Abstract
This paper describes the molecular and metabolic adaptations of Lactococcus lactis during the transition from a growing to a near-zero growth state by using carbon-limited retentostat cultivation. Transcriptomic analyses revealed that metabolic patterns shifted between lactic- and mixed-acid fermentations during retentostat cultivation, which appeared to be controlled at the level of transcription of the corresponding pyruvate dissipation-encoding genes. During retentostat cultivation, cells continued to consume several amino acids but also produced specific amino acids, which may derive from the conversion of glycolytic intermediates. We identify a novel motif containing CTGTCAG in the upstream regions of several genes related to amino acid conversion, which we propose to be the target site for CodY in L. lactis KF147. Finally, under extremely low carbon availability, carbon catabolite repression was progressively relieved and alternative catabolic functions were found to be highly expressed, which was confirmed by enhanced initial acidification rates on various sugars in cells obtained from near-zero-growth cultures. The present integrated transcriptome and metabolite (amino acids and previously reported fermentation end products) study provides molecular understanding of the adaptation of L. lactis to conditions supporting low growth rates and expands our earlier analysis of the quantitative physiology of this bacterium at near-zero growth rates toward gene regulation patterns involved in zero-growth adaptation.
INTRODUCTION
Fundamental knowledge of microbial physiology and cellular regulation is obtained mainly from studies of microorganisms in batch cultures. However, the pace of life and its associated physiological phases in batch cultivation differ strongly from what is found in natural environments (1). During the early phase of batch cultivations, all nutrients, including carbon and energy sources, are usually present in excess and the specific growth rate of the microorganism equals the maximum specific growth rate (2). Thereby, our understanding of microbial energy metabolism originates mostly from microbial population studies performed under laboratory conditions that include rapid growth, high metabolic activity, and high cell density. However, natural microbial communities generally live under relative famine conditions with low specific growth and metabolic rates due to a limited supply of nutrients and energy sources (3). Analogously, under specific industrial fermentation conditions, microorganisms may experience strongly restricted access to nutrients for longer periods of time. For example, lactic acid bacteria (LAB) experience long periods of extremely low nutrient availability during the maturing process of dry sausage (4) and cheese (5) production. Despite these harsh conditions, several LAB survive these processes during months of maturation and may continue to contribute to flavor and aroma formation in the product matrix (4, 6, 7).
Lactococcus lactis is used in food fermentation processes for several products, including cheese, sour cream, and other fermented milk products. In these processes, L. lactis converts the available carbon source into lactic acid, resulting in acidification of the food raw material. In addition, L. lactis is also commonly encountered in diverse natural environments, in particular, in decaying plant materials (8). The strain used in this study, L. lactis KF147, was isolated from mung bean sprouts, and its genome sequence reflects many adaptations to the plant-associated habitat, which are in particular apparent from the repertoire of enzymes and pathways predicted to be involved in the utilization of plant cell wall polysaccharides.
To study the physiological and genome-wide adaptations of microorganisms to near-zero growth rates, retentostat cultivation or recycling fermentor setups have been designed (9). Retentostat cultivation is an adaptation of chemostat cultivation in which a growth-limiting substrate is supplied at a fixed dilution rate while the complete biomass is retained in the bioreactor by removing the spent medium effluent through an external cross-flow filter. Prolonged retentostat cultivation leads to growth rates that approximate zero while the rate of energy transduction (through substrate consumption and conversion) equals the maintenance energy requirements (e.g., osmoregulation, turnover of damaged cellular components) (10, 11). Therefore, retentostat cultivation comprises a gradual transition from a growing to a near-zero growth state under stable environmental conditions, which sustain high cell viability.
Although retentostat cultivation has been performed to study the fundamental physiology of several microorganisms, including Escherichia coli (12), Bacillus polymyxa (13), Paracoccus denitrificans, Bacillus licheniformis (11), Nitrosomonas europaea, and Nitrobacter winogradskyi (14), at low growth rates, these studies were not consistently complemented with detailed molecular analyses. Exceptions are the retentostat studies performed with Lactobacillus plantarum (15), Aspergillus niger (16), and Saccharomyces cerevisiae (2, 17) that included analyses of the metabolic and transcriptome responses.
Previously, we have described how retentostat cultivation allows uncoupling of growth- and non-growth-related processes in L. lactis KF147, allowing investigation of the energy household and quantitative physiology of L. lactis at extremely low growth rates, exemplified by the estimated specific growth rate of 0.0001 h−1, which corresponds to a doubling time of more than 260 days (10). Despite the imposed extremely low growth rate, culture viability was sustained above 90% during prolonged retentostat cultivation (10). This study allowed the accurate calculation of maintenance energy requirements and other quantitative physiology parameters of the strain subjected to retentostat cultivation (10). Furthermore, the calculated maintenance-related substrate coefficient and biomass yield values revealed that the relative distribution of carbon source-derived energy toward maintenance-associated processes increased from approximately 30% to 99% during the transition from growing cultures to prolonged, near-zero-growth retentostat cultivation (10). These energy distribution measurements confirmed that the retentostat cultures had reached a typical near-zero growth state after approximately 14 days. Remarkably, the prolonged retentostat culture of L. lactis displayed significant shifts in central carbon metabolism (CCM), switching between mixed-acid fermentation and lactic acid fermentation (10).
In the present study, we complement our previous study with an in-depth molecular-level analysis of L. lactis KF147 under these retentostat cultivation conditions, including transcriptome, amino acid metabolism, and previously obtained fermentation metabolite (10) analyses. Thereby, this study deciphers the molecular adaptation underlying the previously reported physiological observations. The genome-wide transcriptome analyses were in remarkable agreement with the previously observed oscillation of the culture between lactic acid fermentation and mixed-acid fermentation at extremely low growth rates. Moreover, integrated metabolome and transcriptome analyses created a global view of the interconnected carbon and nitrogen metabolism adaptations under these near-zero growth conditions. Notably, these adaptations included the progressive relief of carbon catabolite repression (CCR), preparing the culture to effectively scavenge alternative carbon sources when they become available. Finally, the transcriptome adaptations are discussed in the context of regulatory circuits that are proposed to govern them, including a predicted role for the central carbon and nitrogen metabolism control proteins CcpA and CodY, respectively.
MATERIALS AND METHODS
Bacterial isolates, media, and cultivation conditions.
L. lactis subsp. lactis strain KF147 originates from mung bean sprouts, and its genome sequence has been determined (8). Precultures for retentostat cultivations (10) were inoculated in 50 ml M17 broth (18) complemented with 0.5% (wt/vol) glucose and grown overnight at 30°C. Overnight cultures were harvested by centrifugation (6,000 × g, 10 min., 4°C) and washed twice with a physiological salt solution (0.9% NaCl in water). Next, the culture was inoculated into a chemically defined medium containing 0.5% (wt/vol) glucose for chemostat cultivation. After a steady state had been achieved with six volume changes in the chemostat, the fermentors were switched to retentostat mode by withdrawing the effluent through the cross-filter, and the pH was controlled at 5.5 through automated 5 M NaOH titration in chemostat and retentostat cultivations (10). To keep the medium composition constant during long-term cultivation, 120-liter batches of medium were prepared, filter sterilized, and used during retentostat cultivations (10).
Two independent, carbon source-limited retentostat cultivations were performed under anaerobic conditions, initiating from chemostat cultivation at a dilution rate of 0.025 h−1 as previously described (10). The retentostat setup was assembled with a 1.5-liter fermentor (Applikon Biotechnology, Schiedam, The Netherlands) and an autoclavable polyethersulfone cross-flow filter (Spectrum Laboratories, CA, USA). As removal of samples could interrupt biomass accumulation, the sample volume and sampling frequency were minimized.
Biomass and amino acid determinations.
During fermentations, culture samples were withdrawn at regular intervals to measure cell dry weights (CDWs) and amino acid concentrations. For CDW determination, 5 ml of culture was passed through preweighed membrane filters with a pore size of 0.45 μm (Merck Millipore, Darmstadt, Germany) with a vacuum filtration unit (Sartorius Stedim Biotech, Göttingen, Germany). Subsequently, membrane filters were dried at 55°C for 24 h and the concentration of the biomass collected on the membranes was determined in g/ml.
The concentrations of amino acids in the culture supernatant and the medium feed were measured with the EZ:fast free amino acid analysis kit (KG0-7165; Phenomenex, CA, USA). According to the manufacturer's instructions, the analysis was performed with a gas chromatograph with a flame ionization detector (Thermo Scientific, MA, USA).
Acidification activity.
