 μg/ml final concentration) was added to all culture wells except for negative control wells. MS or MDSCs from individual mice were re-suspended at 8 × 106 cells/ml and step two dilutions were made starting at a concentration of 4 × 105 cells per well. As a negative control 5×105 OT-I cells were cultured with RPMI only. As a positive control 5 × 105 OT-I cells were cultured with 1 μg/ml SIINFEKL peptide. Proliferation was measured after 72 h by flow cytometry. To evaluate suppression of proliferation, plots were gated on CD8+ cells and the percentage of cells that had diluted CFSE was evaluated using FlowJo. The percentage of proliferating cells was then used to calculate the percent suppression of proliferation as described below. In some experiments, MCL-1 and c-FLIPΔM cells were mixed at varying ratios to obtain a constant number (2 × 105) of input MS cells per well. Percent suppression of proliferation was calculated using the following formula: 1 − (% proliferation with MDSCs / % proliferation without MDSCs) × 100. In some experiments, BrdU incorporation into CD8+ cells was used to measure proliferation. Staining for BrdU and flow analysis was performed with standard techniques.
Polyclonal T cell suppression assays
Lymph node cells from C57BL/6 mice were labeled with 5 μM CFSE, washed and added to all wells at 5 × 105 cells per well. Cells were stimulated with plate bound anti-CD3 (0.5 μg/ml) and soluble anti-CD28 (3 μg/ml) both purchased from ebioscience. Proliferation of CD8+ cells was evaluated by flow cytometry after 72hr. Plots are gated on CD8+ cells.
Tumor models
For EG7 tumor studies, 3 × 106 EG7 cells were injected sub-cutaneously in the rear flank of C57BL/6 or MCL-1ΔM mice. 1 × 106 LLC cells and 1.5 × 106 B16 cells were injected onto the flank of C57BL/6 mice. 12–14 days after tumor implantation mice were sacrificed and spleens and tumors were collected. MCL-1ΔM were crossed to a TH-MYCN+ background (Teitz et al., 2011; Weiss et al., 1997) and further backcrossed to a 129 background. Tumor incidence was measured by ultrasound imaging on mice treated with Nair to remove abdominal hair each week for 7 consecutive weeks. Tumors were scored positive at a minimum size of 10 mm3 and were removed from study if the tumor size equaled or exceeded 500 mm3.
Isolation of myeloid cells from tumor-bearing mice
CD11b+ tumor-associated myeloid cells were isolated from mouse tumors by mincing tumor tissue and placing it in dissociation media (1 mg/ml DNase, 2.5 mg/ml collagenase P, 2.5 mg/ml collagenase/dispase, 20 μl/ml B27, 10 μl/ml N2, 1mL neural basal medium) for 20 minutes at 37°C with gentle shaking for digestion. The cell/tissue suspension was then manually dissociated by re-suspending the cell/tissue pellet in a DNase solution (0.5 mg/ml DNase, 1 μl β-mercaptoethanol, 8 μl glucose 45%), pipetted up and down through a 10 mL pipette and passed through a 70 μM strainer. Cells were washed and collected at the 35% – 60% fraction of a Percoll gradient followed by CD11b+ MACS separation. CD11b+ cells were then stained with antibodies against Ly6G and Ly6C and further purified by cell sorting as described above. To purify spleen MDSCs, single cell suspensions were prepared from spleens of tumor bearing mice. Cells were blocked with 10% normal mouse serum in MACS buffer for 10 min on ice In some experiments MDSCs were purified by first performing a Ly6G+ cell depletion followed by enriching for Ly6C+ cells as described (Dolcetti et al., 2010). For other experiments MDSCs were isolated from spleens by CD11b+ MACS separation followed by staining with antibodies against Ly6G and Ly6C and further purification by cell sorting as described above.
Statistical analysis
Two-tailed Student’s t test was used for evaluating statistical significance between groups. Fisher’s exact test was used to estimate differences in tumor appearance between wild type mice and MCL-1ΔM mice on a TH-MYCN background. Wilcoxon Rank Sum was used for tumor volume analysis.
Supplementary Material
Acknowledgments
We thank Richard Pope (Northwestern University) for the gift of the c-FLIP knockout mice and Richard Cross and his team for cell sorting, and J. Wu and S. Mao for the statistical analysis of tumor formation in TH-MYCN MCL-1ΔM mice. This work was supported by NIH Cancer Center developmental funds (PJM and JTO), The Hartwell Foundation Individual Biomedical Research Award (PJM), Alex’s Lemonade Stand Foundation (PJM), NIH Cancer Center grant P30 CA21765, and the American Lebanese Syrian Associated Charities.