Acidification profiles were determined in triplicate in 200 μl of phosphate-buffered saline (PBS) (initial pH of 6) with a 0.5% (vol/vol) concentration of a chosen carbon source (glucose, ribose, mannitol, sucrose, fructose, and raffinose) at 30°C, inoculated with 5 × 108 CFU/ml of stock cultures of retentostat cultures collected at days 14, 21, 35, and 42, with 96-well microplates (HydroPlate HP96U; PreSens, Regensburg, Germany). These plates have an optical pH sensor at the bottom of each well that can be read out through the bottom of the plate with a fluorescence reader. According to the manufacturer's instructions, pH values of cell suspensions in PBS with the chosen carbon sources were measured every 10 min for 10 h with a microplate fluorescence reader (Safire II; Tecan, Grödig, Austria). The maximum acidification rate (Vmax), expressed in arbitrary and pH-based units (pH units min−1 [10−3]), was calculated on the basis of the slope of the pH-versus-time plot by using at least eight subsequent time points (regression coefficient, >0.99).
RNA isolation and transcriptome analysis.
Total L. lactis RNA was isolated from two independent retentostat cultures harvested at days 0, 2, 7, 14, 21, 28, 35, and 42. RNA extraction, reverse transcription, labeling, hybridization, and data analysis were done as previously described (19). Briefly, following methanol quenching, RNA was phenol-chloroform extracted and purified with the High Pure RNA isolation kit (Roche Diagnostics, Mannheim, Germany). The total RNA was used as the template to synthesize cDNA with the Superscript III reverse transcriptase enzyme (Invitrogen, Carlsbad, CA), followed by purification with the CyScribe GFX purification kit (GE Healthcare, Buckinghamshire, United Kingdom) and Cy3 or Cy5 labeling (Amersham; CyDye postlabeling reactive dye pack; GE Healthcare, Buckinghamshire, United Kingdom). L. lactis KF1471 cDNA was hybridized to OligoWiz (20) designed oligonucleotide DNA microarrays (Agilent Technologies, Santa Clara, CA). After washing (19), slides were scanned at several photomultiplier tube values, optimal scans were selected on the basis of signal distribution, and data were normalized by Lowess and interslide scaling normalization as available in MicroPrep (21). Median intensities of different probes per gene were taken as absolute gene expression intensities per gene for each condition. The microarray hybridization scheme for the transcriptome analyses at retentostat cultivations consisted of a compound loop design with 26 arrays (see Fig. S1 in the supplemental material).
The gene expression intensities were compared and clustered by Short Time-series Expression Miner (STEM, version 1.3.6; http://www.cs.cmu.edu/∼jernst/stem/) (22). The STEM clustering algorithm was used to identify enrichment of Gene Ontology (GO) terms, with Bonferroni correction to determine significance and a maximum of 50 model profiles. The expressions of genes involved in specific pathways (e.g., glycolysis, pyruvate dissipation, and amino acid metabolism) were projected in heat maps with the MultiExperiment Viewer (MeV) (http://www.tm4.org/mev.html) (23). The correlation of the transcriptome data at each time point for the two independent retentostat cultivations was calculated by Pearson correlation analysis and displayed as hierarchical clustering with the MeV tool.
DNA motif mining.
Gene expression data from the significant model profiles identified by STEM were used as a data source to identify transcription factor binding sites (TFBSs) in the genome of L. lactis KF147. Binding site searches were performed by using the 300-bp region upstream of each regulated gene, the algorithm for fitting of a mixture model by expectation maximization (24), and the parameters mod anr (unlimited number of motifs per upstream sequence) and revcomp (allowing motifs to be present on both plus and minus strands) and by allowing maximally three motifs to be found in each upstream region without restricting the total number of motifs. The PePPeR database was used as a source of literature-based regulon clusters (25).
Microarray data accession numbers.
The microarray data obtained in this study and the experimental procedures used to obtain them have been submitted to the NCBI Gene Expression Omnibus under accession numbers GPL17806 and GSE51494 (http://www.ncbi.nlm.nih.gov), respectively.
RESULTS
Transcriptome data analysis.
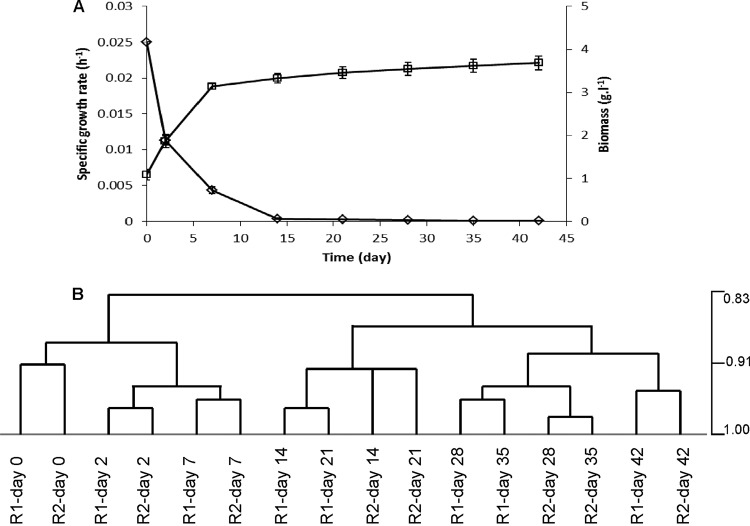
In our earlier study, the metabolic adaptations and physiology of L. lactis KF147 at extremely low growth rates were studied in anaerobic and carbon-limited retentostat cultivations (sustained for 42 days) (10). During retentostat cultivation, biomass accumulated, reaching a plateau level after approximately 14 days, and the growth rate ultimately declined from 0.025 h−1 to approximately 0.0001 h−1 after 42 days of retentostat cultivation (Fig. 1A) (10). To examine the time-resolved transcriptome adaptation of L. lactis KF147 to near-zero growth conditions, samples were taken before starting the retentostat cultivation regimen (t = 0 days; chemostat cultivation conditions at D = 0.025 h−1) and 2, 7, 14, 21, 28, 35, and 42 days after initiation of the retentostat regimen in biologically independent duplicate cultivations. Hierarchical clustering and Pearson correlation analysis illustrated that the transcriptome profiles taken from the two replicates displayed highly similar transcriptome evolutions over time (Pearson correlation coefficient, >0.92), supporting the high reproducibility of the experimental setup (Fig. 1B). In addition, cluster analysis clearly separated the transcriptome patterns of samples taken during the growing stages of the experiment (days 0, 2, and 7) from those where growth was stagnating to eventually reach near-zero growth conditions (days 14, 21, 28, 35, and 42) (Fig. 1B). The discrimination of these two main clusters underpins the separation of the transcriptome signatures related to growth-associated and near-zero-growth-associated processes.
FIG 1.
Growth of L. lactis KF147 and hierarchical clustering linkage of transcriptome profiles in retentostat culture. A steady-state anaerobic chemostat culture was switched to retentostat mode at time zero. (A) Data points represent average values ± standard deviations of measurements of two independent cultures. Shown are the specific growth rate (h−1) (diamonds) and biomass accumulation (g liter−1) (squares) of L. lactis KF147 under retentostat conditions (adapted from reference 10). (B) Hierarchical clustering linkage of retentostat 1 (R1) and 2 (R2) samples. Complete clustering linkage was performed for samples obtained on days 0, 2, 7, 14, 21, 28, 35, and 42 of duplicate retentostat cultivations based on Pearson correlation analysis using MeV.
To identify gene expression patterns during the course of the experiment, the absolute expression levels of all of the genes (2,533 genes in L. lactis KF147) were subjected to expression cluster analysis with the STEM module, which employs a process of statistical clustering of time series data sets into precomposed patterns of expression (22). STEM analysis divided the expression patterns into 50 time-resolved model expression profiles, which were sorted on the basis of the number of genes assigned to the profile. Of the annotated L. lactis KF147 genes in the retentostat duplicates, 66.9 and 64.4% were clustered by STEM into 8 and 11 statistically significant model profiles, respectively (see Fig. S2A and B in the supplemental material). Since the congruence between the transcriptome profiles obtained in the replicate experiments was very high and highly similar STEM profile distributions were obtained for the duplicate retentostat cultivations, the data presented here are those obtained from one of the retentostat cultures, which consistently displayed expression highly congruent with that in the duplicate retentostat cultivation. Since the expression patterns in model profiles 7, 8, 40, and 41 coincided with the carbon metabolism shifts that we described previously (10), we focused on the conserved gene sets in these model profiles (Table 1; see Fig. S2 and S4 in the supplemental material), analyzing the significantly (P ≤ 0.05) enriched GO.
TABLE 1.
GO enrichment analysisa
| Model profile no. | GO category |
|---|---|
| 7 | Membrane |
| Integral to membrane | |
| Membrane part | |
| 8 | Cellular amino acid metabolic process |
| Fatty acid metabolic process | |
| Organic acid metabolic process | |
| Carboxylic acid biosynthetic process | |
| Lipid metabolic process | |
| Primary metabolic process | |
| Glutamine family amino acid metabolic process | |
| Branched-chain family amino acid metabolic process | |
| Arginine metabolic process | |
| 40 | Sequence-specific DNA binding transcription factor activity |
| RNA biosynthetic process | |
| Regulation of RNA metabolic process | |
| Regulation of cellular biosynthetic process | |
| Regulation of gene expression | |
| Regulation of macromolecule metabolic process | |
| 41 | Regulation of transcription, DNA dependent |
| Carbohydrate kinase activity | |
| Regulation of RNA biosynthetic process | |
| Regulation of biological process |
Shown are the GO enrichment results for the set of genes shown in Fig. S3A and B in the supplemental material. Enrichment was computed on the basis of actual size enrichment (P ≤ 0.05).
Cell membrane biosynthesis-related process expression during retentostat cultivation.
In STEM-Model, profile 8 was enriched for a variety of metabolic processes (Table 1). Genes in this cluster were characterized by progressively reducing expression during the growth-associated stages of retentostat cultivation, followed by a period of stable low-level expression and subsequent increasing expression under prolonged near-zero growth conditions (see Fig. S2 and S3 in the supplemental material). This cluster also included the category “fatty acid metabolic process” containing the fabDFGHZ and accABCD genes associated with fatty acid biosynthesis (see Fig. S4A in the supplemental material), the expression of which also remained very low upon prolonged retentostat cultivation. This observation indicates that L. lactis KF147 adapted to near-zero growth by repression of genes related to fatty acid production, thus downregulating the synthesis of one of the main building blocks of the cell membrane. Model profile 7 clusters genes that were expressed at continuously declining levels during retentostat cultivation (see Fig. S2 and S3 in the supplemental material). In this profile, the overrepresented functional categories related to membrane-associated functions (Table 1). These clusters included many ATP-binding cassette (ABC) and phosphotransferase transport systems. In addition, cluster 7 contained genes involved in exopolysaccharide (EPS) synthesis (see Fig. S4B and C in the supplemental material), which is in agreement with the notion that LAB produce and secrete EPSs into their environment only during growth (26). Model profiles 40 and 41 appear to have similar patterns of expression, characterized by initially increasing expression during early stages of retentostat cultivation and remaining stably expressed or showing slightly lower levels of expression during prolonged retentostat cultivation (see Fig. S2 and S3 in the supplemental material). These model profiles were enriched for functional categories related to RNA synthesis and regulation, transcription, and translation (Table 1).
CCM expression during retentostat cultivation.
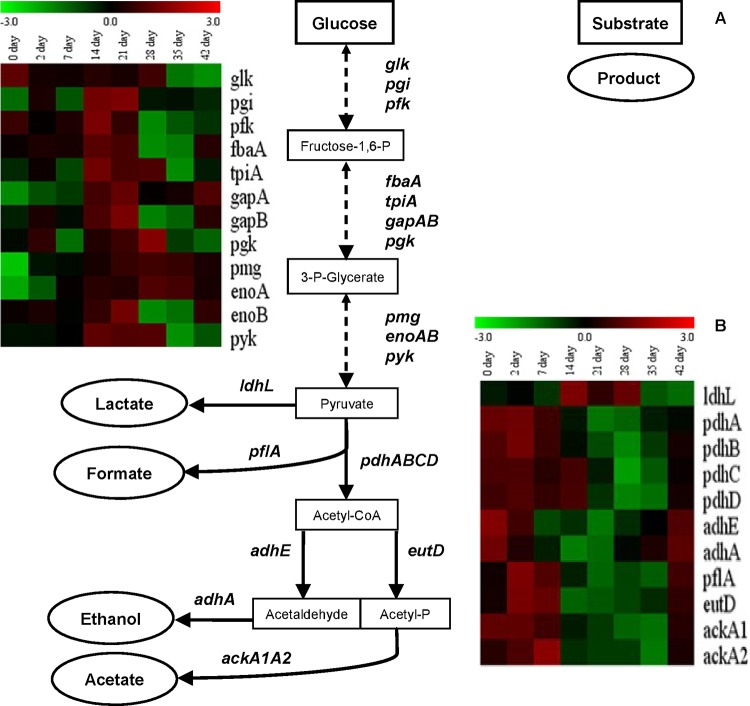
STEM-Model profile 8 was enriched for GO terms associated with “organic acid metabolic process” (Table 1), and the genes in this class displayed progressively lower expression levels during the growth-associated stages of retentostat cultivation (up to day 7), followed by a period of stable low-level expression and a recovery of high expression levels during prolonging retentostat conditions and near-zero growth rates (see Fig. S2 in the supplemental material). Thereby, the expression profiles of these genes followed the timing of the carbon and pyruvate dissipation metabolism fluctuations observed during retentostat fermentation (10), which is in good agreement with the “organic acid metabolic process” enrichment. The expression profiles of the genes associated with these processes were visualized by heat map representation during chemostat cultivation (day 0) and retentostat cultivation (Fig. 2). Pyruvate dissipation-associated genes displayed a remarkably consistent time-dependent transcription profile during retentostat cultivation. At days 0, 2, and 7, the lactate dehydrogenase-encoding gene (ldhL, involved in lactic acid production) was expressed at a low level, whereas the genes encoding pyruvate dehydrogenase (pdhABCD), alcohol dehydrogenase (adhAE), pyruvate formate lyase (pflA), phosphotransacetylase (eutD), and acetate kinase (ackA1A2), involved in mixed-acid fermentation (production of ethanol, formic acid, and acetate), were highly expressed (Fig. 2B). Subsequently, the transcript level of ldhL was elevated on days 14, 21, and 28, while those of pdhABCD, adhAE, pflA, and eutD, and ackA1A2 were decreased during these stages of cultivation. At the last stage of retentostat cultivation (days 35 and 42), the ldhL gene transcription was again repressed, which coincided with a recovery of the higher expression level of genes involved in mixed-acid fermentation (Fig. 2B). These findings indicate that the fluctuations in pyruvate dissipation behavior during retentostat cultivation (10) accurately reflect the transcriptome profiles of the genes involved in these catabolic pathways and could explain the mixed-acid and lactic acid fermentation switches observed (10).
FIG 2.
Overview of CCM in L. lactis KF147. (A) Simple scheme of glycolysis and pyruvate dissipation pathways. Each one-headed arrow represents one metabolic reaction, and each two-headed dashed arrow corresponds to more than one reaction. Genes are indicated beside the arrows. End products are indicated in ellipses. (B) Heat map of L. lactis KF147 glycolysis and pyruvate dissipation genes differentially expressed (on a log2 scale, P ≤ 0.05) during retentostat cultivation over the beginning of chemostat cultivation (day 0) (retentostat 1). Similar transcriptome results obtained in retentostat 2 confirmed the consistency of these results in an independent experiment.
Unlike the pyruvate dissipation-related genes, the genes of the glycolysis pathway (glk, pgi, pfk, fbaA, tpiA, gapAB, pgk, pmg, enoAB, and pyk) were inconsistently regulated in the first week of retentostat cultivation, where all glycolytic genes were repressed or did not change (i.e., <2-fold change), except the glk, pfk, and fbaA genes (Fig. 2B). However, during subsequent days of retentostat cultivation (days 14 and 21), the expression of glk, pgi, pfk, fbaA, tpiA, gapAB, pgk, pmg, enoAB, and pyk was consistently upregulated, whereas their expression was again suppressed during prolonged retentostat cultivation (days 28, 35, and 42), except for the gapA, pmg, and enoA genes (Fig. 2B). Moreover, the progressive reduction of the growth rate (Fig. 1A) (10) appears to be paralleled by reduced glycolytic gene expression, although at the time points that corresponded to increasing lactic acid production, the expression of the glycolytic genes was transiently increased. These findings imply that under retentostat cultivation conditions, the expression pattern of the majority of the glycolytic genes appears to follow the previously described metabolic fluctuations between mixed-acid fermentation and lactic acid fermentation (10) rather than the actual growth rates. The exceptions appear to be gapA, pmg, and enoA, which were continuously expressed at elevated levels after 14 days of retentostat cultivation. Intriguingly, the metabolites associated with these enzymes (substrates and products), also happen to be substrates for the interconversion to amino acid biosynthesis and thereby appear to be consistent with the amino acid production observed at certain stages of fermentation (see below).