Footnotes
Author Contributions
JMH performed the majority of experiments. AMS and JMH established the T cell assay platforms and MDSC isolation and culture. RW, CPD and DRG contributed key reagents and ideas, and contributed to the writing. JEQ and BK did experiments. GN analyzed microarray data using samples purified by YK. MJH contributed key reagents. VB contributed methodologies essential to the overall project. JTO and PJM devised the overall strategy of using death pathways to dissect MDSCs. JMH and PJM wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Condamine T, Kumar V, Ramachandran IR, Youn JI, Celis E, Finnberg N, El-Deiry WS, Winograd R, Vonderheide RH, English NR, et al. ER stress regulates myeloid-derived suppressor cell fate through TRAIL-R-mediated apoptosis. J Clin Invest. 2014;124:2626–2639. doi: 10.1172/JCI74056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon CP, Oberst A, Weinlich R, Janke LJ, Kang TB, Ben-Moshe T, Mak TW, Wallach D, Green DR. Survival function of the FADD-CASPASE-8-cFLIP(L) complex. Cell Rep. 2012;1:401–407. doi: 10.1016/j.celrep.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolcetti L, Peranzoni E, Bronte V. Measurement of myeloid cell immune suppressive activity. Curr Protoc Immunol. 2010;Chapter 14(Unit 14–17) doi: 10.1002/0471142735.im1417s91. [DOI] [PubMed] [Google Scholar]
- Dzhagalov I, St John A, He YW. The antiapoptotic protein Mcl-1 is essential for the survival of neutrophils but not macrophages. Blood. 2007;109:1620–1626. doi: 10.1182/blood-2006-03-013771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI, Bronte V, Chen SH, Colombo MP, Ochoa A, Ostrand-Rosenberg S, Schreiber H. The terminology issue for myeloid-derived suppressor cells. Cancer Res. 2007;67:425. doi: 10.1158/0008-5472.CAN-06-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon J, Angell HK, Bedognetti D, Marincola FM. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity. 2013;39:11–26. doi: 10.1016/j.immuni.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Germano G, Frapolli R, Belgiovine C, Anselmo A, Pesce S, Liguori M, Erba E, Uboldi S, Zucchetti M, Pasqualini F, et al. Role of macrophage targeting in the antitumor activity of trabectedin. Cancer Cell. 2013;23:249–262. doi: 10.1016/j.ccr.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol. 2014;14:392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- Gordy C, Pua H, Sempowski GD, He YW. Regulation of steady-state neutrophil homeostasis by macrophages. Blood. 2011;117:618–629. doi: 10.1182/blood-2010-01-265959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Oberst A, Dillon CP, Weinlich R, Salvesen GS. RIPK-dependent necrosis and its regulation by caspases: a mystery in five acts. Mol Cell. 2011;44:9–16. doi: 10.1016/j.molcel.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp JM, Crist SA, Elzey BD, Cimen C, Ratliff TL. In vivo suppressive function of myeloid-derived suppressor cells is limited to the inflammatory site. Eur J Immunol. 2011;41:749–759. doi: 10.1002/eji.201041069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highfill SL, Rodriguez PC, Zhou Q, Goetz CA, Koehn BH, Veenstra R, Taylor PA, Panoskaltsis-Mortari A, Serody JS, Munn DH, et al. Bone marrow myeloid-derived suppressor cells (MDSCs) inhibit graft-versus-host disease (GVHD) via an arginase-1-dependent mechanism that is up-regulated by interleukin-13. Blood. 2010;116:5738–5747. doi: 10.1182/blood-2010-06-287839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Li B, Li X, Zhao X, Wan L, Lin G, Yu M, Wang J, Jiang X, Feng W, et al. Transmembrane TNF-alpha Promotes Suppressive Activities of Myeloid-Derived Suppressor Cells via TNFR2. J Immunol. 2013;192:1320–1331. doi: 10.4049/jimmunol.1203195. [DOI] [PubMed] [Google Scholar]