Amino acid profiles and amino acid metabolism regulation during retentostat cultivation.
Amino acid concentrations were determined (Fig. 3; see Fig. S5 in the supplemental material) at each time point after the initiation of retentostat growth (day 0). The branched-chain amino acids (BCAAs) valine, leucine, and isoleucine were constantly consumed during the first week of retentostat cultivation, and then their consumption decreased gradually until days 21 and 28 of retentostat cultivation, whereas prolonged retentostat cultivation beyond 28 days led to a gradual increase in BCAA consumption toward the end of the cultivation period (Fig. 3A). Notably, the consumption of the aromatic amino acids (AAAs) phenylalanine, tyrosine, and tryptophan appeared to display a pattern similar to that of BCAA consumption (Fig. 3B).
FIG 3.
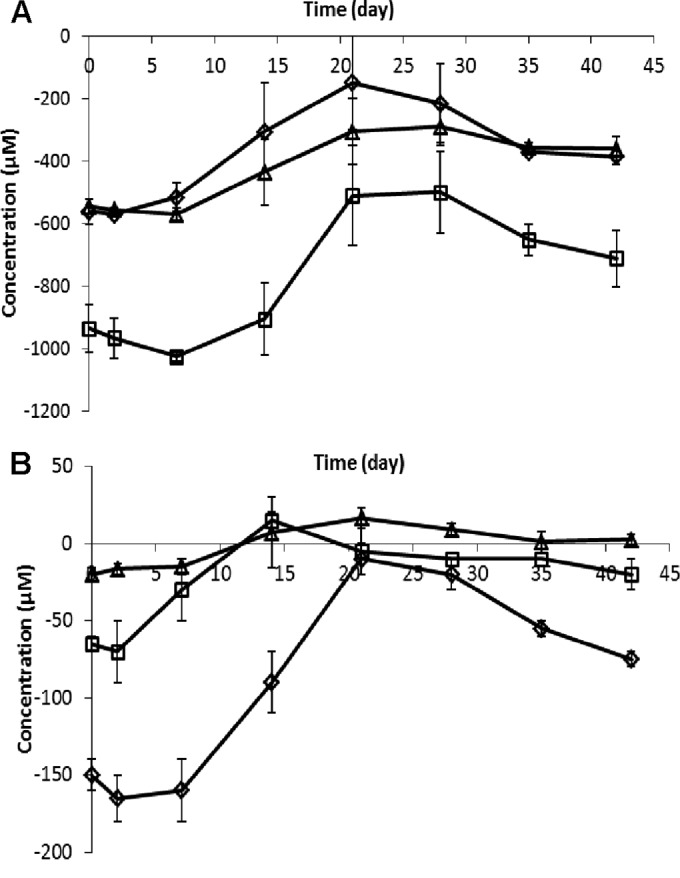
Concentration of BCAAs (A) and AAAs (B) in L. lactis KF147 in retentostat culture. Data points represent average values ± standard deviations of measurements of two independent cultures. (A) Concentrations of Val (diamonds), Leu (squares), and Ile (triangles). (B) Concentrations of Phe (diamonds), Tyr (squares), and Trp (triangles). Each concentration in panels A and B is presented as the difference between the measured concentration in the medium feed and the measured concentration in the filter line efflux sample. Negative values indicate net consumption; positive values indicate net production.
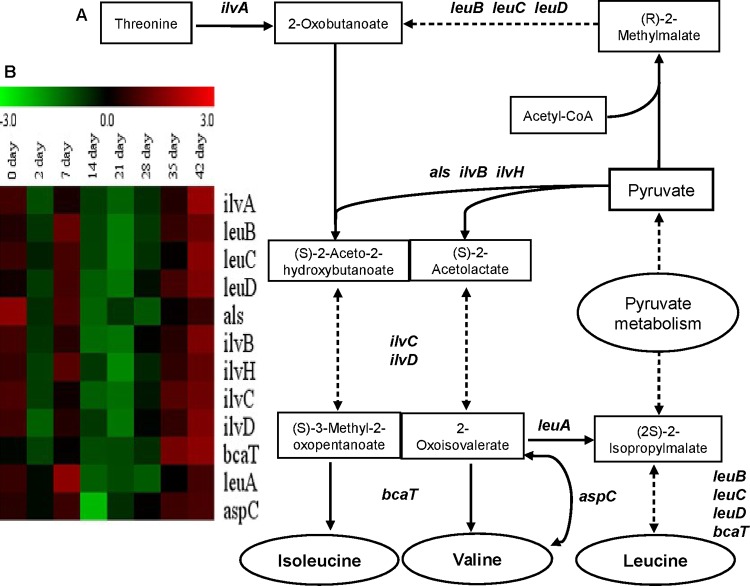
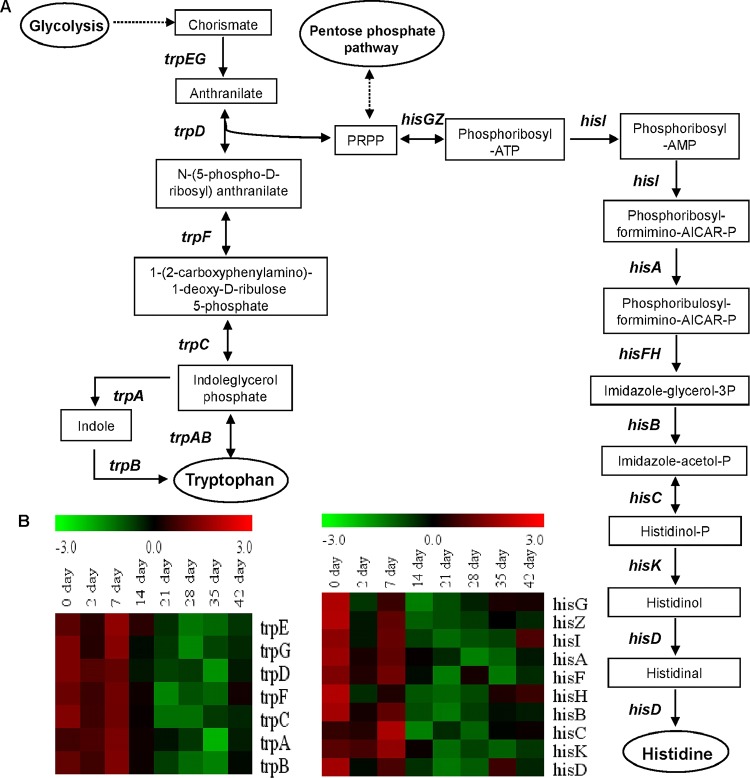
The GO terms “cellular amino acid metabolic process” and “branched-chain family amino acid metabolic process” were overrepresented in transcriptome model profile 8 in both retentostat cultivations (Table 1) and thereby appeared to reflect the pattern of amino acid consumption/production during retentostat cultivation. The ilvABCDH, leuABCD, als, bcaT, and aspC genes, which encode enzymes responsible for BCAAs biosynthesis, displayed similar transcription patterns during retentosat cultivation. Their expression fluctuated during the first week of retentostat cultivation but consistently decreased after 7 days and was followed by increased expression after 35 days of retentostat cultivation (Fig. 4). Notably, genes associated with tryptophan (trpCDFGS) and histidine (hisABDFGHIKZ) synthesis were also clustered in model profile 8 and displayed an expression pattern similar to that of the BCAA-associated genes (Fig. 5). These transcriptional profiles establish a relatively good correspondence between the consumption/production of these amino acids and their biosynthetic-pathway-encoding genes.
FIG 4.
Overview of BCAA biosynthesis in L. lactis KF147. (A) Simple scheme of Ile, Val, and Leu amino acid production pathways. Each one-headed arrow represents one metabolic reaction, and each two-headed dashed arrow corresponds to more than one reaction. Genes are indicated beside the arrows. End products are indicated in ellipses. (B) Heat map of L. lactis KF147 BCAA biosynthesis genes differentially expressed (on a log2 scale, P ≤ 0.05) during retentostat cultivation over the beginning of chemostat cultivation (day 0) (retentostat 1). Similar transcriptome results obtained in retentostat 2 confirmed the consistency of these results in an independent experiment.
FIG 5.
Overview of Trp (AAA) and His biosynthesis in L. lactis KF147. (A) Simple scheme of Trp and His amino acid production pathways. One arrow represents one metabolic reaction, and dashed-line arrows correspond to more than one reaction. Genes are indicated beside the arrows. End products are indicated in ellipses. (B) Heat map of L. lactis KF147 BCAA Trp and His biosynthesis genes differentially expressed (on a log2 scale, P ≤ 0.05) during retentostat cultivation over the beginning of chemostat cultivation (day 0) (retentostat 1). Similar transcriptome results obtained in retentostat 2 confirmed the consistency of these results in an independent experiment.
The other amino acids appeared to display concentration fluctuations similar to those of BCAAs and AAAs during retentostat cultivation (see Fig. S5 in the supplemental material). Notably, at certain time points (days 14 and 21), several amino acids appeared to be net produced by the culture. At these time points, the genes encoding glutamate synthase, a glycerol-3-phosphate transporter, and a glutamate ABC transporter (gltBDPS), as well as cysteine synthase (cysK), a lysine-specific permease (lysQ), and a homoserine kinase and threonine synthase (thrBC), were expressed at low levels, while their expression was higher at stages prior to and after the period of net production (see Fig. S6 in the supplemental material). Conversely, the diaminopimelate decarboxylase-encoding lysA gene displayed an expression profile that is the inverse of that of the related lysQ gene (see Fig. S6). Taken together, the results show that the production of glutamic acid, cysteine, threonine, and lysine coincides with the period that is characterized by enhanced lactic acid fermentation and could be controlled by the gltBDPS, cysK, thrBC, and lysAQ genes, respectively.
Identification of a cis-acting DNA motif potentially involved in near-zero growth gene regulation.
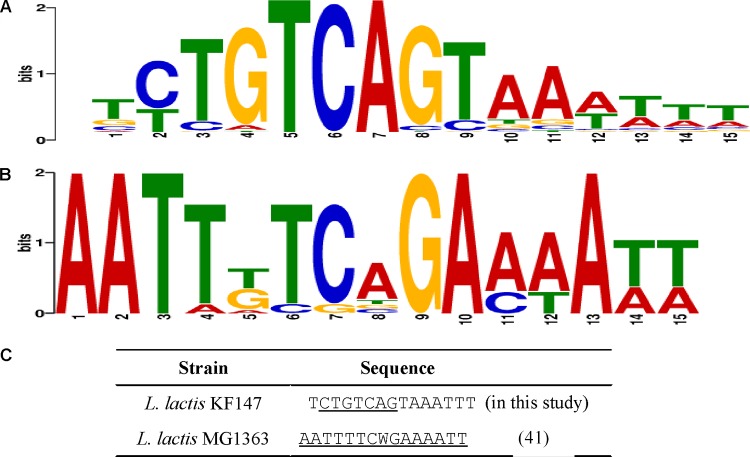
Transcriptional regulators strongly control the expression levels of genes by binding to TFBSs. To identify candidate DNA motifs that are potential TFBSs involved in adaptation to near-zero growth conditions, we searched for DNA sequences overrepresented in the upstream regions of genes that showed correlated expression. As a result, a highly conserved motif encompassing the palindromic sequence element 5′-CTGTCAG-3′ (Fig. 6A) was identified in profiles 7 and 8. The motif is present upstream of genes related to the synthesis of BCAAs (ilvAB, ileS, leuC), histidine (hisB), cysteine (cysK), arginine (argBF, arcD), serine (serS), tryptophan (trpG), and fatty acids (accD, fabZ) and genes related to peptide uptake (dtpT, pepC) (see Fig. S7 in the supplemental material), suggesting that it could play a key role in the adaptation to near-zero growth conditions by its role in the regulation of nitrogen metabolism in L. lactis KF147 (see Discussion).
FIG 6.
WebLogo visualization of the postulated CodY motif in L. lactis KF147 (A) and the experimentally identified CodY motif in L. lactis MG1363 (41) (B) and alignment of the CodY motifs of both strains. (A) The postulated CodY binding sequence found in L. lactis KF147. The CTGTCAG palindrome sequence that forms the core of the motif is positioned at nucleotides 2 to 8. The thymidine at position 5 appears to be conserved as well. (B) The experimentally verified CodY motif in L. lactis MG1363. (C) The consensus CodY motifs identified in L. lactis MG1363 and L. lactis KF147, in which the proposed motif sequence is underlined.
Genome level prediction of enhanced catabolic flexibility of L. lactis KF 147.
Model profiles 40 and 41 also encompass many genes involved in the uptake and metabolism of alternative carbon sources such as ribose, mannitol, galacturonate, raffinose, sucrose, and fructose. These genes were expressed at relatively low levels during the growth-associated stages of retentostat cultivation (up to day 14), and subsequently, their expression levels gradually became higher upon prolonged retentostat cultivation, reaching very high levels under the near-zero growth conditions reached at the end of cultivation (days 35 and 42) (Fig. 7; see Fig. S8 in the supplemental material). For example, the expression of genes associated with the uptake and metabolism of ribose (rbsABCDKR) and mannitol (mtlADFR) was increased more than 10-fold during prolonged retentostat cultivation (Fig. 7). These observations illustrate the transcriptional response to prolonged and severe carbon source limitation that is encountered during prolonged retentostat cultivation, leading to the progressive derepression and/or activation of expression of several genes required for the catabolism of alternative carbon sources in L. lactis KF147. These responses raised the question of whether these retentostat-adapted L. lactis KF147 cultures would display significantly enhanced catabolic flexibility and would more readily and rapidly ferment carbon sources other than glucose. To address this question, the rates of fermentation of various carbohydrates by nongrowing bacterial suspensions of L. lactis KF147 withdrawn from the retentostat culture on days 14, 21, 35, and 42 were determined.
FIG 7.
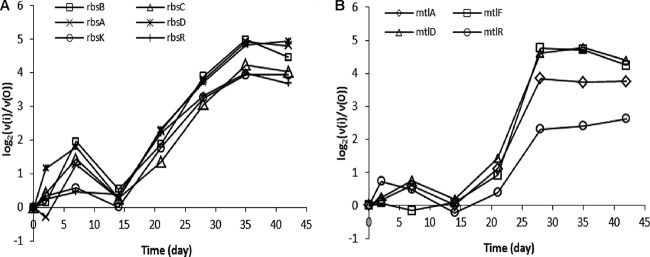
Graphs of the expression of genes involved in ribose (A) and mannitol (B) uptake and metabolism during retentostat cultivation over the beginning of chemostat cultivation (day 0) from model profiles 40 and 41 (retentostat 1). Similar transcriptome results obtained in retentostat 2 confirmed the consistency of these results in an independent experiment.
To this end, the Vmax, which was expressed in arbitrary, pH-based units (pH units min−1 [10−3]; see Materials and Methods), was determined in L. lactis KF147 cell suspensions derived from the retentostat culture on days 14, 21, 35, and 42 by incubation with glucose, ribose, mannitol, sucrose, fructose, or raffinose as the fermentable substrate. Since the culture biomass did not significantly increase after 14 days of retentostat cultivation and at this stage the expression levels of the relevant genes appeared to be unaffected compared to those measured under carbon-limited chemostat growth conditions, this sample was used as a reference in these analyses. The highest Vmax values for all of the carbon sources used were obtained from the sample taken after 42 days of retentostat cultivation (Table 2). Specifically, the Vmax of the cell suspension derived from the retentostat culture on day 42 was severalfold higher than the Vmax of the suspension based on day 14 cultures for the carbon sources ribose (3-fold), mannitol (5-fold), sucrose (2-fold), and raffinose (3-fold), respectively (Table 2). Only the fructose and glucose acidification rates obtained were similar at all of the time points evaluated (days 14, 21, 35, and 42), where glucose served as a positive control in these analyses and was consistently associated with the highest acidification rate in all of the suspensions tested (Table 2). These results confirmed that the induced transcription of genes involved in alternative sugar utilization pathways enabled the cells to rapidly adjust to the utilization of alternative carbon sources when these became available. Notably, the progressive derepression of metabolic pathways dedicated to the use of alternative carbon and energy sources clearly exceeds the basal levels of expression that can be seen under the initial carbon limitation conditions, e.g., during carbon-limited chemostat cultivation.
TABLE 2.