- Huang QQ, Perlman H, Huang Z, Birkett R, Kan L, Agrawal H, Misharin A, Gurbuxani S, Crispino JD, Pope RM. FLIP: a novel regulator of macrophage differentiation and granulocyte homeostasis. Blood. 2010;116:4968–4977. doi: 10.1182/blood-2009-11-252841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowanetz M, Wu X, Lee J, Tan M, Hagenbeek T, Qu X, Yu L, Ross J, Korsisaari N, Cao T, et al. Granulocyte-colony stimulating factor promotes lung metastasis through mobilization of Ly6G+Ly6C+ granulocytes. Proc Natl Acad Sci U S A. 2010;107:21248–21255. doi: 10.1073/pnas.1015855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Dadgostar H, Cheng Q, Shu J, Cheng G. NF-kappaB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc Natl Acad Sci U S A. 1999;96:9136–9141. doi: 10.1073/pnas.96.16.9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz MG, Boettcher S. Emergency granulopoiesis. Nat Rev Immunol. 2014;14:302–314. doi: 10.1038/nri3660. [DOI] [PubMed] [Google Scholar]
- Marigo I, Bosio E, Solito S, Mesa C, Fernandez A, Dolcetti L, Ugel S, Sonda N, Bicciato S, Falisi E, et al. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity. 2010;32:790–802. doi: 10.1016/j.immuni.2010.05.010. [DOI] [PubMed] [Google Scholar]
- McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014;16:717–727. doi: 10.1038/ncb3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motz GT, Coukos G. Deciphering and reversing tumor immune suppression. Immunity. 2013;39:61–73. doi: 10.1016/j.immuni.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movahedi K, Guilliams M, Van den Bossche J, Van den Bergh R, Gysemans C, Beschin A, De Baetselier P, Van Ginderachter JA. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood. 2008;111:4233–4244. doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- Nathan C, Ding A. Nonresolving inflammation. Cell. 2010;140:871–882. doi: 10.1016/j.cell.2010.02.029. [DOI] [PubMed] [Google Scholar]
- Newton K, Dugger DL, Wickliffe KE, Kapoor N, de Almagro MC, Vucic D, Komuves L, Ferrando RE, French DM, Webster J, et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science. 2014;343:1357–1360. doi: 10.1126/science.1249361. [DOI] [PubMed] [Google Scholar]
- Opferman JT, Iwasaki H, Ong CC, Suh H, Mizuno S, Akashi K, Korsmeyer SJ. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science. 2005;307:1101–1104. doi: 10.1126/science.1106114. [DOI] [PubMed] [Google Scholar]
- Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- Ottina E, Grespi F, Tischner D, Soratroi C, Geley S, Ploner A, Reichardt HM, Villunger A, Herold MJ. Targeting antiapoptotic A1/Bfl-1 by in vivo RNAi reveals multiple roles in leukocyte development in mice. Blood. 2012;119:6032–6042. doi: 10.1182/blood-2011-12-399089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peranzoni E, Zilio S, Marigo I, Dolcetti L, Zanovello P, Mandruzzato S, Bronte V. Myeloid-derived suppressor cell heterogeneity and subset definition. Curr Opin Immunol. 2010;22:238–244. doi: 10.1016/j.coi.2010.01.021. [DOI] [PubMed] [Google Scholar]
- Perlman H, Pagliari LJ, Georganas C, Mano T, Walsh K, Pope RM. FLICE-inhibitory protein expression during macrophage differentiation confers resistance to fas-mediated apoptosis. J Exp Med. 1999;190:1679–1688. doi: 10.1084/jem.190.11.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priceman SJ, Sung JL, Shaposhnik Z, Burton JB, Torres-Collado AX, Moughon DL, Johnson M, Lusis AJ, Cohen DA, Iruela-Arispe ML, et al. Targeting distinct tumor-infiltrating myeloid cells by inhibiting CSF-1 receptor: combating tumor evasion of antiangiogenic therapy. Blood. 2010;115:1461–1471. doi: 10.1182/blood-2009-08-237412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restifo NP, Dudley ME, Rosenberg SA. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol. 2012;12:269–281. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SO, Johnson JL, Cobb BA. Neutrophils confer T cell resistance to myeloid-derived suppressor cell-mediated suppression to promote chronic inflammation. J Immunol. 2013;190:5037–5047. doi: 10.4049/jimmunol.1203404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman M, Kanterman J, Ish-Shalom E, Elnekave M, Horwitz E, Baniyash M. Tumor Necrosis Factor-alpha Blocks Differentiation and Enhances Suppressive Activity of Immature Myeloid Cells during Chronic Inflammation. Immunity. 2013;38:541–554. doi: 10.1016/j.immuni.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Schouppe E, Mommer C, Movahedi K, Laoui D, Morias Y, Gysemans C, Luyckx A, De Baetselier P, Van Ginderachter JA. Tumor-induced myeloid-derived suppressor cell subsets exert either inhibitory or stimulatory effects on distinct CD8+ T-cell activation events. Eur J Immunol. 2013;43:2930–2942. doi: 10.1002/eji.201343349. [DOI] [PubMed] [Google Scholar]