Vmax values on different carbon sources of L. lactis KF147 samples taken on days 14, 21, 35, and 42 of the retentostat cultivation period
| Carbon source | Avg Vmax (pH units min−1 [10−3])a ± SD on day: |
|||
|---|---|---|---|---|
| 14 | 21 | 35 | 42 | |
| Glucose | 3.9 ± 0.34 | 2.7 ± 0.69 | 3.7 ± 0.41 | 5.6 ± 0.43 |
| Ribose | 0.2 ± 0.05 | 0.1 ± 0.06 | 0.5 ± 0.05 | 0.6 ± 0.06 |
| Mannitol | 0.2 ± 0.03 | 0.2 ± 0.04 | 0.7 ± 0.03 | 1.0 ± 0.02 |
| Sucrose | 0.5 ± 0.02 | 0.5 ± 0.01 | 1.1 ± 0.04 | 1.1 ± 0.03 |
| Fructose | 2.3 ± 0.05 | 1.8 ± 0.06 | 2.3 ± 0.04 | 2.5 ± 0.05 |
| Raffinose | 0.3 ± 0.01 | 0.3 ± 0.01 | 0.6 ± 0.02 | 0.9 ± 0.02 |
Regression coefficient (R2), >0.99.
DISCUSSION
This paper presents the molecular adaptation of L. lactis to near-zero growth rates induced by carbon-limited retentostat cultivation and thereby expands our previous analysis of the quantitative energy household and physiology of this bacterium under these conditions (10). Genome-wide transcriptional data were integrated with metabolite data sets of organic and amino acid production and consumption under retentostat conditions, allowing the identification of transcription signatures that reflect near-zero growth adaptation inferred by retentostat cultivation. Transcriptome adaptations established the repression of several growth-associated functions, specifically, those related to the biosynthesis of particular macromolecules, i.e., membrane components and extracellular polysaccharides. Notably, the retentostat conditions did not induce stringency-like responses and the genes encoding the components of the machineries for DNA replication, transcription, and translation remained relatively highly expressed.
Our previous study (10) highlighted that L. lactis KF147 retentostat cultures display intriguing metabolic switches within their central carbohydrate and energy metabolism, fluctuating between mixed-acid fermentation and lactic acid fermentation. Notably, in the present study, we show that the fermentation end product analyses (10) are congruent with the transcriptional patterns of the genes involved in the corresponding pathways, implying that the distribution of pyruvate among the different dissipation pathways is not controlled only by allosteric interactions or redox-balance changes (27, 28) but also controlled at the transcriptional level in L. lactis KF147. Intriguingly, carbon-starved batch cultures of L. lactis strongly suppressed carbon metabolism genes during the stationary phase but sustained high expression of glycolytic and pyruvate dissipation functions (29). In the present experiments, genes encoding glycolytic enzymes were transiently expressed at elevated levels during early stages of stagnated growth (days 14 to 28), which are proposed to lead to enhanced glycolytic flux that may drive the changes of pyruvate dissipation from mixed-acid toward lactic acid fermentation, which includes boosted lactate dehydrogenase expression. The transcriptional control of glycolytic and pyruvate dissipation pathways appears to contradict the previously proposed regulation by enzyme level control through allosteric interactions with glycolytic intermediates, ATP demand, and/or carbon source import rates (28–31). Several glycolytic enzymes have been assigned key roles in the regulation of glycolytic flux in L. lactis as a function of the specific strain of this species and its environmental condition, like the pyruvate kinase and phosphofructokinase in L. lactis MG1363 during batch cultivation on maltose (32, 33) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in strain NCDO 2118 during batch growth on lactose (34). Although the regulation of glycolytic flux in L. lactis is not entirely understood, it has been shown that its flux is neither controlled by an individual and rate-limiting glycolytic enzyme nor dictated by sugar import rates (35), although the latter have been proposed to predominantly regulate glycolytic flux under conditions of very low carbon flux (30). Glycolytic flux also appears to be controlled by the cellular energy state, which could elegantly be shown by decreasing ATP levels by increasing F1F0-H+-ATPase expression, which could drive up to 3-fold-increased glycolytic flux but led to declining growth rates and biomass yields (36, 37). Nevertheless, mathematical models using the kinetic parameters of the enzymes that constitute the lactococcal glycolytic pathway demonstrated that glycolytic flux could accurately be predicted with such a model. This is quite remarkable, since the kinetic parameters of enzymes are commonly obtained from in vitro experiments that in many cases neglect many potentially relevant interactions and modifications, including transcriptional regulation, protein phosphorylation, and allosteric modulation (35). The present study revealed that transcriptional regulation contributes to glycolytic flux and pyruvate dissipation control in L. lactis KF147. This could be specific for lactococci isolated from plants, like KF147, which would be supported by the many studies that address metabolic control in dairy L. lactis strains that reach alternative but not necessarily mutually exclusive or consistent control conclusions. Alternatively, this could also be specific for the conditions employed in this study and thus depend on the specific metabolic characteristics induced by near-zero growth conditions.
The response of L. lactis KF147 to extremely low growth rates also included some remarkable fluctuations of amino acid metabolism. During the continuous reduction of the growth rate in the first retentostat cultivation period, the overall declining rate of amino acid consumption is most likely explained by the reduced requirement for these biomass building blocks, which has previously also been shown for batch cultures that enter the stationary phase of growth (29). Intriguingly, the transcription of the genes encoding the glycolytic enzymes GAPDH, phosphoglycerate mutase, and enolase, which are involved in the production of the intermediates 3-phosphoglycerate (3PG), phosphoenolpyruvate (PEP), and pyruvate, were continuously overexpressed after 14 days of retentostat cultivation. These glycolytic intermediates are known to serve as substrates in reactions that link the CCM to amino acid synthesis and are involved in the pathways that lead to the synthesis of Ser, Cys, and Gly; Phe, Trp, and Tyr and Ala, Ile, Leu, Val, and Thr, respectively (38, 39). This possible role of these intermediates is supported by the observation that intermediate stages (days 14 to 28) of retentostat cultivation were associated with the net production of certain amino acids and in parallel expressed their glycolytic genes at elevated levels.
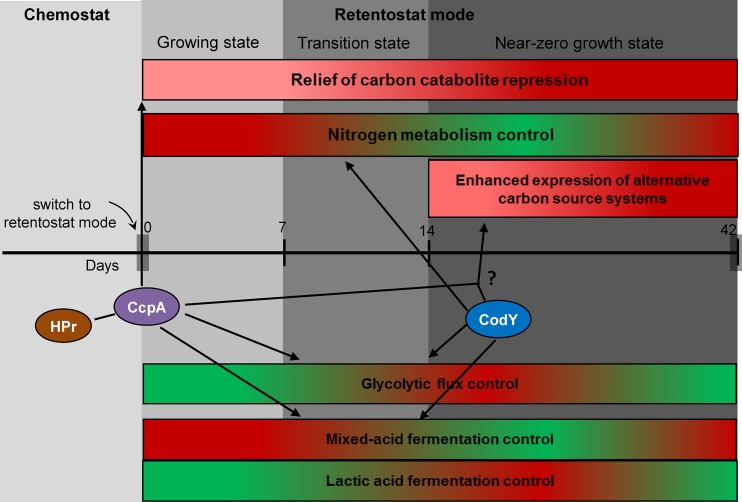
In several low-GC Gram-positive bacteria, including L. lactis, the transcriptional regulator CodY controls the expression of degradation of oligopeptide uptake and metabolism of di/tripeptides and amino acids, especially BCAAs, Asn, Glu, His, and Arg, in response to the availability of amino acids or peptides (40, 41). When lactococcal cultures reach the stationary phase and nutrients become limited, CodY-dependent repression of peptide and amino acid transporter systems is relieved to maintain nitrogen metabolism in the cells (41). Moreover, CodY has also been shown to modulate functions in carbon metabolism since it has been shown to control the expression of citrate synthase (gltA), isocitrate dehydrogenase (icd), and aconitase (citB), which belong to the incomplete Krebs cycle in L. lactis MG1363 (41). Importantly, this incomplete Krebs cycle can support the production of α-ketoglutarate, which can serve as a substrate for glutamate production and as a cosubstrate for the first step of BCAA catabolism (41, 42), illustrating how CodY regulation could connect nitrogen metabolism regulation to carbon metabolism regulation (41). Similarly, the detection of a CodY-like regulatory motif in the near-zero growth regulons of L. lactis KF147 led us to propose CodY-mediated control of nitrogen metabolism under these conditions, although the expression of the codY gene itself appeared to be constitutive under the experimental conditions used in this study. This role could include a connecting regulatory role for CodY in CCM regulation via the glycolytic intermediates 3PG, PEP, and pyruvate (see above). Moreover, the connection of pyruvate metabolism and BCAA biosynthesis (Fig. 4A) also implies that CodY could indirectly influence pyruvate dissipation and thereby may play a role in controlling lactic acid fermentation versus mixed-acid fermentation behavior in L. lactis KF147 under retentostat conditions (Fig. 8).
FIG 8.
Integrated view of adaptive regulation of L. lactis KF147 to near-zero growth rates induced by retentostat cultivation. In rectangular boxes, red and green indicate increased and decreased expression of the functional categories shown, respectively.
In L. lactis MG1363, a 15-bp cis-acting element with the consensus sequence AATTTTCWGAAAATT has been identified as a high-affinity binding site for CodY (41) (Fig. 6B). The proposed CodY binding motif identified in L. lactis KF147 resembles the CodY binding motif of strain MG1363 (41), despite differences between the deduced consensus sequences of the strains (Fig. 6C). Notably, the motif identified in strain KF147 encompasses a palindromic element and is frequently encountered in a head-to-tail tandem orientation upstream of the identified genes, expanding the palindromic nature of the composite cis-acting element. Therefore, we propose that the motif we identified represents the CodY target site in L. lactis KF147 and that CodY plays a prominent role in the regulation of nitrogen metabolism adaptations in L. lactis KF147 at extremely low growth rates and may possibly also control the typical carbon metabolism fluctuations observed under these conditions in a more indirect manner (Fig. 8). The relevance of nitrogen metabolism regulation in the context of industrial applications of L. lactis is obvious, since it has been well established that flavor formation, for example, in cheese ripening, is driven largely by amino acid conversions that involve nitrogen metabolism-associated enzymes (43).
This study also established that genes involved in the import and utilization of alternative carbon sources (other than glucose) were progressively derepressed during prolonged retentostat cultivation, enabling the bacteria to more rapidly switch to alternative energy and carbon resources upon their availability in the environment. This adaptation may reflect the evolutionary benefit of scavenging trace amounts of nutrients in growth-limiting environments, which clearly goes beyond the relief of catabolite control protein A (CcpA)-mediated CCR (44–48), since CCR relief was already effective in the carbon-limited chemostat culture that was used to initiate retentostat cultivation. In LAB, CcpA-mediated CCR includes the repression of catabolic operons in response to the availability of a “preferred” carbon source, which commonly is glucose (45). Intriguingly, in L. lactis, CcpA also acts as an activator of the las operon, which encodes three key enzymes in the glycolytic pathway, phosphofructokinase (pfk), pyruvate kinase (pyk), and lactate dehydrogenase (ldh), creating a CcpA connection to central metabolism control (33, 49, 50). The role of CcpA and/or its coregulators (e.g., HPr, ATP/ADP levels, etc.) in the high-level induction of alternative carbon utilization systems under near-zero growth conditions remains to be determined (Fig. 8).
In conclusion, transcriptome and metabolite adaptations of L. lactis KF147 to near-zero growth rates inferred by retentostat cultivation are clearly distinct from those elicited by starvation or stationary-phase conditions and include particular fluctuating metabolic behavior. The regulation of nitrogen metabolism and, indirectly, possibly also the fluctuating mixed-acid and lactic acid fermentation patterns, might involve CodY (Fig. 8) through the identified cis-acting motif that resembles the previously reported CodY box. Retentostat cultivation also led to the progressive relief of CCR and the activation of pathways associated with the utilization of alternative substrates, which goes beyond the canonical CcpA-mediated carbon catabolite regulation. Intriguingly, recent work with Bacillus subtilis established that CcpA and CodY can form a complex that interacts with RpoA, underpinning the interactions between the gene regulation networks involved in carbon and nitrogen metabolism regulation (51). The gene regulation profiles identified in this study include several CcpA and CodY target genes, and their regulation may involve a similar regulatory complex encompassing both CodY and CcpA.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sacha van Hijum for designing the hybridization scheme, Jan van Riel for technical assistance with gas chromatography, and Marjo Starrenburg for her assistance during hybridization and scanning procedures (NIZO Food Research, Ede, The Netherlands). In addition, we thank our colleagues from the Industrial Microbiology Section, Delft University of Technology, and the Molecular Genetics Group, University of Groningen, in the joint zero-growth project group (Kluyver Centre, The Netherlands) for invaluable discussions.
This project was carried out within the research program of the Kluyver Centre for Genomics of Industrial Fermentation, which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02484-14.
REFERENCES
- 1.Hoehler TM, Jorgensen BB. 2013. Microbial life under extreme energy limitation. Nat Rev Microbiol 11:83–94. doi: 10.1038/nrmicro2939. [DOI] [PubMed] [Google Scholar]
- 2.Boender LGM, de Hulster EAF, van Maris AJA, Daran-Lapujade P, Pronk JT. 2009. Quantitative physiology of Saccharomyces cerevisiae at near-zero specific growth rates. Appl Environ Microbiol 75:5607–5614. doi: 10.1128/AEM.00429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koch AL. 1971. The adaptive responses of Escherichia coli to a famine and feast existence. Adv Microb Physiol 6:147–217. doi: 10.1016/S0065-2911(08)60069-7. [DOI] [PubMed] [Google Scholar]
- 4.Hugas M, Monfort JM. 1997. Bacterial starter cultures for meat fermentation. Food Chem 59:547–554. doi: 10.1016/S0308-8146(97)00005-8. [DOI] [Google Scholar]
- 5.Smit G, Smit BA, Engels WJ M. 2005. Flavour formation by lactic acid bacteria and biochemical flavor profiling of cheese products. FEMS Microbiol Rev 29:591–610. doi: 10.1016/j.fmrre.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Crow VL, Coolbear T, Gopal PK, Martley FG, McKay LL, Riepe H. 1995. The role of autolysis of lactic acid bacteria in the ripening of cheese. Int Dairy J 5:855–875. doi: 10.1016/0958-6946(95)00036-4. [DOI] [Google Scholar]
- 7.Erkus O, de Jager VCL, Spus M, van Alen-Boerrigter IJ, van Rijswijck IMH, Hazelwood L, Janssen PWM, van Hijum SAFT, Kleerebezem M. 2013. Multifactorial diversity sustains microbial community stability. ISME J 7:2126–2136. doi: 10.1038/ismej.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siezen RJ, Bayjanov J, Renckens B, Wels M, van Hijum SAFT, Molenaar D, van Hylckama Vlieg JET. 2010. Complete genome sequence of Lactococcus lactis subsp. lactis KF147, a plant-associated lactic acid bacterium. J Bacteriol 192:2649–2650. doi: 10.1128/JB.00276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbert D. 1961. A theoretical analysis of continuous culture systems, p 21–53. In Henderson DW, Hastings JJH, Southgate BA, Brian PW (ed), Continuous culture of microorganisms (Microbiology Group Symposium papers, University College, London, 31 Mar-1 Apr, 1960). Society of Chemical Industry, London, United Kingdom. [Google Scholar]
- 10.Ercan O, Smid EJ, Kleerebezem M. 2013. Quantitative physiology of Lactococcus lactis at extreme low-growth rates. Environ Microbiol 15:2319–2332. doi: 10.1111/1462-2920.12104. [DOI] [PubMed] [Google Scholar]
- 11.van Verseveld HW, de Hollander JA, Frankena J, Braster M, Leeuwerik FJ, Stouthamer AH. 1986. Modeling of microbial substrate conversion, growth and product formation in a recycling fermenter. Antonie Van Leeuwenhoek 52:325–342. doi: 10.1007/BF00428644. [DOI] [PubMed] [Google Scholar]
- 12.Chesbro W, Evans T, Eifert R. 1979. Very slow growth of Escherichia coli. J Bacteriol 139:625–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arbige M, Chesbro WR. 1982. Very slow growth of Bacillus polymyxa: stringent response and maintenance energy. Arch Microbiol 132:338–344. doi: 10.1007/BF00413386. [DOI] [Google Scholar]