- Srivastava MK, Zhu L, Harris-White M, Kar UK, Huang M, Johnson MF, Lee JM, Elashoff D, Strieter R, Dubinett S, et al. Myeloid suppressor cell depletion augments antitumor activity in lung cancer. PLoS One. 2012;7:e40677. doi: 10.1371/journal.pone.0040677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimer DA, Boyd K, Takeuchi O, Fisher JK, Zambetti GP, Opferman JT. Selective roles for antiapoptotic MCL-1 during granulocyte development and macrophage effector function. Blood. 2009;113:2805–2815. doi: 10.1182/blood-2008-05-159145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer. 2013;13:739–752. doi: 10.1038/nrc3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitz T, Stanke JJ, Federico S, Bradley CL, Brennan R, Zhang J, Johnson MD, Sedlacik J, Inoue M, Zhang ZM, et al. Preclinical models for neuroblastoma: establishing a baseline for treatment. PLoS One. 2011;6:e19133. doi: 10.1371/journal.pone.0019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugel S, Peranzoni E, Desantis G, Chioda M, Walter S, Weinschenk T, Ochando JC, Cabrelle A, Mandruzzato S, Bronte V. Immune tolerance to tumor antigens occurs in a specialized environment of the spleen. Cell Rep. 2012;2:628–639. doi: 10.1016/j.celrep.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Vasquez-Dunddel D, Pan F, Zeng Q, Gorbounov M, Albesiano E, Fu J, Blosser RL, Tam AJ, Bruno T, Zhang H, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest. 2013;123:1580–1589. doi: 10.1172/JCI60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Guttridge DC, Mayo MW, Baldwin AS., Jr NF-kappaB induces expression of the Bcl-2 homologue A1/Bfl-1 to preferentially suppress chemotherapy-induced apoptosis. Mol Cell Biol. 1999;19:5923–5929. doi: 10.1128/mcb.19.9.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bathina M, Lynch J, Koss B, Calabrese C, Frase S, Schuetz JD, Rehg JE, Opferman JT. Deletion of MCL-1 causes lethal cardiac failure and mitochondrial dysfunction. Genes Dev. 2013;27:1351–1364. doi: 10.1101/gad.215855.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Boger R, Vick B, Urbanik T, Haybaeck J, Zoller S, Teufel A, Krammer PH, Opferman JT, Galle PR, et al. Hepatocyte-specific deletion of the antiapoptotic protein myeloid cell leukemia-1 triggers proliferation and hepatocarcinogenesis in mice. Hepatology. 2010;51:1226–1236. doi: 10.1002/hep.23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinlich R, Oberst A, Dillon CP, Janke LJ, Milasta S, Lukens JR, Rodriguez DA, Gurung P, Savage C, Kanneganti TD, et al. Protective roles for caspase-8 and cFLIP in adult homeostasis. Cell Rep. 2013;5:340–348. doi: 10.1016/j.celrep.2013.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. The EMBO journal. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WC, Sun HW, Chen HT, Liang J, Yu XJ, Wu C, Wang Z, Zheng L. Circulating hematopoietic stem and progenitor cells are myeloid-biased in cancer patients. Proc Natl Acad Sci U S A. 2014;111:4221–4226. doi: 10.1073/pnas.1320753111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin B, Ma G, Yen CY, Zhou Z, Wang GX, Divino CM, Casares S, Chen SH, Yang WC, Pan PY. Myeloid-derived suppressor cells prevent type 1 diabetes in murine models. J Immunol. 2010;185:5828–5834. doi: 10.4049/jimmunol.0903636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JI, Collazo M, Shalova IN, Biswas SK, Gabrilovich DI. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. J Leukoc Biol. 2011;91:167–181. doi: 10.1189/jlb.0311177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JI, Collazo M, Shalova IN, Biswas SK, Gabrilovich DI. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. J Leukoc Biol. 2012;91:167–181. doi: 10.1189/jlb.0311177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Rong L, Li X, Liu X, Deng J, Wu H, Xu X, Erben U, Wu P, Syrbe U, et al. TNF signaling drives myeloid-derived suppressor cell accumulation. J Clin Invest. 2012;122:4094–4104. doi: 10.1172/JCI64115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.