- 14.Tappe W, Laverman A, Bohland M, Braster M, Rittershaus S, Groeneweg J, van Verseveld HW. 1999. Maintenance energy demand and starvation recovery dynamics of Nitrosomonas europaea and Nitrobacter winogradskyi cultivated in a retentostat with complete biomass retention. Appl Environ Microbiol 65:2471–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goffin P, van de Bunt B, Giovane M, Leveau JH, Höppener-Ogawa S, Teusink B, Hugenholtz J. 2010. Understanding the physiology of Lactobacillus plantarum at zero growth. Mol Syst Biol 6:413. doi: 10.1038/msb.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jørgensen TR, Nitsche BM, Lamers GE, Arentshorst M, van den Hondel CA, Ram AF. 2010. Transcriptome insights into the physiology of Aspergillus niger approaching a specific growth rate of zero. Appl Environ Microbiol 76:5344–5355. doi: 10.1128/AEM.00450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boender LGM, van Maris AJ A, de Hulster EAF, Almering MJH, van der Klei IJ, Veenhuis M, de Winde JH, Pronk J T, Daran-Lapujade P. 2011. Cellular responses of Saccharomyces cerevisiae at near-zero growth rates: transcriptome analysis of anaerobic retentostat cultures. FEMS Yeast Res 11:603–620. doi: 10.1111/j.1567-1364.2011.00750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terzaghi BE, Sandine WE. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol 29:807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meijerink M, van Hemert S, Taverne N, Wels M, de Vos P, Bron PA, Savelkoul HF, van Bilsen J, Kleerebezem M, Wells JM. 2010. Identification of genetic loci in Lactobacillus plantarum that modulate the immune response of dendritic cells using comparative genome hybridization. PLoS One 5:e10632. doi: 10.1371/journal.pone.0010632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wernersson R, Nielsen HB. 2005. OligoWiz 2.0–integrating sequence feature annotation into the design of microarray probes. Nucleic Acids Res 33(Web Server issue):W611–W615. doi: 10.1093/nar/gki399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Hijum SA, Garcia de la Nava J, Trelles O, Kok J, Kuipers OP. 2003. MicroPreP: a cDNA microarray data pre-processing framework. Appl Bioinformatics 2:241–244. [PubMed] [Google Scholar]
- 22.Ernst J, Bar-Joseph Z. 2006. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. 2006. TM4 microarray software suite. Methods Enzymol 411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 24.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 25.de Jong A, Pietersma H, Cordes M, Kuipers OP, Kok J. 2012. PePPER: a web server for prediction of prokaryote promoter elements and regulons. BMC Genomics 13:299. doi: 10.1186/1471-2164-13-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laws A, Gu Y, Marshall V. 2001. Biosynthesis, characterization, and design of bacterial exopolysaccharides from lactic acid bacteria. Biotech Adv 19:597–625. doi: 10.1016/S0734-9750(01)00084-2. [DOI] [PubMed] [Google Scholar]
- 27.Collins LB, Thomas TD. 1974. Pyruvate kinase of Streptococcus lactis. J Bacteriol 120:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garrigues C, Loubiere P, Lindley ND, Cocaign-Bousquet M. 1997. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J Bacteriol 179:5282–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redon E, Loubiere P, Cocaign-Bousquet M. 2005. Transcriptome analysis of the progressive adaptation of Lactococcus lactis to carbon starvation. J Bacteriol 187:3589–3592. doi: 10.1128/JB.187.10.3589-3592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cocaign-Bousquet MD, Even S, Lindley ND, Loubiere P. 2002. Anaerobic sugar catabolism in Lactococcus lactis: genetic regulation and enzyme control over pathway flux. Appl Microbiol Biotechnol 60:24–32. doi: 10.1007/s00253-002-1065-x. [DOI] [PubMed] [Google Scholar]
- 31.Thomas TD, Turner KW, Crow VL. 1980. Galactose fermentation by Streptococcus lactis and Streptococcus cremoris: pathways, products, and regulation. J Bacteriol 144:672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersen HW, Solem C, Hammer K, Jensen PR. 2001. Twofold reduction of phosphofructokinase activity in Lactococcus lactis results in strong decreases in growth rate and glycolytic flux. J Bacteriol 183:3458–3467. doi: 10.1128/JB.183.11.3458-3467.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solem C, Koebmann B, Yang F, Jensen PR. 2007. The las enzymes control pyruvate metabolism in Lactococcus lactis during growth on maltose. J Bacteriol 189:6727–6730. doi: 10.1128/JB.00902-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Even S, Garrigues C, Loubiere P, Lindley ND, Cocaign-Bousquet M. 1999. Pyruvate metabolism in Lactococcus lactis is dependent upon glyceraldehyde-3-phosphate dehydrogenase activity. Metab Eng 1:198–205. doi: 10.1006/mben.1999.0120. [DOI] [PubMed] [Google Scholar]
- 35.Martinussen J, Solem C, Holm AK, Jensen PR. 2013. Engineering strategies aimed at control of acidification rate of lactic acid bacteria. Curr Opin Biotechnol 24:124–129. doi: 10.1016/j.copbio.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Koebmann BJ, Solem C, Pedersen MB, Nilsson D, Jensen PR. 2002. Expressing of genes encoding F-1-ATPase result in uncoupling of glycolysis from biomass production in Lactococcus lactis. Appl Environ Microbiol 68:4274–4282. doi: 10.1128/AEM.68.9.4274-4282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koebmann BJ, Westerhoff HV, Snoep JL, Solem C, Pedersen MB, Nilsson D, Michelsen O, Jensen PR. 2002. The extent to which ATP demand controls the glycolytic flux depends strongly on the organism and conditions for growth. Mol Biol Rep 29:41–45. doi: 10.1023/A:1020398117281. [DOI] [PubMed] [Google Scholar]
- 38.Ganesan B, Stuart MR, Weimer BC. 2007. Carbohydrate starvation causes a metabolically active but nonculturable state in Lactococcus lactis. Appl Environ Microbiol 73:2498–2512. doi: 10.1128/AEM.01832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novák L, Loubiere P. 2000. The metabolic network of Lactococcus lactis: distribution of 14C-labeled substrates between catabolic and anabolic pathways. J Bacteriol 182:1136–1143. doi: 10.1128/JB.182.4.1136-1143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brinsmade SR, Kleijn RJ, Sauer U, Sonenshein AL. 2010. Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J Bacteriol 192:6357–6368. doi: 10.1128/JB.00937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.den Hengst CD, van Hijum SAFT, Geurts JMW, Nauta A, Kok J. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J Biol Chem 280:34332–34342. doi: 10.1074/jbc.M502349200. [DOI] [PubMed] [Google Scholar]
- 42.Yvon M, Chambellon E, Bolotin A, Roudot-Algaron F. 2000. Characterization and role of the branched-chain aminotransferase (BcaT) isolated from Lactococcus lactis subsp. cremoris NCDO 763. Appl Environ Microbiol 66:571–577. doi: 10.1128/AEM.66.2.571-577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smid EJ, Kleerebezem M. 2014. Production of aroma compounds in lactic fermentations. Annu Rev Food Sci Technol 5:313–326. doi: 10.1146/annurev-food-030713-092339. [DOI] [PubMed] [Google Scholar]
- 44.Egli T. 1995. The ecological and physiological significance of the growth of heterotrophic microorganisms with mixtures of substrates. Adv Microb Ecol 14:305–386. doi: 10.1007/978-1-4684-7724-5_8. [DOI] [Google Scholar]
- 45.Fujita Y. 2009. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci Biotechnol Biochem 73:245–259. doi: 10.1271/bbb.80479. [DOI] [PubMed] [Google Scholar]
- 46.Lendenmann U, Egli T. 1995. Is Escherichia coli growing in glucose-limited chemostat culture able to utilize other sugars without lag? Microbiology 141:71–78. doi: 10.1099/00221287-141-1-71. [DOI] [PubMed] [Google Scholar]
- 47.Sepers AJB. 1984. The uptake capacity for organic compounds of two heterotrophic bacterial strains at carbon limited growth. Z Allg Mikrobiol 24:261–267. doi: 10.1002/jobm.3630240414. [DOI] [Google Scholar]
- 48.Wick LM, Quadroni M, Egli T. 2001. Short- and long-term changes in proteome composition and kinetic properties in a culture of Escherichia coli during transition from glucose-excess to glucose-limited growth conditions in continuous culture and vice versa. Environ Microbiol 3:588–599. doi: 10.1046/j.1462-2920.2001.00231.x. [DOI] [PubMed] [Google Scholar]
- 49.Even S, Lindley ND, Cocaign-Bousquet MD. 2001. Molecular physiology of sugar catabolism in Lactococcus lactis IL1403. J Bacteriol 183:3817–3824. doi: 10.1128/JB.183.13.3817-3824.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luesink EJ, van Herpen RE, Grossiord BP, Kuipers OP, de Vos WM. 1998. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol Microbiol 30:789–798. doi: 10.1046/j.1365-2958.1998.01111.x. [DOI] [PubMed] [Google Scholar]
- 51.Wünsche A, Hammer E, Bartholomae M, Völker U, Burkovski A, Seidel G, Hillen W. 2012. CcpA forms complexes with CodY and RpoA in Bacillus subtilis. FEBS J 279:2201–2214. doi: 10.1111/j.1742-4658.2012.08604.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.